Abstract
The genetic risk factors for chronic obstructive pulmonary disease (COPD) are still largely unknown. To date, genome-wide association studies (GWASs) of limited size have identified several novel risk loci for COPD at CHRNA3/CHRNA5/IREB2, HHIP and FAM13A; additional loci may be identified through larger studies. We performed a GWAS using a total of 3499 cases and 1922 control subjects from four cohorts: the Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints (ECLIPSE); the Normative Aging Study (NAS) and National Emphysema Treatment Trial (NETT); Bergen, Norway (GenKOLS); and the COPDGene study. Genotyping was performed on Illumina platforms with additional markers imputed using 1000 Genomes data; results were summarized using fixed-effect meta-analysis. We identified a new genome-wide significant locus on chromosome 19q13 (rs7937, OR = 0.74, P = 2.9 × 10−9). Genotyping this single nucleotide polymorphism (SNP) and another nearby SNP in linkage disequilibrium (rs2604894) in 2859 subjects from the family-based International COPD Genetics Network study (ICGN) demonstrated supportive evidence for association for COPD (P = 0.28 and 0.11 for rs7937 and rs2604894), pre-bronchodilator FEV1 (P = 0.08 and 0.04) and severe (GOLD 3&4) COPD (P = 0.09 and 0.017). This region includes RAB4B, EGLN2, MIA and CYP2A6, and has previously been identified in association with cigarette smoking behavior.
INTRODUCTION
Chronic obstructive pulmonary disease (COPD) is defined as airflow limitation that is not fully reversible and is usually caused by exposure to noxious particles or gases—predominantly cigarette smoking, though other exposures such as biomass fuels are an important cause worldwide (1). COPD does not reverse with smoking cessation, and despite efforts to curtail cigarette smoking, COPD is a leading and increasing cause of morbidity and mortality. Worldwide, it is projected to rise to rank fifth in disease burden by 2020 (1), and in the USA, it now ranks as the third leading cause of death (2). The development of COPD among smokers is not uniform; a minority of smokers develops the disease (3), and lung function response to similar levels of cigarette smoke exposure varies greatly (4). Numerous studies have demonstrated a genetic component to COPD and smoking-related changes in lung function (5–8). However, the results of many small candidate gene studies have been inconsistent (9,10). Genome-wide association studies (GWASs) of COPD (11–13) have, to date, identified three susceptibility loci that have been well replicated (14–21). We report the results of a follow-up GWAS in four cohorts that identifiy a new COPD susceptibility locus.
RESULTS
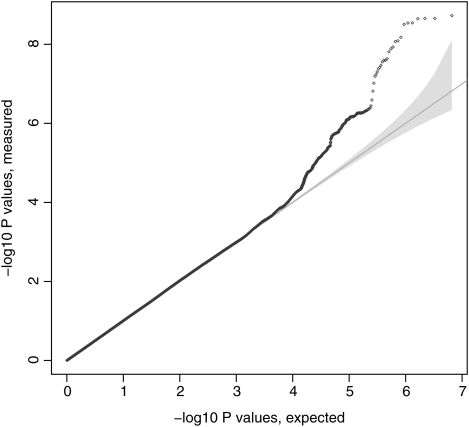
Baseline characteristics for subjects from the four cohorts: the Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints (ECLIPSE); Normative Aging Study (NAS) and National Emphysema Treatment Trial (NETT); Bergen, Norway COPD Cohort (GenKOLS); and the COPDGene study (first 1000 subjects) are shown in Table 1. Fixed-effects meta-analysis across the four cohorts for case–control status showed no evidence of substantial deviation from the null (lambda = 1.01, Fig. 1). The most significantly associated single nucleotide polymorphisms (SNPs) were in the previously identified locus on chromosome 4 in FAM13A (Table 2). However, a new genome-wide significant locus was identified on chromosome 19q13. The top P-value was at rs7937 (OR = 0.74, P-value = 2.88 × 10−9). There was some evidence of heterogeneity (P-value = 0.15 for Cochrane's Q, I2 = 43.8); however, similar results were found using a modified random-effects model (22) (P = 3.36 × 10−9).
Table 1.
