Abstract
Purpose
Tumor-specific antigens of 3-methylcholanthrene (MCA)-induced sarcomas were defined by the narrow immune responses they elicited, which uniquely rejected the homologous tumor, with no cross-reactions between independently-derived syngeneic MCA-induced tumors. This study examines if an autophagosome-enriched vaccine derived from bortezomib-treated sarcomas can elicit an immune response that cross-reacts with other unique sarcomas.
Experimental Design
Mice were vaccinated with either MCA-induced sarcomas or autophagosomes derived from those tumors and later challenged with either homologous or non-homologous sarcomas. Additionally, 293 cells expressing a model antigen were used to understand the necessity of short-lived proteins in this novel vaccine. These findings were then tested in the sarcoma model. Autophagosomes were characterized by western blot and fluorescent microscopy and their ability to generate immune responses was assessed in vitro by CFSE dilution of antigen-specific T cells and in vivo by monitoring tumor growth.
Results
In contrast to a whole cell tumor vaccine, autophagosomes isolated from MCA-induced sarcomas treated with a proteasome inhibitor prime T cells that cross-react with different sarcomas and protect a significant proportion of vaccinated hosts from a non-homologous tumor challenge. Ubiquitinated short-lived proteins (SLiPs), which are stabilized by proteasome blockade and delivered to autophagosomes in a p62/sequestosome-dependent fashion, are a critical component of the autophagosome vaccine as their depletion limits vaccine efficacy.
Conclusion
This work suggests that common short-lived tumor-specific antigens, not physiologically available for cross-presentation, can be sequestered in autophagosomes by p62 and used as a vaccine to elicit cross-protection against independently-derived sarcomas.
Keywords: autophagosome, MCA-induced sarcoma, p62/sequestosome, bortezomib, cancer vaccine, immunotherapy
INTRODUCTION
Cross-presentation was identified as a means by which antigens can be presented by cells in which they were not synthesized, thus obviating direct presentation as the sole mechanism to prime an immune response (1). These findings were expanded by the demonstration that cross-presentation of melanoma antigens during vaccination was essential for the generation of an effective anti-tumor immune response (2). One component of cross-presentation that has been debated and still remains unknown is the source of antigen and the method of its delivery to professional antigen-presenting cells (pAPC). While some groups have shown that the source of antigen is cellular protein, others argue that it is peptides chaperoned by heat-shock proteins (HSPs) (3, 4). To complicate the debate further, Johanthan Yewdell’s group demonstrated that cross-priming results from both the donation of proteasome substrates as well as stable cytosolic peptides in conjunction with HSP90 (5, 6). Recently, we described a pool of antigen used for cross presentation that is dependent on macroautophagy (hereafter referred to as autophagy) (7, 8).
Autophagy is a cellular process in which portions of the cytoplasm are sequestered by double membrane vesicles termed autophagosomes that range in size from 300–900 nm(9). The contents of these autophagosomes are degraded in a lytic compartment, which facilitates the turnover of long-lived proteins and is critical for maintaining the pool of amino acids needed for anabolism. The formation, function and isolation of autophagosomes are distinct from that of exosomes, 100 nm single-membrane vesicles originating from endosomes. Exosomes are actively secreted into the extracellular environment to signal to or otherwise modulate neighboring cells via a variety of stimulatory or inhibitory mechanisms (10–12). Despite their discrete biophysical properties both dendritic cell-derived exosomes and tumor-derived autophagosomes can generate robust immune responses (12). However, unlike tumor-derived autophagosomes, tumor-derived exosomes have been demonstrated by different groups to be immunosuppressive via a variety of mechanisms (9, 10, 13, 14).
A key marker of the induction of autophagy is the conversion of the cytosolic form of microtubule-associated protein 1 light chain 3 (LC3-I) via a series of ubiquitin-like conjugation steps to the lipidated form (LC3-PE or LC3-II) that is tightly associated with autophagosomes. Conversion to LC3-II is not only a definitive marker of autophagy but it can promote curvature and hemifusion of membranes, and is therefore critical to the process (15). A recently described protein, p62/SQSTM1 (sequestosome), binds both polyubiquitin and LC3 and thus facilitates degradation of ubiquitinated proteins via autophagy (16, 17). Interaction of LC3 with p62 has added a layer of complexity to the autophagic network and suggests that this bulk degradation process may be more selective than previously appreciated.
We have shown that autophagy in tumor cells is essential for efficient cross-presentation and subsequent induction of tumor immunity in a B16 melanoma model (8). Cross-presentation, which was measured by proliferation of CFSE-labeled antigen-specific T cells, was significantly inhibited when autophagy was blocked and increased when autophagy was promoted. Interestingly, when cell lysates were fractionated and used as an antigen source, the fraction with the greatest cross-presentation activity also had the highest level of the specific autophagosome marker LC3. By treating cells with the proteasome inhibitor, bortezomib, and the lysosomotropic agent, NH4Cl, which prevents fusion of autophagosomes with lysosomes, autophagosome-containing vesicles could be isolated. These isolated autophagosome-containing vesicles, termed DRibbles (7) served as a potent antigen source in cross-presentation assays and in in vivo vaccine studies. In combination with the results of two recent publications, which demonstrated enhanced antigen presentation related to autophagy (18, 19), our work has further defined the function of autophagy as a means of sequestering antigen for cross-presentation.
To understand better the function of autophagy in cross-presentation, we developed a model that incorporates the DRiP hypothesis (20). A significant proportion of MHC class I binding peptides originate from defective ribosomal products (DRiPs), including misfolded and truncated polypeptides, which are degraded by the proteasome shortly after their translation and loaded onto MHC class I molecules (5). Since DRiPs, as well as other short-lived proteins (SLiPs), are stabilized by proteasome inhibition, we hypothesized that autophagosome-containing vesicles isolated from bortezomib-treated cells would contain DRiPs and SLiPs and thereby provide a unique spectrum of potential tumor rejection antigens. We further hypothesized that using these vesicles to prime an immune response will generate a broader T-cell response.
Prehn and Main established the unique specificity of chemically induced 3-methycholanthrene (MCA) sarcomas, whereby sarcomas generated in genetically identical mice with similar morphology and growth characteristics would only protect vaccinated mice from a challenge with the immunizing tumor but not other syngeneic sarcomas. While there has been a paucity of antigens associated with the unique specificity of this tumor model (21), genetic analysis of a MCA-induced sarcoma after CTL immunoselection revealed a deletion in a region rich with oncogenes and tumor suppressor genes (22). Even though a unique immunodominant antigen results from each MCA treatment, this data demonstrates that specific loci or chromosomal regions are more susceptible to the mutating effects of MCA. Moreover, using CTL immunoselection, a secondary tumor antigen shared by an independent sarcoma cell line was uncovered, demonstrating that the unique rejection antigen is only part of the tumor antigen profile (23). Others have demonstrated cross-reactivity among heterogenic clones of the MCA-106 sarcoma using effector cells primed with the parental MCA-106 line but no cross-reactivity with an “antigenically” distinct MCA-205 sarcoma (24). There are therefore limited examples of common antigens among the MCA-induced sarcomas in the few publications reported.
In this paper, we examine the role of autophagy in tumor immunity by focusing on autophagosomes as the source of antigen for cross-presentation. We find that vaccination with antigens derived from autophagosomes can broaden the T-cell response beyond that seen following whole cell vaccination. Studies using MCA sarcomas as well as HEK 293T cells stably expressing a short-lived model antigen both indicate that short-lived proteins are necessary for this unique autophagosome-mediated immune response. Further, we demonstrate that the ubiquitin/LC3-binding protein p62 (sequestosome) has a key role as a regulator of selective autophagy, as it associates with both ubiquitinated antigen and LC3 and is needed for the sequestration of SLiPs into the autophagosomes. Based on these findings, we propose that the broad array of tumor antigens contained in autophagosomes is dependent upon ubiquitinated SLiPs incorporated by the sequestosome and that cross-presentation of these autophagosomes primes a unique cross-protective immune response.
MATERIALS AND METHODS
Mice and cell lines
Female C57BL/6J (B6) mice were purchased from Charles River (Wilmington, MA). OT-1 breeders transgenic for the TCR that recognizes chicken ovalbumin (peptide sequence SIINFEKL) in the context of H-2Kb were purchased from Jackson Laboratory (Bar Harbor, ME). All mice were maintained and used in accordance with the Earl A. Chiles Research Institute Animal Care and Use Committee and recognized principles of laboratory animal care were followed (Guide for the Care and Use of Laboratory Animals, National Research Council, 1996).
Human embryonic kidney 293T cells were cultured in complete medium (CM), which consisted of DMEM (Lonza) supplemented with 10% FBS (Life Technologies, Grand Island, New York). Cell lines were maintained in T-75 or T-150 culture flasks in a 5% CO2 incubator at 37°C. Stable expression of both the stable OVA (mOVA) and short-lived OVA (rOVA) in HEK 293T cells was achieved with lentiviral transduction as described earlier (8). For the generation of recombinant lentiviruses, HEK 293T cells were transiently transfected with vector plasmid pWPT or pGIPz, virus packaging plasmid pPAX2, envelope plasmid VSV-G MD2, and helper plasmid pAdv. Viral supernatant was used to infect HEK 293T cells and transduced cells were sorted based on GFP expression by flow cytometry.
