Abstract
Doxorubicin-induced cardiomyopathy in cancer patients is well established. The proposed mechanism of cardiac damage includes generation of reactive oxygen species, mitochondrial dysfunction and cardiomyocyte apoptosis. Exposure of adult rat cardiomyocytes to low levels of DOX for 48 h induced apoptosis. Analysis of protein expression showed a differential regulation of several key proteins including the voltage dependent anion selective channel protein 2 and methylmalonate semialdehyde dehydrogenase. In comparison, proteomic evaluation of DOX-treated rat heart showed a slightly different set of protein changes that suggests nuclear accumulation of DOX. Using a new solubilization technique, changes in low abundant protein profiles were monitored. Altered protein expression, modification and function related to oxidative stress response may play an important role in DOX cardiotoxicity.
Keywords: Doxorubicin, cardiomyopathy, proteomics
1. Introduction
The anthracycline antibiotic, doxorubicin (DOX), is an anticancer drug that is used widely in treating leukemias, lymphomas, lung, breast, ovary and uterine cancers. However, DOX chemotherapy causes a cumulative and delayed cardiomyopathy in cancer patients (2% in 2-year versus 5% in 15-year survivors) as a major side effect [1,2]. Human case study reports and preclinical data in animal models indicate that DOX treatment caused mitochondrial damage and decrease in ATP / ADP ratio [3]. The proposed mechanism of cardiotoxicity involves free radical generation [4], lipid peroxidation [5], cardiomyocyte apoptosis [6,7] and somatic mitochondrial alteration and dysfunction [8-10]. DOX undergoes a one-electron redox cycling at the complex I site of the mitochondrial electron transport chain (ETC), forming a semiquinone that reduces molecular oxygen to superoxide that dismutates to form hydrogen peroxide [11-13]. DOX can be reduced by a variety of flavoproteins including cytochrome P450 reductase, mitochondrial NADH (nicotinamide adenine dinucleotide – reduced) dehydrogenase and nitric oxide synthase (reductase domain) [12,14]. In the presence of redox-active metal ions (iron and copper), highly reactive hydroxyl radicals or perferryl iron are formed from DOX redox activation [15]. These oxidants target lipid, protein, and DNA causing extensive modifications [16-18]. Reactive oxygen species formed from DOX redox cycling also affect iron uptake and iron homeostasis [19,20], inhibit aconitase and iron regulatory protein (IRP) function and alter the transferrin / ferritin mRNA (messenger ribonucleic acid) levels. Another mechanism by which aconitase was inactivated is via the interaction with the secondary alcohol derivative of DOX [21,22].
Doxorubicin caused a dose-dependent cell death in isolated adult rat cardiomyocyte [23,24]. Studies with antioxidants [25] and iron chelators [26] suggest that mechanisms other than peroxidative stress contribute to cardiac impairment in DOX chemotherapy [27,28]. In addition, DOX accumulates in the nucleus causing DNA damage. DOX also selectively inhibits gene expression and interferes with the transcription factor's ability to bind to DNA due to formation of GC adducts at the 5’-UTRs (untranslated regions). We have previously demonstrated the critical role of activated transcription factor kappa-B in DOX-mediated apoptosis in endothelial cells and in isolated adult rat cardiomyocyte [29-33]. The DNA repair process in DOX-treated cells is different from that of hydrogen peroxide-treated cells [34-36]. Overall, published data suggest that oxidative stress appears to play a key role in DOX-mediated cardiotoxicity and apoptotic myocyte death. In this study, we report the protein expression changes observed in isolated cardiomyocyte and adult rat heart during DOX treatment and also provide a methodological improvement that identifies changes in non-abundant protein levels.
2. Materials and methods
2.1. Reagents
All reagents of the highest quality commercially available were purchased. TRIS base, glycine, sodium dodecyl sulfate (SDS), doxorubicin-HCl, dithiotheritol (DTT), Urea, 3-[(3-Cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS), ammonium bicarbonate, α-cyano-4-hyroxycinnamic acid (CHCA), deoxyribonuclease (DNAse), bovine serum albumin (fraction V), 80 mesh screen filter, medium 199, iodoacetamide, creatine, taurine, insulin, cytosine β-D-arabinofuranoside, vitamin solution (100x), sodium deoxycholate, Triton x-100, glycerol, urea, thiourea, ethylenediaminetetraacetic acid (EDTA), ethyleneglycoltetraacetic acid (EGTA), diethylene triamine pentaacetic acid (DTPA), phenylmethylsulphonyl fluoride (PMSF), aprotinin, leupeptin, β-glycerophosphate, sodium fluoride, sodium orthovanadate, sodium pyrophosphate, 4-(2-Hydroxyethyl)-1-piperazineethane sulfonic acid (HEPES), sucrose, mannitol, calcium chloride (CaCl2), tween-20, acetonitrile (ACN), dimethyl sulfoxide (DMSO), acetone and HPLC pure water were purchased from Sigma-Aldrich Co. USA. DMEM, fetal bovine serum (FBS), collagenase (type II, the preparation with low collagenase activity of 200-250U/mg) and Dulbecco phosphate buffered saline (DPBS), complete amino acid mixture (50x) and MEM non-essential amino acid solution (100x) were from Gibco, USA. Immobilized IPG (3-10) strips, Bio-lyte 3-10 ampholytes and prestained protein molecular weight marker were purchased from Biorad, USA. 2D-quant kit and advanced ECL kit were purchased from GE Life Sciences. Syringe filters (5 μ) and Zip-Tip C18 was purchased from Millipore, USA. All other general chemicals such as potassium dihydrogen phosphate (KH2PO4), sodium bicarbonate (NaHCO3), sodium chloride (NaCl), potassium chloride (KCl), magnesium sulfate (MgSO4) were purchased from Fisher Scientific, USA.
2.2. Buffers
All buffers involved in cardiomyocyte isolation were prewarmed to 37°C before use. Perfusion buffer contains 25 mM HEPES buffer, 110 mM NaCl, 11 mM glucose, 1 mg/mL BSA, 5 mM creatine, 20 mM taurine, 2.6 mM KCl, 1.2 mM MgSO4, 20 μl/ml of 50x complete amino acid mixture, 10 μl of 100x stock non-essential amino acid mixture, 10 μl/mL of 100x stock of vitamin solution and 1.2 mM KH2PO4 and 5 mM pyruvate sodium (pH 7.4). The pH of the perfusion buffer was adjusted to 7.4 and sterile-filtered using 0.2 μm cellulose acetate filter. Digestion buffer contain perfusion buffer, 25 μM CaCl2 and 200 U/mL of collagenase [37]. Isolation buffer contains digestion buffer, 20 μg/mL of DNAse and 1% BSA. Wash buffers I, II and III are made up of perfusion buffer containing 200 μM, 500 μM and 1 mM CaCl2 , respectively. Isolated cardiomyocytes were cultured in M-199 medium containing 25 mM NaHCO3, 25 mM HEPES, 2 mg/mL BSA, 0.5 mM creatine, 5 mM taurine, 0.1 μM insulin, 10 μM cytosine β-D-arabinofuranoside, 10% FBS, 100 U/mL penicillin and 100 μg/mL streptomycin. Protease inhibitor cocktail contains 5 mM EDTA, 2 mM EGTA, 5 mM DTPA, 2 mM PMSF, 5 μg/mL aprotinin, 5 μg/mL leupeptin, 1 mM β-glycerophosphate, 1 mM sodium fluoride, 1 mM sodium orthovanadate, 2.5 mM sodium pyrophosphate and 5 mM DTT. Radio-immunoprecipitation assay (RIPA) buffer was made with 50 mM Tris pH 7.6, 150 mM NaCl, 1% triton x-100, 1% sodium deoxycholate, 0.1% SDS and protease inhibitor cocktail. Rehydration buffer is made up of 7 M urea, 2 M thiourea, 4% CHAPS, 0.2% ampholytes (3-10) and 100 mM DTT. Equilibration buffer I is made up of 6 M urea, 375 mM Tris-HCl, pH 8.8, 2% SDS, 20% glycerol and 2% DTT. Equilibration buffer II is made up of 6 M urea, 375 mM Tris-HCl, pH 8.8, 20% glycerol and 2.5% idoacetamide. Cytoplasmic extraction buffer is made up of 1x PBS pH 7.0 containing 0.1% digitonin and protease inhibitor cocktail. Membrane fraction I extraction buffer is made up of 1x PBS pH 7.0 containing 0.3% triton x-100 and protease inhibitor cocktail. Membrane fraction II extraction buffer is made up of 1x PBS pH 7.0 containing, 1% triton x-100 and protease inhibitor cocktail. Filamentous protein extraction buffer is made up of 1x PBS pH 7.0 containing 0.5% triton x-100 and 0.6 M KCl and protease inhibitor cocktail.
2.3. Adult rat cardiomyocyte isolation and culture
The ventricular cardiomyocytes were isolated according to a previously described method [38,39] that was modified in our laboratory. Adult male Sprague-Dawley rats weighing approximately 180-220 g were anesthetized with intraperitoneal injections of pentobarbital (60 mg/kg body weight) followed by intraperitonial injection of 250 U/kg of heparin. The heart was excised, mounted on aortic cannula and perfused (in a Langendorff non-recirculating mode) with the oxygenated perfusion buffer supplemented with 1 mM CaCl2. After 15 min of perfusion, it was switched to calcium-free oxygenated buffer in a non-recirculating mode for 5 min followed by recirculation at a constant flow rate for 30 min. At the end of perfusion the ventricular tissue was dissected, minced and incubated with gentle shaking in the isolation buffer at 37°C for 10 min. The single cells released were dispersed from the remaining undigested tissue by mixing the suspension with a 5 ml serological pipette, and the cells were collected using 80 mesh screen filter. The cells were washed by centrifugation at 150 rpm in succession with wash buffers I, II, and III and resuspended in a small volume of wash buffer III. This cell suspension was layered on a 4% BSA - 1 mM CaCl2 cushion and centrifuged at 200 rpm for 2-3 min. Ventricular myocytes settled down and the resulting cell pellet was resuspended in growth medium and plated. Cardiomyocytes were separated from other cells by differential plating at 1.5 × 106 cells / mL on to laminin coated 150 mm dishes. This isolation procedure generally yields 75-80% rod-shaped cardiomyocyte [40] that exclude trypan blue.
