Abstract
The goal of this study is to evaluate the stability of lyophilized siRNA formulations. The gene silencing efficiency of a stored lyophilized siRNA formulation (i.e. siRNA nanosomes) was evaluated in interferon-α (IFN-α) resistant hepatitis C virus (HCV) at different time points up to three months in an in vitro cell culture model and compared with freshly prepared siRNA formulations. Novel siRNA sequences were encapsulated within nanosize liposomes following condensation with protamine sulfate. The siRNA encapsulated nanosomes were lyophilized and stored at 4°C for 3 months, along with liquid liposomes (L) and lyophilized liposome powder (P) which were subsequently used to prepare siRNA nanosomes (L) and siRNA nanosomes (P), respectively at different time points. Physiochemical and biological properties of all three formulations were compared at different time points up to 3 months. The particle size of the stored siRNA nanosomes (642±25 nm) was considerably larger initially in comparison with the liquid liposomes (134±5 nm) and lyophilized liposomes (118±3). However, the particle size gradually became smaller over time (413±128 nm by the third month). The zeta potential of all three formulations was initially very high (> +40 mV), followed by a gradual decrease over time. The amount of siRNA in the stored siRNA nanosomes decreased ~ 18% during the 3 month storage period (1.16±0.03 nmol initially on day 1 vs. 0.95±0.04 nmol after 3 months). With respect to biological potency, all three formulations were significantly effective to knock-down HCV throughout the storage time. The cell viability was well-maintained throughout this period. Thus, this study indicates that the stored lyophilized siRNA formulation is as effective as the fresh preparation and that long-term storage could be a viable option to treat deadly diseases such as cancer and viral infection.
Keywords: siRNA delivery, Hepatitis C virus, Nanosome, Lyophilization, Stability
1. Introduction
Hepatitis C virus (HCV) has been recognized as one of the deadliest killers among viral diseases, with an estimated 170 million carriers worldwide. PEGylated interferon-alpha (IFN-α) in combination with ribavirin has been used to treat HCV with substantial success (Manns et al., 2001; Keating and Plosker, 2005); although, half of the patients failed to develop a sustained virologic response after this therapy, indicating a need for the development of an alternative strategy to improve treatment. Sequence specific gene silencing based on RNA interference (RNAi) is one of the newly evolving fields in biomedical science and may be a potential alternative strategy to treat HCV. Several investigators, including the authors of this paper, have attempted to develop delivery vehicles for siRNA. These delivery systems have been assessed for their ability to knock-down HCV in in vitro cell culture studies, as well as in in vivo animal studies (Chevalier et al., 2007; Wang et al., 2005; Watanabe et al., 2007; Wilson et al., 2003). A number of experimental techniques have recently been attempted for siRNA delivery, such as, hydrodynamic injection (McCaffrey et al., 2002; Lewis et al., 2002), viral genomes (McCaffrey et al., 2003), as well as liposomal and polymeric nanoparticles (Yano et al., 2004; Andersen et al., 2008; Gao and Huang, 2009). In our laboratory, we have successfully developed vehicles to deliver siRNA by applying high pressure homogenization to a cationic liposomal formulation containing DOTAP, cholesterol, protamine sulfate and trehalose. Unlike viral vectors for gene delivery which are associated with high toxicity (Engelhardt et al., 1993) and immunogenicity (Herz and Gerard, 1993), our lipid-based delivery vehicles are comparatively non-toxic, nano-sized and easy to assemble, and they have been shown to completely clear HCV in cell culture studies. The siRNA delivery vehicles used in experiments to date were always freshly prepared. However, long-term use of such formulations would require that they be stored under controlled temperature for long periods of time. During this storage period, the particle size, shape and siRNA entrapment should be maintained in order to retain functional activity. But due to their rapid deterioration by aggregation, aqueous formulations cannot be stored for long periods of time, either at room temperature or at 4°C. This limits their utility as a viable option for therapeutic application on a long-term basis. Moreover, preparation of fresh formulations at the patient’s bedside requires proper training of the health care providers, which is often difficult to fulfill (Caplen et al., 1995). In this context, lyophilization appears to be a feasible approach to solve this problem. Lyophilized powders can be stored at 4° C or even at room temperature for extended time periods and can be reconstituted in a reproducible manner immediately prior to use (Reinisalo et al., 2006). Since siRNA therapeutics could benefit from storable formulations, there is a demonstrable need to develop a stable siRNA formulation to extend siRNA therapy for the widespread treatment of deadly diseases.
The lyophilization of polymeric particles (Andersen et al., 2008) and lipoplex (Yadava et al., 2008) encapsulating siRNAs, as well as the functional characteristics of short-term stored lyophilized nanoparticles (Gao and Huang, 2009; Li et al., 2000), have been reported, but the resiliency and functional aspects of long-term stored siRNA formulations with more extensive therapeutic applications have not been studied. The goal of the current study is to assess the long-term storage effects on the physicochemical characteristics of lyophilized siRNA nanosomes (i.e. siRNA nanosome), such as particle size, morphology, zeta potential and quantity of siRNA, and to correlate them with siRNA’s knock-down efficiency. For comparison, liquid liposomes and lyophilized liposomes powder were also prepared and stored at 4°C, and then they were used to freshly prepare siRNA nanosomes (L) and siRNA nanosomes (P), respectively. They were compared with stored siRNA nanosomes at different time periods to determine the effects of storage on the physicochemical properties as well as on the biological efficiency (i.e. viral inhibition and cell toxicity in an in vitro cell culture system). This study therefore provides valuable information on the effects of storage on siRNA formulations for their potential long-term therapeutic applications.
2. Materials and Methods
2.1. Materials
1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) and cholesterol were purchased from Avanti Polar-lipids Inc. (Birmingham, AL, USA). Protamine sulfate salt Grade X, trehalose dihydrate and HPLC grade chloroform were obtained from Sigma Chemical Co. (St. Louis, MO, USA). si-321 (5′-GGU CUC GUA GAC CGU GCA CTT -3′), si-359 (5′-CCU CAA AGA AAA ACC AAA ATT -3′) and si-74 (5′-GCG CCU AGC CAU GGC GUU ATT -3′) were designed and purchased from Applied Biosystems (Austin, TX). Fetal bovine serum albumin (BSA), Dulbecco’s modified Eagle’s medium (DMEM) and penicillin/streptomycin antibiotics were purchased from Gibco, Invitrogen Corp. (Carlsbad, CA, USA). The Ribogreen assay kit was supplied by Molecular Probes (Eugene, OR, USA). INF-α was purchased from Schering Corporation (Kenilworth, NJ). All other reagents were of analytical grade and were supplied by Sigma Chemical Co. (St. Louis, MO, USA).