3-Methylcholanthrene (MCA)-induced sarcomas were previously generated in our laboratory (25, 26). All tumors aliquots used had been in vivo passaged less than 6 times. Unique MCA sarcomas were passaged in vivo by excising a tumor from a female C57BL/6 mouse followed by triple enzyme digestion of the tumor (incubation at room temperature with hyaluronidase, collagenase and DNAse for 1–3 hours with agitation) and subcutaneous injection into a naïve mouse. Uniqueness of each sarcoma was confirmed by vaccine challenge studies summarized in figure 2G.
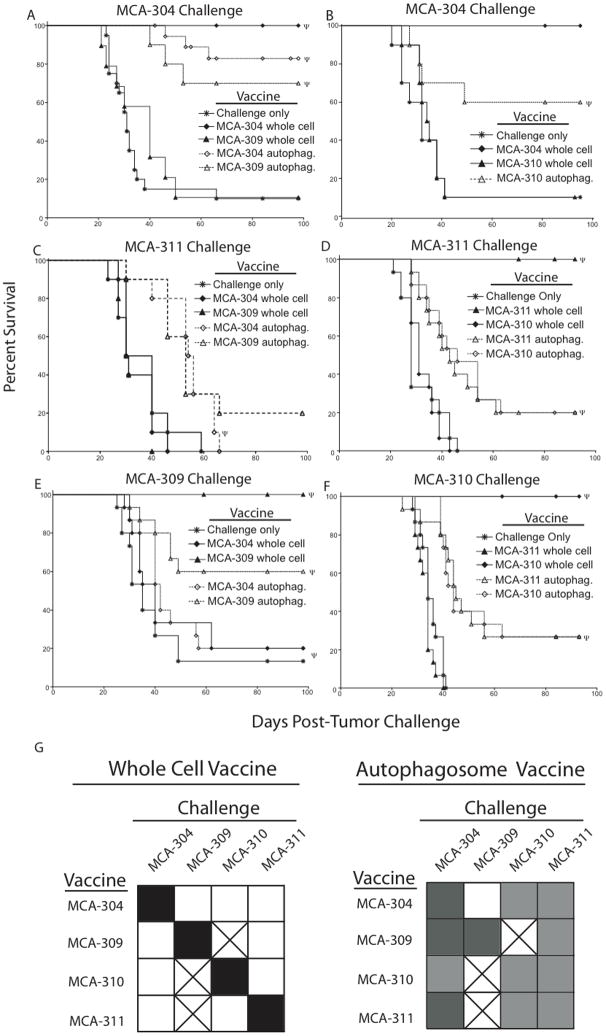
Figure 2.
Autophagosome vaccination protects mice from a challenge against non-homologous sarcomas. (A–F) Kaplan-Meier survival plots comparing protection of mice vaccinated with an irradiated whole cell sarcoma or autophagosomes derived from the sarcomas. The subcutaneous autophagosome vaccination consisted of 3×106 APCs pulsed with 3×106 CE of autophagosomes injected into the lower right flank of the mouse. Whole cell vaccinations were performed as above in Figure 1A. Vaccinated mice were challenged with 30,000 viable (A and B) MCA-304, (C and D) MCA-311, (E) MCA-309 or (F) MCA-310 tumors. Each survival plot represents 3 independent experiments with 5 mice per group (n=15) or (B and C) 2 independent experiments with 5 mice per group (n=10). Survival curves denoted with Ψ represent statistically significant (p>0.05) protection from a tumor challenge compared to no vaccine. (G) Summary of vaccine/challenge experiments. All filled squares represent statistically significant (p<0.05) protection from a tumor challenge compared to no vaccine. Filled black squares represent maximum observed protection from a tumor challenge (p<0.001). After subtracting survival of the challenge only group, black squares represent a survival of 70% or greater, medium grey squares represent a survival of 30–69%, and light grey squares represent a survival of less than 30%. Unfilled squares represent the same protection from a tumor challenge as the no vaccine group (p>0.05). Boxes with an “x” were not determined. Each square represents 2–4 independent experiments with 5 mice per group (n=10–20).
B16BL/6J-D5 (D5), a poorly immunogenic subclone of the spontaneously arising B16BL6 melanoma, was kindly provided by Dr. S. Shu (Earle A. Chiles Research Institute, Portland, Oregon). A stock of D5 cells, screened to be free of mycoplasma, were banked and assayed for immunogenicity and expression of melanoma-associated antigens, gp100 and TRP2. D5 cells from this stock were used within 4–6 weeks of culture. D5 cells were cultured in complete medium (CM), which consisted of RPMI 1640 (BioWhittaker,Walkersville, Maryland) supplemented with 10% FBS (Life Technologies, Grand Island, New York), 50μM 2-mercaptoethanol (Aldrich, Milwaukee, Wisconsin), 0.1mM non-essential amino acids, 1 mM sodium pyruvate, 2mM L-glutamine and 50μg/mL gentamicin sulfate. Cell lines were maintained in T-75 or T-150 culture flasks in a 5% CO2 incubator at 37°C.
Antigen presenting cells (APC) were generated in the spleens of C57BL6 mice via hydrodynamic gene transfer (27). Female C57BL/6J (B6) mice were sequentially injected with 2 mL i.v. containing 2μg of plasmid DNA encoding murine Flt3 ligand or granulocyte macrophage colony stimulating factor. The spleens are typically enlarged and enriched for CD8+/CD11c+ cells (8, 28, 29). Spleens were harvested and processed into a single cell suspension and frozen.
Tumor vaccine and challenge
Female C57BL/6J mice were vaccinated in the lower right flank with a subcutaneous injection of 5×106 irradiated tumor cells from a freshly digested sarcoma or with 3×106 APCs pulsed withautophagosomes (3×106 cell equivalents (CE)) for 6–8 hours. To control for the use of dendritic cells in the autophagosome vaccine, 3×106 APCs pulsed with 5×106 irradiated tumor cells for 6 hours, were washed and used as a vaccine as described above. For the cyclohexamide experiments, 5×106 frozen tumor cells (previously treated with cyclohexamide) were thawed, irradiated and used as vaccine. Mice were challenged 2 weeks after vaccination by subcutaneous injection in the lower left (opposite) flank with 2–3×104 viable digested MCA tumor cells. Tumor growth was assessed by measuring the perpendicular diameters of the sarcoma. Mice were sacrificed when the area of the tumor, determined by the product of the perpendicular diameters, reached 150 mm2 or greater.
Isolation of autophagosome-containing vesicles
Autophagosome-containing vesicles (DRibbles) were harvested from HEK 293 cells or tumor cells after treating the cells with 100nM bortezomib (Velcade) and 10 mM of NH4Cl in CM for 18–24 hours in a 5% CO2 incubator at 37°C. The cells and the supernatant were harvested and spun at 480 x g, as described by Stromhaug et al. (30). The supernatant was then spun at 12,000 x g to harvest the autophagosome-containing pellet. Based on the amount of cells originally seeded, the autophagosome pellet was resuspended at 108 cell equivalents (CE) per mL (typically in the range of 0.5–1 mg/mL total protein by BCA).
DNA Construction and Transfection
Plasmid DNA vector cloning was described earlier (8). Briefly, Ub-X-GFP-expressing plasmids were fused with an OVA antigen by PCR with Vent polymerase and cloned into the lentiviral vector pWPT (kindly provided by Dr. D Trono at the Department of Microbiology, Geneva School of Medicine, Geneva, Switzerland). The LC3 fusion plasmid, pCMV-GFP-LC3 and the p62 fusion plasmid, pCMV-tdTomato-p62 were kindly provided Dr. T Johansen (Biochemistry Department, Institute of Medical Biology, University of Tromso, Tromso, Norway). Both the LC3 and p62 constructs were sub-cloned into the pWPT vector using PCR with Vent polymerase. Transient transfections of HEK 293FT cells with LC3, p62 or OVA expressing vectors were performed using metafectene pro (Biontex Laboratories GmbH).
Measurement of cytokine production by primed LN T cells from vaccinated mice
To measure the priming of naïve lymphocytes, mice were vaccinated in both the fore and hind flanks by subcutaneous injection of 1×106 irradiated sarcoma cells or 2×106 dendritic cells pulsed with 3×106 cell equivalents of autophagosomes. Draining lymph nodes (DLN) were harvested after 11 days and cultured with soluble anti-CD3 (5ug/mL) for 48 hours and then expanded with IL-2 (60 IU/mL) for 72 hours. After this in vitro activation and expansion, effector T cells were washed, resuspended inCM and IL-2 (60 IU/ml), and seeded at 2×106/2 ml/well in a24-well plate. The cells were either cultured without further stimulation or stimulated with 2×105 D5, primary (triple enzyme digested) MCA-304, MCA-309, MCA-310, MCA-311 tumor cells,primary kidney cells or immobilized anti-CD3 (positive control). Supernatants wereharvested after 24 h and assayed for IFN-γ by ELISA using commercially available reagents (IFN-γ, BD Biosciences Pharmingen, San Diego, CA). The concentration of cytokines in the supernatantwas determined by regression analysis.
siRNA knockdown of sequestosome (p62)
siGENOME SMARTpool M-010230-00-0005 against human SQSTM1 (p62) was purchased from Dharmacon (Thermo Scientific). Non-specific control siRNA sc-36869 was purchased from Santa Cruz Biotechnology. 50 nM of either siRNA was transfected into HEK 293 T cells using Invitrogen’s lipofectamine 2000.
