Summary Paragraph
Dramatic changes in socioeconomic status, cultural traditions, population growth, and agriculture are affecting diets worldwide. Understanding how our diet and nutritional status influence the composition and dynamic operations of our gut microbial communities, and the innate and adaptive arms of our immune system, represents an area of scientific need, opportunity and challenge. The insights gleaned should help address a number of pressing global health problems.
The need
A number of reviews have appeared recently about efforts to decipher the interactions between the innate and adaptive immune system and the tens of trillions of microbes that live in our gastrointestinal tracts (‘the gut microbiota’). Here we emphasize how the time is right and the need is great to better understand the interrelationships between diet, nutritional status, the immune system and microbial ecology in humans at different stages of life, living in distinct cultural and socioeconomic settings. Why now? The answer lies in a confluence of forces occurring at the present time that will affect the future. First, there is enormous pressure to devise new ways to feed healthy foods to a human population whose size is predicted to expand to 9 billion by 2050. At the same time, the solutions will have to address the challenges of developing sustainable forms of agriculture in the face of constrained land and water resources 1. Second, there is a great need to develop new translational medicine pipelines for more rigorously defining the nutritional value of foods we consume currently and that we envision creating in the future. These pipelines are required to evaluate health claims made about food ingredients. Third, there is increasing evidence that the nutritional value of food is influenced in part by the structure and operations of a consumer’s gut microbial community, and that food in turn shapes the microbiota and its vast collection of microbial genes (the ‘gut microbiome’) (e.g. ref 2). Therefore, to better define the nutritional value of foods and our nutritional status, we need to know more about our microbial differences and their origins, including how our lifestyles influence the assembly of gut microbial communities in children, and about the transmission of these communities within and across generations of a kinship 3. Fourth, we are learning how our gut microbial communities and immune systems co-evolve during our lifespans and how components of the microbiota impact the immune system. At the same time, we are obtaining more information about how our overall metabolic phenotypes (metabotypes) reflect myriad functions encoded in our human genomes and gut microbiomes. These observations raise the question of how gut microbial community metabolism of the foods we consume affects our immune systems. Fifth, the link between infections that occur within and outside the gut, and the development of nutritional deficiencies has been emphasized for many years. Poor nutrition in turn, increases the risk for infection. Nonetheless, there is still a dearth of mechanistic information that explains these observations. Sixth, only five years remain to achieve the UN’s eight Millennium Development Goals (http://www.undp.org/mdg/). Two of these goals relate to human nutrition: goal 1 seeks to eradicate extreme poverty and hunger while goal 4 aims to reduce by two thirds the under-five mortality rate. Up to one billion people suffer from undernutrition of varying degrees, including ‘silent’ or asymptomatic malnutrition (http://www.fao.org/publications/sofi/en/), making this condition an enormous global health problem. Of the ~10 million children under the age of 5 who die each year, undernutrition contributes in some fashion to >50% of deaths 4. Sadly, children who survive periods of severe undernutrition can suffer long-term sequelae including stunting and neurodevelopmental deficits 5. Moreover, the effects of undernutrition can be felt across generations. Undernourished mothers suffer higher rates of morbidity and mortality and are more likely to give birth to low birth weight children who in turn have increased risk for developing type 2 diabetes, hypertension, dyslipidemia, cardiovascular pathology, and obesity as adults 6. One testable hypothesis is that the gut microbiota may contribute to the risk and pathogenesis of undernutrition through effects on nutrient metabolism and on immune function (Fig. 1). Similarly, does the experience of undernutrition in childhood affect the development of metabolic capacities by this microbial ‘organ’ in ways that result in persistent metabolic dysfunction or inadequate function, thereby contributing to the sequelae of malnutrition? Finally, if we define malnutrition as the inadequate or excessive consumption of dietary ingredients leading to development of disease, then we need to also consider the alarming epidemic of obesity that is sweeping the world and its relationship to the gut microbiome and immune system.
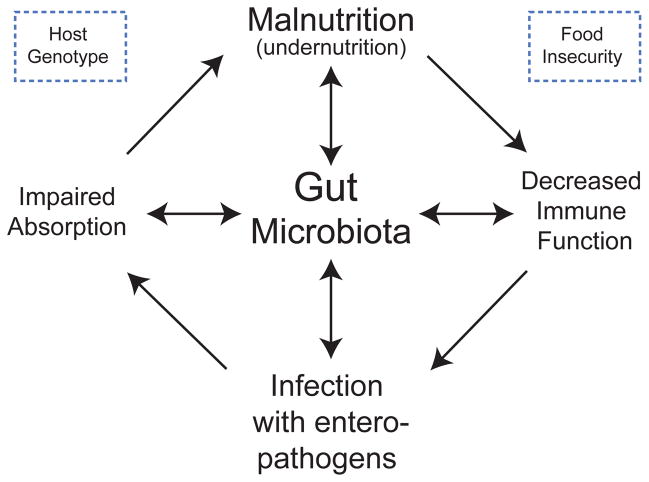
Fig. 1. A schematic of envisioned interrelationships between the gut microbiota, the immune system and diet that underlies the development of malnutrition.
Under-nutrition is associated with a variety of defects in the innate and adaptive immune system that, in turn, are associated with increased predisposition to diarrheal illnesses. Recurrent (enteric) infections predispose to macro- and micronutrient deficiencies, as well as impaired intestinal mucosal barrier function 77. These factors lead to a cycle of further susceptibility to infection and worsening nutritional status. A confounding problem is that vaccines designed to protect children from certain pathogens (including enteropathogens) exhibit poor efficacy in areas of the world where poor nutrition is rampant 74. A testable hypothesis is that the gut microbiota may contribute to disease risk and pathogenesis through effects on nutrient processing and absorption, and on immune function. These interrelationships are illustrated in the Figure. Diet shapes gut microbial community structure and function while the microbiota adapts in ways that promote nutrient processing; the ability of the microbiota to process a given diet affects the nutrient and energetic value of that diet. The microbiota and immune systems co-evolve: malnutrition affects the innate and adaptive immune system as well as the microbiota. The microbiota serves as a barrier to enteropathogen infection; this barrier function may be disrupted by malnutrition as well as by perturbations in immune system function. The microbiota affects nutrient processing and nutrient distribution to the host, including the expression of host genes involved in nutrient transport and metabolism.
The marriage of metagenomics and gnotobiotics
We believe that the ‘marriage’ of two approaches, one involving culture-independent (metagenomic) methods for describing the gut microbiota/microbiome and the other involving gnotobiotics (the rearing of animals under germ-free conditions, with or without subsequent exposure at various stages of postnatal life or adulthood to a microbial species or species consortium) represents a potentially powerful way to address a number of questions related to the interrelationships between diet, nutritional status, the assembly and dynamic operations of gut microbial communities, and the nature of the interkingdom communications between the gut microbiota and host (including host-microbial co-metabolism, and the co-evolution of the immune system2,7,8). Without dismissing caveats related to the use of gnotobiotic models (see below), we describe ways that may be useful for joining gnotobiotics and metagenomic methods to compare and contrast the functional properties of various types of gut microbial communities, to explicitly test or generate hypotheses, and to develop new experimental (and computational) approaches that together inform the design, execution, and interpretation of human studies.
What is changing about what we are eating
Changes in dietary consumption patterns affect many aspects of human biology. A full understanding of the determinants of nutritional status requires that we know what people are eating and how these diets are changing. Unfortunately, accurate information of this type is hard to obtain and when available generally covers a relatively limited time period. As a corollary, searchable databases that effectively integrate information obtained from the surveillance efforts of many international and national organizations (e.g., WHO, the UN Food and Agriculture Organization, the United States Department of Agriculture (USDA) Economic Research Service) are needed to monitor changing patterns of food consumption in different human populations. Analysis of USDA data tracking the availability of over 200 common food items between 1970 and 2000 reveals that diets in the USA have changed both in terms of overall caloric intake and the relative amounts of different food items (http://www.ers.usda.gov/Data/FoodConsumption). Linear regression of total caloric intake over time shows that the average number of kcal consumed per day increased markedly over this 30-year period (R2=0.911, P<10−15). This is consistent with estimates from the US National Health and Nutrition Examination Survey (NHANES), which indicate that adult men and women increased their daily calorie intake by 6.9% and 21.7%, respectively, during the same period 9. If total caloric intake is analogous to ‘primary productivity’ in macro-ecosystems, where primary productivity is used as a proxy for available energy, then increasing the amount of energy input from the diet would be predicted to affect the number of microbial species living in the gut of a single host, as well as the magnitude of the compositional differences that exist between different hosts or even different regions of a single gut (for discussions about the mechanisms underlying productivity-species richness relationships in macro-ecosystems see refs. 10,11). Intriguingly, metagenomic studies of bacterial composition in the fecal microbiota of obese and lean twin pairs living in the USA have shown that obesity is associated with decreased numbers of bacterial species 3. Reductions in diversity could impact community function, resilience to various disturbances, and the host immune system.