2.2. Preparation of cationic liposomes
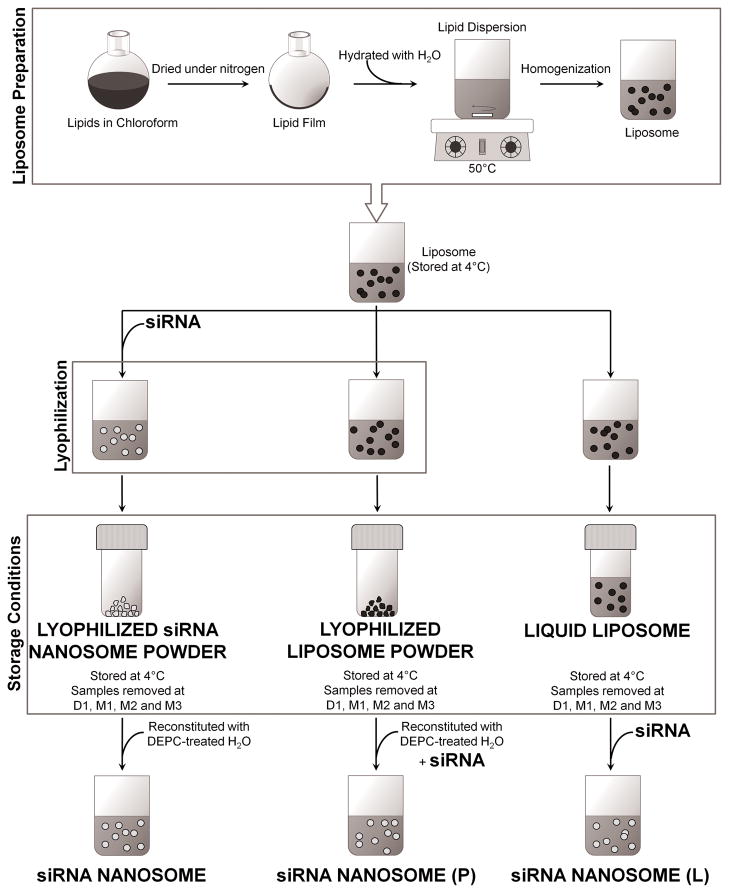
Fig. 1 summarizes the stages of the liposome preparation process and illustrates the difference among the preparation methods for each nanosome sample. Briefly, the liquid liposomes were prepared from a mixture of two lipids: cholesterol and DOTAP at the molar ratio of 1:1. The lipids (containing 50 mg DOTAP and 26.7 mg cholesterol) were dissolved in 15 ml of HPLC-grade chloroform in a round bottom flask and then dried under nitrogen gas and overnight vacuum. The resulting lipid film was hydrated in de-ionized water to give a final concentration of 20 mM. This lipid dispersion was warmed to 50°C in a water bath for 45 minutes by rotation, followed by warming again at 35°C for another 10 minutes. The resultant dispersion was stored at room temperature for 3 hours before it was transferred into a scintillation vial and warmed to 50°C for 10 minutes. The final lipid dispersion was homogenized by using an EmulsiFlex-B3 high pressure homogenizer at 20,000 psi for 5 cycles (Avestin Inc., Ottawa, Canada) (Pupo et al., 2005; Barnadas-Rodriguez and Sabes, 2001; Berger et al., 2001). For each cycle, 2.5 ml of lipid dispersion was subjected to homogenization, and the resultant liposomes were collected in another scintillation vial. The liposomes were kept at room temperature for 1 hour prior to overnight storage at 4°C.
Fig. 1.
Abbreviated illustrated summary of the liposome and nanosome preparation methodology. The liposomes prepared from DOTAP and cholesterol at 1:1 molar ratio using high pressure homogenization were stored as liquid or lyophilized powder form at 4°C. The nanosomes were prepared using siRNA, protamine sulphate, trehalose and differentially stored liposomes. The final nanosomes produced were categorized by their preparation method: siRNA nanosome was prepared from lyophilized siRNA nanosome powder, siRNA nanosome (P) was freshly prepared from lyophilized liposome powder, and similarly siRNA nanosome (L) was freshly prepared from the liquid liposome.
When preparing the lyophilized liposome powder, the refrigerated liposomes were brought to room temperature (approximately 2 hours). The liposomes were then added to a trehalose solution (lyoprotectant) at the mass ratio (in μg) of liposome to trehalose equal to 8.6:100 in DEPC-treated water and vortexed for 15 seconds to obtain a homogeneous mixture. A portion of the mixture was subsequently maintained at −80°C for 2 hours and finally put in a lyophilizer (Labconco freeze dryer; Labconco Corp., Kansas City, MO). The lyophilization protocol was performed as follows: the freeze dryer was cooled to −50°C for 1 hour, and then the chamber pressure was reduced to 0.310 mBar. The frozen sample was dried in a drying chamber at −20°C for 5 days followed by secondary drying at +20°C for 3 hours. After the secondary drying, the lyophilized liposome powder was collected and mixed using a sterilized spatula and stored in a desiccator at 4°C. The remaining portion which had not been used for lyophilization was designated as the “liquid liposomes” and was stored similarly in a desiccator at 4°C.
2.3. Preparation of siRNA nanosomes
Three different preparation methods were used to make three distinct siRNA nanosome samples (Fig. 1). For the first preparation method, freshly prepared protamine sulfate solution in DEPC-treated water was added drop wise to an aqueous solution of siRNA while vortexing the solution at a moderate speed. siRNA and protamine sulfate condensation was continued for 40 minutes at room temperature. Following siRNA-protamine sulfate condensation, freshly-prepared, pre-warmed liposome was added to the mixture. The final preparation was mixed rapidly by pipetting up and down 30 times. Freshly-prepared trehalose solution in DEPC-treated water was added and vortexed to allow thorough mixing of the nanosomes and trehalose. This first prepared sample was designated siRNA nanosomes. The siRNA nanosomes were aliquoted into several tubes; each containing 15 μg of siRNA entrapped in the siRNA nanosomes. The different tubes corresponded to the different storage time periods for the sample. All of these siRNA nanosome samples were kept at −80°C for 2 hours followed by lyophilization for 5 days. After lyophilization, the samples were collected, sealed and kept in a desiccator at 4°C. The stored samples were removed at each storage time: Day 1 (D1), Month 1 (M1), Month 2 (M2), and Month 3 (M3). At each time point, the lyophilized siRNA nanosome powder was reconstituted with DEPC-treated water.
The second nanosome preparation method involved the use of lyophilized liposome powder which has been stored in a desiccator at 4°C. For this preparation method, at each time point (i.e. D1, M1, M2 and M3) a sample of the liposome powder was removed and reconstituted to 20 mM with DEPC-treated water. These reconstituted liposomes were then combined with siRNA and protamine sulfate in the same methodology as described above for the siRNA nanosomes. These nanosome samples were designated siRNA nanosomes (P) since they were made from stored liposome powder.
The third preparation method is the same as the second method except that the nanosomes were prepared from stored liquid liposomes. Liquid liposomes were stored at 4°C for D1, M1, M2 and M3. At each time point, a sample of the liquid liposome was removed; and siRNA and protamine sulfate were added to create liquid nanosomes. The resulting nanosomes were designated siRNA nanosomes (L) since they were made from stored liquid liposomes. Table 1 summarizes the composition of the samples made from the three different preparation methods. In each preparation method, two different siRNA sequences (i.e. si-321 and si-359) were used to make nanosome formulations with the exception of si-74 which was prepared using only liquid liposomes.
Table 1.