2.4. Doxorubicin treatment of cardiomyocyte and isolation of cell lysate
The isolated cardiomyocytes were placed in the incubator at 37°C with 5% CO2 for overnight incubation. DOX was added to the medium at 0.5 μM concentration and the cells were lysed in RIPA buffer at 0, 24, or 48 h after treatment. At these concentrations the cells did not release significant lactate dehydrogenase (marker for cellular damage) [40] but exhibit apoptosis as seen by a large amount of floating cells and positive TUNEL assay [41]. The cell lysate was left to stand on ice for 30 min and centrifuged at 4°C at 13,000 rpm for 30 min. The clear supernatant was transferred to a fresh tube and aliquots in triplicate were analyzed for protein concentration using 2D-quant kit (bicinchoninic acid method) using BSA. Protein concentration equivalent to 600 μg was precipitated by incubating with 8 volumes of ice cold acetone and left to stand at -20°C overnight. The precipitated protein was collected by centrifuging at 4°C at 13,000 rpm for 30 min, and the pellet washed with ice cold acetone twice. The final pellet was dried under vacuum and was stored at -80°C until use.
2.5. Fractionation of cell lysate
For fractionation experiments, DOX-treated (0.5 μM for 48 h and 20 μM for 24 h) and untreated control cells (vehicle for 48 h) were collected in 1x DPBS and washed twice in the same buffer. The cell pellet was resuspended in 2 mL of cytoplasmic extraction buffer, incubated on ice for 1 hr and spun at 16,000 rpm for 15 min at 4°C. The resultant supernatant was spun at 100,000x g for 20 min at 4°C (supernatant collected). The pellets from 16,000 rpm and 100,000x g were pooled and reextracted with membrane fraction I extraction buffer and spun at 16,000 rpm for 20 min at 4°C (supernatant collected). The resultant pellet was extracted with membrane fraction II extraction buffer and spun at 16,000 rpm for 20 min at 4°C (supernatant collected). The final pellet was again extracted with filamentous protein extraction buffer and spun at 16,000 rpm for 20 min at 4°C (supernatant collected). Protein estimation of the extracts was performed as above.
2.6. DOX-treated adult rat
Animal experiments were performed in accordance with the animal protocol approved by the Medical College of Wisconsin Animal Care and Use Committee. Adult male Sprague-Dawley rats were segregated into groups as control (n=16) and DOX-treated (n=11). The control group was injected through the tail vein with saline and the DOX-treated group received 2.5 mg/kg of DOX in saline. Echocardiographic monitoring of cardiac function was performed at various time points and the animals were sacrificed at 8 wks with intraperitoneal injection of sodium pentabarbitol (100 mg/kg body weight). The heart tissue was collected, flash frozen and stored at -80°C for further analysis. Approximately 500 mg of tissue was homogenized in RIPA buffer, protein estimated, and sample processing for 2D electrophoresis was performed as described above.
2.7. 2D gel electrophoresis and silver staining
To each 600 μg of the dried precipitated protein extract, 600 μl of rehydration buffer was added to solubilize the protein, and 300 μl (equivalent to 300 μg protein sample) of this sample was loaded on 17 cm IPG strips (pH 3-10, 0.5 mm thick and 4% T / 3% C composition). Isoelectric focusing was performed using Protean IEF system (Biorad). After initial rehydration at 20°C for 16 h and at 50 V constant, isoelectric focusing was done in three stages. The settings were 20°C, 250 V for 0.5 h (rapid ramping – conditioning step), 10,000 V for 3 h (linear ramping – voltage ramping step) and 10,000 V for 60,000 V-hr (rapid ramping – final focusing step) with maximum current set at 50 μA/gel strip. After the first dimensional run the gels were washed to remove the excess mineral oil, and reduced using equilibration buffer I for 15 min at room temperature, followed by alkylation using equilibration buffer II for 15 min at room temperature in the dark. Finally the gels were washed and equilibrated in the Laemmli SDS-running buffer (Tris / glycine / SDS) and layered on a 12% SDS-PAGE gel. The gel was sealed with 1% agarose containing 0.001% Bromophenol blue dye and ran at 35 mA/gel constant and at 4°C using precooled (4°C) running buffer. Gels were stained with modified silver staining method [42] that is compatible with mass spectroscopic analysis. Briefly, after running the gels, they were marked and fixed overnight at 4°C with shaking in fixative solution. The gels were sensitized with sensitizer containing 30% methanol, 4% sodium thiosulphate solution (stock – 50 mg/mL) and 68 g/L sodium acetate for 30 min at room temperature. The gels were washed with water thrice for 10 min each, followed by silver solution (2.5 g/L silver nitrate) treatment for 25 min at room temperature. This was followed by water wash thrice and developer (25 g/L sodium carbonate and 0.04% formaldehyde solution) until all spots were developed, and the reaction was terminated using 14.6 g/L EDTA for 10 min followed by water wash. The stained gels were documented using a Kodak gel documentation system.
2.8. Mass spectrometry - MALDI-TOF detection
2.8.1. In-gel digestion
Spots of interest were punched out, transferred to fresh tubes and trypsin digested according to [43] with modifications. The gel pieces were destained using 15 mg/mL of sodium thiosulphate and 10 mg/mL of potassium ferricyanide (1:1 ratio) for 30 min at room temperature. The destained slices were washed twice with 50 mM ammonium bicarbonate, reduced, and S-alkylated with 20 mM DTT (20 min at 60°C) and 100 mM iodoacetic acid (20 min at room temperature in the dark) in 50 mM ammonium bicarbonate. The gel slices were washed and dehydrated with ACN and dried under vacuum. The dried gels were rehydrated with 12.5 ηg/μl trypsin and allowed to swell for 1 h on ice. Finally the excess buffer was removed and trypsin-free buffer was added and allowed to digest overnight at 37°C. The digested peptides were eluted with 0.1% TFA followed by 50% ACN in 0.1% TFA and dried down using vacuum. The dried material was resuspended in 0.1% TFA and used for the desalting procedure.
2.8.2. Sample preparation
Matrix solution was prepared using recrystallized α-CHCA at 10 mM to prepare saturated solution in 50% ACN / 0.1% TFA (1:1) by vortexing and centrifuging at 13,000 rpm for 5 min at room temperature. The clear supernatant was transferred to a new amber tube and used for co-crystallizing the peptides. The in-gel digested peptides were desalted using C18 reverse phase Zip-Tips according to manufacturer's protocol. After the final wash, the bound peptides were eluted using the matrix solution and directly deposited onto the MALDI target plate. The plate was dried and immediately used for data collection.
Recrystallization of CHCA was done by dissolving approximately 5 mg/mL in ultrapure ethanol and warmed to 40°C for 2 h with stirring. The bright yellow liquid was filtered using Whatman # 1 paper and diluted to 50% with ultrapure water and left overnight at 4°C. CHCA precipitates out of the solution. The precipitated material was collected by filtering and vacuum dried.
2.8.3. Data collection
Mass spectroscopic data collection was done using Applied Biosystem's PerSeptive voyager DE-PRO mass spectrometer (MALDI-TOF). Data was collected at 3 GHz laser repetition rate, in reflector mode, positive polarity, delayed extraction (resolution = 8000) mode at 25 kV accelerating voltage. The ion signals were recorded under delay time, and grid voltages set depending on the standards for accurate m/z values (routinely the extraction delay time is at 150-180 ηsec, grid voltage at 70-77%, and grid wire voltage set at 0.01-0.02%). Nitrogen laser pulse at 337 nm was used for sample ionization and 200 laser shots / spectrum were averaged. After correcting for background and noise levels the m/z values of the collected spectrum were externally calibrated with bradykinin (monoisotopic [M+H]+ - 1060.57 Da), angiotensin I (monoisotopic [M+H]+ -1296.69 Da), ACTH (1-17) - (monoisotopic [M+H]+ - 2093.08 Da), ACTH (18-39) – (monoisotopic [M+H]+ - 2465.20 Da) and internally calibrated using known trypsin auto digest peptides (3 point calibration, m/z 842.51, m/z 1045.564 and m/z 2211.105). The trypsin that is used in our in-gel digestion protocol is TPCK (N-p-tosyl-L-phenylalanine chloromethyl ketone) treated and reductively methylated to limit auto digestion. An empty gel spot was always digested with excess of trypsin in order to identify the trypsin auto digest peptides under our experimental conditions.
2.8.4. Database search
Database search was done with MASCOT (Matrix Science) peptide mass fingerprint software with peptides in the monoisotopic [M+H]+ mass range of 800-3500 Da. The target protein was identified using NCBI database, taxonomy restricted to Rattus, missed cleavage set at maximum of four (in-gel digestions are often incomplete), enzyme chosen, cysteine modification with iodoacetamide and monoisotopic tolerance at 50 ppm. This protocol had consistently given us correct identification of known standards, and therefore was applied for identifying unknown peptides. Any identification with more than 6 peptides matching, and at least 3 overlapping peptides, were considered positive identification in order to eliminate bias and maintain stringency in data analysis. The masses of those fragments were not included in the search if the isotopic pattern did not follow the typical mass range pattern. Each fragment mass pattern was individually analyzed manually prior to accepting for inclusion in database search, and the known trypsin peptides were manually eliminated from the search.