Baseline characteristics (mean ± SD or percentage)
| COPDGene |
ECLIPSE |
NETT/NAS | GenKOLS |
|||||
|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | |
| n | 499 | 501 | 1764 | 178 | 373 | 435 | 863 | 808 |
| Age | 64.77 (8.12) | 60.2 (8.66) | 63.63 (7.1) | 57.48 (9.44) | 67.47 (5.78) | 69.8 (7.49) | 65.53 (10.03) | 55.62 (9.71) |
| Pack-years | 54.76 (26.69) | 38.87 (21.07) | 50.29 (27.42) | 32.11 (24.84) | 66.43 (30.68) | 40.66 (27.85) | 31.98 (18.46) | 19.66 (13.58) |
| Average cigarettes per day | 27.58 (11.76) | 24.89 (11.18) | 25.54 (12.39) | 21.87 (11.35) | 32.55 (13.47) | 29.27 (14.33) | 15.6 (7.71) | 13.82 (7.44) |
| FEV1, % predicted | 48.73 (18.41) | 97.98 (11.32) | 47.63 (15.62) | 107.83 (13.56) | 28.12 (7.38) | 99.97 (13.2) | 50.63 (17.44) | 94.91 (9.24) |
| Sex (% male) | 49.5 | 50.1 | 67.0 | 57.9 | 63.8 | 100 | 60.1 | 50.1 |
Figure 1.
The Quantile–quantile plot for the four-cohort meta-analysis including 1000 Genomes project imputed data, after adjustment for genetic ancestry using principal components.
Table 2.
Meta-analysis results
| Locus | SNP | Minor allele | MAF | COPDGene |
ECLIPSE |
NETT/NAS | GenKOLS |
Overall |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | I2 | P-value (Q) | ||||
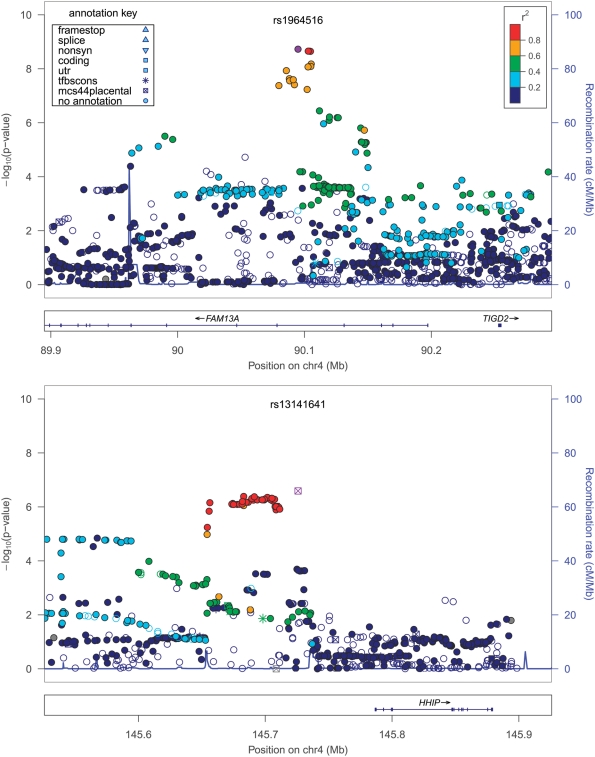
| 4q22 | rs1964516 | C | 0.49 | 0.78a (0.64–0.95) | 1.39 × 10−2 | 0.72a (0.56–0.91) | 7.37 × 10−3 | 0.62a (0.49–0.8) | 2.27 × 10−4 | 0.76a (0.64–0.89) | 7.58 × 10−4 | 0.73 (0.66–0.81) | 1.88 × 10−9 | 0 | 0.56 |
| rs7671167 | C | 0.49 | 0.78 (0.64–0.95) | 1.38 × 10−2 | 0.72 (0.57–0.92) | 7.75 × 10−3 | 0.63 (0.49–0.81) | 2.69 × 10−4 | 0.76 (0.64–0.89) | 7.67 × 10−4 | 0.73 (0.66–0.81) | 2.22 × 10−9 | 0 | 0.58 | |
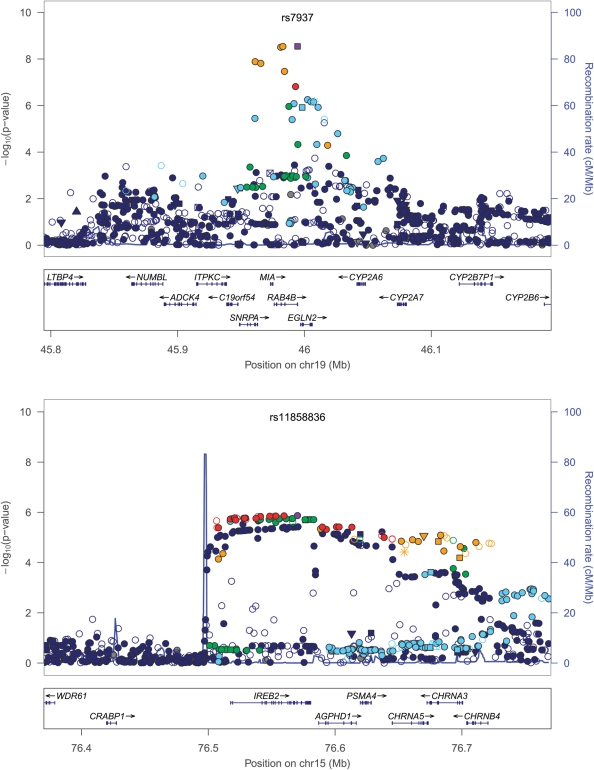
| 19q13 | rs7937 | C | 0.44 | 0.64 (0.52–0.78) | 1.06 × 10−5 | 0.72 (0.57–0.92) | 9.48 × 10−3 | 0.68 (0.54–0.86) | 1.35 × 10−3 | 0.85 (0.72–0.99) | 4.06 × 10−2 | 0.73b (0.63–0.83) | 2.88 × 10−9 | 44 | 0.15 |
