Abstract
Phosphotyrosine-binding domains, typified by the SH2 (Src homology 2) and PTB domains, are critical upstream components of signal transduction pathways. The E3 ubiquitin ligase Hakai targets tyrosine-phosphorylated E-cadherin via an uncharacterized domain. In this study, the crystal structure of Hakai (amino acids 106–206) revealed that it forms an atypical, zinc-coordinated homodimer by utilizing residues from the phosphotyrosine-binding domain of two Hakai monomers. Hakai dimerization allows the formation of a phosphotyrosine-binding pocket that recognizes specific phosphorylated tyrosines and flanking acidic amino acids of Src substrates, such as E-cadherin, cortactin and DOK1. NMR and mutational analysis identified the Hakai residues required for target binding within the binding pocket, now named the HYB domain. ZNF645 also possesses a HYB domain but demonstrates different target specificities. The HYB domain is structurally different from other phosphotyrosine-binding domains and is a potential drug target due to its novel structural features.
Keywords: Hakai, phosphotyrosine, Src substrates, ubiquitin ligases, zinc fingers
Introduction
In eukaryotic cells, phosphorylation events regulate cell signalling by providing docking sites for protein domains, such as the Src homology 2 (SH2) and phosphotyrosine-binding (PTB) domains (Forman-Kay and Pawson, 1999; Yaffe, 2002; Pawson and Nash, 2003). The SH2 was the first signalling domain to be identified and has been extensively characterized (Songyang et al, 1993; Forman-Kay and Pawson, 1999; Yaffe, 2002; Pawson and Nash, 2003; Liu et al, 2006; Filippakopoulos et al, 2009). The SH2 is a dedicated phosphotyrosine-binding domain and plays a critical role in signal transduction, hence making it a target for drug development (Pawson and Nash, 2003; Machida and Mayer, 2005; Taylor et al, 2008; Kasembeli et al, 2009). Binding specificity of SH2 domains is generally conferred by the sequences flanking the C-terminus of the phosphotyrosine (pTyr), and motif recognition is usually relatively inflexible. The other major class of pTyr-binding domain is the PTB domain. The specificity of binding to the PTB domain is conferred typically by residues on the target that are N-terminal to the pTyr. However, the PTB domain also recognizes non-pTyr motifs (Forman-Kay and Pawson, 1999; Yaffe, 2002; Pawson and Nash, 2003; Farooq and Zhou, 2004; Smith et al, 2006). Atypical phosphotyrosine-binding domains have also been detected in PKCδ and the human M2 pyruvate kinase (PKM2) (Benes et al, 2005; Christofk et al, 2008).
In 2002, Fujita et al (2002) discovered a novel ubiquitin E3 ligase protein that targeted pTyr sites on E-cadherin. The E3 ligase, Hakai, possesses three domains: a RING domain, a short pTyr recognition sequence and a proline-rich domain (Fujita et al, 2002). Hakai is involved in the regulation of cell adhesion, cell migration and embryogenesis (Figueroa et al, 2009; Kaido et al, 2009; Gong et al, 2010). Among the reported protein interactions of Hakai, its association with and ubiquitination of E-cadherin upon Src activation is the best characterized (Fujita et al, 2002).
Based on molecular modelling, Fujita et al (2002) assumed the pTyr-binding domain of Hakai to be a derivative SH2 domain. Our previous experience analysing the SH2 domain of a similar E3 ubiquitin ligase, c-Cbl (Ng et al, 2008) led us to examine the nature of the Hakai pTyr-binding domain to provide structural insights into its interaction with E-cadherin. In this study, we report that the Hakai pTyr-binding (HYB) domain consists of a homodimer formed at a structurally novel interface. Each monomer consists of two zinc-finger domains: a RING domain and a minimum pTyr-binding domain that incorporates a novel, atypical zinc coordination motif. Both domains play key roles in dimerization. The HYB domain is therefore composed of four zinc-binding domains cooperating to bind pTyr residues surrounded by acidic amino acids. Whereas the RING domain appears in other proteins, the atypical zinc-binding domain component is a novel protein fold that incorporates an intertwined configuration. In order to obtain its consensus target sequence, we have characterized the recognition motif of the HYB domain and identified several Src substrates that are also targeted. In addition, the HYB domain can also be found in a testis-specific ubiquitin E3 ligase, ZNF645, and possibly Ligand-of-Numb protein X1 and 2 (LNX1 and LNX2).
Results
A novel protein fold in Hakai
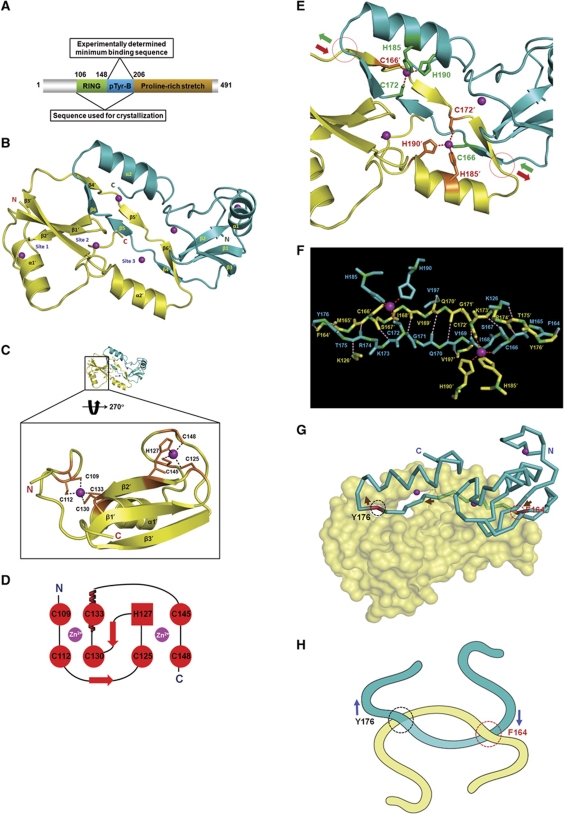
We experimentally established that the minimum E-cadherin phosphotyrosine-binding sequence in Hakai was contained within amino acids 148–206 (aa 148–206) (Supplementary Figure S1A–G). Circular dichroism analysis however revealed that purified Hakai (aa 148–206) was unstructured (Supplementary Figure S1H), and was not suitable for crystallization. Therefore, the Hakai sequence spanning amino-acid residues 106–206 (aa 106–206) that contained both the RING domain and the minimum pTyr-binding domain, as represented schematically in Figure 1A, was purified and crystallized.