Immunoprecipitation/Western blotting
Radioimmunoprecipitation assay (RIPA) buffer with a protease inhibitor cocktail (Roche) was used to lyse autophagosomes isolated from treated 293rGFP-OVA or parental HEK 293T cells. The lysate (50 μg) was initially incubated with 2 μg of goat anti-human IgG (Jackson ImmunoResearch). The pre-cleared lysate (~25 μg) was then incubated overnight with 1 μg of a goat anti-GFP antibody (Rockland). Protein A/G agarose was added to the anti-GFP/lysate mixture and incubated for 4 hours. 60uL of 2X NuPAGE LDS buffer were added directly to beads. The beads were subjected SDS PAGE and western blotting as described below.
For western blots, HEK 293T cells (1×106) or autophagosomes (10×106 cell equivalents) were lysed in 100 μL of radioimmunoprecipitation assay (RIPA) buffer with a protease inhibitor cocktail (Roche). The lysates were mixed with 4X NuPAGE LDS sample buffer and samples were resolved by 4% to 20% SDSPAGE (Invitrogen). Proteins were transferred to a nitrocellulose membrane, incubated with primary antibodies, diluted in blocking buffer (5% dry milk) overnight, and then incubated with horseradish peroxidase (HRP)–conjugated secondary antibodies for 1 h. Protein bands were revealed by using chemiluminescent reagents (Pierce). The primary antibodies included rabbit anti-actin (1:2,000; Sigma), goat anti-GFP (1:1,000; Rockland), rabbit anti-ubiquitin (1:1,000; Upstate), rabbit anti-LC3 (1:1,000; MBL) and goat anti-sequestosome (1:700; santa cruz biotechnology). The secondary antibodies were goat anti-rabbit HRP (1:10,000; Jackson ImmunoResearch), and donkey anti-goat HRP (1:10,000; Jackson ImmunoResearch).
CFSE proliferation assay
CFSE-labeled naïve T cells from ovalbumin-specific OT-1 TCR transgenic mice were added to APCs pulsed with autophagosomes, SIINFEKL peptide (10 μg) or whole soluble ovalbumin (50 μg). Cross-presentation of antigens to labeled OT-I T cells was assessed by measuring the dilution of CFSE by flow cytometry. Splenocytes from OT-I mice were labeled with 5 μM of CFSE according to the manufacturer’s protocol (Invitrogen). T-cell proliferation was measured as loss of CFSE intensity after 3 or 4 days of APC and T-cell co-incubation.
Confocal Microscopy
The images were acquired at the Advanced Light Microscopy Core at The Jungers Center of Oregon Health and Science University on a high resolution wide field Core DV system (Applied Precision™). This system is an Olympus IX71 inverted microscope with a proprietary XYZ stage enclosed in a controlled environment chamber; differential interference contrast (DIC) transmitted light and a short arc 250W Xenon lamp for fluorescence. The camera is a Nikon Coolsnap ES2 HQ. Each image was acquired as Z-stacks in a 1024×1024 format with a 60×1.42 NA Plan Apo N objective in 3 colors: green, red and blue. The pixel size was 0.107 × 0.107 × 0.5 microns. The images were deconvolved with the appropriate OTF (optical transfer function) using an iterative algorithm of 10 iterations. The histogram was optimized for the most positive image and applied to all the other images for consistency before saving the images as 24 bit merged TIFF. A reference DIC image was acquired from the middle of the Z-stack.
Statistical Methods
For the time-to-death endpoint, the survival distribution was estimated using the Kaplan-Meier method. The log-rank test was used to compare the hazard rates of the vaccines. For ELISA data, the log10-transformed IFN-γ values were analyzed using a mixed effects model that appropriately accounted for different sources of variability introduced by the experimental design. This model had fixed effects for the vaccine group, stimulator, and their interaction, and random effects for the experiment (nested within vaccine group), the stimulator pool interaction, and the assay replicate. Different variances were allowed for each vaccine group. Contrasts using least squares means for each stimulator-vaccine group combination were used to test for mean differences. A significance level of α=0.05 between different vaccine groups was limited to individual stimulators. The significance level was α=0.05. All analyses were done using SAS v9.2 using either PROC LIFETEST or PROC MIXED.
RESULTS
Vaccination with autophagosome-containing vesicles derived from 3-MCA-induced sarcomas generates cross-reactive T cells
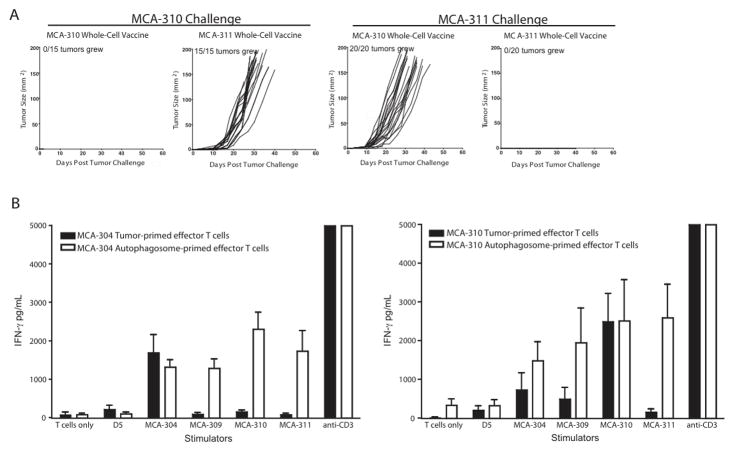
Multiple MCA sarcomas were irradiated and used in an attempt to protect mice from independently generated sarcomas. Similar to what has been described (31), irradiated MCA-310 cells protected mice against a MCA-310 tumor challenge but not a MCA-311 tumor challenge. Conversely, irradiated MCA-311 cells protected mice against a MCA-311 tumor challenge but not against MCA-310 (Figure 1A). Previous work in our lab and others has described how preeffector T cells found in the lymph node draining a tumor could be sequentially activated with anti-CD3 and IL-2 and retain tumor specificity (32, 33). MCA-304 and MCA-310 tumor-draining lymph nodes were harvested, activated in vitro with anti-CD3 and expanded with IL-2. The resultant effector T cells were assayed for IFN-γ secretion after stimulation with different tumor targets. Both MCA-304 and MCA-310 primed effector T cells secreted tumor-specific IFN-γ when stimulated with the immunizing tumor (Figure 1B, black bars). However, both groups of effector T cells failed to secrete appreciable IFN-γ in response to all other targets, including the non-vaccinating MCA-induced sarcomas. Based on our hypothesis that short-lived proteins (SLiPs) or defective ribosomal proteins (DRiPs) incorporated into autophagosomes contain unique antigens that will broaden the T-cell response, autophagosome-containing vesicles were isolated from bortezomib-treated, intact sarcomas and used as a subcutaneous vaccine. Surprisingly, MCA 304-derived autophagosomes pulsed onto APCs primed an immune response to not only MCA-304, but also to other independently derived sarcomas (Figure 1B, grey bars). Similar results were observed with an autophagosome vaccine derived from the MCA-310 sarcoma. These responses were specific because the autophagosome vaccine did not prime T cells to react with the syngeneic, but unrelated, B16 melanoma subclone (D5) or a primary kidney cell line (Figure 1B and data not shown). This suggests that an autophagosome vaccine may prime a more diverse repertoire of T cells to sarcoma-related antigens.
Figure 1.
Isolated autophagosomes prime a broader array of T cells. (A) Tumor growth curves of mice vaccinated with 5×106 irradiated sarcoma cells and challenged 14 days later with 30,000 live sarcoma cells are depicted. Tumor growth was monitored twice weekly. Each growth curve represents either 3 independent experiments with 5 mice per group (n=15 mice) or 4 independent experiments with 5 mice per group (n=20). (B) IFN-γ secretion by effector T cells primed with either a whole cell or an autophagosome vaccine was measured by ELISA. T cells were stimulated with targets for 24 hours, supernatants were harvested and IFN-γ was measured (n=3 independent experiments except for MCA-310 autophagosome-primed effectors stimulated with MCA-311 and MCA-309 sarcomas (n=2 independent experiments)).
Autophagosome vaccination protects against multiple independently derived sarcomas
To determine if the autophagosome vaccine could not only prime a broader T cell response to tumor or tumor-associated antigens, but could also protect a vaccinated host from a non-homologous (non-immunizing) sarcoma challenge, groups of mice were vaccinated with irradiated MCA-induced tumors or autophagosomes derived from these tumors and subsequently challenged with tumor. Vaccination with irradiated whole tumor cells provided complete protection against the homologous tumor. Although the irradiated whole cell vaccine protected more mice than an autophagosome vaccine derived from the same tumor, the autophagosome vaccine provided immunity against not only the homologous sarcoma, but also against other independently derived sarcomas (Figure 2A-F). These results coupled with the previous experiment in Figure 1B demonstrate that autophagosome-containing vesicles derived from MCA-induced sarcomas contain epitopes that can be cross-presented and prime a T-cell population capable of protecting the host from a challenge with sarcomas other than the one used during vaccination.
