Abstract
Genome-wide association studies have identified numerous loci demonstrating genome-wide significant association with Crohn's disease. However, when many single nucleotide polymorphisms (SNPs) have weak-to-moderate disease risks, genetic risk prediction models based only on those markers that pass the most stringent statistical significance testing threshold may be suboptimal. Haplotype-based predictive models may provide advantages over single-SNP approaches by facilitating detection of associations driven by cis-interactions among nearby SNPs. In addition, these approaches may be helpful in assaying non-genotyped, rare causal variants. In this study, we investigated the use of two-marker haplotypes for risk prediction in Crohn's disease and show that it leads to improved prediction accuracy compared with single-point analyses. With large numbers of predictors, traditional classification methods such as logistic regression and support vector machine approaches may be suboptimal. An alternative approach is to apply the risk-score method calculated as the number of risk haplotypes an individual carries, both within and across loci. We used the area under the curve (AUC) of the receiver operating curve to assess the performance of prediction models in large-scale genetic data, and observed that the prediction performance in the validation cohort continues to improve as thousands of haplotypes are included in the model, with the AUC reaching its plateau at 0.72 at ∼7000 haplotypes, and begins to gradually decline after that point. In contrast, using the SNP as predictors, we only obtained maximum AUC of 0.65. Validation studies in independent cohorts further support improved prediction capacity with multi-marker, as opposed to single marker analyses.
INTRODUCTION
Crohn's disease is one subtype of inflammatory bowel disease. It affects as many as 630 000 people in North America, and 850 000 in Europe with similar incidence. Its incidence is lower in Asia and Africa, and higher among Ashkenazi Jews (1). Symptoms include abdominal pain, diarrhea, and among children, growth failure is common (2).
The pathogenesis of Crohn's disease is multifactorial and includes a strong genetic component (2). Individuals with an affected sibling are significantly more likely to develop the disease than the control population. Two studies estimated the ratio of the risk of siblings of patients to the reported population prevalence in white populations resident in the UK. In Oxford, UK, the relative risk for Crohn's disease was estimated at 36.5 (3). Multiple genome-wide association studies (GWAS) have been performed to identify genetic variants associated with Crohn's disease in European ancestry populations (4–10), and meta-analysis of GWAS has identified >30 distinct susceptibility loci for Crohn's disease (11). A number of loci were identified for which the power to detect association was limited, suggesting that presently identified loci represent only a fraction of contributing genetic loci in Crohn's disease. Despite the plethora of genome-wide significant loci identified in Crohn's disease thus far, present association signals account for <25% of the predicted heritability (11). In addition to common variation of modest effects identified through single-point analysis of the GWAS data, it is anticipated that uncommon variation at distinct loci may contribute significantly to overall disease risk (12,13). Taken together, the overall genetic architecture and optimal development of risk models of Crohn's disease may be significantly more complex than previously anticipated.
Two prior papers have considered risk prediction for Crohn's disease. Jakobsdottir et al. (14) constructed a risk prediction model using five single nucleotide polymorphisms (SNPs). Based on the Lu and Elston method (15), they developed a model with an area under the curve (AUC) of 0.66. However, validation of their model was based on theoretical results instead of an independent validation cohort. In another paper, Evans et al. (16) constructed a risk model for Crohn's disease using the disease susceptibility markers identified from the Crohn's disease meta-analysis (11). Using a 10-fold cross-validation scheme, they observed an average AUC of 0.769. However, because the validation cohort used in their analysis was a subset of the discovery cohort that identified the significant markers in the first place, there was a substantial amount of overlap between the marker discovery cohort and training/testing cohort. This is a well-documented problem in the data-mining literature, and could severely upwardly bias estimates of prediction performance (17–19). In fact, in an unpublished work, we found that using the same Crohn's disease cohort for feature selection, model development and testing, prediction accuracy can be inflated by as much as 35%. Therefore, a key component of accurately assessing genetic risk prediction models is the use of independent training and testing cohorts.
When many SNPs have weak-to-moderate disease risks, a genetic risk prediction model based only on those markers that pass the most stringent statistical significance testing threshold may be suboptimal. We found that the best prediction accuracy was often achieved when hundreds or more markers were included in the model, instead of including only those few with highly statistically significant associations (20). Given the enormous complexity of genetic contributions in Crohn's disease, we sought to develop improved prediction risk models based on genome-wide approaches that more comprehensively capture potential contributing genetic variation.