During the past 30-plus years, the American diet has also shifted in terms of the relative contributions of different foods to total energy intake. Since 1970, two dietary ‘epochs’ can be distinguished based on the contribution of grains to overall calories (mean increase in daily carbohydrate intake for men and women during this period, 62.4g and 67.7g, respectively9). Consumption of other food items has also changed: Spearman’s rank correlations between food availability and time, followed by adjustments of p-values to reflect false discovery rates, reveal that the representation of 177 of 214 items tracked by the USDA has either increased or decreased significantly in American diets since 1970. For example, Americans now eat less beef and more chicken, and corn-derived sweeteners have increased at the expense of cane and beet sugars. Additionally, methods of food modification and preparation have changed. Comparable data are needed for other countries with distinct cultural traditions, including countries where people are undergoing dramatic transformations in their socioeconomic status and lifestyles.
We know from metagenomic studies of the human gut microbiota and microbiome that (i) early postnatal environmental exposures play a very important role in determining the overall phylogenetic structure of an adult human gut microbiota, (ii) assembly of the microbiota towards an adult configuration occurs during the first three years of life 12, and (iii) features of the organismal and gene content of gut communities are shared among family members and transmitted across generations of a kinship 3. We also know that dietary habits influence the structure of the human genome. For example, populations that consume diets high in starch have a higher number of copies of the salivary amylase gene (AMY1) than those consuming low-starch diets 13. We know that these habits also affect the gut microbiome. A wonderful illustration of the latter point is the acquisition of a β-porphyranase gene that degrades seaweed-associated glycans from marine microbes associated with non-sterile food consumed by Japanese populations. Zobellia galactanivorans is a marine Bacteroidetes that is able to process porphyran derived from marine red algae belonging to the genus Porphyra. Homologs of porphyranases from Z. galactanivorans are present in the human gut bacterium Bacteroides plebeius and prominently represented in the gut microbiomes of Japanese but not North Americans, leading to the suggestion that porphyranases from Z. galactanivorans or another related bacterium were acquired, perhaps through horizontal gene transfer, by a resident member of the microbiota of Japanese consumers of non-sterile food, and that this organism and gene was subsequently transmitted to others in Japanese society14. Together, these observations lead to the notion that systematic changes in overall dietary consumption patterns across a population might lead to changes in the microbiota/microbiome with consequences for host nutritional status and immune responses.
We also know from work in gnotobiotic mice that have received human fecal microbial community transplants that the relative abundances of different bacterial species and genes in the gut microbiota are highly sensitive to the proportions of different foods in the diet 2. Gnotobiotic mice harboring defined collections of sequenced human gut symbionts or transplanted human fecal microbial communities could provide an approach for modeling the effects of different dietary ‘epochs’ on the gut microbiota and on different facets of host biology. If the desired result is an account of the effects of individual food items or nutrients, then feeding the animals a series of defined diets, each with a different element removed or added might be an appropriate strategy if the food ingredients for the epoch are known and available. If the focus is on the effects of overall differences in dietary habits within or between groups of humans, then diets should reflect the overall nutritional characteristics of the different groups without merely being representative of a single individual. Designing such diets requires detailed accounts of the identity and quantity of each food item consumed, ideally for a large number of people, as well as the methods used for food preparation. The American diet presents a rare opportunity for such an approach, as NHANES datasets (http://www.cdc.gov/nchs/tutorials/Dietary/) provide one-day dietary recall data at multiple timepoints dating back to the early 1970s.
The nexus between nutrient metabolism and the immune system
The nexus between nutrient metabolism and the immune system occurs at many levels, ranging from endocrine signaling to direct sensing of nutrients by immune cells. Leptin provides a case study of features of these complex interrelationships. Leptin serves to regulate appetite and is a pleiotropic cytokine, maintaining thymic output and cellularity, and promoting the dominance of Th1 cells over Th2 cells 15,16 while inhibiting the proliferation of T regulatory cells (Tregs) 17. Low levels of leptin may account for the decreased cellular immunity associated with periods of nutrient deprivation 16. Leptin also impacts innate immune cells, ranging from promotion of neutrophil activation and migration to activation of monocytes and macrophages 15. Elegant experiments using mice deficient in the leptin receptor in different cellular compartments traced a requirement for leptin signaling in intestinal epithelial cells for preventing severe disease following exposure to Entamoeba histolytica. Comparisons of db/db mice that lack a functional leptin receptor and their wild-type littermates revealed that leptin controls infectivity and prevents severe inflammatory destruction of the intestine, thereby impacting mortality 18. These studies were extended to mice with engineered mutations in the leptin receptor that are found in human populations (T1138S and T985L, both of which disrupt signaling), Each of these mutations rendered mice more susceptible to E. histolytica infection 18. Leptin levels are significantly reduced in the sera of germ-free mice 19. Moreover, genetically obese leptin-deficient ob/ob mice have marked differences in the taxonomic and gene content of their gut microbial communities 20. To our knowledge, the effects of leptin-receptor deficiency on the gut microbiota have not been reported. Nonetheless, leptin receptor deficiency and E. histolytica pathogenesis provide a setting where the intersections between the endocrine and immune systems, enteric infection, and gut microbial ecology can be explored.
The ability to use macronutrients is essential for the generation and maintenance of a protective effector immune response. Following TCR stimulation and co-stimulation through CD28, the metabolic needs of T cells are met by a dramatic increase in uptake and utilization of glucose, amino acids and fatty acids 21,22. A deficiency in glucose uptake negatively impacts numerous facets of T cell function with impairment of both proliferation and cytokine expression. Similarly, deficiencies in amino acids such as tryptophan arginine, glutamine and cysteine reduce immune activation. Furthermore, TCR stimulation in the absence of co-stimulation, which leads to T cell anergy, has been linked to a failure to upregulate metabolic machinery associated with amino acid and iron uptake 21,22.
Short chain fatty acids (SCFAs) provide one of the clearest examples of how nutrient processing by the microbiota and host diet combine to shape immune responses. SCFAs are end-products of microbial fermentation of macronutrients, most notably plant polysaccharides that cannot be digested by humans alone because our genomes do not encode the large repertoire of glycoside hydrolases and polysaccharide lyases needed to cleave the varied glycosidic linkages present in these glycans 23. These missing enzymes (‘dining utensils’) are provided by the microbiome. The luminal concentration of intestinal SCFAs can be modified by the amount of fiber in the diet: this in turn affects the composition of the microbiota 24. In addition to acting as an energy source for the host, SCFAs exert significant effects on host immune responses. Butyrate can modify the cytokine production profile of helper T cells 25 and promote intestinal epithelial barrier integrity 26, which in turn can help limit exposure of the mucosal immune system to luminal microbes and prevent aberrant inflammatory responses. Production of another SCFA, acetate, by the microbiota promotes the resolution of intestinal inflammation via the G protein-coupled receptor, Gpr43 27. A recent study highlighted the important role of acetate production in preventing infection with the enteropathogen, E. coli 0157:H7. This effect was linked to its ability to maintain gut epithelial barrier function 28. Intriguingly, acetylation of lysine residues may be regulated by SCFA 29 and appears to affect proteins involved in a variety of signaling and metabolic processes. The role of this covalent modification in modulating the activity of proteins intimately involved in innate and adaptive immune responses needs to be explored. It is tempting to speculate that covalent or non-covalent linkage of a variety products of microbial metabolism to host proteins produced within the intestine, or at extra-intestinal sites, will be discovered and found to have important regulatory effects. These different protein modifications could represent a series of mechanisms by which microbial community metabotype is ‘imprinted’ on the host.