Earlier experiments demonstrated that autophagosomes pulsed onto APCs were more effective than autophagosomes alone at generating a productive vaccine (data not shown). Therefore, to control for the possibility that the addition of antigen-presenting cells (APCs) potentiates the cross-protection observed with the autophagosomal vaccination, APCs were pulsed with irradiated whole cells and used as a vaccine. Similar to what was observed in experiments without APCs, whole tumor cell vaccination with APCs was able to protect vaccinated mice from a challenge with the immunizing tumor (Supplemental Figure 1). However, the inclusion of APCs with the whole cell vaccination could not cross-protect mice from a challenge with a closely related but independently derived sarcoma in any of the vaccines tested (Supplemental Figure 1). This autophagosome-mediated cross-protection demonstrated in both Figure 2 and Supplemental Figure 1 was evident in 8 of the 9 combinations tested whereas the whole cell vaccine provided cross-protection in 0 of 9 combinations tested (Figure 2G).
Autophagosomes contain short-lived proteins that can induce proliferation of naive antigen-specific T cells
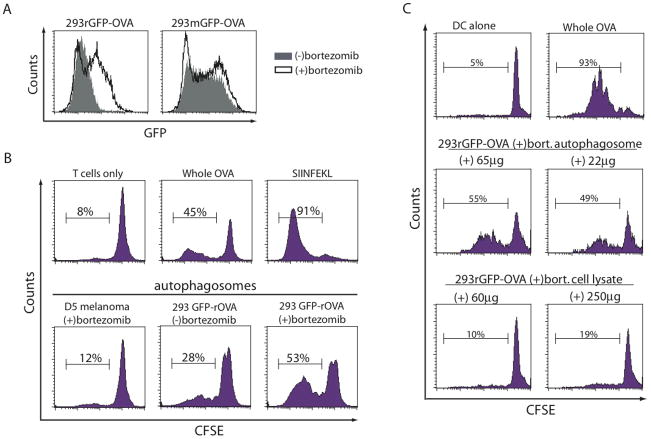
To understand the mechanism of cross-protection with the autophagosome vaccine, we used a model antigen system to test the hypothesis that short-lived proteins (SLiPs) were necessary for autophagosome-induced immunity. HEK 293T cells were transduced to stably express the model antigen ovalbumin (OVA) with a moderate half-life (approximately 8–10 hours) or a shorter half-life (approximately 20 minutes) (8). The half-life of the model proteins was controlled by inserting either a stabilizing methionine residue (mOVA) or a destabilizing arginine residue (rOVA) at the N-terminus of the protein as determined by the N-end rule (34). Additionally, to detect the model antigen more easily, the OVA proteins were fused to an enhanced green fluorescent protein (eGFP) (8). The accumulation of fusion proteins in the HEK 293 T cells was measured by flow cytometry for the presence of eGFP (Figure 3A). The amount of the stable model protein (mGFP-OVA) remained nearly constant when cultured overnight with or without proteasome inhibition. In contrast, the turnover of the short-lived model antigen (rGFP-OVA) was proteasome-dependent; blockade of the proteasome with bortezomib increased the expression of the rGFP-OVA protein.
Figure 3.
Short-lived proteins found in autophagosomes isolated from bortezomib-treated cells stimulate antigen-specific T cells. (A) Expression of the short-lived GFP-OVA fusion protein (rGFP-OVA) or the stable GFP-OVA fusion protein (mGFP-OVA) in HEK 293T cells treated with or without bortezomib was measured by flow cytometry (solid line and filled plot respectively). (B) Proliferation of naïve antigen-specific OT-1 cells measured by CFSE dilution. CFSE-labeled naïve T cells from ovalbumin-specific OT-1 TCR transgenic mice were added to an autophagosome-pulsed APC culture for 3–4 days at 37°C. n=2 experiments. (C) Bortezomib-treated HEK 293rGFP-OVA cells were lysed by sonification and autophagosomes were isolated by differential centrifugation as done previously. CFSE-labeled naïve T cells from ovalbumin-specific OT-1 TCR transgenic mice were added to APC pulsed with different quantities of either purified autophagosomes or total cell lysates and co-cultured for 3 days at 37°C. Proliferation of T cells was measured by CFSE dilution as done previously. n=1 experiment.
Autophagosomes (3×106 cell equivalents (CE)) were isolated from 293rGFP-OVA cells treated with or without the proteasome inhibitor, bortezomib, and pulsed onto antigen-presenting cells for 6 hours, which were used to stimulate naïve OVA-specific, CFSE-labeled T cells. Proliferation of the stimulated T cells was measured by dilution of CFSE. While autophagosomes isolated from untreated 293rGFP-OVA cells caused a significant increase in T cell proliferation, autophagosomes isolated from 293rGFP-OVA cells treated with bortezomib induced substantially more proliferation (Figure 3B). Additionally, autophagosomes isolated from bortezomib-treated 293rGFP-OVA cells were superior at stimulating T cell proliferation when compared to the cell lysate (Figure 3C).
Inhibition of protein synthesis before proteasome blockade prevents accumulation of short-lived proteins
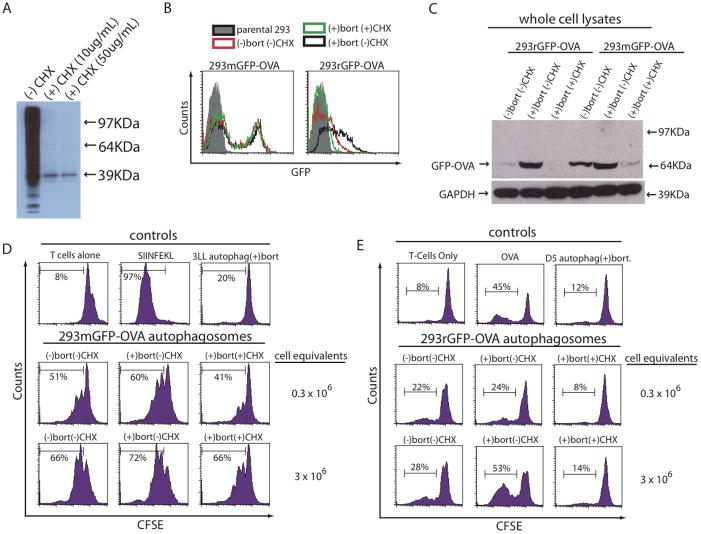
To further define the relationship of short-lived proteins and autophagosomes, cells were pretreated with the protein synthesis inhibitor cyclohexamide (CHX) before the addition of bortezomib to inhibit the production of short-lived proteins and prevent their inclusion into autophagosomes. To determine a dose of CHX capable of preventing protein synthesis, HEK 293T cells were depleted of all methionine and then treated with CHX at 10 μg/mL or 50 μg/mL, or with vehicle alone for 4 hours. Concomitantly, the cells were pulsed with a non-radioactive labeled methionine (L-azidohomoalanine) and visualized using a streptavidin-HRP-enzymatic reaction. The HRP detection system can react with endogenous biotin, such as the biotin carboxylase subunits of acetyl-CoA (39 KDa band in Figure 4A). 10ug/mL in 4 hours of cyclohexamide prevented synthesis of all proteins (Figure 4A).
Figure 4.
Inhibition of protein synthesis before proteasome blockade prevents accumulation of short-lived proteins and limits the ability of autophagosomes to stimulate antigen-specific T cells. (A) HEK 293T cells were pulsed with a non-radioactive-labeled methionine while treated with cyclohexamide or vehicle for 4 hours at the indicated concentrations. Cells were harvested and a western blot was performed to detect any labeled proteins during the 4-hour pulse period. (B and C) HEK 293T cells expressing a stable GFP-OVA fusion protein (mGFP-OVA) or a short-lived GFP-OVA fusion protein (rGFP-OVA) were treated with bortezomib for 4–24 hours or pretreated with cyclohexamide for 4 hours before addition of bortezomib. (B) Expression of GFP-OVA 4 hours after addition of bortezomib measured by flow cytometery. Filled histogram (parental HEK 293T cells), red line (untreated cells), black line (cells treated bortezomib) and green line (cells pretreated with cyclohexamide for 4 hours followed with bortezomib). (C) Expression of GFP-OVA 24 hours after addition of bortezomib or CHX and bortezomib measured by western blot with anti-GFP in total cell lysates from treated cells. (D and E) Autophagosomes were isolated from 293rGFP-OVA or 293mGFP-OVA cells treated as in (C). Non-specific control autophagosomes were isolated from (D) 3LL carcinoma or (E) D5 melanoma. Autophagosomes were pulsed onto APCs and used to stimulate naïve antigen-specific OT-1 T cells. Proliferation was measured by CFSE dilution. (D) 293mGFP-OVA derived autophagosomes n=2 experiments. (E) 293rGFP-OVA derived autophagosomes n=3 experiments.