Figure 1.
A novel protein fold in Hakai. (A) A schematic diagram of the Hakai protein. (B) The crystal structure of Hakai (aa 106–206) reveals a dimer in an anti-parallel configuration. Each monomer contains three zinc coordination sites. Sites 1 and 2 lie in the RING domain. Site 3 is shared between the two monomers. (C) The coordination of zinc ions (purple spheres) by the RING domain of Hakai is shown for one of the monomers. (D) A schematic diagram of the cross-brace arrangement of the Hakai RING domain as shown in (C). (E) The Hakai dimer forms an intertwined configuration spanning the points indicated in circles, with the entry and exit paths shown in green and brown arrows. The zinc-interacting side chains are shown as green and brown sticks. (F) The backbone of the Hakai (aa 106–206) residues involved in intermolecular main-chain H-bonding and the zinc-coordinating side chains of adjacent monomers at the dimer interface are shown in cyan and yellow. The pink dots indicate the main-chain H-bonds; the red dots indicate the zinc coordination bonds. (G) The monomers of the interlinked Hakai dimer are shown in surface representation and Cα trace, respectively. The Cα trace monomer enters and exits the other monomer at the red and black circles, respectively. Brown arrows show its entry and exit path. (H) A schematic diagram of the novel Hakai interlinked arrangement as shown in (G).
The crystal structure of Hakai (aa 106–206) was solved at 1.9 Å resolution (Figure 1B; Supplementary Figure S1I; Supplementary Table SI). The striking feature of the crystal structure was the formation of a dimer from paired, anti-parallel Hakai (aa 106–206) monomers. Each monomer consisted of an N-terminal RING domain, followed by the C-terminal atypical zinc-binding domain that is contained within the experimentally derived minimum pTyr-binding domain. Furthermore, each monomer contained three zinc ions at three distinct sites. One zinc ion coordinated with the atypical zinc-binding domains of both monomers (Figure 1B). The Hakai RING domain (residues 106–148) adopted a typical RING domain fold stabilized by coordinating with two zinc ions, forming a cross-brace arrangement (Figure 1C and D) (Borden, 2000). The zinc-coordinating residues in the RING domain are also indicated in Figure 1D.
The uniqueness of the Hakai (aa 106–206) region was revealed when the structure was compared with other proteins in the PDB (Protein Data Bank) using the DALI server (http://ekhidna.biocenter.helsinki.fi/dali_server/). The results show that only the RING domain of Hakai is structurally similar to RING domains of other proteins. There is, however, no similarity beyond amino-acid residue 159 of the Hakai minimum pTyr-binding domain, which is located on the dimerization interface. The minimal pTyr-binding domain of Hakai adopts a novel, three-dimensional fold and contains three β-strands (β4, β5 and β6) and a C-terminal α-helix. The β-strands β4, β5 and β6 were in an extended configuration and formed anti-parallel β-sheets with the corresponding β-strands of the monomeric partner during homodimerization (Figure 1E). The atypical zinc-finger motif within this region is formed by two histidine residues (H185 and H190) and one cysteine residue (C172) from one monomer and a second cysteine residue (C166) from the adjacent monomer (Figure 1F; Supplementary Figure S2), unlike a classical C2H2 zinc finger (ZnF). Although the Hakai (aa 106–206) region in each monomer fulfils the required criteria of the zinc coordination consensus pattern of cysteine and histidine residues [C-x(5)-C-x(12)-H-x(4)-H], it is not capable of forming the zinc coordination sphere by itself, since a second cysteine residue is required of its anti-parallel monomeric partner (Figure 1F). Therefore, each dimer contains two atypical zinc-finger motifs.
The crystal structure of Hakai also shows that the two Hakai monomers intertwine through a stretch of residues ranging from F164 to Y176 during dimerization, resulting in the formation of three β-sheets based on 12 main-chain hydrogen bonds (Figure 1E–H). This novel interlinked configuration and the two atypical zinc ion interactions at the dimer interface are unique features of this distinctive homodimeric assembly. A surface area of ∼1650 Å2 of each monomer (or ∼21% of each monomer surface) was formed at the dimer interface of Hakai (aa 106–206), with 34 hydrogen bond contacts between the monomers, as analysed by the PISA server (Krissinel and Henrick, 2007).
Hakai forms a dimer in solution
A previous study alluded to the similarity between the Hakai (aa 106–206) polypeptide with the dimerization domain of the V(D)J recombination-activating protein RAG1 (Fujita et al, 2002). In addition to the crystal structure, the intermolecular NOE cross-peaks corresponding to the amides of residues at the dimer interface also indicates that Hakai (aa 106–206) forms a dimer in solution (Figure 2A). Significantly, the structures of the dimerization interface of the RAG1 domain (Bellon et al, 1997) and Hakai (aa 106–206), as described in this study, are completely dissimilar. The formation of Hakai (aa 106–206) dimers in solution is also supported by the results obtained through dynamic light scattering, which show an apparent molecular weight of 24.1 kDa, twice than that of the monomer (Supplementary Figure S3A).
Figure 2.