3. Results
3.1. Proteome changes in DOX-treated adult rat cardiomyocytes
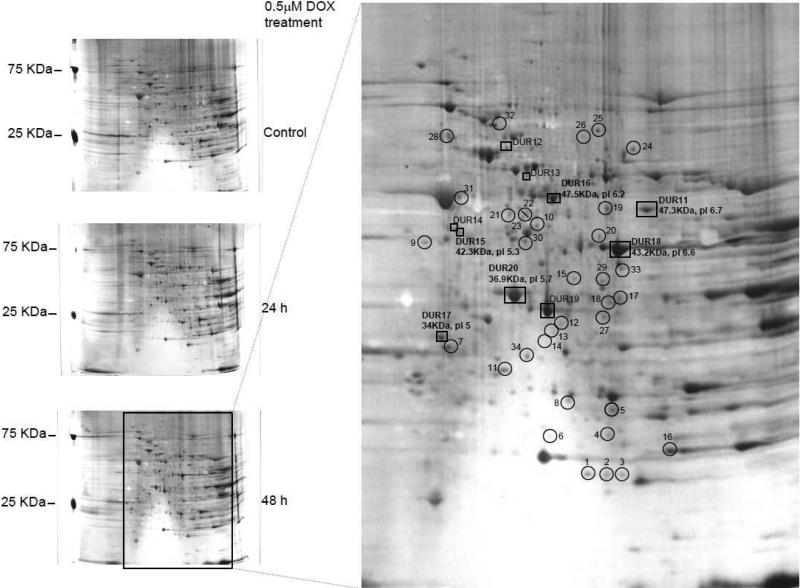
The changes in protein levels in doxorubicin-treated cardiomyocytes were documented by using the 2D gel analysis (Figure 1). We have previously shown that treatment with DOX at clinically relevant concentrations (0.5 μM) induced ROS production and subsequently apoptosis in a caspase-3-dependent manner. We analyzed protein expression changes using 2D gel resolution in isolated adult rat cardiomyocytes under the same conditions. Significant changes occurred after a 48 h DOX treatment, as compared with untreated control and 24 h DOX treatment. Proteins identified include those that are involved in apoptosis, energy metabolism, stress response and signaling. We identified four types of protein changes that were either upregulated, differentially upregulated, differentially downregulated, or downregulated after DOX treatment. The upregulated proteins were detected only in DOX-treated cells but not in control cells, representing treatment response proteins and modified proteins. Differentially upregulated proteins were those that were present at higher quantities in DOX-treated cells; the differentially downregulated were quantitatively less in DOX-treated cells. The differentially regulated proteins could also be representing a group of stress response proteins that are different from the upregulated or downregulated proteins. Finally, the downregulated proteins (minor) were lost due to DOX treatment. These changes were pronounced at 48 h treatment as shown in Figure 1 and to the best of our knowledge, this represents the first documented DOX induced changes in proteome in adult cardiomyocytes.
Figure 1. Upregulated and differentially upregulated proteins in DOX-treated adult rat cardiomyocytes.
Silver stained 12% SDS-PAGE (first dimension pI 3-10) analysis of adult rat cardiomyocyte cultured for 48 h without treatment (control), 0.5 μM DOX treatment for 24 h and 0.5 μM DOX treatment for 48 h. Spots are marked as upregulated and differentially upregulated (DUR) protein. Upregulated proteins are those protein spots that are present in the 48 h DOX treatment and not present in the untreated control gel and DUR are the proteins present in both untreated and treated cells but present quantitatively more in the DOX treatment. Some of the spots were represented in bold to indicate the pI / molecular weight matches across the gel.
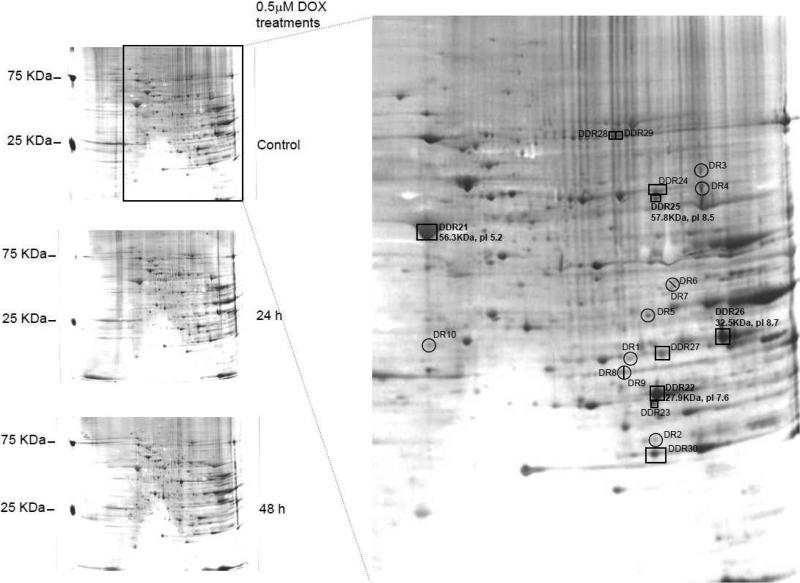
Figure 2 demonstrates differentially downregulated and downregulated proteins. There were several spots that yielded very good mass spectral data (not shown) but did not yield accurate identification. It is likely that the spot represents a complex mixture of modified proteins. Table 1 indicates that the proteins identified are stress responsive (ATP synthase, enolase alpha, Alpha B-crystallin, translocation protein 1, Stress induced phosphoprotein 1) or apoptotic / cell damage markers (p38 alpha, lipocortin, voltage dependent anion selective channel protein 2, creatine kinase, MTUS1). Also, F1-ATP synthase upon DOX treatment was observed in differentially downregulated (P10719), differentially upregulated (AAA40778) and upregulated (AAA40778) list. This is due to the fact that protein P10719 is the precursor of protein AAA40778 that probably undergoes modification (differentially upregulated) and cleavage (differentially downregulated). The amount of the observed protein that in the modified or cleaved form were quantitatively very small as compared to the P10719, indicating the occurrence of a dynamic process. Another differentially downregulated protein, ETF-β (Spot 22, Table 1), might also be downregulated through a putative nitrative modification on the single tyrosine residue (data not shown).
Figure 2. Downregulated / differentially downregulated proteins in DOX-treated adult rat cardiomyocytes.
Silver stained 12% SDS-PAGE demonstrates the differentially downregulated (DDR) and the downregulated (DR) protein changes in DOX-treated cardiomyocytes. DR proteins are proteins present only in the untreated control but not in 48 h DOX treatment and DDR are the proteins present in both untreated and treated cells but present quantitatively more in the control gels.
Table 1. Protein changes in DOX-treated adult rat cardiomyocytes.
Proteins identified to undergo changes due to 48 h DOX treatment. Some of the unknown proteins are included since their mass spectrum is very good yet the data base search does not yield a significant ID.
| Accession # | Molecular function | Matched / Measured peptides | score | ||
|---|---|---|---|---|---|
| Upregulated proteins (DOX treated cardiomyocytes) | |||||
| Spot # 1 | Alpha B-crystallin (19.9KDa, pl 6.8) | CAA42911 | Structural protein / Molecular chaperone | 11 / 46 - 22% coverage | 68 |
| Spot # 2 | Alpha B-crystallin (19.9KDa, pl 6.8) | AAP31996 | Structural protein / Molecular chaperone | 11 / 46 - 22% coverage | 68 |
| Spot # 3 | Potassium dependent sodium-calcium exchanger (NCKX2) (19.7KDa, pl 6.5) | NP_113931 | Transporter | 26/44 - 54% coverage | 212 |
| Spot # 4 | DNA damage inducible transcript 4-like / SMHS1 (Ddit4) (21.9KDa, pl 6.8) | NP_536324 | Energy metabolism / apoptosis regulator | 9 / 33 - 12% coverage | 64 |
| Spot # 9 | Troponin-T (TnnT2) (35.7KDa, pl 5) | NP_036808 | Actin binding motor protein | 16 / 46 -36% coverage | 116 |
| Spot # 11 | Protease (prosome, macropain) 28 subunit, alpha (PSME1) (28.8KDa, pl 5.6) | AAH60574 | Proteosome activator | 6 / 42 - 17% coverage | 74 |
| Spot # 19 | Translocation protein 1 / AB2-292 (Tloc1, 45.9KDa, pl 6.9) | NP_001029301 | Protein transporter activity | 8/28 - 16% coverage | 71 |
| Spot # 22 | MTSG1 346 amino acid isoform (Mtusl, 40.2KDa, pl 5.8) | AAO60218 | Mitochondrial tumor suppressor gene may be involved in regulating cell proliferation | 17 / 42 - 34% coverage | 124 |
| Spot # 23 | MTSG1 346 amino acid isoform (Mtusl, 40.2KDa, pl 5.8) | AAO60218 | Mitochondrial tumor suppressor gene may be involved in regulating cell proliferation | 17 / 43 - 34% coverage | 124 |
| Spot # 24 | Lamin C2 (Lmna, 52.6KDa, pl 6.2) | CAA67641 | Intermediate filament (structural protein) | 9 / 29 - 20% coverage | 194 |
| Spot # 25 | Electron transfer flavoprotein ubiquinone oxidoreductase precursor (ETFDH, 68.1KDa, pl 7.4) | AAQ67364 | Oxidoreductase | 8 / 42 - 16% coverage | 71 |
| Spot # 26 | Stress induced phosphoprotein I (Stipl, 63.2 Kda, pl 6.4) | AAH61529 | Chaperone | 14 / 63 - 22% coverage | 92 |
| Spot # 29 | MAP kinase 14 / p38 alpha (MAPK14, 40KDa, pl 5.9) | NP_112282 | Non receptor serine threonine protein kinase | 12 / 36 - 28% coverage | 107 |
| Spot # 31 | F1-ATPase beta subunit (ATP5b, 38.8KDa, pl 5.1) | AAA40778 | Ion channel hydrogen transporter | 15 / 29 - 45% coverage | 170 |
| Spot # 34 | Lactate dehydrogenase B (Ldhb, 36.9KDa, pl 5.7) | AAH59149 | Dehydrogenase (oxidoreductase) | 10 / 44 - 26% coverage | 64 |
| Differentially upregulated proteins (DOX treated cardiomyocytes) | |||||
| DUR # 11 | Beta enolase (ENO3, 47KDa, pl 7.53) | CAA68788 | Lyase | 19 / 48 - 40% coverage | 173 |
| DUR # 14 | F1- ATPase beta subunit (ATP5b, 56.3KDa, pl 4.9) | AAA40778 | ATP synthase | 9 / 48 - 24% coverage | 104 |
| DUR # 15 | Actin gamma -2 (Actg2, pl 5.2) | NP_037025 | Actin and actin related protein | 7 / 38 - 17% coverage | 97 |
| DUR # 16 | Enolase alpha (Eno1, pl 6.5) | AAH78896 | Lyase | 18 / 38 - 43% coverage | 176 |
| DUR # 17 | Lipocortin V (Anxa5, pl 4.65) | AAC06290 | Transfer/carrier protein | 10 / 32 - 26% coverage | 158 |
| DUR # 18 | Creatine kinase (Ckm, 43KDa, pl 7.1) | AAA40936 | Transferase | 7 / 46 - 14% coverage | 148 |
| DUR # 20 | Lactate dehydrogenase B (Ldhb, pl 5.92) | AAH59149 | Dehydrogenase | 8 / 44 - 22% coverage | 159 |
| Differentially downregulated proteins (DOX treated cardiomyocytes) | |||||
| DDR # 21 | ATP synthase, H+ transporting, mitochondrial F1 complex, beta precursor (Atp5b, 56.3KDa, pl 4.92) | P10719 | ATP synthase | 21 / 45 - 60% coverage | 209 |
| DDR # 22 | Electron transfer flavoprotein beta subunit (ETF-β, pl 7.9) | NP_001004220 | Hydroxylase | 11 / 29 - 44% coverage | 110 |
| DDR # 25 | Methylmalonate semialdehyde dehydrogenase (MMSDH, ALDH6A1, 57.8KDa, pl 7.9) | Q02253 | Aldehyde dehydrogenase member | 16 / 30 - 34% coverage | 163 |
| DDR # 26 | Voltage dependent anion selective channel protein 1 (Vdac1, pl 8.6) | AAH72484 | Anion channel / Voltage gated ion channel | 10 / 29 - 54% coverage | 201 |
| Downregulated proteins (DOX treated cardiomyocytes) | |||||
| DR # 27 | Voltage dependent anion selective channel protein 2 (Vdac2, 31.8KDapl 7.5) | P81155 | Anion channel / Voltage gated ion channel | 8 / 77 - 34% coverage | 74 |
| DR # 28 | Pleckstrin homology domain containing family A (PI binding specific) member 8 (Plekha8, pl 4.7) | NP_001102705 | Nucleic acid binding / Transfer protein | 6 / 41 - 28% coverage | 81 |
Protein score is -10*log(P) where P is the probability that the observed match is a random event.