Haplotype-based association testing may offer several advantages over the standard ‘one-SNP-at-a-time' approach. First, haplotype methods facilitate detection of associations driven by cis-interactions among nearby SNPs that might be missed by methods that consider SNPs one at a time. Several mutations on a haplotype may cause a series of changes in amino acid coding and result in a larger joint effect on the disease trait than the single amino acid changes caused by single mutations. Examples include the lipoprotein lipase-responsible gene in humans (21) and a gene influencing initial lactase activity in humans (22). In this case, haplotypes should reveal more information on disease mechanisms at a candidate gene than single SNPs. Secondly, haplotype-based analysis may be helpful in identifying rare causal variants. Finally, haplotype approaches recognize that variation in populations is inherently structured into genomic blocks and exploit these correlations among SNPs. For all these reasons, using haplotypes in association testing is expected to increase power relative to single-SNP approaches, and studies based on the human haplotype structure have provided support for this (23,24).
In a recent paper by Shim et al. (23), the authors compared the performance of single SNPs versus haplotypes using association data from the North American Rheumatoid Arthritis Consortium. They discovered that some associations were only detected using haplotype-based tests perhaps resulting from the factors cited above. They also found associations using individual SNPs that were not seen in haplotype tests because if there is only a single SNP exhibiting strong linkage disequilibrium with a causal variant, having a long haplotype block with several adjacent SNPs may dilute the strength of the association. In addition, long haplotypes are more difficult to be accurately phased, and the phasing error may worsen the performance of haplotype-based association testing (25,26). Based on these previous findings, analyses based on short haplotype blocks may both provide advantages and mitigate the disadvantages of longer haplotype-based approaches. We hypothesized this because (i) given a reasonably large sample size, short haplotype blocks are easy to phase with the standard expectation maximization algorithm, and the results tend to be stable, and (ii) signal dilution will be less severe in shorter haplotypes, if there is indeed only one causal variant in the entire haplotype block. In this article, we investigate the use of two-marker haplotypes for risk prediction in Crohn's disease, and show that it leads to improved prediction accuracy compared with single-point analyses.
RESULTS
Comparisons of different prediction methods
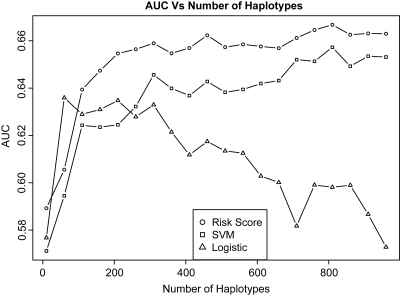
As described above, we considered three classification methods to develop a risk prediction model using the combined National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) and Belgian cohorts as the training data set (n = 2223) and the pediatric data set as the validation cohort (n = 735). For each method, we evaluated the effect of the number of haplotypes included in the risk prediction model. For performance assessment, we calculated the AUCs obtained from the validation cohort as a function of the number of the 1000 most significantly associated haplotypes. The results are summarized in Figure 1.
Figure 1.
AUC as a function of the number of haplotypes (up to 1000) included using risk score, SVM and logistic regression prediction models.
As the number of haplotypes included in the prediction model increases to the range of hundreds or thousands, logistic regression and support vector machine (SVM) methods become more computationally intensive compared with the risk-score method. Furthermore, because logistic regression uses Newton–Raphson to obtain regression coefficients, when the feature set becomes large, algorithm convergence is often not met.
For both SVM and the risk-score methods, we observed the general trend of improved prediction performance with more haplotypes included in the model. The most substantial increase in the AUC occurs when we included around 300 haplotypes in the model (Fig. 1). For the logistic regression method, we notice a gradual decline in the AUC when we included >60 haplotypes in the model. This could be due to the non-convergence of the Newton–Raphson algorithm when the dimensions of the feature set increases.
Comparing the three classification methods' performances on the top 1000 haplotypes, we discovered that the risk-score method not only has the advantage of a much shorter computational time, but also outperformed the other two methods in terms of prediction accuracy. Therefore, in subsequent analyses, we focused on the risk-score method.
The number of haplotypes used in prediction
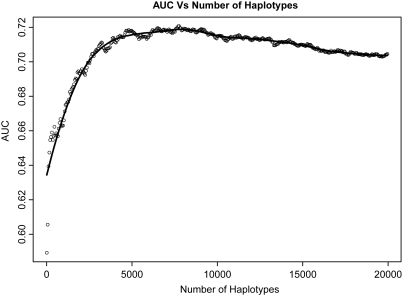
In the next step, we explored whether including >1000 haplotypes in the prediction model would further improve prediction performance, and the results are summarized in Figure 2. To better visualize the relationship between the AUC and the number of haplotypes included in the model, we smoothed the curve using the Loess smoothing technique. The prediction performance in the validation cohort continues to improve as thousands of haplotypes are included in the model. The AUC reaches its plateau at 0.72 when ∼7000 haplotypes are used for prediction, and begins to gradually decline after that point.