Hakai forms a dimer in solution. (A) A 3D 15N-NOESY spectrum showing the intermolecular NOE cross-peaks of amides corresponding to residues of Hakai (aa 106–206). (B) WT Hakai (aa 106–206) and four Hakai (aa 106–206) point mutants were each separately used for gel-filtration chromatography. Their respective elution profiles were overlain and compared. (C) HA- and FLAG-tagged Hakai were overexpressed in the presence of Src in HEK293 cells. FLAG immunoprecipitates were analysed for HA-tagged Hakai. (D) A schematic representation of the point mutations made to C166, C172, H185 and H190 in Hakai. (E) Cell lysates containing WT Hakai or Hakai mutants were used to analyse the effects of Hakai dimerization on E-cadherin recognition.
We next examined whether zinc coordination is necessary for the dimerization of Hakai and its ability to interact with its target. We first investigated whether mutations of the zinc-coordinating residues within the minimum pTyr-binding domain (C166, C172, H185 and H190) would affect the proposed Hakai dimerization. The Hakai (aa 106–206) polypeptides containing point mutations at these residues were separated on a calibrated gel-filtration column. Their gel-filtration elution profiles show that each point mutant had an apparent molecular weight equivalent to a monomeric unit of wild-type (WT) Hakai (aa 106–206) protein (∼12 kDa), whereas WT Hakai eluted as a dimer (Figure 2B). Furthermore, circular dichroism performed using all the Hakai (aa 106–206) mutants ascertained that each one has maintained a well-defined secondary structure (Supplementary Figure S3B). These findings suggest that each zinc-coordinating residue is instrumental in forming the dimer interface.
Having determined that Hakai (aa 106–206) dimerizes in solution, we next examined if this occurs with full-length proteins. Full-length FLAG-tagged Hakai was observed to bind to its HA-tagged counterpart (Figure 2C), indicating that dimerization also occurs between the full-length proteins. To further determine whether Hakai dimerization is required for its function in binding its targets, the full-length proteins containing alanine point mutations of the zinc-coordinating residues (Figure 2D) were tested for their ability to bind to tyrosine-phosphorylated E-cadherin. HEK293 cells were used for such studies as they did not express detectable levels of endogenous E-cadherin, which could have interfered with the mammalian cellular assays used. The evidence presented in Figure 2E indicates that none of the four Hakai point mutants interacted with tyrosine-phosphorylated E-cadherin. The collective results therefore show that zinc coordination is necessary for both dimerization of Hakai and its subsequent function in interacting with its target.
Hakai domain recognizes acidic residues
Having established the novel characteristics of the Hakai zinc-coordinated homodimer, we sought to identify the target motif of this new domain. At this point, the only described target motif was in Src-phosphorylated E-cadherin. Within this motif, two (Y755 and Y756 in mouse; Y753 and Y754 in humans) of three consecutive tyrosine residues were reported to be involved in the interaction with Hakai (Fujita et al, 2002). To analyse the relative contributions of the three tyrosine residues in binding to Hakai, mutations were made to the tyrosine residues (Figure 3A). To determine which of the tyrosines were phosphorylated, we analysed the patterns of the tyrosine phosphorylation of the point mutants after v-Src activation. The pTyr signals shown in Figure 3B indicate that all three adjacent tyrosines were phosphorylated.
Figure 3.
Hakai domain recognizes acidic residues. (A) Y753, Y754 and Y755 (red) of E-cadherin were mutated to phenylalanine (blue) in different combinations. (B) The WT E-cadherin and the mutants shown in (A) were overexpressed in HEK293 cells with v-Src to analyse their pTyr signals. (C) HEK293 cells were co-transfected with WT E-cadherin or its mutants together with Hakai to identify the tyrosine residues recognized by Hakai. (D) The Y754-phosphorylated and non-phosphorylated E-cadherin peptides were titrated against Hakai (aa 106–206) using ITC. The top panels show the heat release profiles after baseline correction and the lower panels indicate the binding isotherms for the interactions. The dissociation constant (Kd) and binding stoichiometry (N) are shown in the table. (E) The E-cadherin aa 747–758 were each substituted with alanine. (F) The E-cadherin mutants from (E) and Hakai were co-transfected into HEK293 cells to identify the target motif on E-cadherin. (G) E-cadherin and Hakai were co-transfected into HEK293 cells. Their interaction was analysed through immunoprecipitation of FLAG-tagged Hakai. (H) Cortactin was co-transfected into HEK293 cells with Hakai. The interaction between cortactin and Hakai was compared with that in (G). (I) Y482 and Y485 (red) were separately substituted with phenylalanine (blue). (J) WT and mutated cortactin were co-transfected into HEK293 cells with Hakai, in the absence or presence of v-Src. The pTyr signal of cortactin and its interaction with Hakai were analysed. (K) An alanine scan of cortactin aa 478–489. Each residue was substituted with alanine (blue). G484 was not mutated as glycine mutations affect the protein structure. (L) The cortactin mutants described in (K) were co-transfected into HEK293 cells with Hakai. The interaction between the cortactin mutants and Hakai was determined using immunoprecipitation.
We next examined the importance of the three tyrosine residues in E-cadherin for its interaction with Hakai. The E-cadherin mutants used in the earlier experiment were analysed for their potential to bind to WT Hakai. The results shown in Figure 3C indicate that Y754 is the only tyrosine significantly involved in binding; each mutant containing a substitution in this position did not bind Hakai, whereas all other mutants showed significant binding. These combined results also show that while Src binds to and phosphorylates most of the E-cadherin mutants, all the mutants with a Y754F substitution do not bind to Hakai, even when phosphorylated. This implies that the interaction between Hakai and E-cadherin depends on the direct recognition of specific pTyr residues on E-cadherin by Hakai. To verify the necessity of Y754 phosphorylation for the interaction between E-cadherin and Hakai, isothermal titration calorimetry (ITC) was performed using phosphorylated and non-phosphorylated E-cadherin peptides corresponding to aa 749–761 with Hakai (aa 106–206). The results in Figure 3D show that binding occurred only with the phosphorylated peptide. Furthermore, the results also indicate that only one E-cadherin peptide binds to the Hakai (aa 106–206) dimer at any one time.