Protein scores greater than 61 are significant (P<0.05).
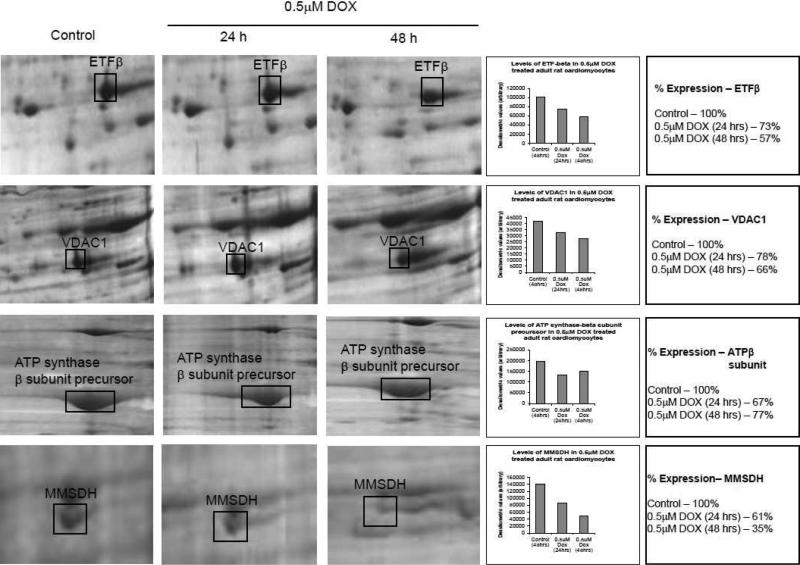
Figure 3 shows differentially downregulated protein displayed on 2D gel, along with the densitometric scan to focus on the level of expression changes (due to the limitation of silver staining technique, the data is to be considered representation of the gels shown here). It is evident from this data that the effect of DOX induced proteome changes were profound at 48 h treatment under the conditions described. The largest drop in the expression is MMSDH involved in valine and pyrimidine catabolic pathway.
Figure 3. Quantitative evaluation of DOX-treated adult rat cardiomyocytes.
Representative changes in protein levels with DOX treatment for 24 h and 48 h as compared to control. The changes are also expressed as densitometric quantification values (average of two independent assays). The spots were quantified from 300 dpi images from the Kodak gel documentation system. Prior to staining, the gels were run in duplicate and stained in an identical manner.
3.2. Proteome changes in DOX-treated adult rat model
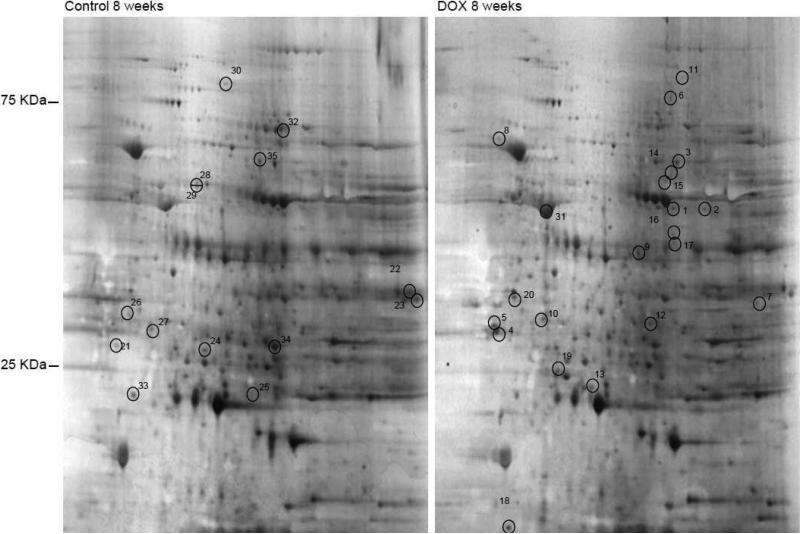
In order to better understand the proteome changes observed in the DOX-treated cardiomyocyte and DOX-treated adult rats, adult male rats were injected with DOX over a period of 8 weeks. The analysis of the heart proteome revealed that although the protein profile are strikingly similar on a 2D gel, the protein changes occurring are somewhat different than those recorded in the cellular model (Figure 4). MALDI-TOF analyses of the proteins indicated that major protein changes involve stress response proteins (Table 2). The upregulated proteins in the animal model included molecular chaperone, hydroxylase, oxidoreductases, cytoskeletal proteins, kinases, phospholipid transporter, endopeptidase and transcription coactivator. The common protein changes between the in vitro cellular model and the in vivo animal model are overexpression of beta enolase, troponin and alpha B-crystallin. Other major protein changes were quite different from those identified in the cell culture model.
Figure 4. Proteome changes in DOX-treated adult rat.
Adult rats were treated with weekly injections of saline or DOX for 8 weeks as described in materials and methods. The heart tissue was collected at the end of experiment and whole tissue extract was prepared and resolved on a 2D gel and silver stained. Protein concentration equivalent to 300 μg was used for each gel.
Table 2. Protein changes in DOX-treated adult rat.
Protein changes in 8 weeks DOX-treated and control animal model. Downregulated proteins were fewer than the upregulated or modified proteins.
| Accession # | Molecular function | Matched / Measured peptides | Score | ||
|---|---|---|---|---|---|
| Upregulaled proteins (DOX treated adult rat) | |||||
| Spot # 1 | Alpha B-crystallin | CAA42911 | Structural protein / Molecular chaperone | 5 / 32 - 28% coverage | 70 |
| Spot # 3 | Similar to β-enolase | CAA68788 | Lyase | 13 / 36 - 28% coverage | 132 |
| Spot # 4 | 14-3-3 protein ε-chain | A60031 | Cytoplasmic chaperones / Activator of tyrosine and tryptophan hydroxylases | 8 / 53 - 36% coverage | 116 |
| Spot # 5 | Tyrosine 3-monooxgenase / tryptophan 5-monooxgenase activation protein, β-polypeptide | AAH76502 | Cytoplasmic chaperones / Activator of tyrosine and tryptophan hydroxylases | 6 / 59 - 24% coverage | 68 |
| Spot # 6 | Catalase | P04762 | Aminoacylase activity / Oxidoreductase activity | 5 / 28 - 22% coverage | 67 |
| Spot # 7 | Actin | NP_062056 | Structural protein | 22 / 47 - 45% coverage | 170 |
| Spot # 8 | H+-transporting two-sector ATPase β-chain, mitochondrial - rat (fragment) | A28701 | ATP-Synthase | 12 / 36 - 35% coverage | 122 |
| Spot # 9 | Aldose reductase (AR) (Aldehyde reductase) | P07943 | Aldehyde reductase / Oxidoreductase | 5 / 55 - 12% coverage | 71 |
| Spot # 10 | Proteasome subunit-α type 3 (Proteasome component C8) | P18422 | Endopeptidase (threonine type) | 5 / 12 - 20% coverage | 84 |
| Spot # 11 | Macropain / Transactivating protein BRIDGE | AAD32925 | Transcription co-activator | 7 / 43 - 26% coverage | 74 |
| Spot # 12 | Macropain / Transactivating protein BRIDGE | AAD32925 | Transcription co-activator | 7 / 47 - 26% coverage | 74 |
| Spot # 13 | Apolipoprotein A-I precursor (Apo-AI) | CAA25224 | Phospholipid transporter activity | 7 / 31 - 25% coverage | 71 |
| Spot # 14 | Creatine kinase, sarcomeric mitochondrial precursor (S-MtCK) | P09605 | Kinase / Nucleotide binding | 9 / 65 - 19% coverage | 63 |
| Spot # 15 | Similar to CCAAT/enhancer binding protein-α | CAA31242 | Transcription factor | 6 / 42 - 22% coverage | 68 |
| Spot # 16 | Lactate dehydrogenase | P42123 | Oxidoreductase | 14 / 44-28% coverage | 98 |
| Spot # 18 | ATP synthase beta subunit | AAB02288 | ATP-Synthase | 12 / 42-35% coverage | 128 |
| Spot # 20 | Neurofilament triplet L protein | A21762 | Cytoskeleton | 10 / 74 - 31% coverage | 104 |
| Spot # 31 | Troponin T2 | NP_036808 | Cytoskeleton / Protein binding | 15 / 32 - 36% coverage | 92 |
| Downregulated proteins (DOX treated adult rat) | |||||
| Spot # 24 | HSP27 | NP_114176 | Heat shock protein / Anti-apoptosis | 7 / 44-11% coverage | 64 |
| Spot # 35 | Phosphocholine cytidylyltransferase A | P19836 | Regulatory rate limiting enzyme | 11 / 38-22% coverage | 86 |
Protein score is -10*log(P) where P is the probability that the observed match is a random event.
Protein scores greater than 61 are significant (P<0 05).