Figure 2.
AUC as a function of the number of haplotypes (up to 20 000) included in the prediction model, using the risk-score method. The Loess-smoothed curve is drawn.
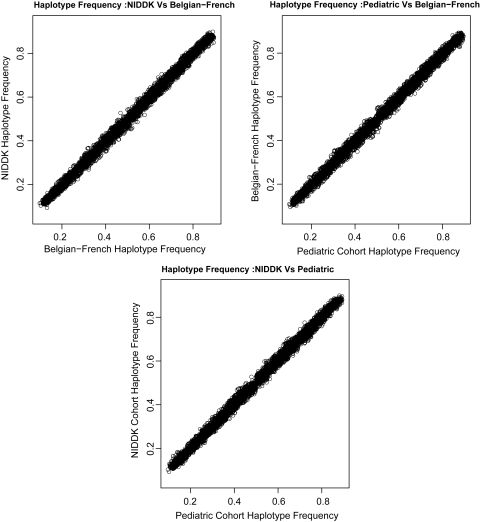
We next examined the frequency distribution in the three cohorts of the top 7000 most significantly associated haplotypes used in the analysis. We found that the haplotype frequencies are highly correlated in the three cohorts (R2 > 0.99), as shown in Figure 3.
Figure 3.
Correlation of the top 7000 associated haplotype frequencies in the three cohorts.
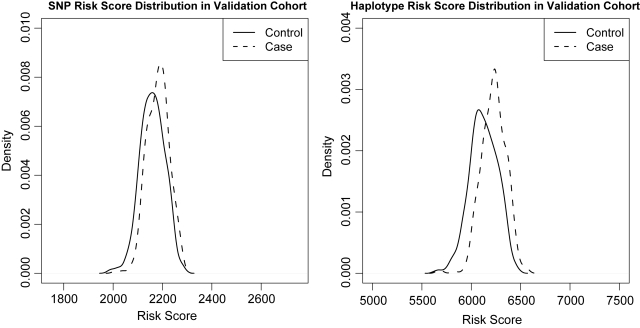
The risk score generated using the risk-score approach provides a proxy for the propensity that someone will develop the disease. In Figure 4, we plot the distribution of the case/control groups' risk scores generated from the 7000 risk haplotypes used in the prediction model in the validation cohort. We find that in the validation cohort, the risk score is capable of separating the cases from the controls, although the separation is not quite profound.
Figure 4.
Risk-score distribution for the most optimal haplotype and SNP models in discovery and validation cohorts.
Comparison with single-SNP-based prediction
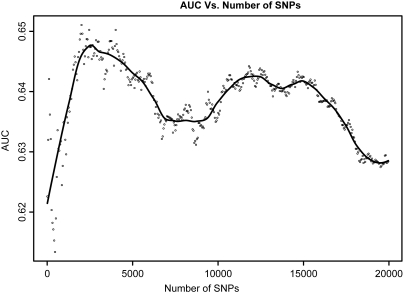
We next investigated how much benefit is gained by using haplotypes instead of single SNPs to construct the risk prediction model for Crohn's disease. Similar to Figure 2, we plotted the AUC generated from the risk-score method as a function of the number of SNPs included in the model in Figure 5. Using directly genotyped SNP as predictors, we obtained a maximum AUC of 0.647 when ∼2000 SNPs were included in the model. When including imputed SNPs into the prediction model, the maximum AUC was improved by <0.01–0.655, which remains significantly lower than the AUC derived using a haplotype-based model.
Figure 5.
The AUC as a function of the number of SNPs (up to 20 000) included in the prediction model, using the risk-score method. The Loess-smoothed curve is drawn.
Comparison with pruned haplotypes
The prediction performance worsened with the pruned set of haplotypes, and the maximum AUC dropped to 0.695.
DISCUSSION
Currently, the main approach adopted by many companies (e.g. 23 and ME, DecodeME, etc.) to carry out direct-to-consumer genetic testing is to test individuals only at well-established loci known to affect the risk of complex diseases. However, for many diseases, the established loci could only collectively explain a small portion of the genetic contribution, which suggests that many more disease-associated genetic variants (especially for those less common variants) are yet to be discovered. Therefore, estimates of risk based upon the known locus associations are likely to change dramatically in the next few years, raising questions on the stability of the current risk estimates. In fact, several papers demonstrate that updating risk factor profile may generate contradictory information about an individual's risk status over time (27,28). In addition, current methods only use single-marker information, whereas haplotypes may offer additional information for risk predictions.