If nutrients and derived metabolites reflect the functional activity of the microbiota, sensors of nutrient/metabolite availability can be considered akin to microbe-associated molecular patterns (MAMPs) that convey information regarding microbes to the host. Several families of innate receptors are involved in recognition of MAMPs: they include Toll-like receptors (TLRs), inflammasomes, C-type lectins such as dectin-1, and RNA-sensing RIG-like helicases such as RIG-I and MDA5. The accompanying review by Maloy and Powrie in this issue provides an overview of this area. Here we would like to emphasize that classical innate immune recognition pathways have evolved to assess the nutrient environment. TLR4 can sense the presence of free fatty acids 30 while ATP is in important activator of the inflammasome 31. A variety of other immune cell-associated ‘sensors’ serve to couple information about the local nutrient/metabolite environment to the co-ordination of local immune responses. Examples include mTOR (mammalian Target Of Rapamycin), a serine/threonine kinase32, PKR (double stranded RNA-activated protein kinase) 33, the aryl hydrocarbon receptor (AhR) 34, and various nuclear hormone receptors such as liver-X-receptor (LXR) and peroxisome-proliferator activated receptors (PPAR-α, –β, –γ) 35 (Fig. 2 and Table 1). The mTOR pathway represents an example of how energy availability impacts immune responses. mTOR is activated by PI3 kinase and AKT activity and is inhibited by AMP-activated protein kinase (AMPK), which is a sensor of cellular energy resources. Genetic and pharmacologic approaches (the latter using rapamycin) indicate that mTOR-signaling affects both the innate and adaptive arms of the immune system, including maturation and effector activity of dendritic cells (DCs), inhibition of Treg development, promotion of the differentiation of Th1, Th2 and Th17 cells, regulation of CD8+ T cell trafficking, and inhibition of memory T cell formation 32,36. PKR couples the presence of free fatty acids to immune activation and has been implicated in the pathogenesis of obesity in mice fed a high fat diet, including their immunoinflammatory and insulin-resistant phenotypes 33 (see below). AhR is activated by a variety of agonists, including kynurenine, a product of tryptophan metabolism by indolamine-2,3-dioxygenase (IDO) 37,38. AhR modulates the differentiation of DCs 39 as well as promoting Th17 and Treg differentiation and effector activity 40,41. Withdrawal of tryptophan and arginine controls immune responses 42,43. The presence of an intact amino acid starvation (AAS) response in T cells is essential for the immunosuppressive activity of tryptophan depletion by IDO 44. This example illustrates how the ability of T-cells to sense levels of a nutrient (tryptophan) in its local environment, rather than using the nutrient solely as a fuel source, is an important determinant of cell fate. If assessment of local nutrient levels or metabolites is an important feature in the immune decision-making process, and if the products of microbial metabolism represent heretofore unappreciated agonists or antagonists of immune cell receptors, then an important challenge is to devise in vitro and in vivo models, including genetically manipulatable gnotobiotic animals (e.g., mice or zebrafish) to identify the array of metabolites produced by a microbiota (and host) as a function of different defined diets.
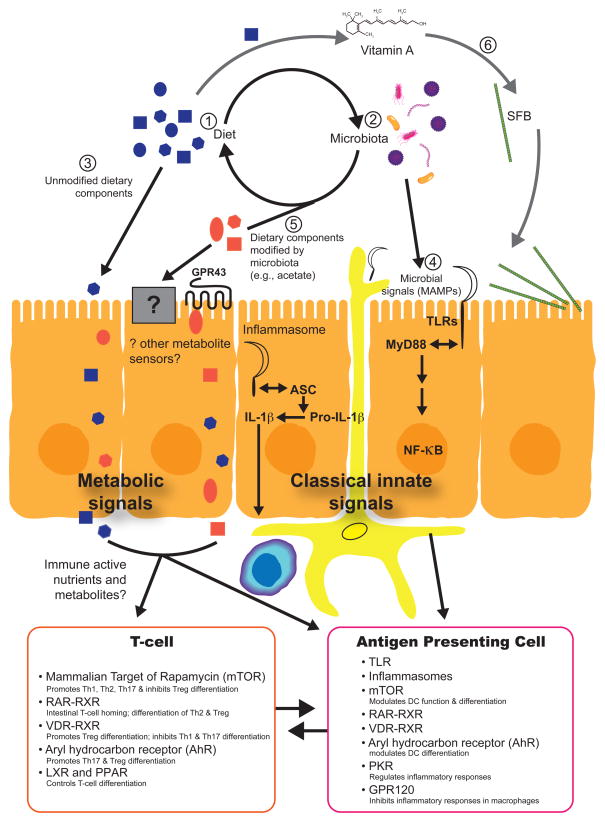
Fig. 2. Metabolic sensors that help co-ordinate immune responses.
(1) Dietary intake of macro- and micro- nutrients shape (2) microbial community structure which, in turn, changes the nutritional value of the consumed food. (3) Unmodified dietary components are directly absorbed in the intestine where they can interact with a variety of immune cells (e.g., omega 3-fatty acids). (4) Microbial signals in the form of Microbe Associated Molecular Patterns (MAMPs) modify local mucosal immune responses through innate signaling pathways such as the inflammasome or TLRs. (5) Microbe-modified dietary components (e.g. acetate produced from fermentation of polysaccharides) provide signals by which the immune system can monitor the metabolic activities of the microbiota. (6) An example of micronutrients directly modifying intestinal microbial ecology: vitamin A can modify the representation of segmented filamentous bacterium (SFB) in the mouse gut microbiota; SFB induce differentiation of Th17 cells.
Table 1.
| Sensor | Agonist | Immune Response Affected | |
|---|---|---|---|
| Mammalian Target of Rapamycin (mTOR) | Sphingosine-1-phosphate (S1P) | Inhibition of Treg differentiation and maintenance88 | |
| Leptin | Promotion of Th1 differentiation 16 | ||
| Leptin | Inhibition of Treg proliferation and function 89,17 | ||
|
| |||
| Aryl Hydrocarbon Receptor (AhR) | (6,12-diformylindolo[3,2-b]carbazole) | Th17 differentiation and IL-22 production by Th17 cells 40, 41 | |
| 2,3,7,8-tetrachlorodibenzo-p-dioxin | Promotes Treg induction 40 | ||
| Kynurenine | Promotes Treg induction 38 | ||
|
| |||
| Double stranded RNA-dependent protein kinase (PKR) | Free fatty acids; palmitic acid | Promotes Insulin Resistance through inhibitory phosphorylation of IRS-1 33 | |
|
| |||
| RAR-RXR | Retinoic Acid | Promotes intestinal T cell homing 90 | |
| Promotes Treg generation 91 | |||
| Promotes T cell proliferation 92 | |||
| Promotes Th2 | |||
| differentiation over Th1 93 | |||
|
| |||
| VDR-RXR | 1,25(OH)2 vitamin D3 | Inhibits lymphocyte | |
| proliferation 94 | |||
| Inhibits IFN-γ, IL-17 and IL-2 expression 95 | |||
| Promotes emergence of Treg 96 | |||
| Drives anti-microbial peptide expression 97 | |||
| Promotes T cell expression of CCR10 98 | |||
|
| |||
| GPR120 | Omega-3-fatty acids | Inhibits inflammatory cytokine production and chemotaxis in macrophages 99 | |
|
| |||
| GPR43 | Acetate | Promotes resolution of intestinal inflammation 27 | |
|
| |||
| P2X Receptors | ATP | Th17 generation 100 | |
The case of micronutrients
The intestinal microbiota has the capacity to synthesize a variety of vitamins involved in myriad aspects of microbial and host metabolism, including cobalamin (vitamin B12), pyridoxal phosphate (active form of vitamin B6), a cofactor in a variety of enzymatic interconversions involved in amino acid metabolism, pantothenic acid (vitamin B5), niacin (vitamin B3), biotin, tetrahydrofolate (generated from dietary forms of folate) and vitamin K. In addition to vitamin B12, gut microbes produce a range of related molecules (corrinoids) with altered ‘lower ligands’ including methyladenine, p-cresol, and other analogs. Over 80% of non-absorbed dietary vitamin B12 is converted to these alternate corrinoids 45,46. There is preliminary evidence that syntrophic relationships among members of the human microbiota, and the fitness of some taxa, may be based on the ability to generate, utilize, or further transform various corrinoids46,47.
The ability of the gut microbiota to produce folate and cobalamin could affect host DNA methylation patterns, while acetate produced from microbial fermentation of polysaccharides could modify chromatin structure and gene transcription via histone acetylation. Thus, inheritance of a mammalian genotype, intergenerational transmission of a microbiome, together with a complex dynamic where the microbiome is viewed both as an epigenome per se and as a modifier of the host epigenome during the postnatal period when host, host diet and microbial community co-evolve, could together shape human physiological phenotypes that are manifest during childhood or later in life.
Numerous observational studies indicate that deficiencies in vitamins A, D, E and zinc can adversely impact immune function, particularly T-cell responses. Although a significant body of work exists detailing the myriad effects of vitamin A, D and E on host immune responses, to date there is little evidence for a role of the microbiota in the biosynthesis or metabolism of these vitamins. However, stimulation of dendritic cells via TLR2 increases the expression of host genes associated with generation of the immunoactive form of vitamin A (retinoic acid) while enteric infection has been linked to vitamin A deficiency 48,49. Intriguingly, a recent study demonstrated that vitamin A deficiency leads to a complete loss of Th17 cells in the small intestine of specified pathogen-free mice, and an associated significant reduction in the abundance of segmented filamentous bacteria (SFB) 50, a member of the Clostridiaceae that drives intestinal Th17 responses in mice 51,52. Thus, vitamin A has the potential to modulate immune responses through direct interactions with immune cells, or indirectly by modulating the composition of the microbiota.