Composition of different siRNA nanosomes
| Sample Preparation | siRNA |
|---|---|
| siRNA nanosome | si-321, si-359 |
| siRNA nanosome (P) | si-321, si-359 |
| siRNA nanosome (L) | si-321, si-359 |
| si-74 nanosome (L) | si-74 |
siRNA concentration 50 pmol; Lipid (DOTAP and cholesterol at 1:1 Molar) amount 8.6 μg; Protamine sulphate 2 μg; Trehalose 100 μg
2.4. Measurement of particle size, morphology and zeta potential
The particle size of different samples (i.e. liquid liposome and lyophilized liposome and siRNA nanosomes) at different time points was determined by dynamic laser light scattering method at room temperature by using a Delsa Nano C Particle Analyzer (Beckman Coulter Inc., Fullerton, CA, USA). Each measurement was performed four times (n=4). The result was reported as 70th percentile (i.e., seventy percentile cut-off points) and 90th percentile (i.e., ninety percentile cut-off points), respectively, and corresponding standard deviation.
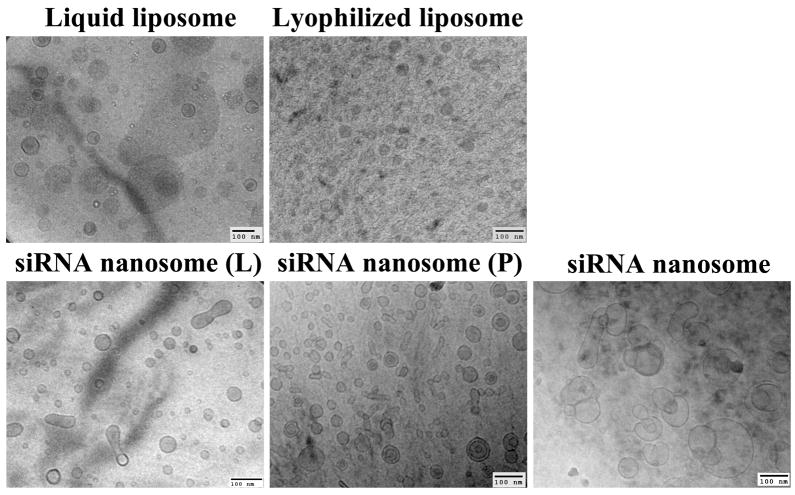
The morphology of blank liposomes (i.e. liquid and lyophilized liposomes), siRNA nanosome, siRNA nanosome (L) and siRNA nanosome (P) was examined by using Transmission Electron Microscope (JEOL 2010, Gatan Inc., Pleasanton, CA). A 6 μl drop of the different formulations was placed on a holey carbon grid and rapidly vitrified in liquid ethane. The sample was then transferred under the protection of liquid nitrogen to the cryo-TEM sample holder and inserted into the cryo-TEM. The temperature of the sample grids was maintained at −175°C during the course of imaging.
Analysis of the charge density of different formulations at various time points was performed by examining their zeta potential with a Malvern Zetasizer 2000 (Malvern Instruments, Malvern, UK). The system was initially calibrated with standards. All experimental samples were prepared in 1mM KCl.
2.5. Measurement of siRNA amount in the stored siRNA nanosomes
The amount of siRNA, either complexed or encapsulated, in siRNA nanosomes was determined at different time periods. Briefly, following reconstitution into water, the stored siRNA nanosomes were centrifuged at 14,000 rpm (Allegra Centrifuge, Beckman Coulter Inc., Fullerton, CA) for 15 minutes at 4°C. Supernatants containing the free siRNA were separated from the pellets. 500 μl of a 1% sodium dodecyl sulfate (SDS) solution was added to the pellets, and to the supernatants. Samples were then incubated at 37°C for 18 hours with gentle agitation (50 rpm). The siRNA amount was then measured by using Ribogreen assay, following the manufacturer’s protocol.
2.6. Cell culture and cell lines
The Huh-7 cells obtained from the laboratory of James Wilson, Wistar Institute of Human Gene Therapy, Pennsylvania and R4GFP HCV (+) replicon cells were previously developed in our laboratory (Hazari et al., 2010). The continuous replication of HCV in R4GFP HCV (+) replicon cells was monitored by GFP expression. The cells were routinely cultured and expanded at 37°C in DMEM, supplemented with 2 mM l-glutamine, nonessential amino acids, 100 U/ml of penicillin, 100 μg/ml of streptomycin and 5% fetal bovine serum in an atmosphere of 5% CO2.
2.7. In vitro functional assessment of HCV inhibition by different siRNA nanosomes at various time points
GFP expression
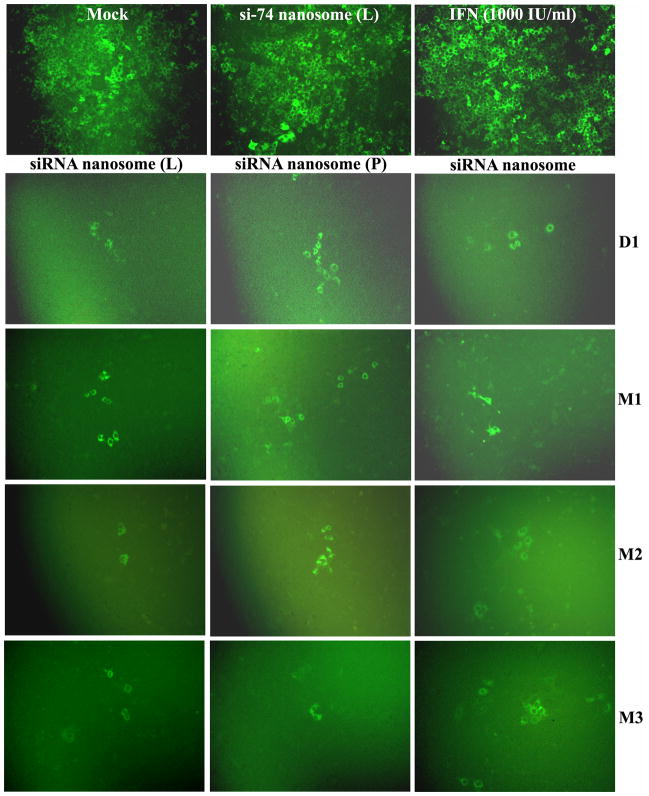
In vitro transfection efficiency and inhibition of HCV by different nanosome formulations at several time points were monitored by both qualitative (GFP expression) and quantitative (FACS) analysis. Briefly, R4GFP HCV (+) replicon cells were cultured in 6-well tissue culture plates (TCPs) for 24 hours in 5% FBS containing DMEM, followed by replenishing the media with 2% FBS containing DMEM. The cells were then transfected with different formulations and kept at 37°C for 48 hours. The cells were subsequently scraped by using trypsin-EDTA, and replated and cultured again for 72 hours. After this time period, the cells were again transfected with the same nanosome formulations, entrapping the same amount of siRNA for another 48 hours. Simultaneously in another well, cells were treated with 1000 IU/ml IFN for both time periods. Following the second dose, the GFP expression was monitored by a fluorescence microscope (Olympus America, Inc., NY), and photographs were taken at 10X magnification.
FACS analysis
After capturing GFP images by fluorescence microscope, flow cytometric analysis (FACS) (BD Biosciences, San Hose, CA, USA) was performed in order to quantify the GFP positive cells post treatment.