| rs2604894 | A | 0.44 | 0.66a (0.53–0.81) | 9.28 × 10−5 | 0.72a (0.56–0.92) | 9.38 × 10−3 | 0.68a (0.53–0.88) | 2.67 × 10−3 | 0.85a (0.72–1) | 5.25 × 10−2 | 0.74b (0.65–0.84) | 3.41 × 10−8 | 30 | 0.23 | |
| 4q31 | rs13141641 | C | 0.43 | 0.91a (0.75–1.1) | 3.35 × 10−1 | 0.7a (0.55–0.89) | 3.00 × 10−3 | 0.62a (0.48–0.79) | 1.39 × 10−4 | 0.78a (0.66–0.92) | 3.06 × 10−3 | 0.76b (0.65–0.88) | 2.56 × 10−7 | 54 | 0.09 |
| rs13118928 | G | 0.40 | 0.92a (0.76–1.12) | 3.95 × 10−1 | 0.74 (0.58–0.93) | 1.11 × 10−2 | 0.6 (0.47–0.78) | 8.04 × 10−5 | 0.78 (0.66–0.92) | 2.45 × 10−3 | 0.76b (0.65–0.89) | 5.97 × 10−7 | 57 | 0.07 | |
| 15q25 | rs11858836 | A | 0.37 | 1.25a (1.02–1.53) | 2.98 × 10−2 | 1.05a (0.82–1.34) | 7.13 × 10−1 | 1.49a (1.16–1.92) | 1.98 × 10−3 | 1.38a (1.16–1.64) | 2.33 × 10−4 | 1.29b (1.13–1.47) | 1.35 × 10−6 | 35 | 0.20 |
| rs13180 | C | 0.36 | 0.78 (0.64–0.95) | 1.52 × 10−2 | 0.78 (0.61–0.99) | 4.26 × 10−2 | 0.74 (0.58–0.95) | 1.75 × 10−2 | 0.79 (0.67–0.94) | 7.59 × 10−3 | 0.78 (0.7–0.86) | 2.02 × 10−6 | 0 | 0.98 | |
The top result at each locus as well as additional genotyped SNPs at that locus is shown.
aImputed genotypes; note that imputed SNPs in this table had imputation quality r2 > 0.9.
bEffect estimates given for random-effects model.
We genotyped rs7937 and another nearby genome-wide significant SNP in linkage disequilibrium (LD) (rs2604894, r2 = 0.74) in 983 probands and 1876 siblings from the family-based International COPD Genetics Network study (ICGN). P-values for the COPD affection status for the same risk allele were 0.28 and 0.11 for rs7937 and rs2604894, respectively. More significant associations were demonstrated for pre-bronchodilator FEV1 (P = 0.08 and 0.04, respectively) and after limiting cases to Global Initiative for Chronic Obstructive Lung Disease (GOLD) severity classifications 3 and 4 COPD (P = 0.09 and 0.017, respectively).