The microbiota also affects the absorption of key minerals. Perhaps the best characterized micronutrient in terms of its interaction with both the microbiota and immune system is iron. Iron-deficient mice are resistant to the development of experimental autoimmune encephalomyelitis, and have reduced delayed type hypersensitivity responses and lower levels of IgM and IgG. Iron deficiency also impairs innate immune responses, as it is required for the respiratory burst 53. Likewise, iron is an essential micronutrient for bacteria. Given the low solubility of Fe3+, microbes have evolved the capacity to produce a variety of high affinity iron-binding siderophores.. Microbes take up soluble Fe3+ siderophore complexes via a variety of active transporters. Early studies in gnotobiotic animals revealed a link between the gut microbiota and development of iron deficiency. Germ-free but not conventionally-raised rats become anemic when fed a low iron diet. Germ-free rats also exhibit increased loss of iron in their feces compared to their conventionally-raised counterparts 54. The iron balance that exists between host and microbiota is disturbed in a mouse model of Crohn’s disease where there is dysregulation of TNF-α expression: oral (but not parenteral) iron supplementation in these animals causes a shift in gut microbial community composition, as defined by 16S rRNA-based surveys, and exacerbates their ileitis 55.
Metagenomic methods need to be applied to further delineate the role of the microbiota in iron and other forms of micronutrient deficiency. For example, what is the impact of developing iron deficiency on the configuration of the gut microbiota and microbiome, including its content of siderophores? Does iron repletion return the microbiota/microbiome to a ‘normal’ pre-deficient state, or are there persistent structural and functional perturbations that require continued nutritional supplementation to correct? Do particular configurations of the microbiota/microbiome predispose the host to iron or other types of micronutrient deficiency? How does the iron content of mother’s milk during post-natal life impact the assembly and metabolic operations of the microbiota? In principle, these questions can be first addressed in a variety of gnotobiotic mouse models, and also extended to macronutrient-deficient states.
The microbiota and immune system in obesity
Obesity, metabolic syndrome and diabetes illustrate the role the diet-microbiota-immune axis plays in shaping human systems biology. Although the dramatic increase in obesity worldwide can be linked to an ever-growing trend towards excessive calorie intake, the microbiota has also been implicated in this disorder. Studies of a cohort of twins living in the USA indicate that the bacterial phylogenetic composition of the fecal microbiota and the representation of microbial genes involved in several aspects of nutrient metabolism in the fecal microbiome are different in lean versus obese twin pairs 3. Different groups applying different primers for amplifying bacterial 16S rRNA genes for culture-independent analyses of gut microbial ecology, and studying different human populations consuming different diets have reported differing results concerning the bacterial phylogenetic composition of the microbiota in lean versus obese individuals 56.
Evidence that a link exists between the microbiota and obesity comes from transplant experiments in gnotobiotic mice: gut communities from leptin-deficient ob/ob mice or mice with diet-induced obesity produce a greater increase in adiposity when transferred to germ-free recipients than do communities from wild-type littermates or mice that have been given a healthy calorically less dense diet 20,57. Germ-free mice are resistant to diet-induced obesity. Additional studies have revealed that the gut microbial community regulates expression of genes that affect fatty acid oxidation and fat deposition in adipocytes. For example, production of the secreted lipoprotein lipase inhibitor angipoietin-like protein 4 (Angptl4; also known as fasting-induced adipose factor) is suppressed by the microbiota: studies of germ-free and conventionalized wild-type and Angptl4−/− animals established that microbiota-mediated suppression of gut epithelial expression of this secreted LPL inhibitor results in increased LPL activity and fat storage in white adipose tissue 19,58. TLR5-deficient mice harbor a gut microbiota with a configuration distinct from that encountered in littermate controls. Moreover, when their gut microbiota is transplanted to wild-type germ-free recipients, food intake is increased compared to recipients of microbiota transplants from wild-type mice: increased adiposity and hyperglycemia ensue 59. The mechanism underlying the increase in food consumption remains to be defined although the authors of this study speculate that inflammatory signaling may desensitize insulin signaling in ways that lead to hyperphagia.
Obesity in mice and humans is associated with infiltration of adipose tissue by macrophages, CD8+ T cells 60, and CD4+ T cells 61,62 expressing inflammatory cytokines and chemokines such as TNF-α, CCL2, IL-6, IFN-γ and IL-17 60,62,63. In contrast, adipose tissue in lean mice is home to a population of immunosuppressive regulatory T cells (Treg) that serve to prevent inflammation 64. Mice deficient in the chemokine receptor CCR2 and with obesity induced by consumption of a high-fat diet have reduced macrophage infiltration of the adipose tissue and improved glucose tolerance relative to CCR2-sufficient controls 60, highlighting the role played by factors recruiting inflammatory immune cells and their associated pro-inflammatory products in the pathogenesis of metabolic abnormalities associated with obesity. Blockade of TNF-α 65 or expanding Tregs using anti-CD3 mAbs 62 serves to prevent the onset of obesity-associated insulin resistance in a mouse model of diet-induced obesity.
Inflammation drives development of insulin resistance through phosphorylation of insulin receptor 1 (IRS1) via TNF-α activated JNK, IKK-β, protein kinase C (PKC) or mTOR activity 60. Although MyD88 signals promote development of type 1 diabetes in specified pathogen-free NOD (non-obese diabetic) mice, germ-free MyD88 deficient NOD animals are susceptible to this disorder 66. These findings suggest that particular intestinal microbial configurations can promote or prevent inflammatory immune responses that drive metabolic dysfunction.
Mice fed a high fat diet have increased serum LPS 67. Furthermore, genetically obese mice deficient in leptin or its receptor have reduced intestinal barrier function 68. As noted above, SCFA produced by microbial fermentation affect barrier function. Thus, it will be important to assess whether or not obese humans display similar reductions in barrier function: one scenario is that a high fat diet alters the structure of the intestinal microbiota leading to a reduction in intestinal barrier integrity, enhanced translocation of microbes and/or their antigens resulting in increased microbial antigen load at extra-intestinal sites, enhanced immune stimulation, and the development of insulin-resistance. Furthermore, nutrients are known to directly activate inflammatory arms of the immune system 69. The capacity of the intestinal microbiota to shape immune responses outside of intestine is well documented. Studies have highlighted the ability of the microbiota and specifically SFB to support the development of autoimmune arthritis 70 and experimental allergic encephalomyelitis 71, both of which have been linked to excessive Th17 responses.
Unfortunately, we have scant knowledge of the spatial relationships between members of the microbiota as well as their proximity to elements of the gut-associated immune system in healthy individuals, or individuals with mucosal barrier dysfunction. Gnotobiotic mouse models of obesity may help provide important insights about the biogeography of microbial communities along the length and width of the gut, including whether microbial consortia occupy ectopic sites that could impact the development and perpetuation of barrier dysfunction (e.g., in the crypts of Lieberkuhn where multipotential gut stem cells reside as described in the accompanying article by Medema and Vermeulen). Newer methods, such as CLASI-FISH 72, offer a great deal of promise for characterizing the spatial features of microbe-microbe and microbe-host cell interactions in the gut mucosa, especially if they are applied to gnotobiotic models.
Undernutrition and environmental enteropathy
Undernutrition can have a variety of clinical manifestations ranging from mild asymptomatic micronutrient deficiencies to severe, life-threatening conditions such as kwashiorkor or marasmus. Estimates are that implementing current ‘best practice’ interventions, including lengthening the time of breastfeeding, supplementing diets with zinc and vitamins, improving handwashing and other hygiene measures, and optimizing treatment of acute severe malnutrition, could reduce mortality during the first three years of life by only 25%, even if there is near perfect compliance 5. While a variety of environmental and genetic factors have long been postulated to influence the development of moderate to severe forms of malnutrition 73, the underlying mechanisms remain poorly defined. Food availability, while certainly a major factor, is not the only contributor. For example, in Malawi, the concordance for severe malnutrition between twins within the same household and fed similar diets is only 50% (M. Manary, personal communication). This observation raises a number of questions. Do different configurations of the microbiota predispose one co-twin to kwashiorkor or marasmus? What is the impact of nutrient deficiency, in either the mother or her child, on the configuration of the gut microbiota and microbiome in the developing gut? Does nutrient deficiency in the mother impact the assembly of the microbiota via changes in the mother’s gut microbiota or in the nutrient and immune content of her breast milk: both the microbiota and milk are ‘transmitted’ to the infant yet we have much to learn about how the biochemical and immunologic features of breast-milk change and how breast milk and infant microbiota ‘co-evolve’ during the suckling period when a mother is healthy or when she is malnourished (see below). If malnutrition delays the maturation of the gut’s microbial metabolic organ or skews it towards a different and persistent configuration that either lacks necessary functions for health or that expresses functions that may increase the risk for disease, including immunoinflammatory disorders, does nutrient repletion return the microbiota/microbiome to a ‘normal’ pre-deficient state, or are there persistent structural and functional perturbations that require continued nutritional supplementation to correct? Are there microbiome configurations that correlate with vaccine responsiveness 74?