2.8. Colony assay
The inhibition of HCV replication by different formulations at various time points was analyzed by colony formation assay. Briefly, R4GFP HCV (+) replicon cells were cultured and transfected with different formulations and with IFN at several time points following the same procedure described earlier. Following the second dose in 8 days, the cells were scraped with trypsin-EDTA and washed twice with sterile PBS. Subsequently, about 1x104 cells were plated in a 100 mm dish in a DMEM selection media containing 5% FBS and 1 mg/ml G-418. The cell culture was continued for 3 weeks with media being replenished once a week. Finally, the cells were washed and stained with Giensa Dye (Sigma Chemical Co., St. Louis, USA), and the colonies were counted by using a manual colony counter.
2.9. Measurement of cell viability
The viability of R4GFP HCV (+) replicon cells transfected with different siRNA nanosomes was determined at specific time points by MTT assay in accordance with the manufacturer’s protocol (Sigma Chemical Co., MO, USA). Briefly, the cells (1x104) were cultured on 24-well TCP in 5% FBS containing DMEM media for 24 hours. After that, the DMEM media was replaced with fresh 2% FBS media. The cells were then transfected with different formulations for an additional 48 hours. For comparison, cells were simultaneously treated with 1000 IU/ml IFN or transfected with si-74 nanosomes (L) encapsulating 100 pmol si-74 siRNA. Finally, the cells were incubated with MTT solution at 37°C for 3 hours, and cell viability was measured by reading the absorbance at 570 nm.
2.10. Statistical analysis
The statistical significance of particle size differences between liquid liposomes and lyophilized liposomes at both the 70th percentile and 90th percentile was compared by the Student-Newman-Keul’s nonparametric test, using the GraphPad Prism 5 software. A p value of <0.05 was considered as evidence of a significant difference.
3. Results
3.1. Storage effects on physical and morphological properties of different formulations
3.1.1. Particle size and zeta potential
The particle size of liposomes (liquid vs. lyophilized) and siRNA nanosomes at different time periods was measured and the results of particle size at a 70th percentile are presented in Table 2. The particle sizes of the liquid and lyophilized liposomes at D1 were 134±6 nm and 118±4 nm, respectively. With increasing storage time periods, both the liquid and lyophilized liposomes increased slightly in size (Table 2). The particle sizes of liquid liposomes at M1, M2 and M3 were 150±4 nm, 170±5 nm and 179±9 nm, respectively, whereas the particle sizes for lyophilized liposomes at M1, M2 and M3 were 114±4 nm, 127±4 nm and 123±11 nm, respectively. The significant difference in average particle size between liquid and lyophilized liposomes was observed with a majority of the particles (i.e. 90th percentile) being compared. The size difference was observed to increase with increasing length of storage times. For example, the lyophilized liposome particle size was shown to be 205±10 nm at M2 and 219±12 nm at M3, respectively, which is significantly lower than that of the liquid liposomes at M2 (311±84 nm) (p=0.0011) and M3 (341±78 nm) (p< 0.001). Moreover, a more heterogeneous population of particles was observed in liquid liposomes at the two and three month mark, unlike the more homogeneous distribution in the lyophilized liposomes. In contrast, the particle size (70th percentile) of the stored siRNA nanosomes was initially found to be 642±25 nm at D1. Their particle sizes at M1, M2 and M3 were determined to be 572±49 nm, 435±22 nm and 413±128 nm, respectively.
Table 2.
Measurement of particle size (70th percentile) and zeta potential at different time periods (n=4)
| Samples | Day 1 | Month 1 | Month 2 | Month 3 | ||||
|---|---|---|---|---|---|---|---|---|
| Particle size (nm) | Zeta potential (mV) | Particle size (nm) | Zeta potential (mV) | Particle size (nm) | Zeta potential (mV) | Particle size (nm) | Zeta potential (mV) | |
| Liquid liposome | 134±5 | 56±3 | 150±6 | 47±2 | 170±26 | 36±3 | 179±31 | 16±2 |
| Lyophilized liposome powder | 118±3 | 40±6 | 114±5 | 30±10 | 127±11 | 29±4 | 123±18 | 11±3 |
| siRNA nanosome | 642±25 | 48±4 | 572±49 | 30±3 | 435±35 | 19±2 | 413±128 | 17±1 |
The results of zeta potential measurement are listed in Table 2. Initially these formulations had a higher charge density on their surface (liquid liposome ~ 56±3 mV, lyophilized liposome ~ 40±6 mV and siRNA nanosome ~ 48±4 mV), which gradually decreased over time. The surface charge at M1, M2 and M3 was determined to be 47±2 mV, 36±3 mV, and 16±2 mV for liquid liposomes; 30±10 mV, 29±4 mV, and 11±3 mV for lyophilized liposomes; and 30±3 mV, 19±2 mV, and 17±1 mV for siRNA nanosomes, respectively.
3.1.2. Particle morphology
The morphology of various liposomes and nanosomal formulations encapsulating siRNAs, as assessed by cryo-TEM, reveals that most of the particles of the liquid and lyophilized liposomes were discrete and spherical, although the particles of the lyophilized liposomes were smaller and highly homogenous compared to their liquid counterpart (Fig. 2). The siRNA nanosome (L) and siRNA nanosome (P) had a large number of smaller spherical particles, but also had a small percentage of elongated and larger particles. The particles of the stored siRNA nanosome were relatively larger compared to the freshly prepared siRNA nanosome (L) and siRNA nanosome (P); and though most of the particles in stored siRNA nanosome were round and spherical in shape, some were larger and oval shape due to fusion of some particles.
Fig. 2.
Cryo-TEM pictures of liposomes (liquid and lyophilized) and different siRNA nanosomes. Scale bar represents 100 nm.
3.2. Inhibition of HCV replication in an in vitro cell culture study
3.2.1. Measurement of GFP expression and FACS analysis
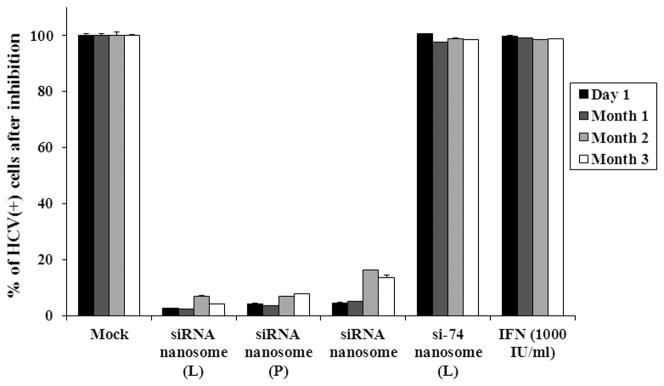
A comparison of the results of GFP expression of R4GFP HCV (+) replicon cells transfected with different nanosomal formulations at various time points showed that the freshly prepared siRNA nanosome (L) and siRNA nanosome (P) as well as the stored siRNA nanosomes almost completely cleared the viruses, irrespective of time periods. This was evident by the presence of only a few HCV positive cells in the culture plate after transfection (Fig. 3A). Quantification of the HCV (+) cells by FACS analysis (Fig. 3B) indicated that all three formulations were significantly effective in clearing the viruses (> 95%) at early time points (D1 and M1). With increasing storage periods, the HCV inhibition was insignificantly diminished in freshly prepared siRNA nanosomes (L) (~4%) and siRNA nanosomes (P) (~6%) at M2 and M3. However, when the siRNA nanosome was stored for 2 months (M2) and 3 months (M3), the knock-down efficiency of HCV was determined to be ~ 84% at M2 and ~ 87% at M3 compared to > 95% on the first day. Despite this slight decrease of knock-down efficiency during long-term storage, the stored siRNA nanosome was still significantly effective in clearing IFN-resistant HCV compared to non-targeted siRNA formulations (i.e. si-74 nanosome) or IFN treatment.