The 19q13 locus includes the genes RAB4B, EGLN2 and CYP2A6, and SNPs in this locus have recently been identified in a large GWAS of smoking behavior (23,24) as associated with average number of cigarettes smoked per day; rs7937 was the second-ranked association to cigarettes per day at this locus in the study by Thorgeirsson et al. (23), with a combined P-value of 2.4 × 10−9. To explore whether our findings were due to associations with cigarette smoking behavior, we examined the relationship of rs7937 and rs2604894 with both pack-years and average number of cigarettes smoked per day separately in the case and control groups. In none of these analyses were any significant associations found; the strongest association (P = 0.27) was with pack-years of smoking in controls. These results are consistent with the results reported for rs7937 in the Bergen, Norway GenKOLS cohort, as this cohort was included in the GWAS by Thorgeirsson et al. (23). While these main effects were not significant, we also examined whether there was any evidence of a gene-by-cigarettes per day or pack-years interaction; none of these analyses were significant (P > 0.1).
At chromosome 19q13, and at the three previously demonstrated genome-wide associated loci (11–13) (4q22—FAM13A; 4q31—HHIP; and 15q25—CHRNA3/CHRNA5/IREB2), we used the imputed marker data from the 1000 Genomes study to attempt to further refine previously determined association signals (Table 2 and Fig 2). At 4q22 and 4q31, imputed SNPs in high LD (r2 of 1.0 and 0.9, respectively) with the most significant genotyped association had only marginally smaller P-values. At 15q25, the top hit was rs11858836; this SNP is in strong LD with the previously reported rs8034191 and rs1051730 (r2 of ∼0.75–0.8), both part of locus 1 of Saccone et al. (25), but also in moderate LD (r2 ∼0.4) with rs13180, our previously reported most significant SNP in this region. We also attempted to assess whether any known non-synonymous SNPs could account for these association signals. At only one locus, 15q25, was there any non-synonymous SNP at an r2 of >0.5 with the most significant SNP in the region; this was the CHRNA5 SNP rs16969968, which has been described as part of locus 1 above. To evaluate the potential effect of rarer SNPs—those with low r2 but high D′—we additionally evaluated SNPs with D′ > 0.8 within 500 kb of the top four loci. Two other imputed SNPs had D′ ∼ 1 with rs7937 at 19q13 and nominal associations with COPD: rs1801272 in CYP2A6 [P = 0.03, minor allele frequency (MAF) = 0.04, imputation r2 0.3–0.4 using HapMap2 data] previously described in this cohort as associated with lifetime average cigarettes per day (26), and rs36012476 in ADCK4 (P = 0.02, MAF = 0.07, and imputation r2 0.6–0.7). Inclusion of these SNPs only modestly attenuated the signal for rs7937 (P after adjustment = 8.3 × 10−8). In addition to single SNP analyses, we also performed analyses conditioning on the top SNP in each of the four GWAS regions, using markers 250 kb on either side and a threshold of P < ∼5 × 10−4 to reflect an approximate adjustment for a 500 kb interval flanking the most significant SNP. None of the loci demonstrated evidence of other independent SNP effects.
Figure 2.
Association plots at genome-wide significant loci. R2 values are given in relation to the most significant, labeled and highlighted SNP. Plots created using LocusZoom (83).
Finally, we attempted to estimate the relative contribution of these four risk loci to the sibling relative risk of COPD. We used odds ratios aggregated over the four cohorts and assumed independence of the four loci under a standard multiplicative model; acknowledging that the resulting estimates using the discovery cohorts are likely to be inflated. These four loci accounted for ∼5% of the total variance explained using either pseudo-R2 or a liability threshold model. Alternatively, using estimates of sibling relative risk from family-based studies of 2–3 (5,6), these four loci account for ∼8–15% of the sibling relative risk.
DISCUSSION
While a familial component to COPD has been recognized for at least the last 40 years (27)—aside from the nearly- contemporaneous identification of alpha-1 antitrypsin deficiency (28)—the genetic determinants of COPD have been generally elusive. Our analysis adds an additional cohort to three prior GWASs and identifies a new genome-wide significant locus associated with COPD susceptibility at chromosome 19q13.