In a similar manner, we analysed the importance of the amino-acid residues flanking the tyrosines for the interaction between the two proteins via an alanine scan (Figure 3E). The immunoprecipitation results in Figure 3F show that there were profound contributions from the aspartic acid D756 and glutamic acid E757, and significant contributions from valine V752 and aspartic acid D750. Consequently, a cluster of negative charges from the acidic amino acids is formed around the centrally binding tyrosine 754 of E-cadherin.
To verify the motif recognized by Hakai, additional target proteins were identified. This identification was accomplished by analysing Hakai-binding proteins phosphorylated by Src using mass spectrometry. A list of the proteins obtained from a typical experiment is appended in Supplementary Figure S4A and B, in which cortactin was identified as a potential target. Mouse cortactin is phosphorylated by Src primarily on Y482 and Y485 (Ren et al, 2009). Interestingly, these two tyrosines are also surrounded by several acidic residues. We investigated whether Hakai binds to Src-phosphorylated cortactin, as well as the importance of Y482, Y485 and their flanking residues in this interaction. In addition to the WT proteins, phenylalanine substitution and alanine scan experiments were also performed on cortactin as described for E-cadherin. The results for cortactin mirror those obtained for E-cadherin. Like E-cadherin, cortactin interacts with Hakai only when phosphorylated by Src (Figure 3G and H). Phenylalanine substitutions of the Src-phosphorylated tyrosine residues revealed that Y482 is the main tyrosine residue involved in Hakai binding (Figure 3I and J). Furthermore, the acidic residues (E478, D480 and E486) surrounding Y482 contributed profoundly to the interaction between Hakai and E-cadherin, and significant contribution was also observed from S487 (Figure 3K and L).
To further verify that Hakai targets pTyrs of Src substrates with surrounding acidic residues, another Src substrate was selected: DOK1, which also contains pTyrs with adjacent acidic groups (Luo et al, 2008). One such particular tyrosine residue was found to be a primary phosphorylation site of Src (Luo et al, 2008) (Figure 4A). The results show that DOK1 interacts with Hakai. Furthermore, DOK1 competed with endogenous cortactin for Hakai, implying that DOK1 and cortactin bind Hakai on the same site (Figure 4A–C).
Figure 4.
DOK1 interacts with Hakai. (A) The sequence alignment of the different Src phosphorylation target sites in E-cadherin, cortactin and DOK1. The acidic amino-acid residues flanking the phosphorylated tyrosine are shown in blue. (B) Hakai and DOK1 were overexpressed in HEK293 cells in the absence or presence of Src. FLAG immunoprecipitates were analysed for DOK1 interaction. (C) DOK1 was co-transfected into HEK293 cells with Hakai to study its competition with endogenous cortactin for binding to Hakai. FLAG immunoprecipitates were immunoblotted for cortactin.
Target-binding amino acids of Hakai
To identify the residues in Hakai within aa 106–206 necessary for its interaction with E-cadherin, 2D 1H–15N-HSQC spectra of 15N-labelled Hakai (aa 106–206) were acquired in the absence and presence of a pTyr peptide derived from amino-acid residues 749–761 of E-cadherin. Based on the changes in the NMR spectrum of Hakai (aa 106–206), several residues underwent perturbations in chemical shift (Figure 5A and B). A minimum criterion of a chemical shift difference Δδp.p.m.>0.15 p.p.m. was applied. Five residues were identified (Y176, H185, N187, H188 and R189), which resided in the Hakai minimum pTyr-binding domain, whereas a sixth residue (H127) was from the RING domain (Figure 5C). However, the crystal structure shows that only four of these six residues reside in a close three-dimensional spatial proximity. While H127, Y176, H185 and R189 face the interior of the E-cadherin-binding site (Figure 5C) and form a positively charged pocket, N187 and H188 face outward and are not part of the pocket (Figure 5D).
Figure 5.
Target-binding amino acids of Hakai. (A) An overlay of the 1H–15N-HSQC spectra of Hakai (aa 106–206) in the absence (green) or the presence (red) of an tyrosine-phosphorylated E-cadherin peptide. (B) A graphical representation of the combined chemical shift perturbation (p.p.m.) plotted against all Hakai (aa 106–206) residues, with the cutoff at the combined chemical shift perturbation of 0.15 p.p.m. (C) The six potential E-cadherin-interacting residues in Hakai (aa 106–206) are highlighted as sticks in the ribbon representation of the crystal structure. (D) An electrostatic surface potential representation of Hakai (aa 106–206) shows that H127, Y176, H185 and R189 form part of the positively charged pocket. (E) The interaction between E-cadherin and the Hakai mutants of the residues identified in (C) was analysed by immunoprecipitating FLAG-tagged Hakai. (F) HEK293 cells were transfected with the identified Hakai mutants, and their interaction with endogenous cortactin was studied. (G) Immunoprecipitates of either WT Hakai or the Hakai zinc-coordinating mutants were tested for interaction with endogenous cortactin. (H) A schematic representation of the Hakai dimer and the HYB domain.
These residues identified through in vitro peptide-domain binding assays were then further tested by expressing full-length point mutants in HEK293 cells. The immunoprecipitation results shown in Figure 5E indicate that the residues identified in the NMR analysis also abrogated binding when mutated, with the exception of residues N187 and H188. As expected, the required residues were situated on the interior of the target-binding domain, whereas the non-binding residues, N187 and H188, were on the exterior. Similar results were obtained with experiments using cortactin (Figure 5F), demonstrating the importance of these Hakai residues. Furthermore, dimerization of Hakai is also required, as with E-cadherin, as cortactin was unable to bind to Hakai containing mutations to its zinc-coordinating residues (Figure 5G).