We hypothesized that since mOVA is a stable protein, its expression would be detected after a 4 hour pretreatment with CHX prior to either a 4-hour or overnight culture with bortezomib whereas there would be diminished expression of rOVA because of its short half-life. Expression of GFP in HEK 293mGFP-OVA cells was essentially unchanged after 4 hours regardless of whether they were treated with bortezomib or CHX (Figure 4B, left panel). However, the expression of GFP in HEK 293rGFP-OVA cells was diminished in the absence of proteasome inhibition and absent following CHX pretreatment (Figure 4B, right panel). The reduced expression of the model short-lived protein was not due to decreased cell viability or a secondary effect of bortezomib or CHX as both 293rGFP-OVA cells and the 293mGFP-OVA cells treated with CHX and bortezomib or bortezomib alone had similar morphology (slightly more rounded cells with CHX), growth patterns and were able to exclude trypan blue even after an overnight culture (data not shown).
The expression of both mGFP-OVA and rGFP-OVA in HEK 293T cell lysates was also examined by western blot after a 24 hour treatment. Similar to what was observed by flow cytometry, bortezomib caused a modest increase in the more stable mOVA protein. However, HEK 293mGFP-OVA cells cultured overnight with CHX and bortezomib exhibited a substantial reduction in the levels of the 64 KDa fusion protein (Figure 4C). Bortezomib increased the half-life of the rGFP-OVA fusion protein in HEK 293rGFP-OVA cells only in the absence of CHX.
Autophagosomes derived from cells that lack short-lived proteins have diminished capacity to stimulate antigen-specific T cells
Autophagosomes derived from cells expressing either the short-lived rGFP-OVA protein or the stable mGFP-OVA protein were isolated, used to pulse APCs and drive naïve antigen-specific OT-1 T-cell proliferation. Autophagosomes from cells expressing the stable mGFP-OVA protein caused OT-1 T cells to proliferate at nearly the same level whether they were generated in the presence or absence of bortezomib, with or without CHX (Figure 4D). These data indicate that CHX pretreatment does not prevent stable proteins from being sequestered in autophagosomes nor does it prevent the formation of “functional” autophagosomes. However, autophagosomes from CHX-treated HEK 293rGFP-OVA cells were unable to induce antigen-specific T-cell proliferation (Figure 4E). The diminished proliferation was more apparent when higher doses of autophagosomes were used to stimulate the naïve T cells. These experiments demonstrate that stable proteins, but not short-lived proteins are incorporated into autophagosomes when protein synthesis is inhibited.
Autophagosomes that lack short-lived proteins fail to protect hosts from MCA tumor challenge
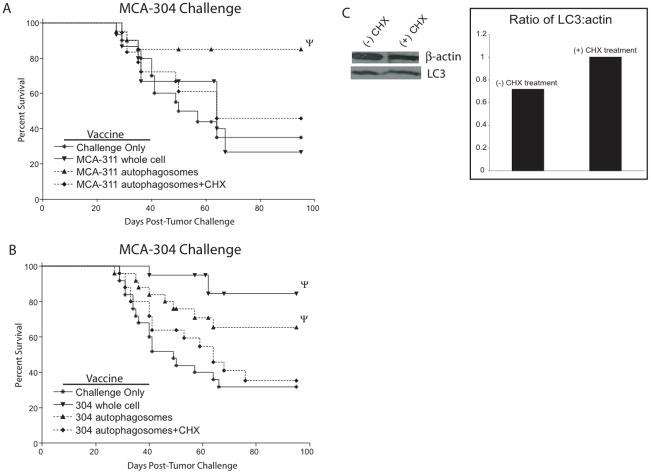
Based on data from the HEK 293 model antigen system, which suggests that SLiPs are incorporated into autophagosomes and can be necessary for their efficacy as a vaccine, MCA-induced sarcomas were treated with CHX to inhibit protein synthesis before the addition of bortezomib. Autophagosomes isolated from MCA-311 cells treated with or without CHX were used to vaccinate mice, which were then challenged with MCA-304 to assess the ability of MCA-311 autophagosomes to cross-protect (Figure 5A). Vaccination with autophagosomes isolated from bortezomib-treated cells (complete autophagosomes) protected 85% (17 of 20) of the mice. In contrast, vaccination with autophagosomes isolated from cells that were pretreated with CHX before the addition of bortezomib (CHX autophagosomes) protected only 47% (9 of 19 mice) of the mice, which was not significantly different than non-vaccinated mice (8 of 20 mice protected) or mice vaccinated with non-homologous MCA-311 whole cells (4 of 10 mice protected). These results suggest that the presence of short-lived proteins in the autophagosome vaccine was necessary to generate cross-protection to an independently-derived sarcoma.
Figure 5.
Autophagosomes isolated from MCA-induced sarcomas lacking short-lived proteins lose the ability to protect hosts from viable sarcoma challenge. (A and B) Kaplan-Meier survival plots comparing protection of mice vaccinated with a whole cell sarcoma or autophagosomes derived from the sarcomas with or without CHX pretreatment and challenged with MCA-304. Survival curves denoted with Ψ were significantly different than the corresponding no vaccine group of mice (p<0.05). Each survival plot used 4 independent experiments with 5 mice per group (n=20). (C) Detection of β-actin and LC3 in autophagosomes isolated from MCA-304 treated with or without CHX determined by western blot and the relative ratio of β-actin and LC3 determined by densitometry.
To understand further the role that SLiPs may play in generating a protective immune response against a homologous tumor challenge, autophagosomes isolated from CHX-treated MCA-304 cells were pulsed onto APCs and used to vaccinate groups of mice. Whereas only 20% (4 of 20) of mice vaccinated with a MCA-304 whole cells and 32% (8 of 25) with MCA-304 autophagosome vaccine grew tumor, 64% (16 of 25) of mice vaccinated with CHX-treated MCA-304 autophagosome grew tumor (Figure 5B). This incidence of tumor growth was nearly the same as the non-vaccinated mice (17 of 25).
While CHX can have dramatic biological effects, MCA-304 cells treated overnight with bortezomib and with or without CHX displayed similar viability and the autophagosomes isolated from CHX-treated tumors had similar protein concentrations (1.97 mg/mL and 2.15 mg/mL respectively). To assess the potential negative effects of CHX on autophagy, which could limit more than just the incorporation of SLiPs and DRiPs into autophagosomes, LC3, a critical component necessary for the induction of autophagy and thus autophagosome formation, was measured by western blot. The ratio of LC3 to total protein was the same or even greater with CHX pretreatment in both HEK293mGFP-OVA (Figure 6A) and MCA-304 autophagosomes (Figure 5C).
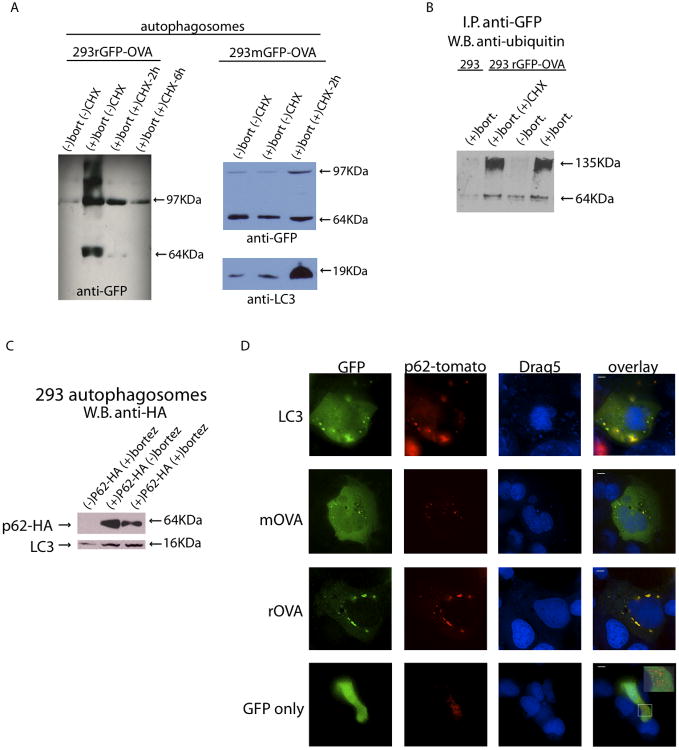
Figure 6.
Autophagosomes contain ubiquitinated short-lived proteins and the ubiquitin/LC-3 binding protein p62 that co-localize in HEK 293 cells. (A) Western blot measuring the expression of GFP-OVA in autophagosomes isolated 24 hours after addition of bortezomib or CHX and bortezomib to either HEK 293rGFP-OVA cells or HEK 293mGFP-OVA cells. (B) Autophagosomes produced from parental HEK 293T cells or HEK 293rGFP-OVA cells were used for an immunoprecipitation with anti-GFP antibody. The eluate from the i.p. was used for a western blot with anti-ubiquitin. (C) HEK 293 FT cells were mock transfected or transfected with an HA-tagged p62. After 48 hours autophagosomes were isolated and used for a western blot with anti-HA. (D) HEK 293 FT cells were co-transfected with either GFP-tagged LC3 or rOVA and tomato-tagged p62. After 48 hours the cells were treated with bortezomib and analyzed by fluorescent microscopy after 24 hours. Scale bar, 10μM. Inset box in lower right panel is a 2.2x magnification.