Fig. 3.
Qualitative and quantitative assessment of the inhibition of HCV replication by different siRNA nanosomes at different time points. The HCV (+) replicon cells were cultured in 6-well plates for 24 hours followed by transfection with different formulations, or with IFN for 48 hours. The cells were scraped and re-cultured for 72 hours before they were transfected for a second time with the same formulations, or with IFN for 48 hours. A. Fluorescence photographs of GFP expression using a fluorescence microscope at 10X magnification and B. Quantitative measurement of GFP expression by FACS analysis. The results represent mean ± standard deviation (n=3).
3.2.2. Colony selection of HCV (+) cells after nanosomal transfection
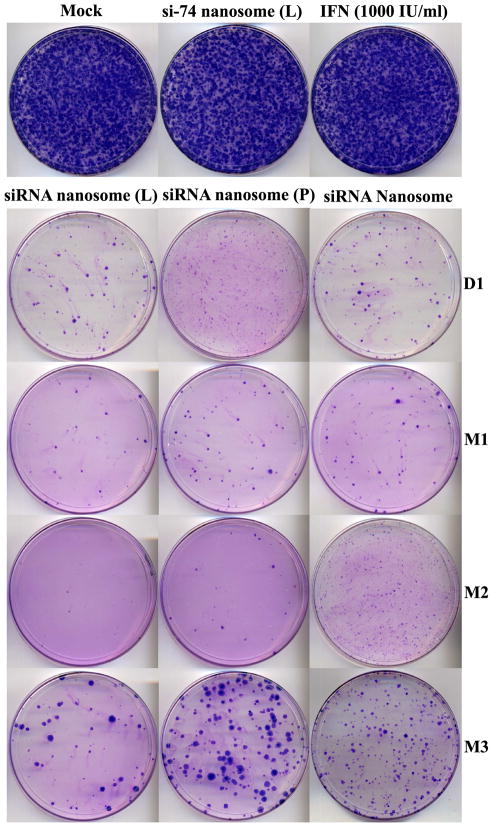
The inhibition of HCV replication by different formulations was also monitored by colony selection assay. The results shown in Fig. 4 indicate that neither the non-targeted siRNA formulation (i.e. si-74 nanosomes), nor IFN was successful in prohibiting HCV replication. This was evident by the generation of a vast population of HCV colonies apparent in the petri dish. In comparison, when the cells were transfected with different nanosomal formulations encapsulating si-359 and si-321, the level of colony formation was significantly downgraded up to the three month time mark. However, the inhibition of HCV was not as pronounced as it was on day one (D1) for each formulation. At 3 month time mark, the number of colonies for freshly prepared siRNA nanosome (L) (~32±12) and siRNA nanosome (P) (~128±23) as well as stored siRNA nanosome (~169±34) was significantly lower than non-transfected control (~740±18), non-targeted si-74 nanosome transfection (~718±26) and IFN inhibition (~733±15). These results clearly indicate that the stored siRNA nanosome maintains its effectiveness to clear the virus as like as the fresh preparation during the long storage time period.
Fig. 4.
Colony selection of HCV (+) replicon cells transfected with different siRNA nanosomes and with IFN at specific time points. After 24 hours culture in 6-well plates, HCV (+) replicon cells were transfected with different formulations or with IFN for 48 hours. The cells were scraped and re-cultured for 72 hours and then were transfected for a second time with the same formulations, or with IFN for 48 hours. Finally the cells were scraped and re-plated in a 100 mm dish and cultured in the presence of G-418 to select HCV (+) cell colonies that were not inhibited by siRNA nanosomes.
3.3. Effect of storage on siRNA stability within the siRNA nanosomes
The siRNA stability within the stored siRNA nanosomes was measured at D1 and at M3. The results indicate no loss of siRNA amount at D1. Following 3 months of storage at 4°C, the siRNA amount was reduced from 1.16±0.03 nmol to 0.95±0.04 nmol which was nearly a reduction of 15–18% from its actual amount at D1. This drop in siRNA content in stored siRNA nanosome was possibly due to the degradation of the siRNA or degradation of the lipid/siRNA complex (i.e. siRNA nanosome) over time. This would also explain the slight decline of functional inhibition of this formulation at later time points, particularly at M3.
3.4. Cell viability
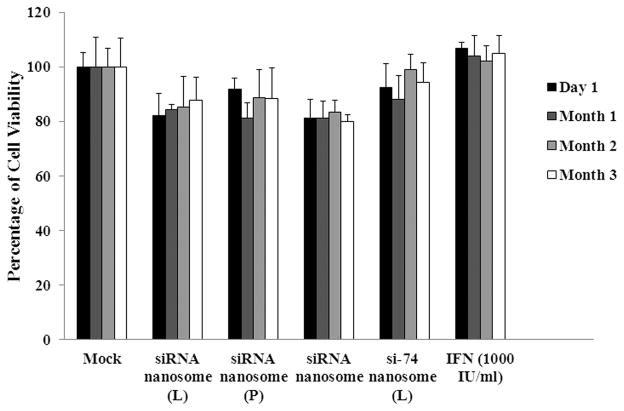
The results of the MTT assay (Fig. 5) indicate that freshly prepared siRNA nanosome (L) maintained cell viability of 82±7%, 84±2%, 85±11%, 87±8% at D1, M1, M2 and M3, respectively; whereas the freshly prepared siRNA nanosome (P) maintained cell viability of 91±4%, 81±6%, 88±10%, 88±11% at the same time points, respectively. Similarly, the stored siRNA nanosomes maintained cell viability of 81±7%, 81±6%, 83±4% and 80±3% at D1, M1, M2 and M3, respectively. All the formulations maintained cell viability greater than 80% at all time points. However, freshly prepared siRNA nanosome (L) and siRNA nanosome (P) showed slightly superior cell viability compared to the stored siRNA nanosomes.
Fig. 5.
Cell viability of R4GFP HCV (+) replicon cells transfected with different siRNA nanosomes and IFN at specific time points. After 24 hours culture in 24-well TCPs, the cells were transfected with different formulations, or with IFN, for another 48 hours. Subsequently, the cells were incubated with MTT solution at 37°C for 3 hours, and the absorbance reading was taken at 570 nm. The percentage of cell viability was determined by comparing viable cells to the untreated cells during transfection at different time points. The results represent mean ± standard deviation (n=5).