Based on the evidence obtained, it can be concluded that two Hakai monomers interact in an anti-parallel manner to form a dimer via the interlinked zinc-coordinating domain. This domain binds pTyrs flanked by acidic amino acids in Src substrates. The target-binding domain resulting from this dimerization process represents the functional Hakai phosphotyrosine-binding domain, henceforth referred to as the HYB (Hakai pY-binding) domain (Figure 5H; Supplementary Figure S5).
The HYB domain in other proteins
We next investigated whether the HYB domain is found in other proteins. Literature and database searches revealed that the testis-specific ubiquitin E3 ligase ZNF645 exhibited high-sequence homology with Hakai (Liu et al, 2010), as shown in Figure 6A. We therefore questioned whether ZNF645 could also interact with E-cadherin and cortactin. The results in Figure 6B show that ZNF645 bound to v-Src-phosphorylated E-cadherin but not to cortactin (Figure 6C). This result implies that although there is significant homology between Hakai and ZNF645, they are likely to have their own sets of targets due to the differences in their sequences between the key zinc-coordinating residues.
Figure 6.
The HYB domain in other proteins. (A) A comparison of the Hakai protein from amino-acid residues 127–191 and the equivalent sequence in ZNF645. (B) E-cadherin and ZNF645 were analysed for their interaction using immunoprecipitation. Hakai was used as a positive control. The dotted arrow indicates a non-specific band; the solid arrow indicates the ZNF645 band. (C) ZNF645 was overexpressed in HEK293 cells and its interaction with endogenous cortactin was analysed using immunoprecipitation. (D) Sequence alignment of LNX1 and LNX2 with Hakai and ZNF645 based on the conserved zinc-coordinating residues from Hakai aa 106–206.
Based on the key amino-acid residues involved in zinc coordination and binding in HYB, we searched the NCBI database to analyse gene origins and protein homologies (Supplementary Table SII). Two interesting results emerged. First, a comparison of the species distribution of the Hakai and ZNF645 gene products indicated that the latter, found only in primates, is most likely a recent copy of the former. Second, ZNF645 is an intronless gene, implying that it is a retrotransposed copy of Hakai.
Further database searches based on the conserved zinc-coordinating cysteine and histidine residues within the HYB domain showed that a similar series of residues is present in LNX1 and LNX2 (Figure 6D). This implies that the HYB domain may be distributed in other proteins, although the latter observation requires experimental confirmation.
Novel structure of the HYB domain
The structures of the five pTyr-binding domains that have been discovered to date are illustrated in Figure 7A and B. All of the domains, except for the HYB domain, are contained within one monomer. The HYB domain consists of a pair of monomers arranged in an anti-parallel configuration and is composed of two RING and two atypical zinc-coordinating domains. From this comparison, it is apparent that all five of these pTyr domains have completely different structures, with different strategies to recognize tyrosine phosphorylation.
Figure 7.
Novel structure of the HYB domain. (A) Representative structures of SH2 (PDB code 1SHB), PTB (PDB code 1SHC), PKCδ C2 (PDB code 1YRK) and PKM2 (PDB code 3BJF) in ligand-free forms are compared with the HYB domain. (B) The corresponding topologies of the domains in (A).
Discussion
ZnFs consist of a diverse group of protein domains that are able to bind to zinc ions. Initially thought to bind only nucleic acids, ZnFs were later shown to mediate protein–protein interactions (Gamsjaeger et al, 2007; Klug, 2010). The common structural feature in the different ZnF classes, where determined, is that the zinc-coordinating cysteine and histidine residues are all situated on the same monomer. The evidence in this study has shown that Hakai, in addition to having a typical RING domain, possesses an atypical ZnF with a novel structural fold. In contrast to previously reported structures, the formation of this atypical ZnF requires amino-acid residues from two intertwining Hakai monomers. Three of the four residues are contributed by one monomer, whereas one cysteine residue comes from another monomer, implying that the formation of this ZnF is accompanied by dimerization. Although this particular ZnF is novel by itself, it is made more intriguing by the fact that this domain lies within a pTyr-binding region. The results from our study demonstrate that in conjunction with the RING domain of Hakai, dimerization of Hakai through the formation of the atypical ZnF creates a novel pTyr-binding domain, called the HYB domain. Originally postulated to recognize multiple consecutive pTyrs, it was instead found that the HYB domain recognizes a single pTyr residue flanked by acidic amino acids. Interestingly, this may indicate a bias towards Src substrates because Src kinase has a preferred, but not strict, recognition sequence of Y(p)-E-E-I/V (Yeatman, 2004).
The zinc-coordinating residues of each Hakai monomer consist of a conserved series of nine cysteine and three histidine residues that facilitate Hakai dimerization. Based on sequence analysis, LNX1 and LNX2 also harbour such a sequence motif, although the last conserved histidine is replaced by cysteine. While the full structures of the LNX proteins are currently unknown, it is possible that, based on sequence homology, they also contain the HYB domain. As the HYB domain contains a RING domain, its occurrence would be predominantly in E3 ubiquitin ligases, which has implications in the way these proteins recognize their targets and effect downstream signalling.
Although the zinc-coordinating residues within the ZnFs of the HYB domain are critical, the protein folding of the primary sequences surrounding the cysteine and histidine residues are also important in defining its characteristics. This chelation-derived folding provides the necessary three-dimensional structure while specific amino acids within the sequences between the zinc-coordinating residues are instrumental in recognizing and binding phosphorylated tyrosines of target proteins. In the case of Hakai, four residues were identified through NMR studies to bind to E-cadherin, H127, Y176, H185 and R189. It is noteworthy that there is strict conservation of these residues between Hakai and ZNF645. On the other hand, the differences in their binding targets are most likely dictated by amino acids that are not common to both proteins, although this aspect requires further study. It would also be interesting to observe if LNX1 or LNX2 binds phosphorylated tyrosine residues. Based on sequence homology, both LNX1 and LNX2 harbour most of the zinc-coordinating residues of the HYB domain. However, only LNX2 has the equivalent of R189 of Hakai. Neither of the LNX proteins has the equivalent of Y176. It is possible that these two proteins bind a different set of tyrosine-phosphorylated proteins by utilizing different residues within their sequences. Thus, while the basic sequence required to provide a unique pTyr-binding structure is apparent within the four proteins, a number of questions regarding the exact pTyr-binding requirements of these proteins remain unanswered.