In the light of these issues, including a large number of predictors in the model (e.g. both nominally significant variants and established risk loci) seems to be an attractive alternative to using only the most highly associated loci. It has been shown that the prediction performance can be improved when there are a large number of weak predictors, each of which is merely nominally significant, especially for diseases such as bipolar disorder and coronary heart disease (16). In this paper, we showed that the prediction model for Crohn's disease also follows this trend. In our Crohn's disease prediction model, the prediction performance steadily improves until the number of predictors reaches the scale of thousands, and this is observed for both haplotype and single-SNP predictors. When there are far more predictors than sample size (n<<P), traditional classification methods such as logistic regression and SVM may become inadequate both in terms of algorithm convergence and computational efficiency. An alternative way that has been considered in the literature to summarize large-scale genotype data is to use the risk-score method. Although the risk-score approach procedure does not incorporate information about effect size of each individual predictor, we found that it may outperform methods that do utilize this information (e.g. logistic regression, SVM) when the power of the study is low. This observation agrees with the results from simulation studies conducted by Kang et al. (20), and can probably be accounted for by the difficulties of accurate effect-size estimates in the high-dimensional data setting.
The risk-score method has been adopted in a number of previously published studies for complex diseases, such as diabetes, bipolar disease and Crohn's disease. In our analysis, we constructed Crohn's disease risk prediction model using both SNPs and two-marker haplotypes as predictors. Although the model evaluation criteria are different (e.g. external evaluation versus cross-validation), the performance of the single-SNP prediction model in our analysis seems to closely match the results presented by Evans et al. (16), in which they also used the risk-score method to perform Crohn's disease risk prediction. However, recognizing that for some diseases, haplotypes might be a better unit of capturing the genetic information, we also constructed our risk prediction model for Crohn's disease using haplotypes as predictors. For Crohn's disease, we observe that the risk model established with haplotypes has a better performance models built with single-SNP information; compared to the single-marker approach, the multi-marker approach increased variance explained from 0.08 to 0.14. It is worthwhile noting that although the risk-score method uses a summary statistic to build the model and may not be sensitive to the problem of correlated predictors, we compared the prediction performance of the model with and without pruning (e.g. with and without removing those highly correlated haplotypes from the predictor set), and it appears that the prediction performance worsens with pruning.
MATERIALS AND METHODS
Cohort description
We included three European ancestry Crohn's disease cohorts in our analyses. The North American NIDDK IBD Genetics Consortium cohorts were genotyped on the Illumina HumanHap300 platform. Only the non-Jewish NIDDK subjects were included in the analysis. The Belgian–French cohort was also genotyped on the Illumina HumanHap300 platform, while the pediatric cohort was genotyped on the Illumina HumanHap550 Genotyping BeadChip. More details about the individual cohorts can be found in their original publications (6–8).
We combined the NIDDK and the Belgian–French cohorts into one cohort as our discovery cohort, and use the pediatric cohort as the validation cohort. Although the discovery cohort was primarily ascertained through adult gastroenterology practices, it is believed that there is very little difference in the genetic component between the adult-onset and pediatric-onset cohorts (9); therefore, the pediatric cohort is a suitable independent replication cohort.
Data cleaning
Genotype data of each individual cohort were subjected to rigorous quality control (QC) measures to remove poor-quality SNPs. First, we excluded any SNP in Hardy–Weinberg disequilibrium (Chi-square test P-value <0.0001 in cases or controls), SNPs that have >5% missing data and any SNP with a minor allele frequency <2%. Using these filters, 3% of the SNPs in the NIDDK Non Jewish cohort, 2% of the SNPs in the Belgium–France cohort and 6.5% of the SNPs in the pediatric cohort were removed. We then included an intersection of the post-QC genotypes from the three cohorts; 284 941 SNPs shared by all of the three cohorts. In the next step, we aligned the strands of the three cohorts to the Hapmap CEU population to facilitate subsequent merging.
At the subject level, individuals having >5% missing SNPs were removed, and one, zero and four people were removed from the NIDDK, the Belgian–French and the pediatric cohorts, respectively.