4. Discussion
RNAi offers great clinical potential as a therapeutic modality for a variety of viral diseases, especially HCV (Chevalier et al., 2007). Being a single-stranded RNA virus, the HCV becomes a suitable candidate for siRNA therapy. Given the poor performance of IFN-α and ribavirin for the clearance of IFN-resistant HCV genotype, an alternative therapeutic approach for chronic hepatitis C is required. To meet this need, we have designed some novel siRNAs that can target the highly conserved sequences in 5′ UTR of IFN-resistant HCV, which render complete clearance of IFN-resistant HCV in the in vitro cell culture study. A novel gene delivery vehicle (i.e. nanosomes) was also developed in our laboratory by applying high pressure homogenization to a liposomal formulation, which was then used to carry multiple siRNAs simultaneously to treat IFN-resistant HCV in cell culture studies. The complete clearance of IFN-resistant HCV was accomplished by freshly preparing siRNA encapsulated nanosomes just prior to use. However, in order to enable their use in actual clinical settings, a storable siRNA formulation is required, one which will maintain both physicochemical and biological potency over time. In this context, several recent reports highlighted the effects of lyophilization of the siRNA encapsulated liposomal and polymeric nanoparticles (Andersen et al., 2008; Yadava et al., 2008; Li et al., 2000; Gao et al., 2010). However, none of these reports have provided detailed information concerning the characterization and biological significance of the lyophilized siRNA nanoparticles. It is expected that such information will have enormous therapeutic potential in treating deadly viruses such as HCV.
The current study compares three sets of siRNA nanosomes with regard to their physical state and biological potency at different storage time periods. In the available scientific literature, the particle size has exclusively been used to determine the deterioration of lyophilized nanoparticles. Yadava et al. (2008) and other investigators (Li et al., 2000) have reported that lyophilization itself causes an increase in particle size, which can be prevented to some extent by the use of proper lyoprotectants. Sucrose and trehalose are two main lyoprotectants that are often used for lyophilization of nanoparticles (Andersen et al., 2008; Yadava et al., 2008; Christensen et al., 2007; Maitani et al., 2008). A higher liposome to trehalose ratio (~ 8.6:100) has been used in our study, since a higher amount of trehalose seems to preserve liposomal activity (Christensen et al., 2007). The lyophilization helps to keep lyophilized liposomes powder small and uniform in a majority of the particles. Our results show that the liquid liposomes deteriorate upon long-term storage, especially at the three month time points (M3).
Our results also show that the stored siRNA nanosomes are 2–5 times larger in size than the liquid and lyophilized liposomes. Since lyophilization apparently has no significant influence on the size of lyophilized liposomes over time, it may be the siRNA entrapment which causes the increase in particle size of the stored siRNA nanosomes. It was also observed that the entrapment of siRNA within stored siRNA nanosomes generated larger and elongated particles that were absent in the freshly prepared siRNA nanosomes (P) (Fig. 2). This further confirms the fact that it is the siRNA entrapment, rather than the lyophilization step which is largely responsible for the increase in particle size of the stored siRNA nanosomes. In our preliminary studies, we have observed that sonication for a period of 2 to 4 minutes can significantly reduce the particle size of freshly prepared siRNA nanosomes (L) (100 nm vs. 211nm) (Kundu et al., personal communication). Thus, sonication of the siRNA nanosomes just prior to lyophilization may be attempted to reduce particle size.
A published report by Li et al. (2000) mentions that the nanoparticle size remained almost unchanged when frozen in liquid nitrogen, whereas when the nanoparticles were kept at −80°C for 60 minutes prior to lyophilization, aggregation was observed. In a separate note, it was reported that addition of even a small quantity (1%) of sucrose could help preserve particle size. Since our nanosomal formulations were prepared utilizing a high ratio of liposome to trehalose, freezing the nanosomes at −80°C for 2 hours just prior to lyophilization should not affect the particle size in this study. Nevertheless, future studies will be designed to observe the effects of sonication of the siRNA nanosomes and to observe the effects of freezing them in liquid nitrogen instead of maintaining at −80°C for 2 hours prior to lyophilization. These approaches may be worthwhile in attempting to minimize the particle size.
In the stored siRNA nanosomes, it was also noticed that the particles were gradually getting smaller over time. In the last few years, several articles have highlighted possible reasons for the degradation of lipid/DNA or lipid/siRNA complex during a longer storage time period. Andersen et al. (2008) have assumed that the decrease of the knockdown efficiency of lyophilized siRNA nanoparticles during storage was possibly due to the breakdown of the lipid or siRNA or lipid/siRNA complex. In this context, a series of studies done by Molina et al. (2004), Molina and Anchordoquy (2008a), and Molina and Anchordoquy (2008b) clearly revealed that the degradation of the lipid/DNA complexes during long-term storage resulted from the generation of reactive oxygen species (ROS) and thiobarbituric reactive substances (TBARS) which are highly reactive to lipid formulations, especially to unsaturated lipids (i.e. DOTAP). The formation of free radicals and the peroxidation of lipids in lyophilized formulations during long-term storage are enhanced by lipid contamination (such as lipid hydroperoxides) present in commercially available lipids, by the presence of sugars or exposure to oxygen in the air (Breen and Murphy, 1995), and even by sonication (Almog et al., 1991). Similar to Molina et al. (2004) and Molina and Anchordoquy (2008a), we have also assumed that the degradation of lipids or siRNA in the stored siRNA nanosomes during long-term storage leads to the generation of smaller particles over time. Similarly, the generation of ROS can lead to the oxidation of siRNA which ultimately degrades siRNA and lowers the amount of siRNA in the stored siRNA nanosomes. The slim reduced amount of siRNA observed in stored siRNA nanosomes at the 3 month mark in our experiment also confirms this hypothesis.
The effect of long-term storage on the surface charge (i.e. zeta potential) of nanoparticles has not been studied adequately. Our results show that the zeta potential of all three formulations was initially very high on day one (D1). The zeta potential subsequently tended to decrease with increasing storage time periods, irrespective of formulation type. It is known that a zeta potential measurement less positive than +30 mV, or less negative than −30 mV, is a possible indication of instability of the particles in solution. It is possible that the zeta potential of nanoparticles might gradually be getting altered over time. Ohshima et al. (2009) have recently shown that the zeta potential of particles declined with decreasing particle size. Huang et al. (2008) have shown that addition of different lipids in a nanoparticle formulation generated different degrees of zeta potential after storage for 28 days. Also, lack of addition of lipids caused a slight decrease in the zeta potential of the nanoparticles over a period of time. In our study, the zeta potential of both liquid and lyophilized liposomes was maintained at levels ≥ +30 mV until the measurements done at three month time point (M3). This finding reveals that storage of liposomes for longer time periods in excess of three months does not guarantee the stability even in the dry powder form. On the other hand, the zeta potential of stored siRNA nanosomes was maintained at levels ≥ +30 mV until M1and then gradually reduced to ≤ +20 mV during extended storage period. Similar to the observation by Ohshima et al. (2009), we also assumed that the gradual decrease of particle size in stored siRNA nanosomes with time caused a reduction in their surface charge. Also, it is possible that a decrease in siRNA amount over time is partially responsible for the reduction of surface charge of stored siRNA nanosomes.