In conclusion, we have identified a novel HYB domain, made up of two structurally atypical ZnF domains, which recognize a previously unidentified pTyr motif. Our structural data indicate that the HYB domain forms a positively charged pocket, implying that it represents a highly suitable drug target. As dysfunctional E3 ubiquitin ligases are often associated with diseases (Burger et al, 2006; Lohr et al, 2010), the presence of the HYB domain in these proteins offers a specific target for directed therapies.
Materials and methods
Plasmids
Mouse Hakai, human E-cadherin and avian v-Src were gifts from W Birchmeier (Max-Delbrück-Center for Molecular Medicine, Germany), W Hunziker (IMCB, Singapore) and XM Cao (IMCB, Singapore), respectively. Mouse cortactin was from Addgene (Cambridge, MA) (plasmid 26722, deposited by A Weaver). Human ZNF645 and DOK1 were from Origene (Rockville, MD). Where necessary, the genes were cloned into pXJ40–HA or pXJ40–FLAG. For structural studies, Hakai constructs were cloned into pGEX6P-1 (GE Healthcare, UK). Point mutants and truncates were generated using the proofreading Pfu DNA polymerase.
Antibodies and reagents
Mouse anti-FLAG M2, rabbit anti-FLAG and anti-HA and agarose-conjugated anti-FLAG M2 beads were obtained from Sigma-Aldrich (St Louis, MO). Rabbit GST, cortactin, E-cadherin and DOK1 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Protein-A-conjugated agarose beads were from Roche Molecular Biochemicals (Germany). Mouse anti-E-cadherin and HRP-conjugated anti-pTyr PY20 were from BD Transduction Laboratories (Lexington, KY). Mouse anti-β-actin was obtained from Abcam (Cambridge, MA).
Cell lines and transfection
HEK293 cells were purchased from ATCC (Manassas, VA) and maintained as described (Yusoff et al, 2002). Transfections were performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions.
Immunoprecipitation and immunoblotting
Immunoprecipitation and immunoblotting were carried out as described (Yusoff et al, 2002) with the following modifications. HEK293 cells were harvested 24 h post-transfection with a lysis buffer containing protease inhibitors (Roche) and 1 mM Na3VO4. Immunoprecipitations were performed using agarose-conjugated anti-FLAG or protein-specific antibodies followed by incubation with protein-A-conjugated agarose beads at 4°C.
Liquid chromatography–mass spectrometry/mass spectrometry
Immunoprecipitates were separated by SDS–PAGE, and stained with Coomassie Blue G250. Protein bands were excised and washed with 25 mM ammonium bicarbonate (ABB) in 50% acetonitrile (ACN) buffer thrice. The proteins in the gel were reduced with 10 mM DTT in 25 mM ABB buffer, alkylated with 5 mM iodoacetamide, dehydrated and digested with trypsin overnight. After in-gel digestion, the solution was transferred to a clean tube and sonicated for 30 min in the presence of 50 μl 50% ACN and 5% acetic acid for protein extraction. This extraction procedure was repeated three times; the pooled extracts were dried with a vacuum concentrator. The samples were processed and analysed as described (Zhang et al, 2010) using a LTQ-FT ultra mass spectrometer.
For each experiment, MS/MS (dta) spectra were extracted from the raw data files using the extract_msn program in Biowork 3.3 (ThermoFinnigan). The extracted dta files were combined into a single file in the Mascot generic file (mgf) format. Except for the conversion of precursor mass from MH+ in dta to m/z in mgf, the fragment ion m/z and intensity values were used as determined. Proteins were identified by searching the combined data against the IPI human database (downloaded on 25 November 2009, including 86 845 sequences and 35 122 444 residues) via an in-house Mascot server (version 2.2.07). Two missing cleavages were allowed. Precursor ion and MS/MS fragment ion error tolerances were set to <10 p.p.m. and <0.8 Da, respectively. A protein was accepted as a true positive if it had a significant score (P<0.05) and at least two unique peptides.
Protein purification and gel-filtration chromatography
The GST-tagged Hakai (aa 106–206) constructs were expressed in Escherichia coli BL21 (DE3) and purified using glutathione-conjugated sepharose (GE Healthcare). The GST-tag was cleaved using GST-PreScission Protease (GE Healthcare) and the proteins were applied to a Superdex 75 size-exclusion column (GE Healthcare) equilibrated using 10 mM Bis-Tris, pH 6.5, 250 mM NaCl and 5 mM DTT and pre-calibrated using a gel-filtration standard (Bio-Rad).
NMR spectroscopy and chemical shift perturbation analysis
15N/13C-labelled Hakai (aa 106–206) was obtained from cultures grown in M9 media supplemented with 15N-labelled ammonium chloride and 13C-labelled glucose as the sole nitrogen and carbon sources, respectively. The labelled proteins were purified as described above. NMR spectra were acquired at 298 K in an 800-MHz NMR spectrometer (Bruker, Karlsruhe, DE). The backbone assignment was obtained using standard 15N-edited HSQC, HNCACB and CBCA (CO)NH experiments; side chains were assigned using standard 3D-TOCSY, 3D-NOESY and HCCH-TOCSY experiments. NMR data were processed using NMRPipe (Delaglio et al, 1995) and analysed by NMRView (Johnson and Blevins, 1994).
For the chemical shift perturbation analysis, the 2D 1H–15N-HSQC spectra for the 15N-labelled Hakai (aa 106–206) were acquired in the absence or presence of the phosphorylated E-cadherin peptide. Perturbed residues on Hakai were assigned by superimposing the two HSQC spectra.