Recognizing the potential confounding that may be created from combining two samples into a single discovery cohort, before combining the NIDDK and Belgian cohorts, SNPs having significantly different minor allele frequencies (Chi-square proportion test P-value <0.01) between the two cohorts were removed from the analysis (Table 1). A total of 21 959 SNPs were removed following this scheme, and 262 982 SNPs were retained in the proceeding analysis. The genomic inflation factor of the post-QC discovery cohort was 1.08. Note that there is no overlap between the discovery and the validation cohorts in terms of their geographic locations, therefore the false association signals resulting from the population structure in the discovery cohort are less likely to be replicated in the validation cohort. And because our main goal for this work is to perform prediction instead of association detection, we did not adjust for the population structure in the subsequent analysis. Next, we coded the individual cohort membership difference in the merged cohort as a binary variable (e.g. 0 = Belgian–French, 1 = NIDDK), and tested its significance by including it in the logistic regression model of marker association testing. In the discovery cohort, there were 35 markers passing a lenient genome-wide threshold (5 × 10−5), and the membership variable is not significant (>0.05) for all the top markers. Furthermore, because false association signals resulting from the population structure are less likely to be replicated in an independent validation cohort, we think including false positives from the discovery cohort will more likely hurt the prediction performance of the prediction model instead of inflating it. Therefore, combining all of the above findings, we believe that merging the two data sets to form the discovery cohort does not impose a serious problem in the risk prediction analysis.
Table 1.
Samples used in this study
| NIDDK | Belgian–French | Pediatric | |
|---|---|---|---|
| Cases | 547 | 534 | 335 |
| Controls | 548 | 594 | 400 |
Genotype imputation
Recognizing that prediction performance using single SNPs may be improved using imputed instead of directly genotyped markers, we utilize PLINK to perform the imputation, with the HapMap III CEU population as the reference panel. The required confidence threshold for making a genotype call was set at 0.8. The same QC filters as described previously for the directly genotyped markers were applied.
Haplotype construction
The entire genome was scanned with a sliding window of size 2, i.e. two SNPs were considered one at a time. Haplotypes were phased using the standard EM algorithm implemented in PLINK (29). Assuming an additive genetic model, each possible two-SNP haplotype in the sliding window is represented as a three-level categorical variable, corresponding to the number of copies each haplotype presents within each study participant. Following this recoding scheme, haplotypes can be treated in a similar way as their single-SNP counterparts, and methods developed for single-SNP association testing can be applied to the haplotypes. To avoid spurious association signals resulting from systematic missing patterns between the case and the control groups, missing haplotypes due to missing SNPs were imputed using the K-nearest neighbor algorithm. Specifically, a block containing 1000 haplotypes centered at the missing haplotype is extracted from the genome, and the genotype of the missing haplotype is imputed based on a vote of its K-nearest neighbor haplotypes (Table 2). We chose k = 3 in our imputation because it has been shown to work well when the block size is small (30).
Table 2.
Top significant haplotypes identified from the discovery cohort
| Hap | CHR | P-value | Gene |
|---|---|---|---|
| rs5743289|rs2076756 | 16 | 2.35E−17 | NOD2 |
| rs2076756|rs5743291 | 16 | 2.70E−17 | NOD2 |
| rs10521209|rs5743289 | 16 | 1.51E−16 | NOD2 |
| rs2066843|rs10521209 | 16 | 2.40E−15 | NOD2 |
| rs11647841|rs2066843 | 16 | 5.24E−15 | NOD2 |
| rs8057341|rs11647841 | 16 | 4.56E−14 | NOD2 |
| rs7194886|rs8057341 | 16 | 9.87E−14 | |
| rs9302752|rs7194886 | 16 | 5.28E−13 | |
| rs790631|rs7517847 | 1 | 1.47E−10 | IL23R |
| rs1004819|rs790631 | 1 | 1.78E−10 | IL23R |
| rs7143973|rs9285572 | 14 | 1.38E−09 | |
| rs1343151|rs10889675 | 1 | 1.78E−09 | IL23R |
| rs7530511|rs10489629 | 1 | 4.75E−09 | IL23R |
| rs10889675|rs10889677 | 1 | 3.68E−08 | IL23R |
| rs10489629|rs2201841 | 1 | 4.51E−08 | IL23R |
| rs2241880|rs3792106 | 2 | 7.81E−08 | ATG16L1 |
| rs2289476|rs2241880 | 2 | 7.81E−08 | ATG16L1 |
| rs2201841|rs11804284 | 1 | 7.94E−08 | IL23R |
| rs3818562|rs4970779 | 1 | 1.08E−07 | EPS8L3 |
| rs2302759|rs4785452 | 16 | 1.13E−07 | CYLD |
Haplotype selection
For each haplotype, a standard logistic regression is performed to obtain its marginal association significance with the disease phenotype. Haplotypes are then ranked by the significance of their disease association, measured by P-values. Assuming that those haplotypes most significantly associated with disease are also good classifiers, a significance cut-off threshold is applied, and features (i.e. haplotypes) with more significant associations than the cut-off level are included in the prediction model. As part of the QC, we removed those haplotypes occurring at frequencies <10%, and those haplotypes having significantly different frequencies between the discovery and the validation cohorts (Chi-square proportion test P-value <0.05). Following this QC scheme, among the 197 851 haplotypes we analyzed, 20.4% of them were removed, and 157 490 haplotypes were kept in the following analysis.