3.3. Proteome analysis of DOX-treated cardiomyocytes with prefractionation of the cell lysate
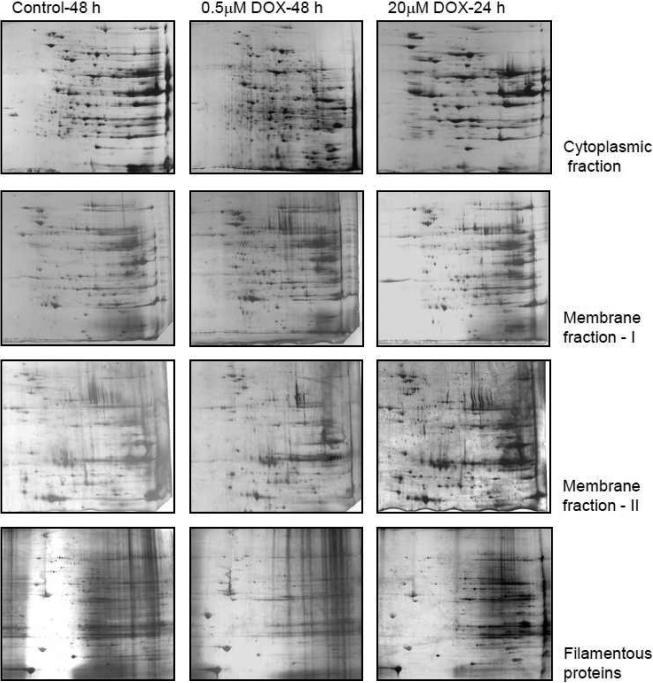
The above models indicate that there are significant differences in proteome changes in cellular and in vivo models of DOX cardiomyopathy. We investigated fractionation of protein by solubilization technique in order to enrich less abundant proteins which might be difficult to identify by traditional methods. In our solubilization methods and 2D gel analysis of our cellular model, we were able to resolve many more proteins that were not easily detected before (Figure 5). This is the result of enrichment of the protein; we are able to load sufficient quantities of those proteins that were previously below the detection limit. In addition, the changes on protein profile are quite dramatic for 0.5 μM DOX treatment for 48 h as compared to untreated control; however, DOX treatment at 20 μM concentration for 24 h had quantitatively higher level of a small number of proteins and this difference is more pronounced in the cytoplasmic fraction than in other fractions.
Figure 5. Prefractionated samples of DOX-treated adult rat cardiomyocytes – 2D gel analysis.
Two dimensional gel analysis of different solubilization extracts. Protein extract of 300 μg was resolved and silver stained. The controls were 48 h treated with DMSO and DOX treatment were for the indicated period.
4. Discussion
The cytotoxic drugs such as DOX are capable of causing cardiotoxicity due to generation of free radicals, lipid peroxidation, immune modulation and apoptosis. DOX also binds to several cellular and plasma proteins, modulating their functions [44,45]. Several models including isolated cardiomyocyte culture [40,46], mitochondrial preparation [47] and animal model [9] have been used to study the mechanism of DOX-mediated cardiotoxicity. Protein changes in any model may depend on the kind of secondary metabolites it generates [48]. Also, DOX accumulates in the nucleus besides mitochondria, thus interfering with nuclear functions [49]. The excessive amount of oxidant generated caused oxidative stress that is beyond the cellular antioxidant capacity, and therefore damages cell membranes due to lipid peroxidation and alters the signaling pathways and protein expression. As a consequence of this process, several proteins undergo changes that are critical for understanding DOX-induced damage. This study, using proteomic analysis, attempts to give insight into this process.
Among the proteins identified, lipocortin V (annexin V) is an annexin family protein that is found on the cytosolic side of the plasma membrane, and it is known to exhibit anti-inflammatory effects by inhibiting the cytosolic phospholipase A2 activity [50]. Annexin V (along with other isoform II and VI) is overexpressed in end stage heart failure [51], and it binds to phosphatidylserine (PS) exposed on the apoptotic cell surface [52]. Alpha B-crystallin, a 22 KDa protein found in rat heart, is a stress inducible molecular chaperone and has been shown to salvage cardiomyocyte apoptosis [53]. It is actually a functional small heat-shock protein induced by heat and other physiological stress and hyper-induced in neurodegenerative disease such as Alzheimer's, Creutzfeldt-Jacob, and Parkinson's diseases [54].
The ETF-QO / ETFDH couple accepts electrons (2 × 1e-) from five acyl Co-A dehydrogenases and four amino acid catabolism products through ETF-β. DOX treatment is known to inhibit long chain fatty acid oxidation and transport across mitochondrial membrane [55,56]. Loss of ETF-β in the DOX-treated samples and low levels of ETFDH overexpression / modification detected in the 48 h sample could be a defense mechanism, as ETFDH is a nuclear encoded mitochondrial protein and is regulated by signaling from AMP-activated protein kinase that detect the low level ATP / AMP involved in fatty acid oxidation and amino acid oxidation. Both of these are catabolic processes that result in generation of energy to compensate for the loss of mitochondrial function [57]. MMSDH is an aldehyde dehydrogenase in the valine and pyrimidine catabolic pathway. Fatty acid acylation via myristate (C14:0) is a covalent modification of the active site cysteine that inhibits the enzyme activity, and the level of inactivity varies with the metabolic state of the mitochondria [58]. MMSDH has been identified as a marker for aging heart where it is nitrated [59]. Deregulation of this protein was not previously reported, and thus the large extent of downregulation reported here by DOX is a novel finding. The significance of its downregulation might be consistent with the problems with fatty acid metabolism involving ETF-β / ETFDH, deregulation of ETC, and loss of mitochondria that is damaged with DOX treatment. The alternative explanation is that the modified (acylated / nitrated) protein migrated differerently than the original protein. The role of nitrative stress in DOX-induced cardiotoxicity has been reported (60,61). Taken together, protein modification by nitration appears to play a crucial role in DOX mediated cardiomyocyte cell death.
The cardiomyopathy results also from a loss of F-actin network (due to depolymerization) which coordinates troponin (F-actin binding protein) involved in controlling contraction. Thus the modifications of troponin T such as phosphorylation could be a manifestation of DOX-induced injury. DOX treatment suppresses alpha actin, troponin I (resulting in myofibrillar loss) [29], sarcoplasmic reticulum Ca2+ ATPase, calcium gated Ca2+ release channel (resulting in impaired Ca2+ handling and perturbation of contraction and relaxation cycle) [62], Rieske Fe-S protein, ADP / ATP translocase, phosphofructokinase, creatine kinase M isoform (resulting in energy impairment) [31]. It is worth noting that the animals treated for 10 wks with DOX did not survive well with the treatment plan. In this animal model, the anti-apoptotic protein HSP27 that confer resistance to DOX and other anticancer drugs was downregulated [63,64]. Suppression of this protein in the animal model in the presence of DOX is of interest with respect to the apoptotic mechanism. In our cardiomyocyte model, p38 is highlighted in the overexpressed protein list, and the, blot analysis shows that this protein gets phosphorylated with time upon DOX treatment (data not shown). Phosphorylation activates p38 kinase, and it in turn phosphorylates many substrates including HSP27 thus regulating apoptosis [65,66].
Doxorubicin-treated cardiomyocyte mitochondrial preparation showed a small but significant decrease in the complex III activity, while the complex I activity remained unaffected (data not shown). It is possible that under our experimental conditions, superoxide generated could be at both complexes I and III, but complex III is also affected by DOX effects on ETFDH and ETF-β. We attempted to confirm if the change in ETFDH levels are true overexpression of ETFDH or some modification by testing the untreated control and DOX-treated sample on a Western blot (data not shown) using an ETFDH antibody (kindly donated by Rikke K.J. Olsen, Aarhus University Hospital and Faculty of Health Sciences, Denmark). We could not detect any marked modification. There is, however, a quantitative increase in ETFDH levels in DOX-treated samples. Therefore it is possible that the ETF-QO protein levels and loss of ETF-β protein levels drastically affect complex III ability to function properly, thus increasing the chances of superoxide generated at this site. In addition, generation of superoxide at complex I site could be due to redox cycling of DOX. It would be interesting to obtain a direct evidence to prove this point, as superoxide generated at complex I and complex III (Qi center) is released towards the matrix where glutathione and other oxidant protection machinery are present, while the superoxide generated from complex III (Qo center- quinol oxidase) are released to the inner membrane side [67,68]. DOX also affects highly unsaturated fatty acids, desaturating and elongating enzymes that are involved in the biosynthesis of essential fatty acids [69]. Increased ROS formation in the mitochondria triggers the intrinsic pathway that leads to the opening of transition pores, and the process is favored by oxidation of glutathione and other sulfhydryls [70]. Events such as activation of BAK (BCL-2 antagonist / Killer 2) by BCL2-Homology (BH3)-only proteins translocate to mitochondrial outer membrane. This results in the translocation of cytochrome C to cytoplasm where it triggers the formation of apoptosome in association with Apaf1 (apoptotic protease activating factor 1) protein. Apoptosome activates caspase 9 which then activates the effector caspase 3 resulting in apoptosis.
Another important downregulated protein in response to DOX treatment was VDAC2. It is well known that VDAC2 plays a critical role in mitochondrial apoptosis. VDAC2 is a member of anion channel proteins (porins) residing in the outer mitochondrial membrane. These proteins are involved in regulation of metabolic interactions and solutes exchange between mitochondria and cytosol, and in regulating the permeabilization of outer mitochondrial membrane during apoptosis. Permeabilization of the membrane is known to initiate the release of cytochrome c into the cytosol and initiate mitochondrial apoptotic pathway. VDAC2 is an isoform that is present in low abundance in the outer mitochondrial membrane and specifically interacts with a multidomain proapoptotic member of Bcl-2 family of proteins BAK (71). This interaction prevents oligomerization of BAK, the event that leads to the formation of outer mitochondrial membrane pores and leaking of cytochrome c into the cytosol (71). Genetic depletion of VDAC2 resulted in excessive BAK oligomerization and apoptotic cell death, while overexpression of this protein prevented BAK activation and inhibited mitochondrial pathway (71,72). On the other hand, other studies performed on mouse embryonic fibroblasts and other cell types, have shown that VDAC2 recruits BAK to the mitochondrial membrane and is required for BAX- and BID-induced apoptosis (73,74). The role of VDAC2 in the regulation of mitochondrial apoptotic pathway is cardiomyocytes is not known. Interestingly, another proteomic study reported upregulation of VDAC2 abundance in hepatoma cells as a result of the action of hepatocarcinogenic dioxin compound (75). The fate of the regulation of VDAC2 expression in cardiomyocytes has not been described previously. It has been known from our previous publication that doxorubicin induces an abundant release of cytochrome c into the cytosol in both adult and neonatal cardiomyocytes (76). Changed expression of this protein by doxorubicin could clearly be an important factor regulating the mitochondrial apoptotic pathway in cardiomyocytes.