Crystallization and structure determination
SelMet-substituted Hakai (aa 106–206) was expressed in a methionine auxotroph (Doublie, 1997) and purified as described above. SelMet incorporation was verified by MALDI-TOF.
SelMet Hakai (aa 106–206) crystals were grown at 289 K by the hanging drop vapour diffusion method. The protein (30 mg/ml) was mixed with an equal volume of reservoir solution (140 mM Li2SO4; 100 mM Tris, pH 7.8; 15 mM Na2S2O3; 20–22% PEG 5000 MME; 1% isopropanol and 20–25% ethylene glycol). A complete SAD data set was collected to 1.9 Å resolution at the synchrotron beamlines (NSLS, Brookhaven National Laboratory and the National Synchrotron Radiation Research Center [NSRRC], Taiwan) using a Quantum4-CCD detector (Area Detector Systems Corp., Poway, CA). The data sets were processed and scaled using HKL2000 (Otwinowski and Minor, 1997). The crystals belonged to space group P6222 with a=64.66 Å, b=64.66 Å, c=121.04 Å, and contained one molecule in the asymmetric unit.
All four expected selenium sites in the asymmetric unit were located by SOLVE (Terwilliger and Berendzen, 1999). Initial phases were developed by RESOLVE (Terwilliger, 2003); the overall figure of merit was improved to 0.83, and over 90% of the molecule was built automatically. The remaining parts of the model were built manually using COOT (Emsley and Cowtan, 2004) and alternatively refined by CNS (Brunger et al, 1998) and PHENIX (Adams et al, 2002). The final model was refined to a 1.9-Å resolution with an R-factor of 0.2175 (Rfree=0.2396) and analysed using PROCHECK (Laskowski et al, 1993). All structure-related figures were prepared using PyMOL (DeLano, 2002).
Circular dichroism spectrometry
Far UV spectra (260–190 nm) of Hakai (aa 148–206) and Hakai (aa 106–206) and its mutants were measured using a Jasco J-810 spectropolarimeter in phosphate buffer (pH 7.5) at room temperature using a 0.1-cm path length and stoppered cuvettes. Six scans were recorded, averaged and the baseline subtracted.
Dynamic light scattering
Dynamic light scattering studies were carried out on a DynaPro Light Scattering instrument (Protein Solutions, USA) at a protein concentration of 2 mg/ml, in a buffer containing 10 mM Bis-Tris pH 6.5, 250 mM NaCl and 5 mM DTT.
Isothermal titration calorimetry
Phosphorylated and non-phosphorylated peptides of E-cadherin corresponding to amino-acid residues 749–761 were titrated at a molar concentration of 800 μM against 100 μM of Hakai (aa 106–206) dimer in a VP-ITC microcalorimeter (Microcal, Northhampton, UK) at 293 K. The titrations were carried out using 30 10-μl injections of the appropriate peptide into the sample cell containing Hakai (aa 106–206) and the data were analysed with a one-site binding model using the Origin software package v7.0 supplied by Microcal. All measurements were repeated twice.
Accession number
The coordinates and structure factors have been deposited at the Protein Data Bank (PDB) with the accession code 3VK6.
Supplementary Material
Acknowledgments
This work was partly supported by the Agency of Science Technology and Research and AcRF Grant (R154000438112) to J Sivaraman, Singapore. MM is a PhD student in receipt of a research scholarship from the National University of Singapore (NUS).
Author contributions: MM performed the cloning, protein expression, purification, crystallization, NMR spectroscopy and chemical shift perturbation analysis. Data collection, structure determination and analysis of crystal structure were done by J Sivaraman, MM and J Seetharaman. CD and ITC experiments were performed by MM and XZ. Mutations of Hakai (aa 106–206) for structural studies were done by MM. Determination of the minimum binding region of Hakai was performed by RAJ, JY and PY. All the mutational analyses performed in mammalian cells and biochemical assays for Hakai, E-cadherin and cortactin were done by SYC, PY, CN, SS, NFMA, DL, DY, AI and YPL. Experiments with DOK1 were done by XWK. PY prepared the samples for mass spectrometry and LC–MS/MS and the subsequent processing and analysis were performed by JQ and SKS. Sequence analysis and supervision was provided by GRG and J Sivaraman. Manuscript preparation was done by GRG and SYC with input from J Sivaraman and MM.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 [DOI] [PubMed] [Google Scholar]
- Bellon SF, Rodgers KK, Schatz DG, Coleman JE, Steitz TA (1997) Crystal structure of the RAG1 dimerization domain reveals multiple zinc-binding motifs including a novel zinc binuclear cluster. Nat Struct Biol 4: 586–591 [DOI] [PubMed] [Google Scholar]
- Benes CH, Wu N, Elia AE, Dharia T, Cantley LC, Soltoff SP (2005) The C2 domain of PKCdelta is a phosphotyrosine binding domain. Cell 121: 271–280 [DOI] [PubMed] [Google Scholar]
- Borden KLB (2000) RING domains: master builders of molecular scaffolds? J Mol Biol 295: 1103–1112 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Burger A, Amemiya Y, Kitching R, Seth AK (2006) Novel RING E3 ubiquitin ligases in breast cancer. Neoplasia 8: 689–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC (2008) Pyruvate kinase M2 is a phosphotyrosine binding protein. Nature 452: 181–186 [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhe G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6: 277–293 [DOI] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOL Molecular Graphics System, Version 0.99. USA: Schrödinger, LLC http://www.pymol.org [Google Scholar]