Autophagosomes contain ubiquitinated proteins and the sequestosome/p62, both of which co-localize with LC3
To examine the effects of CHX on protein incorporation into autophagosomes, expression of mGFP-OVA and rGFP-OVA in isolated autophagosomes was determined by western blot. Autophagosomes isolated from HEK 293mGFP-OVA cells had nearly the same expression of mGFP-OVA regardless of treatment with bortezomib or CHX (Figure 6A). Interestingly, the autophagosomal marker LC3 was overexpressed in the CHX-treated cells relative to the +/− bortezomib groups (Figure 6A). This increase in LC3 has been demonstrated to occur when cells undergo stress, such as inhibition of protein synthesis (9). It is also of note that while the cellular expression of mGFP-OVA pre-treated with CHX was reduced (Figure 4C), its expression in the autophagosome-containing vesicles was not (Figure 6A). This suggests that mOVA-containing autophagosomes blebbed from the cells, reducing the cellular expression of this fusion protein while maintaining its expression in the autophagosomes. The model short-lived protein, rGFP-OVA, was detected only if the proteasome was inhibited in cells with normal protein synthesis; it was absent in autophagosomes produced from cells treated with CHX. Therefore, by inhibiting protein synthesis before blocking the proteasome, isolated autophagosomes contain minimal amounts of short-lived protein.
Since SLiPs were a critical component of the autophagosome vaccine (Figures 4E and 5), we were interested in how they were delivered to the autophagosomal pathway. Our hypothesis predicts and others have shown (8) that treating cells with bortezomib increases the pool of ubiquitinated proteins, which may provide a means of delivery to the autophagosomal pathway. Interestingly, higher molecular weight bands were detected with anti-GFP in HEK 293rGFP-OVA autophagosomes isolated from bortezomib-treated cells (Figure 6A), but not in the cells used to produce the autophagosomes (Figure 4B). Although these high weight bands were also present in autophagosomes isolated from cells expressing the more stable mGFP-OVA protein, they were less abundant. The N-end rule predicts that the model short-lived protein will be quickly degraded in a ubiquitin-proteasome-dependent fashion (34). We therefore hypothesized that these larger proteins were a ubiquitinated species of the fusion protein that escaped degradation by the proteasome and were enriched in the autophagosomes isolated from the cells. To test this hypothesis, autophagosomes from HEK 293rGFP-OVA cells were immunoprecipitated with anti-GFP and western blotted with anti-ubiquitin. The anti-ubiquitin antibody bound only to the higher molecular weight proteins found in autophagosomes isolated from cells treated with bortezomib (+/− CHX) (Figure 6B) but not in autophagosomes from untreated 293rGFP-OVA cells or bortezomib-treated parental HEK 293T cells. This observation demonstrates that ubiquitinated short-lived proteins were present in autophagosomes.
Recent work has demonstrated that the ubiquitin-binding protein sequestosome/p62 can interact with both ubiquitinated proteins and autophagy-related initiator LC3, potentially delivering these proteins to the autophagy pathway (16, 17, 35). Western blots showed clearly that p62 was present in the autophagosomes (Figure 6C). Co-localization of LC3 and p62 in HEK 293 T cells was seen with confocal microscopy using GFP-tagged LC3 and tomato-tagged p62. The short-lived protein, rGFP-OVA, also co-localized with tomato-tagged p62 in transfected HEK 293 T cells but not with a control GFP vector (Figure 6D). These results demonstrate that ubiquitinated proteins are packaged into autophagosomes with ubiquitin/LC3 binding protein p62, especially when cells are treated with bortezomib.
p62 is necessary for the delivery of ubiquitinated proteins to autophagosomes
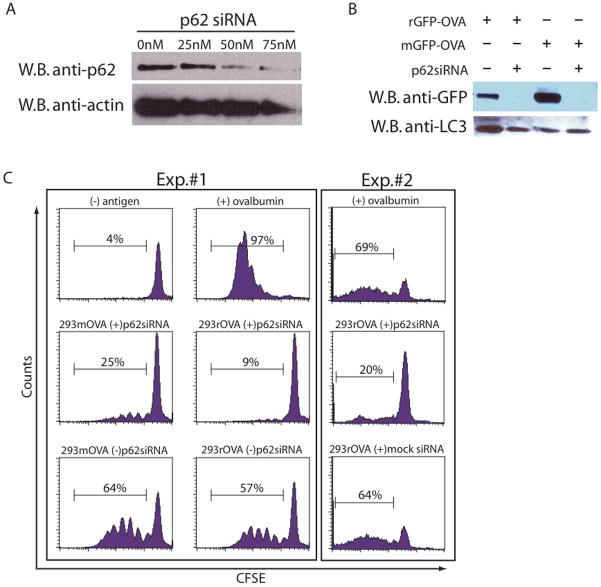
To understand how the sequestosome may influence trafficking of specific proteins to autophagosomes, p62 expression was knocked down in HEK 293T cells. A titration of p62 siRNA showed that protein expression was greatly reduced with 50 nM of the p62 smart pool (Figure 7A). HEK 293T cells were then transfected simultaneously with both p62 siRNA and a plasmid that expressed either rGFP-OVA or mGFP-OVA. After 24-hours, bortezomib was added and the cells were cultured for an additional 24 hours before isolating the autophagosome-containing vesicles. A portion of the autophagosomes isolated from the transfected cells was used to detect the fusion protein by western blot (Figure 7B). The knockdown of p62 in both rGFP-OVA and mGFP-OVA transfected cells greatly reduced the level of the GFP-OVA. This knockdown did not affect the levels of LC3 in the autophagosomes, suggesting that autophagosome formation was independent of p62 expression (Figure 7B).
Figure 7.
p62 in necessary for delivery of ubquitinated proteins to autophagosomes. (A) HEK 293T cells were transfected with a pool of p62 siRNA at various concentrations. After 48 hours the cells were collected and lysates were probed for the presence of p62 by western blot. (B and C) HEK 293T cells were cotransfected with either mGFP-OVA or rGFP-OVA and p62siRNA or control siRNA. After 24 hours bortezomib was added to the cells and cultured for an additional 24 hours before harvesting the autophagosomes. (B) An anti-GFP and anti-LC3 western blot of the autophagosome preparation. (C) Autophagosome-pulsed APCs were washed and added to CFSE-labeled naïve OT-1 cells to measure T-cell proliferation as in Figure 3. (B and C) n=2 experiments.
Based on the co-localization experiments and the presence of the ubiquitinated protein species in isolated autophagosomes, we hypothesized that knockdown of the sequestosome would greatly diminish the ability of autophagosomes to stimulate naive T cell proliferation. To test this hypothesis, autophagosomes generated from the co-transfection described above were pulsed onto APCs and used to stimulate CFSE-labeled naive antigen specific T cells as previously described. T cells stimulated with autophagosomes generated from 293 cells with the p62 knockdown proliferated less than those stimulated without the knockdown (Figure 7C). While the p62 knockdown diminished the proliferation of T cells stimulated with mGFP-OVA autophagosomes, the inhibition of proliferation was even greater with rGFP-OVA autophagosomes knocked down for p62. These results coupled with the presence of ubiquitinated proteins in the rGFP-OVA autophagosomes and the co-localization of the LC3 and p62 demonstrated that delivery of short-lived proteins to autophagosomes is sequestosome dependent.
DISCUSSION
We have demonstrated that an autophagosome vaccine can prime a repertoire of T cells with a broader tumor-specific reactivity than the intact sarcoma from which it was derived. In contrast to the irradiated whole cell vaccine, vaccination with tumor-derived autophagosomes protected mice from a challenge with independently-derived MCA sarcomas. Experiments using MCA sarcomas, or the OVA model antigen system, both suggest that short-lived proteins incorporated into autophagosomes are necessary in generating a robust immune response. Additionally, these data suggest that the sequestering of SLiPs into autophagosomes is dependent on the ubiquitin binding protein sequestosome/p62. This novel vaccine strategy has redefined the tumor rejection antigens associated with MCA-induced tumors and has created the potential for new vaccine strategies.
While MCA-induced sarcomas can share similar histology, growth patterns, sensitivity to chemotherapeutics and are generated in syngeneic mice using the same carcinogen, each sarcoma induces a unique tumor-specific immune response. This observation has been made by many researchers (31, 33, 36–38), and it is also evident in our studies by the failure of our MCA-induced sarcoma whole cell vaccine to cross-protect against independently derived but related MCA-induced sarcomas (Figure 1A). While this failure to cross-protect has been attributed to each sarcoma’s unique antigenic profile (38), it seems unlikely that a large panel of sarcomas of similar etiology will not have common antigens. A more plausible description of this unique specificity could be described by the concept of a tumor rejection antigen, which functionally relates to how well an immune response directed against a tumor antigen can limit tumor growth (39, 40). The hierarchy of a tumor rejection antigen, which ranges from poor to strong, can be influenced by the ability of a specific antigen to prime an immune response. Thus while there may be a common pool of antigens shared between independently-derived sarcomas, individual tumors can express unique antigen(s) that may dominate the immune response, effectively limiting the priming of shared epitopes. This immune dominance may relate to multiple factors. Pion and colleagues have demonstrated that immune dominance is determined by not only the affinity of the TCR and peptide-MHC complex (p:MHC) but also the surface density of p:MHC on the APC (41), which is directly proportional to an epitope’s MHC binding affinity, linking the establishment of immune dominance to antigen presentation. This observation coupled with previous findings which demonstrate the necessity of cross-presentation during tumor immunity (2) illustrates how cross-presentation may determine immune dominance and consequently the tumor specificity seen with MCA-induced sarcomas. Thus a possible tumor rejection antigen must not only be stable enough to be transferred to an APC, it must also possess an epitope that can compete for binding of the APC’s MHC molecules as well as have an affinity for the TCR. Due to their transient nature, SLiPs are poorly cross-presented and are therefore removed from the pool of possible rejection antigens.