In the in vitro study, all the nanosomal formulations were initially highly effective to knock-down HCV. The gene silencing activity of freshly prepared siRNA nanosome (L) and siRNA nanosome (P) was insignificantly diminished during long-term storage until the three month mark. The particles of the lyophilized liposomes were smaller and more uniform compared to the liquid liposomes at M2 and M3; however, this did not result in superior transfection with the siRNA nanosome (P) at M2 and M3 compared to the siRNA nanosome (L). One likely reason is that a majority of the particles for both liquid and lyophilized liposomes were < 200 nm, which was small enough to carry all the siRNAs into the cells in the cell culture study. Thus, the transfection efficiency was not affected by the presence of a small percentage of larger particles in the liquid formulation. Similarly, the stored siRNA nanosomes showed significant gene silencing activity throughout the storage period compared to non-transfected control or non-targeted (siRNA) transfected cells. A slight decrease of knockdown efficiency at the end of month two and month three was probably due to the decrease of siRNA amount over time. Moreover, the instability of stored siRNA nanosomes at this time period, as represented by their reduced surface charge ( +20 mV), was also partially responsible for this slight decrease of gene silencing activity during extended storage time periods. The degradation of siRNA or lipids in this stored lyophilized formulation (i.e. siRNA nanosomes) as well as their instability over time could be minimized by using appropriate anti-oxidant such as α-tocopherol (Molina and Anchordoquy, 2008b) in future studies.
The delivery of nanoparticles to the liver hepatocytes can be enhanced by conjugating the particles with liver specific target molecules which can specifically recognize and bind to the receptors on the hepatocyte cell surface, such as asyloglycoprotein receptors (ASGP-R) (Fumoto et al., 2003; Kawakami et al., 2000). Watanabe et al. (2007) have examined lactosylated cationic liposomes for their ability to carry siRNA to knock-down HCV core protein in liver hepatocytes and reported that HCV core protein targeted siRNAs have better access to the liver hepatocytes when they are complexed with lactosylated cationic liposomes compared to the conventional cationic liposomes (without lactose conjugation). The freshly prepared liquid nanosomes of our study have also been examined for their ability to knock down GAPDH gene (a house keeping gene) specifically in the liver hepatocytes of a mouse model and we have seen a significant reduction of GAPDH (~70%) in the liver hepatocytes (data not reported). In this context, Kawakami et al. (1998, 2000) have reported that cationic liposomes which contained galactose conjugated cholesterol (i.e. chol-gal) instead of pure cholesterol could be recognized by ASGP-R and were therefore able to enhance nucleic acid delivery to liver hepatocytes. This receptor mediated endocytosis of such formulations might constitute an effective approach for targeted delivery to liver hepatocytes with minimal non targeted deposition to other organs and reduced toxicity. Further modification of the siRNA encapsulated nanosomes by conjugating galactose moieties on the particle surface will be examined in our future studies designed to achieve targeted delivery to the liver hepatocytes and complete clearance of HCV.
In conclusion, the delivery of siRNA for the clearance of HCV in our study could be performed by either freshly prepared or stored formulations. In general, stored formulation will be comparatively easy to use, does not require extensive training of the health care providers and could be easily accessible to a vast majority of people on a long-term basis. This study clearly indicates that the HCV knockdown efficiency of our stored siRNA formulation is as effective as the freshly prepared formulations during long-term storage compared to non-transfected control, as well as non-targeted siRNA transfection and IFN inhibition. A slight decrease of gene silencing efficiency of stored formulations was shown after two months and three months which was possibly due to the degradation of siRNA and particle instability in solution. A further optimization of this formulation to minimize siRNA degradation and instability over time will therefore meet the demand of a storable, stable and active siRNA delivery vehicle for the long-term treatment of cancer and certain viral diseases.
Table 3.
Measurement of siRNA amount in siRNA nanosomes over time
| Nanosome | siRNA amount (nmol) (Day 1) | siRNA amount (nmol) (Month 3) |
|---|---|---|
| siRNA nanosome | 1.16±0.03 | 0.95±0.04 |
Acknowledgments
This work was funded in part by Louisiana Board of Regents RC/EEP (2007–11), LEQSF(2007–12)-ENH-PKSFI-PRS-02, DoD W81XWH-07-1-0136, NIH grants 1R01CA127481, R21CA129776, 1G12RR026260-01, P20CA118768-02 and LCRC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Almog R, Forward R, Samsonoff C. Stability of sonicated aqueous suspensions of phospholipids under air. Chem Phys Lipids. 1991;60:93–99. doi: 10.1016/0009-3084(91)90031-6. [DOI] [PubMed] [Google Scholar]
- Andersen MO, Howard KA, Paludan SR, Besenbacher F, Kjems J. Delivery of siRNA from lyophilized polymeric surfaces. Biomaterials. 2008;29:506–512. doi: 10.1016/j.biomaterials.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Barnadas-Rodriguez R, Sabes M. Factors involved in the production of liposomes with a high-pressure homogenizer. Int J Pharm. 2001;213:175–186. doi: 10.1016/s0378-5173(00)00661-x. [DOI] [PubMed] [Google Scholar]
- Berger N, Sachse A, Bender J, Schubert R, Brandl M. Filter extrusion of liposomes using different devices: comparison of liposome size, encapsulation efficiency, and process characteristics. Int J Pharm. 2001;223:55–68. doi: 10.1016/s0378-5173(01)00721-9. [DOI] [PubMed] [Google Scholar]
- Breen AP, Murphy JA. Reactions of oxyl radicals with DNA. Free Radic Biol Med. 1995;18:1033–1077. doi: 10.1016/0891-5849(94)00209-3. [DOI] [PubMed] [Google Scholar]
- Caplen NJ, Alton EW, Middleton PG, Dorin JR, Stevenson BJ, Gao X, Durham SR, Jeffery PK, Hodson ME, Coutelle C, et al. Liposome-mediated CFTR gene transfer to the nasal epithelium of patients with cystic fibrosis. Nat Med. 1995;1:39–46. doi: 10.1038/nm0195-39. [DOI] [PubMed] [Google Scholar]
- Chevalier C, Saulnier A, Benureau Y, Flechet D, Delgrange D, Colbere-Garapin F, Wychowski C, Martin A. Inhibition of hepatitis C virus infection in cell culture by small interfering RNAs. Mol Ther. 2007;15:1452–1462. doi: 10.1038/sj.mt.6300186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen D, Foged C, Rosenkrands I, Nielsen HM, Andersen P, Agger EM. Trehalose preserves DDA/TDB liposomes and their adjuvant effect during freeze-drying. Biochim Biophys Acta. 2007;1768:2120–2129. doi: 10.1016/j.bbamem.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Engelhardt JF, Simon RH, Yangm Y, Zepeda M, Weber-Pendleton S, Doranz B, Grossman M, Wilson JM. Adenovirus-mediated transfer of the CFTR gene to lung of nonhuman primates: biological efficacy study. Hum Gene Ther. 1993;4:759–769. doi: 10.1089/hum.1993.4.6-759. [DOI] [PubMed] [Google Scholar]