It is interesting to note that the proteomic changes in DOX-treated cardiomyocyte and rat heart are not similar. The exact reasons for this difference are not clear. However, the cultured myocytes were treated with DOX for 24 or 48 h, whereas in vivo cardiac tissues were harvested 7 days after the last injection. Under these conditions, no DOX was present in the cardiac tissues isolated from rats. In contrast, DOX was continuously present during in vitro cardiomyocyte cell culture experiments (thus generating oxy-radicals continuously). Thus, the difference between DOX-induced effects in cardiomyocyte culture and in vivo animals could be due to differences in the amount and duration of DOX exposure. Another reason for the observed difference might also be possibly due to the fact that the adult rat heart tissue has more protein in the whole tissue extract. Therefore, we investigated other methods of sample preparation to improve protein identification. The subcellular fractionation is a useful technique, but isolated fractions are often contaminated with other subcellular organelles. Charge fractionations have protein overlaps, and chromatographic fractionation requires a large amount of sample. On the contrary, solubilization techniques are simple, highly reproducible, and enrich low abundant proteins in an inexpensive and reproducible manner. It is also advantageous to further separate them on narrow-range pH strips to further resolve the proteins. Our experimental results in Figure 5 clearly document the advantage of such a technique which will help in identifying the changes occurring in the low abundant proteins.
5. Conclusions
The global proteomic analyses of DOX-treated cardiomyocytes and heart tissues isolated from DOX-treated adult rat indicate different changes in proteome. The common protein changes between the in vitro cellular model and the in vivo animal model are overexpression of beta enolase, troponin and alpha B-crystallin. Other major protein changes detected in tissues were much different from those identified in the cell culture model.
Table 3.
| Upregulated proteins (Doxorubicin-treated adult rat) | Accession # | Molecular function | Matched / Measured peptides | Score | |
|---|---|---|---|---|---|
| Spot # 1 | Alpha B-crystallin | CAA42911 | Structural protein / Molecular chaperone | 5 / 32 - 28% coverage | 70 |
| Spot # 3 | Similar to β-enolase | CAA68788 | Lyase | 13 / 36 - 28% coverage | 132 |
| Spot # 4 | 14-3-3 protein ε-chain | A60031 | Cytoplasmic chaperones / Activator of tyrosine and tryptophan hydroxylases | 8 / 53 - 36% coverage | 116 |
| Spot # 5 | Tyrosine 3-monooxgenase / tryptophan 5-monooxgenase activation protein, β-polypeptide | AAH76502 | Cytoplasmic chaperones / Activator of tyrosine and tryptophan hydroxylases | 6 / 59 - 24% coverage | 68 |
| Spot # 6 | Catalase | P04762 | Aminoacylase activity / Oxidoreductase activity | 5 / 28 - 22% coverage | 67 |
| Spot # 7 | Actin | NP_062056 | Structural protein | 22 / 47 - 45% coverage | 170 |
| Spot # 8 | H+-transporting two-sector ATPase β-chain, mitochondrial - rat (fragment) | A28701 | ATP-Synthase | 12 / 36 - 35% coverage | 122 |
| Spot # 9 | Aldose reductase (AR) (Aldehyde reductase) | P07943 | Aldehyde reductase / Oxidoreductase | 5 / 55 - 12% coverage | 71 |
| Spot # 10 | Proteasome subunit-α type 3 (Proteasome component C8) | P18422 | Endopeptidase (threonine type) | 5 / 12 - 20% coverage | 84 |
| Spot # 11 | Macropain / Transactivating protein BRIDGE | AAD32925 | Transcription co-activator | 7 / 49 - 26% coverage | 74 |
| Spot # 12 | Macropain / Transactivating protein BRIDGE | AAD32925 | Transcription co-activator | 7 / 47 - 26% coverage | 74 |
| Spot # 13 | Apolipoprotein A-I precursor (Apo-AI) | CAA25224 | Phospholipid transporter activity | 7 / 31 - 25% coverage | 71 |
| Spot # 14 | Creatine kinase, sarcomeric mitochondrial precursor (S-MtCK) | P09605 | Kinase / Nucleotide binding | 9 / 65 - 19% coverage | 63 |
| Spot # 15 | Similar to CCAAT/enhancer binding protein-α | CAA31242 | Transcription factor | 6 / 42 - 22% coverage | 68 |
| Spot # 16 | Lactate dehydrogenase | P42123 | Oxidoreductase | 14 / 44-28% coverage | 98 |
| Spot # 18 | ATP synthase beta subunit | AAB02288 | ATP-Synthase | 12 / 42-35% coverage | 128 |
| Spot # 20 | Neurofilament triplet L protein | A21762 | Cytoskeleton | 10 / 74 - 31% coverage | 104 |
| Spot # 31 | Troponin T2 | NP_036808 | Cytoskeleton / Protein binding | 15 / 32 - 36% coverage | 92 |
| Downregulated proteins (Doxorubicin-treated adult rat) | |||||
| Spot # 24 | HSP27 | NP_114176 | Heat shock protein / Anti-apoptosis | 7 / 44-11% coverage | 64 |
| Spot # 35 | Phosphocholine cytidylyltransferase A | P19836 | Regulatory rate limiting enzyme | 11 / 38-22% coverage | 86 |
Protein score is -10*log(P) where P is the probability that the observed match is a random event.
Protein scores greater than 61 are significant (P<0.05)
Acknowledgements
This work was made possible with the help of a NIH grants (R01CA152810) and (1UL1RR031973).
Abbreviations
- ACTH
adrenocorticotropic hormone
- ADP
adenosine diphosphate
- AMP
adenosine monophosphate
- ATP
adenosine triphosphate
- DOX
doxorubicin
- ETC
electron transport chain
- ETF
electron transfer flavoprotein
- ETF-QO / ETFDH
electron transfer protein dehydrogenase
- HSP27
heat shock protein 27 KDa
- MALDI-TOF
matrix assisted laser desorption and ionization – time of flight
- MMSDH
methylmalonate semialdehyde dehydrogenase
- PBS
phosphate buffered saline
- p38 alpha
Mitogen activated protein kinase p38 alpha
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Singal PK, Iliskovic N. Doxorubicin induced cardiomyopathy. N Engl J Med. 1998;339:900–5. doi: 10.1056/NEJM199809243391307. [DOI] [PubMed] [Google Scholar]
- 2.Kremer LC, Van Dalen EC, Offringa M, Ottenkamp J, Voute PA. Anthracycline induced clinical heart failure in a cohort of 607 children: long term follow up study. J Clin Oncol. 2001;19:191–6. doi: 10.1200/JCO.2001.19.1.191. [DOI] [PubMed] [Google Scholar]
- 3.Kawasaki N, Lee JD, Shimizu H, Ueda T. Long term 1-carnitine treatment prolongs the survival in rats with adriamycin induced heart failure. J Card Fail. 1996;2:293–9. doi: 10.1016/s1071-9164(96)80016-9. [DOI] [PubMed] [Google Scholar]
- 4.Powis G. Free radical formation by anti tumor quinones. Free Radic Biol Med. 1989;6:63–101. doi: 10.1016/0891-5849(89)90162-7. [DOI] [PubMed] [Google Scholar]
- 5.Myers CE, McGuire RH, Liss RH, Ifrim I, Grotzinger K, Young RC. Adriamycin: the role of lipid peroxidation in cardiac toxicity and tumor response. Science. 1977;197:165–7. doi: 10.1126/science.877547. [DOI] [PubMed] [Google Scholar]
- 6.Andrieu-Abadie N, Jaffrezou J, Hatem S, Laurent G, Levade T, Mercadier J. L-carnitine prevents doxorubicin induced apoptosis of cardiac myocytes: role of inhibition of ceramide generation. FASEB J. 1999;13:1501–10. doi: 10.1096/fasebj.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 7.Konorev EA, Kotamraju S, Zhao H, Kalivendi S, Joseph J, Kalyanaraman B. Paradoxical effects of metalloporphyrins on doxorubicin induced apoptosis. scavenging of reactive oxygen species versus induction of heme oxygenase-1. Free Radic Biol Med. 2002;33:988–97. doi: 10.1016/s0891-5849(02)00989-9. [DOI] [PubMed] [Google Scholar]
- 8.Adachi K, Fujiura Y, Mayumi F, Nozuhara A, Sugiu Y, Sakanashi T, Hidaka T, Toshima H. A deletion of mitochondrial DNA in murine doxorubicin induced cardiotoxicity. Biochem Biophys Res Commun. 1993;195:945–51. doi: 10.1006/bbrc.1993.2135. [DOI] [PubMed] [Google Scholar]
- 9.Zhou S, Starkov A, Froberg MK, Leino RL, Wallace KB. Cumulative and irreversible cardiac mitochondrial dysfunction induced by doxorubicin. Cancer Res. 2001;61:771–7. [PubMed] [Google Scholar]
- 10.Lebrecht D, Setzer B, Ketelsen UP, Haberstroh J, Walker UA. Time dependent and tissue specific accumulation of mtDNA and respiratory chain defects in chronic doxorubicin cardiomyopathy. Circulation. 2003;108:2423–9. doi: 10.1161/01.CIR.0000093196.59829.DF. [DOI] [PubMed] [Google Scholar]
- 11.Davies KJ, Doroshow JH. Redox cycling of anthracyclines by cardiac mitochondria. I. Anthracycline radical formation by NADH dehydrogenase. J Biol Chem. 1986;261:3060–7. [PubMed] [Google Scholar]
- 12.Jung K, Reszka R. Mitochondria as sub cellular targets for clinically useful anthracyclines. Adv Drug Deliv Rev. 2001;49:87–105. doi: 10.1016/s0169-409x(01)00128-4. [DOI] [PubMed] [Google Scholar]