The rules governing cross-presentation of tumor or tumor-associated antigens with a cellular vaccine may not apply to an autophagosome vaccine as short-lived proteins can be stabilized by proteasome blockade, isolated in autophagosomes and pulsed onto APCs ex vivo or even injected directly into a lymphnode (intranodal). Consequently, we have focused on the repertoire of antigen, specifically the short-lived proteins sequestered, as a means of altering the tumor specificity with an autophagosome vaccine. Based on the observation that both protection and cross protection from an MCA sarcoma challenge were reduced with elimination of SLiPs from the autophagosome vaccine (Figure 5), antigens common to independently derived MCA-induced sarcomas are likely to be SLiPs. These SLiPs may provide epitopes with a high affinity for MHC molecules allowing them to compete with the dominant whole cell rejection antigen for surface density on the APC. Alternatively, the dominant rejection antigen may not be sequestered into autophagosomes, providing subdominant, neo or cryptic antigens access to MHC molecules for cross presentation.
While the potential of a varied repertoire of antigen in autophagosomes is a novel and significant finding, further studies are needed to augment the efficacy of an autophagosome vaccine. The secretion of IFN-γ by autophagosome-primed effector T cells indicates that cross-reactive tumor immune responses are generated (Figure 1B) yet complete protection of vaccinated mice was not achieved. The lack of a memory T cell response during the autophagosome vaccination may explain why tumor growth curves were only shifted in a proportion of mice vaccinated with autophagosomes. A vaccine strategy that incorporates an OX-40 agonist could boost the generation of memory T cells, providing a further delay in tumor progression and lead to a larger frequency of protected mice. Alternatively, the difference between delayed tumor growth versus complete protection could relate to the initial magnitude of primed T cells. The addition of a TLR agonist during the vaccine could increase critical co-stimulatory molecules during cross-presentation, leading to a stronger immune response. Additionally, our data suggests that increasing the concentration SLiPs or their delivery to the autophagosomes would be a means of increasing the potency of the vaccine.
The targeting of SLiPs into autophagosomes appeared to be coordinated, as cell lysates could not stimulate antigen specific T cells as effectively as could autophagosomes, suggesting a relative concentration of SLiPs in autophagosomes. This observation was supported by the remarkable increase in the proliferation of antigen-specific T cells when simulated with isolated autophagosomes versus whole cell lysate (Figure 3C). Western blot analysis suggests that the short-lived OVA protein was readily detected at much lower total protein concentrations in autophagosomes than in cell lysates (Figures 4C and 6A and data not shown). Additionally, higher molecular weight fusion proteins were detected only in autophagosomes and not in cell lysates. These higher molecular weight proteins were more obvious in autophagosomes derived from bortezomib treated cells and especially those containing the model SLiP antigen which is rapidly ubiquitinated versus the more stable antigen (Figures 6A). The higher weight band, confirmed to be ubiquitinated by western blot analysis after co-immunoprecipitation (Figure 6B), was likely responsible for directing SLiPs into autophagosomes based on recent publications that established a link between autophagy and ubiquitinated proteins via the LC3 and ubiquitin binding protein p62/sequestosome (42–44). Indeed HA-tagged p62 was readily detected in autophagosomes isolated from transfected cells and fluorescent microscopy demonstrated co-localization of p62 with both the model SLiP as well as LC3 (Figures 6C and 6D), all supporting the hypothesis that p62 regulates the sequestering of ubiquitinated proteins into autophagosomes. While using siRNA to knockdown expression of p62 may destabilize or limit expression of the GFP-OVA construct, these experiments corroborate sequestosome’s role in antigen delivery to autophagosomes as reduced expression of p62 limited the model antigen found in the autophagosomes and diminished proliferation of antigen-specific T cell stimulated with these autophagosomes. Autophagosomes isolated from HEK293rGFP-OVA cells treated with p62 siRNA had a greater impairment in their ability to stimulate T-cell proliferation than autophagosomes isolated from p62 siRNA-treated cells expressing the more stable mGFP-OVA antigen (Figure 7C), suggesting a correlation between the stability or state of ubiquitination of the protein and the necessity of p62 to transfer the protein into the autophagosome.
Collectively the data points to a model whereby SLiPs or other ubiquitinated proteins are sequestered into autophagosomes in a p62 dependent fashion when the cell is treated with a proteasome inhibitor. Furthermore, data from the MCA-induced sarcoma cross-protection experiments suggest that these SLiPs include antigenic determinants common to multiple independently-derived sarcomas. Several possibilities may explain how a whole cell vaccine does not afford the cross-reactivity observed with an isolated autophagosome vaccine. The most simple explanation is that tumors rapidly turn-over SLiPs in a proteasome-dependant fashion thereby preventing their accumulation. This explanation makes the assumption that MCA-induced tumors do not directly prime an immune response since rapidly degraded proteins fill a large proportion of MHC class I molecules on the cell surface (20). Alternatively, SLiPs may accumulate at low levels but are not stable enough to be efficiently cross-presented which is supported by the observation that superior T cell proliferation occurs with much less autophagosomes than cell lysates. If both SLiPs and whole cell tumor rejection antigens are equally cross-presented, the rejection antigens may assert immune dominance by binding MHC class I more efficiently or in the context of MHC class I, have a higher affinity for the TCR (41). Addressing these possibilities will provide a greater understanding of not only this novel finding but also a potential class of novel tumor antigens.
The implication of these studies from a translational point of view is that autophagosomes may be a way to uncover common antigenic determinants created during the transformation process. Evidence supporting this concept in this model comes from two studies. The first study identified a region of the mouse genome rich in tumor-modifier genes and associated with MCA-induced tumor antigens (22). A second study showed that mice vaccinated with a MCA-induced tumor that had lost the dominant antigen could generate CTL, which recognized a shared antigen expressed not only by the immunizing cell line, but also by independent sarcoma cell lines (23). The idea that tumor-derived autophagosomes may contain common tumor-specific or tumor-associated antigens is clinically appealing. These autophagosomes could have the potential of an “off the shelf” vaccine in an allogeneic setting, particularly if an autologous tumor is unavailable to the patient. Additionally, this strategy could be useful in priming immune responses against tumor-antigen loss variants seen with a progressively growing tumor and may even serve as an alternative vaccine to treat patients coming out of remission. This strategy has the potential to augment immunity in clinical applications and positively contribute to the field of immunotherapy.
Supplementary Material
Supplemental Figure 1: Irradiated whole cell sarcoma-pulsed APCs do not protect mice from a challenge with non-homologous sarcomas. Kaplan-Meier survival curves of mice vaccinated with irradiated whole cell sarcoma-pulsed APCs or autophagosome-pulsed APCs. Autophagosome vaccination was performed as in Figure 2. Subcutaneous whole cell vaccinations consisted of 3×106 antigen-presenting cells pulsed with 5×106 irradiated tumor cells injected into the lower right flank of the mouse. Mice were challenged with 30,000 viable MCA-304 sarcomas. Tumor growth was monitored at least twice weekly. Each survival plot represents 2 independent experiments with 5 mice per group (n=10). Survival curves denoted with Ψ represent statistically significant (p>0.05) protection from a tumor challenge compared to no vaccine.
TRANSLATIONAL RELEVANCE.
For more than 50 years, the basic paradigm that vaccination with chemically induced tumor cells is effective at protecting mice with a challenge from that specific tumor, but not other tumors induced by the same carcinogen, has been repeatedly documented, is well accepted and has driven numerous autologous cancer vaccine trials. The importance of the current paper is that it identifies a methodology that improves vaccine efficacy sufficiently enough to prime immunity against shared antigens in models where whole tumor vaccines fail to do so. It also provides the mechanisms responsible for augmented priming and insights to further enhancements. Better vaccines will help, but generating therapeutic immune responses in patients with advanced cancer will likely require a multimodality combination approach. A clinical study of this strategy is underway.