- Fumoto S, Nakadori F, Kawakami S, Nishikawa M, Yamashita F, Hashida M. Analysis of hepatic disposition of galactosylated cationic liposome/plasmid DNA complexes in perfused rat liver. Pharm Res. 2003;20:1452–1459. doi: 10.1023/a:1025766429175. [DOI] [PubMed] [Google Scholar]
- Gao J, Sun J, Li H, Liu W, Zhang Y, Li B, Qian W, Wang H, Chen J, Guo Y. Lyophilized HER2-specific PEGylated immunoliposomes for active siRNA gene silencing. Biomaterials. 2010;31:2655–2664. doi: 10.1016/j.biomaterials.2009.11.112. [DOI] [PubMed] [Google Scholar]
- Gao K, Huang L. Nonviral methods for siRNA delivery. Mol Pharm. 2009;6:651–658. doi: 10.1021/mp800134q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazari S, Chandra PK, Poat B, Datta S, Garry RF, Foster TP, Kousoulas G, Wakita T, Dash S. Impaired antiviral activity of interferon alpha against hepatitis C virus 2a in Huh-7 cells with a defective Jak-Stat pathway. Virol J. 2010;7:36. doi: 10.1186/1743-422X-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz J, Gerard RD. Adenovirus-mediated transfer of low density lipoprotein receptor gene acutely accelerates cholesterol clearance in normal mice. Proc Natl Acad Sci USA. 1993;90:2812–2816. doi: 10.1073/pnas.90.7.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZR, Hua SC, Yang YL, Fang JY. Development and evaluation of lipid nanoparticles for camptothecin delivery: a comparison of solid lipid nanoparticles, nanostructured lipid carriers, and lipid emulsion. Acta Pharmacol Sin. 2008;29:1094–1102. doi: 10.1111/j.1745-7254.2008.00829.x. [DOI] [PubMed] [Google Scholar]
- Kawakami S, Yamashita F, Nishikawa M, Takakura Y, Hashida M. Asialoglycoprotein receptor-mediated gene transfer using novel galactosylated cationic liposomes. Biochem Biophys Res Commun. 1998;252:78–83. doi: 10.1006/bbrc.1998.9602. [DOI] [PubMed] [Google Scholar]
- Kawakami S, Fumoto S, Nishikawa M, Yamashita F, Hashida M. In vivo gene delivery to the liver using novel galactosylated cationic liposomes. Pharm Res. 2000;17:306–313. doi: 10.1023/a:1007501122611. [DOI] [PubMed] [Google Scholar]
- Keating GM, Plosker GL. Peginterferon alpha-2a (40KD) plus ribavirin: a review of its use in the management of patients with chronic hepatitis C and persistently ‘normal’ ALT levels. Drugs. 2005;65:521–536. doi: 10.2165/00003495-200565040-00006. [DOI] [PubMed] [Google Scholar]
- Lewis DL, Hagstrom JE, Loomis AG, Wolff JA, Herweijer H. Efficient delivery of siRNA for inhibition of gene expression in postnatal mice. Nat Genet. 2002;32:107–108. doi: 10.1038/ng944. [DOI] [PubMed] [Google Scholar]
- Li B, Li S, Tan Y, Stolz DB, Watkins SC, Block LH, Huang L. Lyophilization of cationic lipid-protamine-DNA (LPD) complexes. J Pharm Sci. 2000;89:355–364. doi: 10.1002/(SICI)1520-6017(200003)89:3<355::AID-JPS7>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Maitani Y, Aso Y, Yamada A, Yoshioka S. Effect of sugars on storage stability of lyophilized liposome/DNA complexes with high transfection efficiency. Int J Pharm. 2008;356:69–75. doi: 10.1016/j.ijpharm.2007.12.033. [DOI] [PubMed] [Google Scholar]
- Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, Goodman ZD, Koury K, Ling M, Albrecht JK. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- McCaffrey AP, Meuse L, Pham TT, Conklin DS, Hannon GJ, Kay MA. RNA interference in adult mice. Nature. 2002;418:38–39. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- McCaffrey AP, Nakai H, Pandey K, Huang Z, Salazar FH, Xu H, Wieland SF, Marion PL, Kay MA. Inhibition of hepatitis B virus in mice by RNA interference. Nat Biotechnol. 2003;21:639–644. doi: 10.1038/nbt824. [DOI] [PubMed] [Google Scholar]
- Molina MDC, Anchordoquy TJ. Degradation of lyophilized lipid/DNA complexes during storage: The role of lipid and reactive oxygen species. Biochim Biophys Acta. 2008a;1778:2119–2126. doi: 10.1016/j.bbamem.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Molina MDC, Anchordoquy TJ. Formulation strategies to minimize oxidative damage in lyophilized lipid/DNA complexes during storage. J Pharm Sci. 2008b;97:5089–5105. doi: 10.1002/jps.21365. [DOI] [PubMed] [Google Scholar]
- Molina MDC, Armstrong TK, Zhang Y, Patel MM, Lentz YK, Anchordoquy TJ. The stability of lyophilized lipid/DNA complexes during prolonged storage. J Pharm Sci. 2004;93:2259–2273. doi: 10.1002/jps.20138. [DOI] [PubMed] [Google Scholar]
- Ohshima H, Miyagishima A, Kurita T, Makino Y, Iwao Y, Sonobe T, Itai S. Freeze-dried nifedipine-lipid nanoparticles with long-term nano-dispersion stability after reconstitution. Int J Pharm. 2009;377:180–184. doi: 10.1016/j.ijpharm.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Pupo E, Padron A, Santana E, Sotolongo J, Quintana D, Duenas S, Duarte C, de la Rosa MC, Hardy E. Preparation of plasmid DNA-containing liposomes using a high-pressure homogenization--extrusion technique. J Control Release. 2005;104:379–396. doi: 10.1016/j.jconrel.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Reinisalo M, Urtti A, Honkakoski P. Freeze-drying of cationic polymer DNA complexes enables their long-term storage and reverse transfection of post-mitotic cells. J Control Release. 2006;110:437–443. doi: 10.1016/j.jconrel.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Wang Q, Contag CH, Ilves H, Johnston BH, Kaspar RL. Small hairpin RNAs efficiently inhibit hepatitis C IRES-mediated gene expression in human tissue culture cells and a mouse model. Mol Ther. 2005;12:562–568. doi: 10.1016/j.ymthe.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Umehara T, Yasui F, Nakagawa S, Yano J, Ohgi T, Sonoke S, Satoh K, Inoue K, Yoshiba M, Kohara M. Liver target delivery of small interfering RNA to the HCV gene by lactosylated cationic liposome. J Hepatol. 2007;47:744–750. doi: 10.1016/j.jhep.2007.06.015. [DOI] [PubMed] [Google Scholar]
- Wilson JA, Jayasena S, Khvorova A, Sabatinos S, Rodrigue-Gervais IG, Arya S, Sarangi F, Harris-Brandts M, Beaulieu S, Richardson CD. RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. Proc Natl Acad Sci USA. 2003;100:2783–2788. doi: 10.1073/pnas.252758799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadava P, Gibbs M, Castro C, Hughes JA. Effect of lyophilization and freeze-thawing on the stability of siRNA-liposome complexes. AAPS PharmSciTech. 2008;9:335–341. doi: 10.1208/s12249-007-9000-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano J, Hirabayashi K, Nakagawa S, Yamaguchi T, Nogawa M, Kashimori I, Naito H, Kitagawa H, Ishiyama K, Ohgi T, Irimura T. Antitumor activity of small interfering RNA/cationic liposome complex in mouse models of cancer. Clin Cancer Res. 2004;10:7721–7726. doi: 10.1158/1078-0432.CCR-04-1049. [DOI] [PubMed] [Google Scholar]