Classification algorithms
We considered three commonly used classification methods: logistic regression, risk-score logistic regression and SVM. For each haplotype selected, we standardize its direction of risk by choosing the reference haplotype as the one that gives an odds ratio ≥1 such that haplotype coding represents the copies of risk haplotypes present. In the risk-score logistic regression, the risk score is calculated as the number of risk haplotypes an individual carries, both within and across loci:
where xi is the number of the two-SNP risk haplotypes (0, 1, 2) at ith selected haplotype window. The overall risk score serves as a proxy for the risk of a subject developing disease, and it is then treated as the single predictor in the logistic regression framework for model training and validation. It is worth noting that this method essentially assumes that all risk haplotypes contribute equally to disease risk.
For both the SVM regression and logistic regression methods, the binary disease trait (or the logit of the binary trait) is constructed as a linear combination of the predictor haplotypes. Note that these two procedures incorporate information about effect-size estimates of each individual haplotype.
Model evaluation
Janssens et al. (31) advocated that the AUC of the receiver operating characteristic (ROC) curve should be used to assess the performance of prediction models in the large-scale genetic data. And for the published genetic prediction models of many diseases, it has become a standard practice to report the AUC as the measurement of the prediction quality (32–34). Briefly, the ROC curve represents the combination of sensitivity and specificity for each possible cut-off value of the continuous test result that can be considered to define positive and negative test outcomes. It is the probability that given a random pair of individuals, between whom one will develop the disease and the other will not; the classifier will assign the former a positive test result and the latter a negative result. Theoretically, the AUC can take values between 0 and 1, where a perfect classifier will take the value of 1. However, the practical lower bound for random classification is 0.5, and classifiers with an AUC significantly >0.5 have at least some ability to discriminate between cases and controls. Here, we evaluated our prediction model for Crohn's disease using the AUC obtained from the independent validation cohort.
Haplotype pruning
Pruning was achieved via a sliding window approach, with window size of 50, and the number of haplotypes to shift the window at each step is 5. Haplotypes within each sliding window are pruned at the pair-wise correlation threshold of 0.7.
Conflict of Interest statement. Neither this manuscript nor any similar manuscript, in whole or in part, other than an abstract, has been or will be submitted to or published in any other scientific journal by the named authors. All authors are aware of and agree to the content of the paper and their being listed as authors on the paper. There are no financial or other interests with regard to the submitted manuscript that might be construed as a conflict of interest.
FUNDING
This work was supported by the National Institutes of Diabetes and Digestive and Kidney Diseases (NIDDK) Grants (U01 DK062429, U01 DK062422 to J.H.C.), the National Institute of General Medical Sciences (NIGMS) Grant (R01 GM059507 to H.Z.) and the Bohmfalk Foundation at Yale University (to J.H.C.).