Studies of severe forms of malnutrition indicate that these patients often have many characteristics of environmental enteropathy 75. Environmental enteropathy, also known as tropical sprue or tropical enteropathy, is a poorly characterized chronic inflammatory disease that primarily affects the small intestine. This disorder afflicts individuals who reside for relatively long periods of time in areas with poor sanitation and who have high exposure to fecal-contaminated water and food. As an example, Peace Corps volunteers returning to the USA from such areas would report a history of diarrheal disease and have signs and symptoms of chronic malabsorption and nutritional deficiencies 76. The malabsorption associated with environmental enteropathy is often subtle, manifesting itself clinically only as stunting due to chronic undernutrition 76. The breakdown in intestinal mucosal barrier function in this disorder can lead to increased susceptibility to enteropathogen infections. Recurrent infections predispose to nutritional deficiencies and further compromise of barrier function, leading to a vicious cycle of further susceptibility to infection and worsening nutritional status77.
Efforts to break this cycle have focused on vaccines that could prevent infection. However, there is significant heterogeneity in the responses to vaccination between children living in highly Westernized societies and children living in certain developing countries. Oral rotavirus vaccine elicits responses in >95% of children living in Westernized societies but only 49% in Malawi 78. Lower oral polio vaccine (OPV) efficacy has been reported in populations with greater enteric disease burden 79. Studies in Chilean children have demonstrated a negative correlation between oral cholera vaccine responses and small bowel bacterial overgrowth 80. In addition, patients with celiac disease, which as noted below, shares phenotypic features with environmental enteropathy, can have a blunted response to parenteral hepatitis B vaccination, but only when their disease is active 81.
Traditionally, the most definitive test for environmental enteropathy has been small intestinal biopsy. Biopsies typically show reductions in small intestinal villus height, increased numbers of intraepithelial lymphocytes, and increased infiltration of the underlying lamina propria by T cells with a predominant Th1 phenotype 75. Some of these features are found in patients with celiac disease, where a luminal antigen (gliadin) drives a T-cell response that, in turn, results in epithelial destruction, reduced absorptive surface area, and malabsorption 76. Unlike celiac disease, the antigens that drive the host immune response in environmental enteropathy are unknown, but there may be an association with certain HLA alleles (e.g., Aw-31 82).
The pathologic events that lead to the development of environmental enteropathy are poorly understood, in part because of the absence of a robust set of readily assayed biomarkers that would improve the ability to diagnose, classify and potentially subcategorize individuals that exhibit the broadly defined clinical manifestations which define this disorder. Epidemiologic data showing a strong association of environmental enteropathy in areas with poor sanitation, occasional epidemic spread of the disease and its responsiveness to antibiotic treatment reinforce the long-standing belief that there is an ‘infectious’ etiology. While cultures of jejunal aspirates from individuals with environmental enteropathy have suggested ‘contamination’ of the proximal small bowel by aerotolerent Gram-negative bacteria 83, no single pathogen or set of pathogens has been identified in the gut microbiota of the majority of affected individuals. There is a distinct possibility that this enteropathy is not the result of a single pathogen but rather the result of colonization with microbial consortia that are inflammogenic in the context of a susceptible host. In fact, what constitutes a ‘normal’ immune repertoire in a healthy gut likely varies considerably depending upon environmental exposures and the configuration of a microbiota. Moreover, most metagenomic studies of the microbiota have focused on members of the domain Bacteria that dominate these communities. Additional tools need to be developed so that they can be extended to viral and eukaryotic components. The latter include parasites that compete for nutrients within the intestines of infected individuals. Parasites can interact directly with bacterial members of the microbiota during their life cycle in ways that promote hatching of parasite eggs, and can shape immune function through factors such as excretory-secretory (ES) products which have been shown to modulate cytokine production, basophil degranulation, immune cell recruitment and interference with TLR signaling 84.
It seems reasonable to posit that individuals living in regions with high oral exposures to fecal contaminated water and foods, and/or with a eukaryotic component of their gut community that includes parasites, will have gut associated-immune systems with significantly different structural and functional configurations than those without these exposures. In this sense, including the term ‘environmental’ together with enteropathy is logical and emphasizes the need to place a host’s immune and gut microbiome phenotypes in the context of their various exposures.
Comparative metagenomic studies could provide important new diagnostic tools in the form of microbial taxa, and microbiome gene functions whose representation in the gut communities of affected individuals versus healthy controls correlates with environmental enteropathy. In addition, they could provide pathophysiological insights about relationships between host diet, enteropathogen representation in the microbiota, and microbiome gene composition and expression (including expressed metabolic functions). A major challenge will be to correlate this data with the results of quantitative phenotyping of the human gut’s innate and adaptive immune system. This will require new and safe approaches for sampling system components, especially in the gut mucosa. Similarly, as noted above, we have scant knowledge of the spatial relationships between members of the microbiota, as well as their proximity to elements of the gut-associated immune system in healthy individuals or in individuals with mucosal barrier dysfunction.
Microbiota assembly and breast milk
Breast milk is known to protect newborns from infection, in part because of the copious amount of maternally generated antibodies that it contains. While these antibodies have specificity for components of the microbiota, the microbial targets are not well defined for given maternal- infant dyads, or as a function of time after delivery. In addition to antibodies, breast milk contains other immunoactive compounds including cytokines (e.g., IL-10), growth factors (e.g., EGF) and antimicrobial enzymes such as lysozyme. The impact of maternal nutritional status on the glycan, protein, lipid and cytokine landscape of breast milk needs to be defined further. This analysis should have a temporal axis that explores co-evolution of the immunological/nutrient properties of mother’s milk and the postnatal assembly and maturation of the infant gut microbiota and of the innate and adaptive immune system. Important feedback systems may be revealed. Similarly, knowledge of the vaginal and cutaneous microbiota of mothers prior to and following birth, as a function of their nutritional status could be very informative. For example, are there common configurations of microbial communities occupying these body habitats that correlate with the development of environmental enteropathy in mothers and their offspring?
Personalized gnotobiotics and culture collections
As noted above, studies have demonstrated the ability of intestinal microbial communities to rapidly re-shape themselves in response to changes in diet. These observations raise the question of whether and how malnourished states impact (i) the spatial/functional organization of the microbiota and the niches (professions) of its component members; (ii) the capacity of the community to respond to changes in diet; (iii) the ability of components of the microbiota to adaptively forage on host-derived mucosal substrates, and (iv) the physical and functional interactions that occur between the changing microbial communities and the intestinal epithelial barrier (including its overlying mucus layer). One way of developing the experimental and computational tools and concepts needed to examine these challenging questions in humans is to turn to gnotobiotic mice who have been ‘humanized’ by transplantation of gut communities from human donors with distinct physiological phenotypes and to feed these mice diets that are representative of those of the microbiota donor.
Personalized gnotobiotic mouse models
We have used metagenomic methods to show that gut (fecal) communities can be efficiently transplanted into germ-free mice and the mice then fed diets that resemble those consumed by the human microbiota donors, or diets whose ingredients are deliberately manipulated in various ways 2. Transplanted human gut microbial communities can be transmitted from gnotobiotic mothers to their pups. In principle, by using mice humanized with microbiota from individuals residing in different regions of the world, and giving them diets that are representative of their cultural traditions, proof-of-principle global ‘clinical trials’ of the nutritional value of foods and their impact on the microbiota and immune system can be performed.
Transplantation of a human fecal microbiota into germ-free mice can be viewed as capturing an individual’s microbial community at a moment in time and replicating it in multiple recipient gut ecosystems. The humanized mice can be followed over time under highly controlled conditions where potentially confounding variables can be constrained in ways that are not achievable in human studies. This type of ‘personalized gnotobiotics’ also provides an opportunity to determine the degree to which human phenotypes can be transmitted via the gut microbiota as a function of diet. Moreover, the documented responses of microbial lineages and genes encoding metabolic pathways in the transplanted, replicated communities may provide mechanistic insights about differences in the adaptations of healthy versus diseased gut microbiomes (and host immune system) to changes in diets, plus new biomarkers of nutritional status and the impact of various therapeutic interventions, including those based on dietary manipulations. Putative microbial biomarkers obtained from studies of these mice can in turn be used to query datasets generated directly from the human donor(s).