- 13.Vasquez-Vivar J, Martasek P, Hogg N, Masters BS, Pritchard KA, Kalyanaraman B. Endothelial nitric oxide synthase-dependent superoxide generation from adriamycine. Biochemistry. 1997;36:11293–7. doi: 10.1021/bi971475e. [DOI] [PubMed] [Google Scholar]
- 14.Minotti G, Cairo G, Monti E. Role of iron in anthracycline cardiotoxicity : new tunes for an old song? FASEB J. 1999;13:199–212. [PubMed] [Google Scholar]
- 15.Kalyanaraman B, Joseph J, Kalivendi S, Wang S, Konorev E, Kotamraju S. Doxorubicin induced apoptosis: Implications in cardiotoxicity. Mol Cell Biochem. 2002:234–235. 119–24. [PubMed] [Google Scholar]
- 16.Doroshow JH, Davis KJ. Redox cycling of anthracyclines by cardiac mitochondria. II. Formation of superoxide anion, hydrogen peroxide and hydroxyl radical. J Biol Chem. 1986;261:3068–74. [PubMed] [Google Scholar]
- 17.Winterbourn CC, Gutteridge JMC, Halliwell B. Doxorubicin-dependent lipid peroxidation at low partial pressure of O2. J. Free Radic Biol Med. 1985;1:43–9. doi: 10.1016/0748-5514(85)90028-5. [DOI] [PubMed] [Google Scholar]
- 18.Gille L, Nohl H. Analyses of the molecular mechanism of adriamycin-induced cardiotoxicity. Free Radic Biol Med. 1997;23:775–82. doi: 10.1016/s0891-5849(97)00025-7. [DOI] [PubMed] [Google Scholar]
- 19.Torti FM, Torti SV. Regulation of ferritin genes and protein. Blood. 2002;99:3505–16. doi: 10.1182/blood.v99.10.3505. [DOI] [PubMed] [Google Scholar]
- 20.Brazzolotto X, Gaillard J, Pantopoulos K, Hentze MW, Moulis JM. Human cytoplasmic aconitase (Iron regulatory protein 1) is converted into its [3Fe-4S] form by hydrogen peroxide in vitro but is not activated for iron responsive element binding. J Biol Chem. 1999;274:21625–30. doi: 10.1074/jbc.274.31.21625. [DOI] [PubMed] [Google Scholar]
- 21.Minotti G, Recalcati S, Liberi G, Calafiore AM, Mancuso C, Preziosi P, Cairo G. The secondary alcohol metabolite of doxorubicin irreversibly inactivates aconitase / iron regulatory protein-1 in cytosolic fractions from human myocardium. FASEB J. 1998;12:541–52. doi: 10.1096/fasebj.12.7.541. [DOI] [PubMed] [Google Scholar]
- 22.Minotti G, Ronchi R, Salvatorelli E, Menna P, Cairo G. Doxorubicin irreversibly inactivates iron regulatory protein 1 and 2 in cardiomyocytes. Evidence for distinct metabolic pathways and implications for iron mediated cardiotoxicity of antitumor therapy. Cancer Res. 2001;61:8422–8. [PubMed] [Google Scholar]
- 23.Tobin TP, Abbot BC. A stereological analysis of the effect of adriamycin on the ultrastructure of rat myocardial cells in culture. J Mol Cell Cardiol. 1980;12:1207–25. doi: 10.1016/0022-2828(80)90066-8. [DOI] [PubMed] [Google Scholar]
- 24.Kumar D, Kirshenbaum L, Li T, Danelisen I, Singal P. Apoptosis in isolated adult cardiomyocytes exposed to adriamycin. Annals NY Acad Sci. 1999;874:156–68. doi: 10.1111/j.1749-6632.1999.tb09233.x. [DOI] [PubMed] [Google Scholar]
- 25.Li T, Danelisen I, Bello-Klein A, Singal PK. Effect of probucol on changes of antioxidant enzymes in adriamycin induced cardiomyopathy in rats. Cardiovasc Res. 2000;46:523–30. doi: 10.1016/s0008-6363(00)00039-0. [DOI] [PubMed] [Google Scholar]
- 26.Heon S, Bernier M, Servant N, Dostanic S, Wang C, Kirby GM, Alpert L, Chalifour LE. Dexrazoxane does not protect against doxorubicin induced damage in the young rat. Am J Physiol Heart Circ Physiol. 2003;285:H499–506. doi: 10.1152/ajpheart.00047.2003. [DOI] [PubMed] [Google Scholar]
- 27.Lawenda BD, Kelly KM, Ladas EJ, Sagar SM, Vickers A, Blumberg JB. Should supplemental antioxidant administration be avoided during chemotherapy and radiation therapy? J Natl Cancer Inst. 2008;100:773–83. doi: 10.1093/jnci/djn148. [DOI] [PubMed] [Google Scholar]
- 28.Block KI, Koch AC, Mead MN, Tothy PK, Newman RA, Gyllenhaal C. Impact of antioxidant supplementation on chemotherapeutic toxicity: A systematic review of the evidence from randomized controlled trials. Int J Cancer. 2008;123:1227–39. doi: 10.1002/ijc.23754. [DOI] [PubMed] [Google Scholar]
- 29.Ito H, Miller SC, Billingham ME, Akimoto H, Torti SV, Wade R, Gahlmann R, Lyons G, Kedes L, Torti FM. Doxorubicin selectivity inhibits muscle gene expression in cardiac muscle cells in vivo and in vitro. Proc Natl Acad Sci. 1990;87:4275–9. doi: 10.1073/pnas.87.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeyaseelan R, Poizat C, Baker RK, Abdishoo S, Isterabadi LB, Lyons GE, Kedes L. A novel cardiac-restricted target for doxorubicin. CARP, a nuclear modulator of gene expression in cardiac progenitor cells and cardiomyocytes. J Biol Chem. 1997;272:22800–8. doi: 10.1074/jbc.272.36.22800. [DOI] [PubMed] [Google Scholar]
- 31.Jeyaseelan R, Poizat C, Wu HY, Kedes L. Molecular mechanisms of doxorubicin induced cardiomyopathy. Selective suppression of Reiske iron sulfur protein, ADP / ATP translocase and phosphofructokinase genes is associated with ATP depletion in rat cardiomyocytes. J Biol Chem. 1997;272:5828–32. doi: 10.1074/jbc.272.9.5828. [DOI] [PubMed] [Google Scholar]
- 32.Cutts SM, Parsons PG, Strum RA, Phillips DR. Adriamycin induced DNA adducts inhibit DNA interactions of transcription factors and RNA polymerase. J Biol Chem. 1996;271:5422–9. doi: 10.1074/jbc.271.10.5422. [DOI] [PubMed] [Google Scholar]
- 33.Wang S, Kotamraju S, Konorev E, Kalivendi S, Joseph J, Kalyanaraman B. Activation of nuclear factor-kappaB during doxorubicin induced apoptosis in endothelial cells and myocytes is proapoptotic: the role of hydrogen peroxide. Biochem J. 2002;367:729–40. doi: 10.1042/BJ20020752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gigli M, Doglia SM, Millot JM, Valentini L, Manfait M. Quantitative study of doxorubicin in living cell nuclei by microspectrofluorometry. Biochim Biophys Acta. 1988;950:13–20. doi: 10.1016/0167-4781(88)90068-1. [DOI] [PubMed] [Google Scholar]
- 35.L'Ecuyer T, Sanjeev S, Thomas R, Novak R, Das L, Cambell W, Heide RV. DNA damage is an early event in doxorubicin induced cardiac myocyte death. Am J Physiol Heart Circ Physiol. 2006;291:H1273–80. doi: 10.1152/ajpheart.00738.2005. [DOI] [PubMed] [Google Scholar]
- 36.Ellis CN, Ellis MB, Blakemore WS. Effect of adriamycin on heart mitochondrial DNA. Biochem J. 1987;245:309–12. doi: 10.1042/bj2450309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piper HM, Volz A, Schwartz P. Adult ventricular rat heart cells. In: Piper HM, editor. Cell culture techniques in heart and vessel research. Springer Verlag, Heidelberg: 1990. pp. 36–60. [Google Scholar]
- 38.Hohl CM, Altschuld RA, Brierley GP. Effects of calcium on the permeability of isolated adult rat heart cells to sodium. Arch Biochem Biophys. 1982;221:197–205. doi: 10.1016/0003-9861(83)90136-4. [DOI] [PubMed] [Google Scholar]
- 39.Armstrong SC, Ganote CE. Effects of 2,3-butanedione monoxime (BDM) on contracture and injury of isolated rat myocytes following metabolic inhibition and ischemia. J Mol Cell Cardiol. 1991;23:1001–14. doi: 10.1016/0022-2828(91)91636-6. [DOI] [PubMed] [Google Scholar]
- 40.Konorev EA, Kennedy MC, Kalyanaraman B. Cell-permeable superoxide dismutase and glutathione peroxidase mimetics afford superior protection against doxorubicin induced cardiotoxicity. The role of reactive oxygen and nitrogen intermediates. Arch Biochem Biophys. 1999;368:421–28. doi: 10.1006/abbi.1999.1337. [DOI] [PubMed] [Google Scholar]
- 41.Kotamraju S, Konorev EA, Joseph J, Kalyanaraman B. Doxorubicin induced apoptosis in endothelial cells and cardiomyocytes is ameliorated by nitrone spin traps and ebselen. Role of reactive oxygen and nitrogen species. J Biol Chem. 2000;275:33585–92. doi: 10.1074/jbc.M003890200. [DOI] [PubMed] [Google Scholar]
- 42.Gharahdaghi F, Weinberg CR, Meagher DA, Imai BS, Mische SM. Mass spectrometric identification of proteins from silver stained polyacrylamide gels: A method for the removal of silver ions to enhance sensitivity. Electrophoresis. 1999;20:601–5. doi: 10.1002/(SICI)1522-2683(19990301)20:3<601::AID-ELPS601>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 43.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver stained polyacrylamide gels. Anal Chem. 1996;68:850–8. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 44.Nagi MN, Mansour MA. Protective effect of thymoquinone against doxorubicin-induced cardiotoxicity in rats: a possible mechanism of protection. Pharmacol Res. 2000;41:283–9. doi: 10.1006/phrs.1999.0585. [DOI] [PubMed] [Google Scholar]
- 45.Kim YK, Lee WK, Jin Y, Lee KJH, Yu YG. Doxorubicin binds to unphosphorylated form of hNopp140 and reduces protein kinase CK2-dependent phosphorylation of hNopp140. J Biochem Mol Biol. 2006;39:774–81. doi: 10.5483/bmbrep.2006.39.6.774. [DOI] [PubMed] [Google Scholar]