References
- 1.Bevan MJ. Cross-priming for a secondary cytotoxic response to minor H antigens with H-2 congenic cells which do not cross-react in the cytotoxic assay. J Exp Med. 1976;143:1283–8. doi: 10.1084/jem.143.5.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang AY, Golumbek P, Ahmadzadeh M, Jaffee E, Pardoll D, Levitsky H. Role of bone marrow-derived cells in presenting MHC class I-restricted tumor antigens. Science. 1994;264:961–5. doi: 10.1126/science.7513904. [DOI] [PubMed] [Google Scholar]
- 3.Shen L, Rock KL. Cellular protein is the source of cross-priming antigen in vivo. Proc Natl Acad Sci U S A. 2004;101:3035–40. doi: 10.1073/pnas.0308345101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kunisawa J, Shastri N. Hsp90alpha chaperones large C-terminally extended proteolytic intermediates in the MHC class I antigen processing pathway. Immunity. 2006;24:523–34. doi: 10.1016/j.immuni.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 5.Norbury CC, Basta S, Donohue KB, Tscharke DC, Princiotta MF, Berglund P, et al. CD8+ T cell cross-priming via transfer of proteasome substrates. Science. 2004;304:1318–21. doi: 10.1126/science.1096378. [DOI] [PubMed] [Google Scholar]
- 6.Lev A, Takeda K, Zanker D, Maynard JC, Dimberu P, Waffarn E, et al. The exception that reinforces the rule: crosspriming by cytosolic peptides that escape degradation. Immunity. 2008;28:787–98. doi: 10.1016/j.immuni.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, Wang LX, Pang P, Twitty C, Fox BA, Aung S, et al. Cross-presentation of tumor associated antigens through tumor-derived autophagosomes. Autophagy. 2009;5:576–7. doi: 10.4161/auto.5.4.8366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Wang LX, Yang G, Hao F, Urba WJ, Hu HM. Efficient cross-presentation depends on autophagy in tumor cells. Cancer Res. 2008;68:6889–95. doi: 10.1158/0008-5472.CAN-08-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abeliovich H, Dunn WAJ, Kim J, Klionsky DJ. Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J Cell Biol. 2000;151:1025–34. doi: 10.1083/jcb.151.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szajnik M, Czystowska M, Szczepanski MJ, Mandapathil M, Whiteside TL. Tumor-derived microvesicles induce, expand and up-regulate biological activities of human regulatory T cells (Treg) PLoS One. 2010;5:e11469. doi: 10.1371/journal.pone.0011469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–93. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 12.Zitvogel L, Mayordomo JI, Tjandrawan T, DeLeo AB, Clarke MR, Lotze MT, et al. Therapy of murine tumors with tumor peptide-pulsed dendritic cells: dependence on T cells, B7 costimulation, and T helper cell 1-associated cytokines. J Exp Med. 1996;183:87–97. doi: 10.1084/jem.183.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huber V, Filipazzi P, Iero M, Fais S, Rivoltini L. More insights into the immunosuppressive potential of tumor exosomes. J Transl Med. 2008;6:63. doi: 10.1186/1479-5876-6-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iero M, Valenti R, Huber V, Filipazzi P, Parmiani G, Fais S, et al. Tumour-released exosomes and their implications in cancer immunity. Cell Death Differ. 2008;15:80–8. doi: 10.1038/sj.cdd.4402237. [DOI] [PubMed] [Google Scholar]
- 15.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–78. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 16.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–14. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 18.English L, Chemali M, Duron J, Rondeau C, Laplante A, Gingras D, et al. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat Immunol. 2009;10:480–7. doi: 10.1038/ni.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jagannath C, Lindsey DR, Dhandayuthapani S, Xu Y, Hunter RLJ, Eissa NT. Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat Med. 2009;15:267–76. doi: 10.1038/nm.1928. [DOI] [PubMed] [Google Scholar]
- 20.Yewdell JW, Nicchitta CV. The DRiP hypothesis decennial: support, controversy, refinement and extension. Trends Immunol. 2006;27:368–73. doi: 10.1016/j.it.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 21.Ikeda H, Ohta N, Furukawa K, Miyazaki H, Wang L, Kuribayashi K, et al. Mutated mitogen-activated protein kinase: a tumor rejection antigen of mouse sarcoma. Proc Natl Acad Sci U S A. 1997;94:6375–9. doi: 10.1073/pnas.94.12.6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akilesh S, Dudley ME, Eden PA, Roopenian DC. Efficient chromosomal mapping of a methylcholanthrene-induced tumor antigen by CTL immunoselection. J Immunol. 2001;167:5143–9. doi: 10.4049/jimmunol.167.9.5143. [DOI] [PubMed] [Google Scholar]
- 23.Dudley ME, Roopenian DC. Loss of a unique tumor antigen by cytotoxic T lymphocyte immunoselection from a 3-methylcholanthrene-induced mouse sarcoma reveals secondary unique and shared antigens. J Exp Med. 1996;184:441–7. doi: 10.1084/jem.184.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsumura T, Krinock RA, Chang AE, Shu S. Cross-reactivity of anti-CD3/IL-2 activated effector cells derived from lymph nodes draining heterologous clones of a murine tumor. Cancer Res. 1993;53:4315–21. [PubMed] [Google Scholar]
- 25.Winter H, Hu HM, Poehlein CH, Huntzicker E, Osterholzer JJ, Bashy J, et al. Tumour-induced polarization of tumour vaccine-draining lymph node T cells to a type 1 cytokine profile predicts inherent strong immunogenicity of the tumour and correlates with therapeutic efficacy in adoptive transfer studies. Immunology. 2003;108:409–19. doi: 10.1046/j.1365-2567.2003.01596.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winter H, Hu HM, Urba WJ, Fox BA. Tumor regression after adoptive transfer of effector T cells is independent of perforin or Fas ligand (APO-1L/CD95L) J Immunol. 1999;163:4462–72. [PubMed] [Google Scholar]
- 27.Suda T, Liu D. Hydrodynamic gene delivery: its principles and applications. Mol Ther. 2007;15:2063–9. doi: 10.1038/sj.mt.6300314. [DOI] [PubMed] [Google Scholar]
- 28.Chen Q, Khoury M, Chen J. Expression of human cytokines dramatically improves reconstitution of specific human-blood lineage cells in humanized mice. Proc Natl Acad Sci U S A. 2009;106:21783–8. doi: 10.1073/pnas.0912274106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He Y, Pimenov AA, Nayak JV, Plowey J, Falo LDJ, Huang L. Intravenous injection of naked DNA encoding secreted flt3 ligand dramatically increases the number of dendritic cells and natural killer cells in vivo. Hum Gene Ther. 2000;11:547–54. doi: 10.1089/10430340050015734. [DOI] [PubMed] [Google Scholar]
- 30.Stromhaug PE, Berg TO, Fengsrud M, Seglen PO. Purification and characterization of autophagosomes from rat hepatocytes. Biochem J. 1998;335:217–24. doi: 10.1042/bj3350217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prehn RT, Main JM. Immunity to methylcholanthrene-induced sarcomas. J Natl Cancer Inst. 1957;18:769–78. [PubMed] [Google Scholar]
- 32.Hu HM, Winter H, Urba WJ, Fox BA. Divergent roles for CD4+ T cells in the priming and effector/memory phases of adoptive immunotherapy. J Immunol. 2000;165:4246–53. doi: 10.4049/jimmunol.165.8.4246. [DOI] [PubMed] [Google Scholar]
- 33.Yoshizawa H, Chang AE, Shu S. Specific adoptive immunotherapy mediated by tumor-draining lymph node cells sequentially activated with anti-CD3 and IL-2. J Immunol. 1991;147:729–37. [PubMed] [Google Scholar]
- 34.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–86. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 35.Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci U S A. 2008;105:20567–74. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Old LJ, Boyse EA. Immunology of experimental tumors. Annu Rev Med. 1964;15:167–86. doi: 10.1146/annurev.me.15.020164.001123. [DOI] [PubMed] [Google Scholar]
- 37.Klein G, Sjogren HO, Klein E, Hellstrom KE. Demonstration of resistance against methylcholanthrene-induced sarcomas in the primary autochthonous host. Cancer Res. 1960;20:1561–72. [PubMed] [Google Scholar]
- 38.Basombrio MA. Search for common antigenicities among twenty-five sarcomas induced by methylcholanthrene. Cancer Res. 1970;30:2458–62. [PubMed] [Google Scholar]
- 39.Van Pel A, Boon T. Protection against a nonimmunogenic mouse leukemia by an immunogenic variant obtained by mutagenesis. Proc Natl Acad Sci U S A. 1982;79:4718–22. doi: 10.1073/pnas.79.15.4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gilboa E. The makings of a tumor rejection antigen. Immunity. 1999;11:263–70. doi: 10.1016/s1074-7613(00)80101-6. [DOI] [PubMed] [Google Scholar]
- 41.Pion S, Christianson GJ, Fontaine P, Roopenian DC, Perreault C. Shaping the repertoire of cytotoxic T-lymphocyte responses: explanation for the immunodominance effect whereby cytotoxic T lymphocytes specific for immunodominant antigens prevent recognition of nondominant antigens. Blood. 1999;93:952–62. [PubMed] [Google Scholar]
- 42.Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell. 2009;33:517–27. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–75. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–16. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Irradiated whole cell sarcoma-pulsed APCs do not protect mice from a challenge with non-homologous sarcomas. Kaplan-Meier survival curves of mice vaccinated with irradiated whole cell sarcoma-pulsed APCs or autophagosome-pulsed APCs. Autophagosome vaccination was performed as in Figure 2. Subcutaneous whole cell vaccinations consisted of 3×106 antigen-presenting cells pulsed with 5×106 irradiated tumor cells injected into the lower right flank of the mouse. Mice were challenged with 30,000 viable MCA-304 sarcomas. Tumor growth was monitored at least twice weekly. Each survival plot represents 2 independent experiments with 5 mice per group (n=10). Survival curves denoted with Ψ represent statistically significant (p>0.05) protection from a tumor challenge compared to no vaccine.