Despite the potential power of using humanized mice to study interactions between the host immune and metabolic systems and the intestinal microbiota under highly controlled conditions, this approach has caveats. Recent work on Th17 responses suggests that unlike the mouse microbiota, which contains SFB, a fecal microbiota from a human donor is not sufficient to drive immune-gene expression in the small intestine of ex-germ free mice52. This raises the possibility that humanization may not fully recapitulate the capacity of a mouse microbiota to mature the intestinal immune system in mice. However, earlier studies on the effects of human microbiota on the mouse immune system revealed that the ability of E. coli heat labile enterotoxin (LT) to break oral tolerance to ovalbumin in germ-free mice can be inhibited by transplantation of either a human or mouse microbiota during the neonatal period 85. Further, a single component of a human gut symbiont, the polysaccharide A component of B. fragilis, is able to mature components of the CD4+ T cell response in mice 86. Finally, we have observed a similar increase in the frequency of TCR-β+ cells in the mesenteric lymph nodes of gnotobiotic recipients of a human or mouse microbiota, when compared to germ-free controls (P. Ahern, V. Ridaura and J. Gordon, unpublished observations). This suggests that although not all components of the immune system will be matured by a human gut microbiota, the immune system is not likely to remain ignorant of these communities. In addition, any differences detected in direct comparisons of the effects of two different human communities may represent responses relevant to the human immune system.
Personalized bacterial culture collections
We have recently shown that the human fecal microbiota consists largely of bacteria that can readily be cultured87. Metagenomic analysis suggests that the majority of predicted functions in a human’s microbiome are represented in its cultured members. In gnotobiotic mice, both complete and cultured communities exhibit similar properties and responses to dietary manipulations. By changing the diet of the host, the community of cultured microbes can be shaped so that it becomes enriched for taxa suited to that diet. These culture collections of anaerobes can be clonally arrayed in multi-well formats: this means that personalized, taxonomically defined culture collections can be created from donors representing different human populations and physiologic phenotypes, and where the microbes have co-evolved and co-existed together within a single human being’s gut habitat.
Together, these advances yield a translational medicine pipeline for examining the interplay between food and food ingredients, the microbiota, the immune system and health. Goals for such a human translational medicine pipeline are to (i) identify individuals with interesting phenotypes, (ii) assess transmissibility of their phenotypes via human microbiota transplants into gnotobiotic animals, (iii) select candidate disease-modifying taxa (retrieved from clonally-arrayed, taxonomically defined personal bacterial culture collections), (iv) sequence selected taxa and (v) reunite them in various combinations in gnotobiotic mice as defined model gut communities, so that their interactions with one another and their impact on host biology can be further explored, using a variety of methods [e.g., RNA-Seq, mass-spec based proteomics and metabolomics, multi-label FISH (for biogeographical studies of the microbiota), whole genome transposon mutagenesis (to identify fitness factors for microbes under various dietary contexts 46), immune profiling and other measurements of mucosal barrier function]. Knowing the degree to which tractable bacterial taxa are able to influence host physiology, and how dietary components can be used to affect specific organisms in the microbiota in ways that provide benefit to the host may be very useful for discovering new generations of pro- and prebiotics.
Looking ahead
With massive prospective national surveys planned and being implemented, such as the NIH’s National Children’s Study that will follow a representative sample of 100,000 children from before birth to age 21, the time is right for an initiative to evaluate the interrelationships between our diets, nutritional status, microbiomes and immune systems. Many components could comprise this initiative. We can readily envision several of these.
Dietary databases
As noted above, there is a need to create more and improved databases for monitoring changing patterns of food consumption that integrates the surveillance efforts of a number of organizations. This tool and other interdisciplinary approaches could be used to define a set of study populations representative of established and emerging food consumption patterns in distinct cultural and socioeconomic settings. An emphasis could be placed on comparing humans living in Westernized societies versus those living in developing countries undergoing marked transitions in lifestyles/cultural traditions. New, reliable, cost-effective and generalizable methods will be needed for acquiring quantitative data about the diets consumed by individual humans in these study populations, and the resulting data deposited in searchable databases together with defined annotation standards. Moreover, guidelines need to be further developed related to ethical and legal aspects of human subjects research involving observational and interventional nutritional studies of pregnant women and their offspring.
New biomarkers of nutritional status
Readily procured human biospecimens could be used together with high throughput, targeted and nontargeted (quantitative) profiling of metabolites in comprehensive time series studies to define the relationship between diet, nutritional status, and microbiome configuration in ‘healthy’ individuals at various stages of life (e.g., in women before, during and after pregnancy and in their children during the first 5 years after birth). This could be accompanied by studies of malnourished individuals before, during and after well justified, defined nutritional interventions. In addition to these data, genomes (genotypes), epigenomes and microbiomes could be characterized in these study cohorts together with a variety of clinical parameters (e.g., vaccine responses) and environmental parameters (e.g., water sanitation). The resulting datasets would be deposited in annotated searchable databases. A translational medicine pipeline that includes relevant cellular and animal models would help guide the design and interpretation of these human studies.
Quantitative phenotyping of the immune system
As noted above, a major challenge is to obtain cellular and molecular biomarkers for quantitative profiling of the innate and adaptive immune system, including biomarkers of mucosa-associated barrier function. Given the small quantities of biomaterials available from some body sites, this initiative should help advance enabling ‘miniaturizing technology’ for quantitative measurements of cells and biofluids. Non-invasive imaging-based biomarkers are also needed.
Aspirational goals include identifying new host and microbial biomarkers and mediators of nutritional status, the nutritional value of various foods, the functioning of the human adaptive/innate immune system (including mucosal barrier integrity and mucosal immunity), and the dynamic operations of the microbiota. This information would be used for ‘demonstration projects’ that rigorously define ‘nutritional health’ and test preventive or therapeutic recommendations for micro- and macronutrient consumption, for example in pregnant women and infants/children, and their impact on the assembly and operations of their immune systems. The ‘microbiome’ component could also help define a previously uncharacterized axis of human genetic evolution (our ‘microbiome-evolution’) reflecting in part our changing dietary habits. It could also produce testable hypotheses about unappreciated aspects of the pathophysiology of ‘Western diseases’, and yield new microbiome-based strategies for disease prevention or treatment.
Supplementary Material
Acknowledgments
We are grateful to members of our lab, plus our colleagues Clay Semenkovich and Andrey Shaw, for many helpful discussions. Work cited from our laboratory was supported by grants from the NIH (DK30292, DK70977, DK078669), the Crohn’s and Colitis Foundation of America, and the Bill and Melinda Gates Foundation.