- 46.Green PS, Leeuwenburgh C. Mitochondrial dysfunction is an early indicator of doxorubicin induced apoptosis. Biochim Biophys Acta. 2002;1588:94–101. doi: 10.1016/s0925-4439(02)00144-8. [DOI] [PubMed] [Google Scholar]
- 47.Tokarska-Schlattner M, Dolder M, Gerber I, Speer O, Wallimann T, Schlattner U. Reduced creatine stimulated respiration in doxorubicin challenged mitochondria: particular sensitivity of the heart. Biochim Biophys Acta. 2007;1767:1276–84. doi: 10.1016/j.bbabio.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 48.Salvatorelli E, Guarnieri S, Menna P, Liberi G, Calafiore AM, Mariggio MA, Mordente A, Gianni L, Minotti G. Defective one- and two- electron reduction of the anticancer anthracycline epirubicin in human heart. Relative importance of vesicular sequestration and impaired efficiency of electron addition. J Biol Chem. 2006;281:10990–1001. doi: 10.1074/jbc.M508343200. [DOI] [PubMed] [Google Scholar]
- 49.Zhou S, Palmeira CM, Wallace KB. Doxorubicin induced persistent oxidative stress to cardiac myocytes. Toxicol Lett. 2001;121:151–7. doi: 10.1016/s0378-4274(01)00329-0. [DOI] [PubMed] [Google Scholar]
- 50.Romisch J, Grote M, Weithmann KU, Heimburger N, Amann E. Annexin protein PP4 and PP4-X: comparative characterization of biological activities of placental and recombinant proteins. Biochem J. 1990;272:223–9. doi: 10.1042/bj2720223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strauss HW, Narula J, Blankenberg F. Radioimaging to identify myocardial cell death and probably injury. Lancet. 2000;356:180–1. doi: 10.1016/S0140-6736(00)02474-0. [DOI] [PubMed] [Google Scholar]
- 52.Andree HA, Reutelingsperger CP, Hauptmann R, Hemker HC, Hermens WT, Willems GM. Binding of vascular anticoagulant (VAC) to planar phospholipid bilayers. J Biol Chem. 1990;265:4923–8. [PubMed] [Google Scholar]
- 53.Ray PS, Martin JL, Swanson EA, Otani H, Dillmann WH, Das DK. Transgene over expression of alpha B crystallin confers simultaneous protection against cardiomyocyte apoptosis and necrosis during myocardial ischemia and reperfusion. FASEB J. 2001;15:393–402. doi: 10.1096/fj.00-0199com. [DOI] [PubMed] [Google Scholar]
- 54.Klemenz R, Frohli E, Steiger RH, Schafer R, Aoyama A. Alpha B-crystallin is a small heat shock protein. Proc Natl Acad Sci. 1991;88:3652–6. doi: 10.1073/pnas.88.9.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abdel-aleem S, el-Merzabani MM, Syed-ahmed M, Taylor DA, Lowe JE. Acute and chronic effects of adriamycine on fatty acid oxidation in isolated cardiac myocytes. J Mol Cell Cardiol. 1997;29:789–97. doi: 10.1006/jmcc.1996.0323. [DOI] [PubMed] [Google Scholar]
- 56.Hong YM, Kim HS, Yoon HR. Serum lipid and fatty acid profiles in adriamycine treated rats after administration of L-carnitine. Pediatr Res. 2002;51:249–55. doi: 10.1203/00006450-200202000-00020. [DOI] [PubMed] [Google Scholar]
- 57.Zong H, Ren JM, Young LH, Pypaert M, Mu J, Birnbaum J, Shulman GI. AMP kinase is required for mitochondrial biogenesis in skeletal muscle in response to chronic energy deprivation. Proc Natl Acad Sci. 2002;99:15983–7. doi: 10.1073/pnas.252625599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Berthiaume L, Deichaite I, Peseckis S, Resh MD. Regulation of enzymatic activity by active site fatty acylation. A new role for long chain fatty acid acylation of proteins. J Biol Chem. 1994;269:6498–505. [PubMed] [Google Scholar]
- 59.Kanski J, Behring A, Pelling J, Schoeneich C. Proteomic identification of 3-nitrotyrosine containing rat cardiac proteins. Effects of biological aging. Am J Physiol Heart Circ Physiol. 2005;288:H371–81. doi: 10.1152/ajpheart.01030.2003. [DOI] [PubMed] [Google Scholar]
- 60.Pacher P, Liaudet L, Bai P, Virag L, Mabley JG, Hasko G, Szabo C. Activation of poly(ADP-ribose) polymerase contributes to development of doxorubicin-induced heart failure. J Pharmacol Exp Therap. 2002;300:862–7. doi: 10.1124/jpet.300.3.862. [DOI] [PubMed] [Google Scholar]
- 61.Mukhopadhyay P, Rajesh M, Batkai S, Kashiwaya Y, Hasko G, Liaudet L, Szabo C, Pacher P. Role of superoxide, nitric oxide, and peroxynitrite in doxorubicin-induced cell death in vivo and invitro. Am J Physiol Heart Circ Physiol. 2009;296:H1466–83. doi: 10.1152/ajpheart.00795.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arai M, Tomaru K, Takizawa T, Sekiguchi K, Yokoyama T, Suzuki T, Nagai RJ. Sarcoplasmic reticulum genes are selectively downregulated in cardiomyopathy produced by doxorubicin in rabbits. J Mol Cell Cardiol. 1998;30:243–54. doi: 10.1006/jmcc.1997.0588. [DOI] [PubMed] [Google Scholar]
- 63.Turakhia S, Venkatakrishnan CD, Dunsmore K, Wong H, Periannan K, Zweier JL, Ilangovan G. Doxorubicin-induced cardiotoxicity: Direct correlation of cardiac fibroblasts and H9c2 cells survival and aconitase activity with heat shock protein 27. Am J Physiol Heart Circ Physiol. 2007;293:H3111–21. doi: 10.1152/ajpheart.00328.2007. [DOI] [PubMed] [Google Scholar]
- 64.O'Callaghan-Sunol C, Gabai VL, Sherman MY. Hsp27 modulates p53 signaling and suppresses cellular senescence. Cancer Res. 2007;67:11779–88. doi: 10.1158/0008-5472.CAN-07-2441. [DOI] [PubMed] [Google Scholar]
- 65.Arrigo AP, Landry J. Expression and function of the low molecular weight heat shock proteins. In: Morimoto RI, Tissieres A, Georgopoulos C, editors. The biology of heat shock proteins and molecular chaperones. Vol. 26. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1994. pp. 335–73. [Google Scholar]
- 66.Venkatakrishnan CD, Tewari AT, Moldovan L, Cardounel AJ, Zweier JL, Kuppuswamy P, Ilangovan G. Heat shock protects cardiac cells from doxorubicin induced toxicity by activating p38 MAPK and phosphorylation of small heat shock protein 27. Am J Physiol Heart Circ Physiol. 2006;291:H2680–91. doi: 10.1152/ajpheart.00395.2006. [DOI] [PubMed] [Google Scholar]
- 67.Chen Q, Vazquez EJ, Moghaddas S, Hoppel CL, Lesnefsky EJ. Production of reactive oxygen species by mitochondria. Central role of complex III. J Biol Chem. 2003;278:36027–31. doi: 10.1074/jbc.M304854200. [DOI] [PubMed] [Google Scholar]
- 68.Young TA, Cunningham CC, Bailey SM. Reactive oxygen species production by the mitochondrial respiratory chain in isolated rat hepatocytes and liver mitochondria : studies using myxothiazol. Arch Biochem Biophys. 2002;405:65–72. doi: 10.1016/s0003-9861(02)00338-7. [DOI] [PubMed] [Google Scholar]
- 69.Bordoni A, Biagi P, Hrelia S. The impairment of essential fatty acid metabolism as a key factor in doxorubicin induced damage in cultured rat cardiomyocytes. Biochim Biophys Acta. 1999;1440:100–6. doi: 10.1016/s1388-1981(99)00113-4. [DOI] [PubMed] [Google Scholar]
- 70.Chernyak BV. Redox regulation of the mitochondrial permeability transition pore. Biosci Rep. 1997;17:293–302. doi: 10.1023/a:1027384628678. [DOI] [PubMed] [Google Scholar]
- 71.Cheng EH, Sheiko TV, Fisher JK, Craigen WJ, Korsmeyer SJ. VDAC2 inhibits BAK activation and mitochondrial apoptosis. Science. 2003;301:513–7. doi: 10.1126/science.1083995. [DOI] [PubMed] [Google Scholar]
- 72.Ren D, Kim H, Tu HC, Westergard TD, Fisher JK, Rubens JA, Korsmeyer SJ, Hsieh JJ, Cheng EH. The VDAC2-BAK rheostat controls thymocyte survival. Sci Signal. 2009;2:ra48. doi: 10.1126/scisignal.2000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roy SS, Ehrlich AM, Craigen WJ, Hajnoczky G. VDAC2 is required for truncated BID-induced mitochondrial apoptosis by recruiting BAK to mitochondria. EMBO Rep. 2009;10:1341–7. doi: 10.1038/embor.2009.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yamagata H, Shimizu S, Nishida Y, Watanabe Y, Craigen WJ, Tsujimoto Requirement of voltage-dependent anion channel 2 for pro-apoptotic activity of Bax. Oncogene. 2009;28:3563–72. doi: 10.1038/onc.2009.213. [DOI] [PubMed] [Google Scholar]
- 75.Sarioglu H, Brandner S, Haberger M, Jacobsen C, Lichtmannegger J, Wormke M, Andrae U. Analysis of 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced proteome changes in 5L rat hepatoma cells reveals novel targets of dioxin action including the mitochondrial apoptosis regulator VDAC2. Mol Cell Proteomics. 2008;7:394–410. doi: 10.1074/mcp.M700258-MCP200. [DOI] [PubMed] [Google Scholar]
- 76.Konorev EA, Vanamala S, Kalyanaraman B. Differences in doxorubicin-induced apoptotic signaling in adult and immature cardiomyocytes. Free Radic Biol Med. 2008;45:1723–8. doi: 10.1016/j.freeradbiomed.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]