REFERENCES
- 1.Loftus E.V., Jr. Clinical epidemiology of inflammatory bowel disease: incidence, prevalence, and environmental influences. Gastroenterology. 2004;126:1504–1517. doi: 10.1053/j.gastro.2004.01.063. doi:10.1053/j.gastro.2004.01.063. [DOI] [PubMed] [Google Scholar]
- 2.Abraham C., Cho J.H. Inflammatory bowel disease. N. Engl. J. Med. 2009;361:2066–2078. doi: 10.1056/NEJMra0804647. doi:10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Satsangi J., Jewell D.P., Bell J.I. The genetics of inflammatory bowel disease. Gut. 1997;40:572–574. doi: 10.1136/gut.40.5.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. doi:10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hampe J., Franke A., Rosenstiel P., Till A., Teuber M., Huse K., Albrecht M., Mayr G., De La Vega F.M., Briggs J., et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 2007;39:207–211. doi: 10.1038/ng1954. doi:10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 6.Libioulle C., Louis E., Hansoul S., Sandor C., Farnir F., Franchimont D., Vermeire S., Dewit O., de Vos M., Dixon A., et al. A novel susceptibility locus for Crohn's disease identified by whole genome association maps to a gene desert on chromosome 5p13.1 and modulates the level of expression of the prostaglandin receptor EP4. PLoS Genet. 2007;3 doi: 10.1371/journal.pgen.0030058. e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rioux J.D., Xavier R.J., Taylor K.D., Silverberg M.S., Goyette P., Huett A., Green T., Kuballa P., Barmada M.M., Datta L.W., et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 2007;39:596–604. doi: 10.1038/ng2032. doi:10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kugathasan S., Baldassano R.N., Bradfield J.P., Sleiman P.M., Imielinski M., Guthery S.L., Cucchiara S., Kim C.E., Frackelton E.C., Annaiah K., et al. Loci on 20q13 and 21q22 are associated with pediatric-onset inflammatory bowel disease. Nat. Genet. 2008;40:1211–1215. doi: 10.1038/ng.203. doi:10.1038/ng.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imielinski M., Baldassano R.N., Griffiths A., Russell R.K., Annese V., Dubinsky M., Kugathasan S., Bradfield J.P., Walters T.D., Sleiman P., et al. Common variants at five new loci associated with early-onset inflammatory bowel disease. Nat. Genet. 2009;41:1335–1340. doi: 10.1038/ng.489. doi:10.1038/ng.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raelson J.V., Little R.D., Ruether A., Fournier H., Paquin B., Van Eerdewegh P., Bradley W.E., Croteau P., Nguyen-Huu Q., Segal J., et al. Genome-wide association study for Crohn's disease in the Quebec Founder Population identifies multiple validated disease loci. Proc. Natl Acad. Sci. USA. 2007;104:14747–14752. doi: 10.1073/pnas.0706645104. doi:10.1073/pnas.0706645104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barrett J.C., Hansoul S., Nicolae D.L., Cho J.H., Duerr R.H., Rioux J.D., Brant S.R., Silverberg M.S., Taylor K.D., Barmada M.M., et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat. Genet. 2008;40:955–962. doi: 10.1038/NG.175. doi:10.1038/ng.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johansen C.T., Wang , J., Lanktree M.B., Cao H., McIntyre A.D., Ban M.R., Martins R.A., Kennedy B.A., Hassell R.G., Visser M.E., et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat. Genet. 2010;42:684–687. doi: 10.1038/ng.628. doi:10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azzopardi D., Dallosso A.R., Eliason K., Hendrickson B.C., Jones N., Rawstorne E., Colley J., Mokvina V., Frye C., Sampson J.R., et al. Multiple rare nonsynonymous variants in the adenomatous polyposis coli gene predispose to colorectal adenomas. Cancer Res. 2008;68:358–363. doi: 10.1158/0008-5472.CAN-07-5733. doi:10.1158/0008-5472.CAN-07-5733. [DOI] [PubMed] [Google Scholar]
- 14.Jakobsdottir J., Gorin M.B., Conley Y.P., Ferrell R.E., Weeks D.E. Interpretation of genetic association studies: markers with replicated highly significant odds ratios may be poor classifiers. PLoS Genet. 2009;5:e1000337. doi: 10.1371/journal.pgen.1000337. doi:10.1371/journal.pgen.1000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu Q., Elston R.C. Using the optimal receiver operating characteristic curve to design a predictive genetic test, exemplified with type 2 diabetes. Am. J. Hum. Genet. 2008;82:641–651. doi: 10.1016/j.ajhg.2007.12.025. doi:10.1016/j.ajhg.2007.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans D.M., Visscher P.M., Wray N.R. Harnessing the information contained within genome-wide association studies to improve individual prediction of complex disease risk. Hum. Mol. Genet. 2009;18:3525–3531. doi: 10.1093/hmg/ddp295. doi:10.1093/hmg/ddp295. [DOI] [PubMed] [Google Scholar]