REFERENCES (also see Supplemental References)
- 1.Whitacre PT, Fagen AP, Husbands JL, Sharples FE. Implementing the New Biology: Decadal Challenges Linking Food, Energy, and the Environment. National Research Council of The National Academies of Science; Washington, D.C.: 2010. [Google Scholar]
- 2.Turnbaugh PJ, et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. This report highlights the utility of gnotobiotic mice harboring a transplanted human gut microbiome in studying the dynamic interplay between diet and the microbial community. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryce J, Boschi-Pinto C, Shibuya K, Black RE. WHO estimates of the causes of death in children. Lancet. 2005;365:1147–1152. doi: 10.1016/S0140-6736(05)71877-8. [DOI] [PubMed] [Google Scholar]
- 5.Bhutta ZA, et al. What works? Interventions for maternal and child undernutrition and survival. Lancet. 2008;371:417–440. doi: 10.1016/S0140-6736(07)61693-6. [DOI] [PubMed] [Google Scholar]
- 6.Barker DJ. Adult consequences of fetal growth restriction. Clin Obstet Gynecol. 2006;49:270–283. doi: 10.1097/00003081-200606000-00009. [DOI] [PubMed] [Google Scholar]
- 7.Wikoff WR, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Nat Acad Sci USA. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin FP, et al. Probiotic modulation of symbiotic gut microbial-host metabolic interactions in a humanized microbiome mouse model. Mol Syst Biol. 2008;4:157. doi: 10.1038/msb4100190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wright JE, Willis GJ, Edwards MS. Nutritional content of hospital diets. JAMA. 2004;291:2194–2196. doi: 10.1001/jama.291.18.2194. [DOI] [PubMed] [Google Scholar]
- 10.Chase JM. Stochastic community assembly causes higher biodiversity in more productive environments. Science. 2010;328:1388–1391. doi: 10.1126/science.1187820. [DOI] [PubMed] [Google Scholar]
- 11.Mittelbach GG, et al. What is the observed relationship between species richness and productivity? Ecology. 2001;82:2381–2396. [Google Scholar]
- 12.Koenig JE, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Nat Acad Sci USA. 2011;108:4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perry GH, et al. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 2007;39:1256–1260. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hehemann JH, et al. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464:908–U123. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- 15.La Cava A, Matarese G. The weight of leptin in immunity. Nat Rev Immunol. 2004;4:371–379. doi: 10.1038/nri1350. [DOI] [PubMed] [Google Scholar]
- 16.Lord GM, et al. Leptin modulates the T-cell immune response and reverses starvation-induced immunosuppression. Nature. 1998;394:897–901. doi: 10.1038/29795. [DOI] [PubMed] [Google Scholar]
- 17.De Rosa V, et al. A key role of leptin in the control of regulatory T cell proliferation. Immunity. 2007;26:241–255. doi: 10.1016/j.immuni.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 18.Guo X, et al. Leptin signaling in intestinal epithelium mediates resistance to enteric infection by Entamoeba histolytica. Mucosal Immunol. 2010 doi: 10.1038/mi.2010.76. Demonstrates the role of signaling via the leptin receptor in protecting the intestinal epithelium against infection and damage by this enteropathogen. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Backhed F, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 21.Fox CJ, Hammerman PS, Thompson CB. Fuel feeds function: energy metabolism and the T-cell response. Nat Rev Immunol. 2005;5:844–852. doi: 10.1038/nri1710. [DOI] [PubMed] [Google Scholar]
- 22.Michalek RD, Rathmell JC. The metabolic life and times of a T-cell. Immunol Rev. 2010;236:190–202. doi: 10.1111/j.1600-065X.2010.00911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lupton JR. Microbial degradation products influence colon cancer risk: the butyrate controversy. J Nutr. 2004;134:479–482. doi: 10.1093/jn/134.2.479. [DOI] [PubMed] [Google Scholar]
- 25.Bird JJ, et al. Helper T cell differentiation is controlled by the cell cycle. Immunity. 1998;9:229–237. doi: 10.1016/s1074-7613(00)80605-6. [DOI] [PubMed] [Google Scholar]
- 26.Peng L, He Z, Chen W, Holzman IR, Lin J. Effects of butyrate on intestinal barrier function in a Caco-2 cell monolayer model of intestinal barrier. Pediatr Res. 2007;61:37–41. doi: 10.1203/01.pdr.0000250014.92242.f3. [DOI] [PubMed] [Google Scholar]
- 27.Maslowski KM, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fukuda S, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. Refs 27 and 28 demonstrate how microbiota-derived short chain fatty acids help modulate immune responses and susceptibility to enteropathogen invasion. [DOI] [PubMed] [Google Scholar]
- 29.Kim GW, Gocevski G, Wu CJ, Yang XJ. Dietary, metabolic, and potentially environmental modulation of the lysine acetylation machinery. Int J Cell Biol. 2010:632739. doi: 10.1155/2010/632739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen MT, et al. A subpopulation of macrophages infiltrates hypertrophic adipose tissue and is activated by free fatty acids via Toll-like receptors 2 and 4 and JNK-dependent pathways. J Biol Chem. 2007;282:35279–35292. doi: 10.1074/jbc.M706762200. [DOI] [PubMed] [Google Scholar]
- 31.Mariathasan S, et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 32.Thomson AW, Turnquist HR, Raimondi G. Immunoregulatory functions of mTOR inhibition. Nat Rev Immunol. 2009;9:324–337. doi: 10.1038/nri2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakamura T, et al. Double-stranded RNA-dependent protein kinase links pathogen sensing with stress and metabolic homeostasis. Cell. 2010;140:338–348. doi: 10.1016/j.cell.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stockinger B. Beyond toxicity: aryl hydrocarbon receptor-mediated functions in the immune system. J Biol. 2009;8:61. doi: 10.1186/jbiol170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glass CK, Ogawa S. Combinatorial roles of nuclear receptors in inflammation and immunity. Nat Rev Immunol. 2006;6:44–55. doi: 10.1038/nri1748. [DOI] [PubMed] [Google Scholar]
- 36.Araki K, Youngblood B, Ahmed R. The role of mTOR in memory CD8 T-cell differentiation. Immunol Rev. 2010;235:234–243. doi: 10.1111/j.0105-2896.2010.00898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Esser C, Rannug A, Stockinger B. The aryl hydrocarbon receptor in immunity. Trends Immunol. 2009;30:447–454. doi: 10.1016/j.it.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 38.Mezrich JD, et al. An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J Immunol. 2010;185:3190–3198. doi: 10.4049/jimmunol.0903670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Platzer B, et al. Aryl hydrocarbon receptor activation inhibits in vitro differentiation of human monocytes and Langerhans dendritic cells. J Immunol. 2009;183:66–74. doi: 10.4049/jimmunol.0802997. [DOI] [PubMed] [Google Scholar]
- 40.Quintana FJ, et al. Control of Treg and TH17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008;453:65–71. doi: 10.1038/nature06880. [DOI] [PubMed] [Google Scholar]
- 41.Veldhoen M, et al. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- 42.Bronte V, Zanovello P. Regulation of immune responses by L-arginine metabolism. Nat Rev Immunol. 2005;5:641–654. doi: 10.1038/nri1668. [DOI] [PubMed] [Google Scholar]
- 43.Mellor AL, Munn DH. IDO expression by dendritic cells: tolerance and tryptophan catabolism. Nat Rev Immunol. 2004;4:762–774. doi: 10.1038/nri1457. [DOI] [PubMed] [Google Scholar]
- 44.Munn DH, et al. GCN2 kinase in T cells mediates proliferative arrest and anergy induction in response to indoleamine 2,3-dioxygenase. Immunity. 2005;22:633–642. doi: 10.1016/j.immuni.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 45.Allen RH, Stabler SP. Identification and quantitation of cobalamin and cobalamin analogues in human feces. Am J Clin Nutr. 2008;87:1324–1335. doi: 10.1093/ajcn/87.5.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goodman AL, et al. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anderson PJ, et al. One pathway can incorporate either adenine or dimethylbenzimidazole as an alpha-axial ligand of B12 cofactors in Salmonella enterica. J Bacteriol. 2008;190:1160–1171. doi: 10.1128/JB.01386-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Curtale F, Pokhrel RP, Tilden RL, Higashi G. Intestinal helminths and xerophthalmia in Nepal. A case control study. J Trop Pediatr. 1995;41:334–337. doi: 10.1093/tropej/41.6.334. [DOI] [PubMed] [Google Scholar]
- 49.Sommer A, Tarwotjo I, Katz J. Increased risk of xerophthalmia following diarrhea and respiratory disease. Am J Clin Nutr. 1987;45:977–980. doi: 10.1093/ajcn/45.5.977. [DOI] [PubMed] [Google Scholar]
- 50.Cha HR, et al. Downregulation of Th17 cells in the small intestine by disruption of gut flora in the absence of retinoic acid. J Immunol. 2010;184:6799–6806. doi: 10.4049/jimmunol.0902944. Demonstrates how a single micronutrient, vitamin A, modulates host immune responses through its effects on the composition of the intestinal microbiota. [DOI] [PubMed] [Google Scholar]
- 51.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gaboriau-Routhiau V, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. Refs 51 and 52 represent seminal studies identifying a single member of the intestinal microbiota that drives differentiation of intestinal Th17 cells. [DOI] [PubMed] [Google Scholar]
- 53.Schaible UE, Kaufmann SH. Iron and microbial infection. Nat Rev Microbiol. 2004;2:946–953. doi: 10.1038/nrmicro1046. [DOI] [PubMed] [Google Scholar]