- Doublie S (1997) Preparation of selenomethionyl proteins for phase determination. Methods Enzymol 276: 523–530 [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Farooq A, Zhou MM (2004) PTB or not to be: promiscuous, tolerant and Bizarro domains come of age. IUBMB Life 56: 547–556 [DOI] [PubMed] [Google Scholar]
- Figueroa A, Kotani H, Toda Y, Mazan-Mamczarz K, Mueller EC, Otto A, Disch L, Norman M, Ramdasi RM, Keshtgar M, Gorospe M, Fujita Y (2009) Novel roles of Hakai in cell proliferation and oncogenesis. Mol Biol Cell 20: 3533–3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Müller S, Knapp S (2009) SH2 domains: modulators of nonreceptor tyrosine kinase activity. Curr Opin Struct Biol 19: 643–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman-Kay JD, Pawson T (1999) Diversity in protein recognition by PTB domains. Curr Opin Struct Biol 9: 690–695 [DOI] [PubMed] [Google Scholar]
- Fujita Y, Krause G, Scheffner M, Zechner D, Leddy HE, Behrens J, Sommer T, Birchmeier W (2002) Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E-cadherin complex. Nat Cell Biol 4: 222–231 [DOI] [PubMed] [Google Scholar]
- Gamsjaeger R, Liew CK, Loughlin FE, Crossley M, Mackay JP (2007) Sticky fingers: zinc-fingers as protein-recognition motifs. Trends Biochem Sci 32: 63–70 [DOI] [PubMed] [Google Scholar]
- Gong EY, Park E, Lee K (2010) Hakai acts as a coregulator of estrogen receptor alpha in breast cancer cells. Cancer Sci 101: 2019–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA, Blevins RA (1994) NMRView: a computer program for the visualization and analysis of NMR data. J Biomol NMR 4: 603–614 [DOI] [PubMed] [Google Scholar]
- Kaido M, Wada H, Shindo M, Hayashi S (2009) Essential requirement for RING finger E3 ubiquitin ligase Hakai in early embryonic development of Drosophila. Genes Cells 14: 1067–1077 [DOI] [PubMed] [Google Scholar]
- Kasembeli MM, Xu X, Tweardy DJ (2009) SH2 domain binding to phosphopeptide ligands: potential for drug targeting. Front Biosci 14: 1010–1022 [DOI] [PubMed] [Google Scholar]
- Klug A (2010) The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu Rev Biochem 79: 213–231 [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Liu BA, Jablonowski K, Raina M, Arcé M, Pawson T, Nash PD (2006) The human and mouse complement of SH2 domain proteins-establishing the boundaries of phosphotyrosine signaling. Mol Cell 22: 851–868 [DOI] [PubMed] [Google Scholar]
- Liu YQ, Bai G, Zhang H, Su D, Tao DC, Yang Y, Ma YX, Zhang SZ (2010) Human RING finger protein ZNF645 is a novel testis-specific E3 ubiquitin ligase. Asian J Androl 12: 658–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr N, Molleston JP, Strauss KA, Torres-Martinez W, Sherman EA, Squires RH, Rider NL, Chikwara KR, Cummings OW, Morton DH, Puffenberger EG (2010) Human ITCH E3 ubiquitin ligase deficiency causes syndromic multisystem autoimmune disease. Am J Human Gen 86: 447–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Slebos RJ, Hill S, Li M, Brábek J, Amanchy R, Chaerkady R, Pandey A, Ham AJ, Hanks SK (2008) Global impact of oncogenic Src on a phosphotyrosine proteome. J Proteome Res 7: 3447–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida K, Mayer BJ (2005) The SH2 domain: versatile signaling module and pharmaceutical target. Biochim Biophys Acta 1747: 1–25 [DOI] [PubMed] [Google Scholar]
- Ng C, Jackson RA, Buschdorf JP, Sun Q, Guy GR, Sivaram J (2008) Structural basis for a novel intrapeptidyl H-bond and reverse binding of c-Cbl-TKB domain substrates. EMBO J 27: 804–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Pawson T, Nash P (2003) Assembly of cell regulatory systems through protein interaction domains. Science 300: 445–452 [DOI] [PubMed] [Google Scholar]
- Ren G, Crampton MS, Yap AS (2009) Cortactin: coordinating adhesion and the actin cytoskeleton at cellular protrusions. Cell Motil Cytoskeleton 66: 865–873 [DOI] [PubMed] [Google Scholar]
- Smith MJ, Hardy WR, Murphy JM, Jones N, Pawson T (2006) Screening for PTB domain binding partners and ligand specificity using proteome-derived NPXY peptide arrays. Mol Cell Biol 26: 8461–8474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songyang Z, Shoelson SE, Chaudhuri M, Gish G, Pawson T, Haser WG, King F, Roberts T, Ratnofsky S, Lechleider RJ, Neel BG, Birge RB, Fajardo JE, Chou MM, Hanafusa H, Schaffhausen B, Cantley LC (1993) SH2 domains recognize specific phosphopeptide sequences. Cell 72: 767–778 [DOI] [PubMed] [Google Scholar]
- Taylor JD, Ababou A, Fawaz RR, Hobbs CJ, Williams MA, Ladbury JE (2008) Structure, dynamics, and binding thermodynamics of the v-Src SH2 domain: implications for drug design. Proteins 73: 929–940 [DOI] [PubMed] [Google Scholar]
- Terwilliger TC (2003) Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr D Biol Crystallogr 59: 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J (1999) Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr 55: 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB (2002) Phosphotyrosine-binding domains in signal transduction. Nat Rev Mol Cell Biol 3: 177–186 [DOI] [PubMed] [Google Scholar]
- Yeatman TJ (2004) A renaissance for SRC. Nat Rev Cancer 4: 470–480 [DOI] [PubMed] [Google Scholar]
- Yusoff P, Lao DH, Ong SH, Wong ES, Lim J, Lo TL, Leong HF, Fong CW, Guy GR (2002) Sprouty2 inhibits the Ras/MAP kinase pathway by inhibiting the activation of Raf. J Biol Chem 277: 3195–3201 [DOI] [PubMed] [Google Scholar]
- Zhang H, Guo T, Li X, Datta A, Park JE, Yang J, Lim SK, Tam JP, Sze SK (2010) Simultaneous characterization of glyco- and phosphoproteomes of mouse brain membrane proteome with electrostatic repulsion hydrophilic interaction chromatography. Mol Cell Proteomics 9: 635–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.