- 17.Ambroise C., McLachlan G.J. Selection bias in gene extraction on the basis of microarray gene-expression data. Proc. Natl Acad. Sci. USA. 2002;99:6562–6566. doi: 10.1073/pnas.102102699. doi:10.1073/pnas.102102699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simon R., Radmacher M.D., Dobbin K., McShane L.M. Pitfalls in the use of DNA microarray data for diagnostic and prognostic classification. J. Natl Cancer Inst. 2003;95:14–18. doi: 10.1093/jnci/95.1.14. doi:10.1093/jnci/95.1.14. [DOI] [PubMed] [Google Scholar]
- 19.Reunanen J. Overfitting in making comparisons between variable selection methods. J. Mach. Learn. Res. 2003;3:1371–1382. doi:10.1162/153244303322753715. [Google Scholar]
- 20.Kang J., Cho J., Zhao H. Practical issues in building risk-predicting models for complex diseases. J. Biopharm. Stat. 2010;20:415–440. doi: 10.1080/10543400903572829. doi:10.1080/10543400903572829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clark A.G., Weiss K., Nickerson D.A., Taylor S.L., Buchanan A., Stengård J., Salomaa V., Vartiainen E., Perola M., Boerwinkle E., et al. (1998) Haplotype structure and population genetic inferences from nucleotide-sequence variation in human lipoprotein lipase. Am. J. Hum. Genet. 63:595–612. doi: 10.1086/301977. doi:10.1086/301977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaid D.J., Rowland C.M., Tines D.E., Jacobson R.M., Poland G.A. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am. J. Hum. Genet. 2002;70:425–434. doi: 10.1086/338688. doi:10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shim H., Chun H., Engelman C.D., Payseur B.A. Genome-wide association studies using single-nucleotide polymorphisms versus haplotypes: an empirical comparison with data from the North American Rheumatoid Arthritis Consortium. BMC Proc. 2009;3(Suppl. 7):S35. doi: 10.1186/1753-6561-3-s7-s35. doi:10.1186/1753-6561-3-s7-s35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Bakker P.I., Yelensky R., Pe'er I., Gabriel S.B., Daly M.J., Altshuler D. Efficiency and power in genetic association studies. Nat. Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. doi:10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 25.Andres A.M., Clark A.G., Shimmin L., Boerwinkle E., Sing C.F., Hixson J.E. Understanding the accuracy of statistical haplotype inference with sequence data of known phase. Genet. Epidemiol. 2007;31:659–671. doi: 10.1002/gepi.20185. doi:10.1002/gepi.20185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong A., Masson G., Frigge M.L., Gylfason A., Zusmanovich P., Thorleifsson G., Olason P.I., Ingason A., Steinberg S., Stefansson K., et al. Detection of sharing by descent, long-range phasing and haplotype imputation. Nat. Genet. 2008;40:1068–1075. doi: 10.1038/ng.216. doi:10.1038/ng.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraft P., Hunter D.J. Genetic risk prediction—are we there yet. N. Engl. J. Med. 2009;360:1701–1703. doi: 10.1056/NEJMp0810107. doi:10.1056/NEJMp0810107. [DOI] [PubMed] [Google Scholar]
- 28.Mihaescu R., van Hoek M., Sijbrands E.J., Uitterlinden A.G., Witteman J.C., Hofman A., van Duijn C.M., Janssens A.C. Evaluation of risk prediction updates from commercial genome-wide scans. Genet. Med. 2009;11:588–594. doi: 10.1097/GIM.0b013e3181b13a4f. doi:10.1097/GIM.0b013e3181b13a4f. [DOI] [PubMed] [Google Scholar]
- 29.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. doi:10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberts A., McMillan L., Wang W., Parker J., Rusyn I., Threadgill D. Inferring missing genotypes in large SNP panels using fast nearest-neighbor searches over sliding windows. Bioinformatics. 2007;23:i401–i407. doi: 10.1093/bioinformatics/btm220. doi:10.1093/bioinformatics/btm220. [DOI] [PubMed] [Google Scholar]
- 31.Janssens A.C., Gwinn M., Valdez R., Narayan K.M., Khoury M.J. Predictive genetic testing for type 2 diabetes. BMJ (Clin. Res. Ed.) 2006;333:509–510. doi: 10.1136/bmj.38953.598947.80. doi:10.1136/bmj.38953.598947.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyssenko V., Almgren P., Anevski D., Orho-Melander M., Sjogren M., Saloranta C., Tuomi T., Groop L. Genetic prediction of future type 2 diabetes. PLoS Med. 2005;2:e345. doi: 10.1371/journal.pmed.0020345. doi:10.1371/journal.pmed.0020345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lango H., Palmer C.N., Morris A.D., Zeggini E., Hattersley A.T., McCarthy M.I., Frayling T.M., Weedon M.N. Assessing the combined impact of 18 common genetic variants of modest effect sizes on type 2 diabetes risk. Diabetes. 2008;57:3129–3135. doi: 10.2337/db08-0504. doi:10.2337/db08-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meigs J.B., Shrader P., Sullivan L.M., McAteer J.B., Fox C.S., Dupuis J., Manning A.K., Florez J.C., Wilson P.W., D'Agostino R.B., Sr., et al. Genotype score in addition to common risk factors for prediction of type 2 diabetes. N. Engl. J. Med. 2008;359:2208–2219. doi: 10.1056/NEJMoa0804742. doi:10.1056/NEJMoa0804742. [DOI] [PMC free article] [PubMed] [Google Scholar]