- 54.Reddy BS, Pleasants JR, Wostmann BS. Effect of intestinal microflora on iron and zinc metabolism, and on activities of metalloenzymes in rats. J Nutr. 1972;102:101–107. doi: 10.1093/jn/102.1.101. [DOI] [PubMed] [Google Scholar]
- 55.Werner T, et al. Depletion of luminal iron alters the gut microbiota and prevents Crohn’s disease-like ileitis. Gut. 2011;60:325–333. doi: 10.1136/gut.2010.216929. [DOI] [PubMed] [Google Scholar]
- 56.Ley RE. Obesity and the human microbiome. Curr Opin Gastroenterol. 2010;26:5–11. doi: 10.1097/MOG.0b013e328333d751. [DOI] [PubMed] [Google Scholar]
- 57.Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mandard S, et al. The fasting-induced adipose factor/angiopoietin-like protein 4 is physically associated with lipoproteins and governs plasma lipid levels and adiposity. J Biol Chem. 2006;281:934–944. doi: 10.1074/jbc.M506519200. [DOI] [PubMed] [Google Scholar]
- 59.Vijay-Kumar M, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. Links changes in the configuration of the intestinal microbiota in TLR5 deficient mice to inflammation and development of metabolic syndrome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gregor MF, Hotamisligil GS. Inflammatory mechanisms in obesity. Ann Rev Immunol. 2011;29:415–445. doi: 10.1146/annurev-immunol-031210-101322. [DOI] [PubMed] [Google Scholar]
- 61.Kintscher U, et al. T-lymphocyte infiltration in visceral adipose tissue: a primary event in adipose tissue inflammation and the development of obesity-mediated insulin resistance. Arterioscler Thromb Vasc Biol. 2008;28:1304–1310. doi: 10.1161/ATVBAHA.108.165100. [DOI] [PubMed] [Google Scholar]
- 62.Winer S, et al. Normalization of obesity-associated insulin resistance through immunotherapy. Nat Med. 2009;15:921–929. doi: 10.1038/nm.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zuniga LA, et al. IL-17 regulates adipogenesis, glucose homeostasis, and obesity. J Immunol. 2010;185:6947–6959. doi: 10.4049/jimmunol.1001269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Feuerer M, et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat Med. 2009;15:930–939. doi: 10.1038/nm.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Uysal KT, Wiesbrock SM, Marino MW, Hotamisligil GS. Protection from obesity-induced insulin resistance in mice lacking TNF-alpha function. Nature. 1997;389:610–614. doi: 10.1038/39335. [DOI] [PubMed] [Google Scholar]
- 66.Wen L, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cani PD, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 68.Brun P, et al. Increased intestinal permeability in obese mice: new evidence in the pathogenesis of nonalcoholic steatohepatitis. Am J Physiol Gastrointest Liver Physiol. 2007;292:G518–525. doi: 10.1152/ajpgi.00024.2006. [DOI] [PubMed] [Google Scholar]
- 69.Shi H, et al. TLR4 links innate immunity and fatty acid-induced insulin resistance. J Clin Invest. 2006;116:3015–3025. doi: 10.1172/JCI28898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu HJ, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experiment autoimmune encephalomyelitis. Proc Natl Acad Sci USA. 2011;108:4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Valm AM, et al. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc Natl Acad Sci. 2011;108:4152–4157. doi: 10.1073/pnas.1101134108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Golden MH. Oedematous malnutrition. Br Med Bull. 1998;54:433–444. doi: 10.1093/oxfordjournals.bmb.a011699. [DOI] [PubMed] [Google Scholar]
- 74.Ferreira RB, Antunes LC, Finlay BB. Should the human microbiome be considered when developing vaccines? PLoS Pathog. 2010;6:e1001190. doi: 10.1371/journal.ppat.1001190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Campbell DI, et al. Chronic T cell-mediated enteropathy in rural west African children: relationship with nutritional status and small bowel function. Pediatr Res. 2003;54:306–311. doi: 10.1203/01.PDR.0000076666.16021.5E. [DOI] [PubMed] [Google Scholar]
- 76.Humphrey JH. Child undernutrition, tropical enteropathy, toilets, and handwashing. Lancet. 2009;374:1032–1035. doi: 10.1016/S0140-6736(09)60950-8. An excellent review of the relationship between environmental enteropathy and malnutrition. [DOI] [PubMed] [Google Scholar]
- 77.Guerrant RL, Oria RB, Moore SR, Oria MO, Lima AA. Malnutrition as an enteric infectious disease with long-term effects on child development. Nutr Rev. 2008;66:487–505. doi: 10.1111/j.1753-4887.2008.00082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Meeting of the immunization Strategic Advisory Group of Experts April 2009--conclusions recommendations. Wkly Epidemiol Rec. 2009;84:220–236. [PubMed] [Google Scholar]
- 79.Grassly NC, et al. Mucosal immunity after vaccination with monovalent and trivalent oral poliovirus vaccine in India. J Infect Dis. 2009;200:794–801. doi: 10.1086/605330. [DOI] [PubMed] [Google Scholar]
- 80.Lagos R, et al. Effect of small bowel bacterial overgrowth on the immunogenicity of single-dose live oral cholera vaccine CVD 103-HgR. J Infect Dis. 1999;180:1709–1712. doi: 10.1086/315051. [DOI] [PubMed] [Google Scholar]
- 81.Nemes E, et al. Gluten intake interferes with the humoral immune response to recombinant hepatitis B vaccine in patients with celiac disease. Pediatrics. 2008;121:e1570–1576. doi: 10.1542/peds.2007-2446. [DOI] [PubMed] [Google Scholar]
- 82.Menendez-Corrada R, Nettleship E, Santiago-Delpin EA. HLA and tropical sprue. Lancet. 1986;2:1183–1185. doi: 10.1016/s0140-6736(86)92195-1. [DOI] [PubMed] [Google Scholar]
- 83.Ghoshal UC, et al. Tropical sprue is associated with contamination of small bowel with aerobic bacteria and reversible prolongation of orocecal transit time. J Gastroenterol Hepatol. 2003;18:540–547. doi: 10.1046/j.1440-1746.2003.03006.x. [DOI] [PubMed] [Google Scholar]
- 84.Hayes KS, et al. Exploitation of the intestinal microflora by the parasitic nematode Trichuris muris. Science. 2010;328:1391–1394. doi: 10.1126/science.1187703. Demonstrates the co-evolution of bacterial and eukaryotic components of the microbiota and its impact on host immunity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gaboriau-Routhiau V, Raibaud P, Dubuquoy C, Moreau MC. Colonization of gnotobiotic mice with human gut microflora at birth protects against Escherichia coli heat-labile enterotoxin-mediated abrogation of oral tolerance. Ped Res. 2003;54:739–746. doi: 10.1203/01.PDR.0000086902.52137.C9. [DOI] [PubMed] [Google Scholar]
- 86.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 87.Goodman AL, et al. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci. 2011 doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu G, Yang K, Burns S, Shrestha S, Chi H. The S1P(1)-mTOR axis directs the reciprocal differentiation of TH1 and Treg cells. Nat Immunol. 2010;11:1047–1056. doi: 10.1038/ni.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Procaccini C, et al. An oscillatory switch in mTOR kinase activity sets regulatory T cell responsiveness. Immunity. 2010;33:929–941. doi: 10.1016/j.immuni.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Iwata M, et al. Retinoic acid imprints gut-homing specificity on T cells. Immunity. 2004;21:527–538. doi: 10.1016/j.immuni.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 91.Siddiqui KR, Powrie F. CD103+ GALT DCs promote Foxp3+ regulatory T cells. Mucosal Immunol. 2008;1 (Suppl 1):S34–38. doi: 10.1038/mi.2008.43. [DOI] [PubMed] [Google Scholar]
- 92.Ertesvag A, Engedal N, Naderi S, Blomhoff HK. Retinoic acid stimulates the cell cycle machinery in normal T cells: involvement of retinoic acid receptor-mediated IL-2 secretion. J Immunol. 2002;169:5555–5563. doi: 10.4049/jimmunol.169.10.5555. [DOI] [PubMed] [Google Scholar]
- 93.Iwata M, Eshima Y, Kagechika H. Retinoic acids exert direct effects on T cells to suppress Th1 development and enhance Th2 development via retinoic acid receptors. Int Immunol. 2003;15:1017–1025. doi: 10.1093/intimm/dxg101. [DOI] [PubMed] [Google Scholar]
- 94.Lemire JM, Adams JS, Sakai R, Jordan SC. 1 alpha,25-dihydroxyvitamin D3 suppresses proliferation and immunoglobulin production by normal human peripheral blood mononuclear cells. J Clin Invest. 1984;74:657–661. doi: 10.1172/JCI111465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mora JR, Iwata M, von Andrian UH. Vitamin effects on the immune system: vitamins A and D take centre stage. Nat Rev Immunol. 2008;8:685–698. doi: 10.1038/nri2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Daniel C, Sartory NA, Zahn N, Radeke HH, Stein JM. Immune modulatory treatment of trinitrobenzene sulfonic acid colitis with calcitriol is associated with a change of a T helper (Th) 1/Th17 to a Th2 and regulatory T cell profile. J Pharmacol Exp Ther. 2008;324:23–33. doi: 10.1124/jpet.107.127209. [DOI] [PubMed] [Google Scholar]
- 97.Wang TT, et al. Cutting edge: 1,25-dihydroxyvitamin D3 is a direct inducer of antimicrobial peptide gene expression. J Immunol. 2004;173:2909–2912. doi: 10.4049/jimmunol.173.5.2909. [DOI] [PubMed] [Google Scholar]
- 98.Sigmundsdottir H, et al. DCs metabolize sunlight-induced vitamin D3 to ‘program’ T cell attraction to the epidermal chemokine CCL27. Nat Immunol. 2007;8:285–293. doi: 10.1038/ni1433. [DOI] [PubMed] [Google Scholar]
- 99.Oh DY, et al. GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell. 2010;142:687–698. doi: 10.1016/j.cell.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Atarashi K, et al. ATP drives lamina propria TH17 cell differentiation. Nature. 2008;455:808–812. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.