The 19q13 locus has demonstrated clear associations with smoking behavior, and has been shown to be associated with cigarettes per day in two large meta-analytic GWASs (23,24). An association operating through cigarette smoking is clearly the most parsimonious and most likely explanation. However, we cannot rule out the possibility that other mechanisms may also be responsible for the association to COPD that we observed. At 15q25, mediation approaches suggest that pack-years explain only ∼1/4 of the association between these SNPs and COPD (29). Another gene at this locus, IREB2, was identified independently in part through gene expression and genetic association (30) as a candidate gene for COPD. In our data set, we were unable to show a significant association between rs7937 and cigarettes per day or pack-years in any of our study populations, and our finding persisted after inclusion of a non-synonymous SNP associated with cigarettes per day in this cohort. The 19q13 locus includes several other genes of potential interest expressed in developing animal or human lung: RAB4B (31), MIA (32) and EGLN2 (33). LTBP4 is ∼150 kb away, and variants in this gene have been associated with functional outcomes in subjects with emphysema (34), and disruption of this gene leads to emphysema in mice (35) and in humans, as part of the Urban-Rifkin-Davis Syndrome (OMIM #613177) (36).
While future studies (both statistical analyses, such as causal modeling and mediation analysis, and functional studies) may help determine whether other genes at this locus play a role in the 19q13 association, it is highly likely that this association occurs through cigarette smoking. CYP2A6 variants have additionally been associated with another major smoking-related disease, lung cancer (37). CYP2A6 and, to a lesser extent, CYP2B6 are involved in nicotine metabolism (38,39); the most significant associations in these studies are in LD with sequence variants that have been shown to reduce CYP2A6 enzyme activity, and rs7937 was associated with the levels of the major nicotine metabolite cotinine in a subset of subjects from the European Network for Genetic and Genomic Epidemiology (23). While we were able to identify a nominally significant association of cigarettes per day with a non-synonymous SNP in CYP2A6 in our COPD cohorts (26), CYPA26 activity is affected by a number of other demonstrated functional variants—both common and rare (40–43). While we failed to demonstrate an association with cigarette smoking behavior and rs7937 within our case or control groups, both the 15q25 and 19q13 loci have been associated with cotinine levels in other studies that did not find an association with cigarettes per day (44,45), suggesting that standard measures of smoking behavior are incomplete. Issues with measuring the complexity of cigarette smoke exposure such as recall bias (46), as well as differential intensity of smoking per cigarette and effects of cigarette metabolism, may be responsible for the lack of association with pack-years or cigarettes per day in our case and control groups.
The identification of this 19q13 COPD locus, in conjunction with 15q25, highlights the critical contribution of variants affecting the major behavioral risk factor for COPD, cigarette smoking. This is in contrast to coronary artery disease, where despite the fact that cigarette smoking is a major risk factor (47), none of the identified genome-wide association loci to date has identified variants known to affect smoking, and a minority of the identified loci have been associated with traditional risk factors (48). Ongoing cigarette smoking causes accelerated lung function decline; conversely, smoking cessation attenuates this decline, improves respiratory symptoms and reduces overall mortality (49–52). Decreasing cigarette consumption is essential to reducing the risk of COPD, and though a subset of our subjects carry lower risk alleles, our data do not suggest that there are smokers unlikely to benefit from smoking cessation. Whether genetic testing may aid a subset of more susceptible individuals in smoking cessation efforts is unclear (53,54).
While our GWAS is the largest reported to date for COPD, our sample size is still substantially smaller—by a factor of 10 or more—than some other complex diseases; a recent coronary artery disease GWAS included 22 233 cases (48). Thus, if the genetic architecture of COPD is similar to other complex common diseases, our study is underpowered to detect many of the likely other common variants contributing to COPD susceptibility. This lack of power may also explain why we were unable to discern independent associations at the 15q25 locus, as others have demonstrated (25); our family-based replication results may have also suffered from power limitations, or, less likely, reflect subtle stratification in our original case–control analysis (55,56). Our study also did not directly assess for rare genetic variation, which may be an important contributor to COPD susceptibility (57), nor did we assess for copy number variation. Finally, we did not attempt to address issues of COPD heterogeneity, for example, through radiographic phenotypes (58).
Our GWAS increases the number of genome-wide significant loci associated with COPD to four. Our analysis at these risk loci, using publicly available dense sequencing data and imputation, helps refines these signals for further study and replication. These loci account for only a small fraction of the observed effect of all genetic variants in COPD risk, and we anticipate analysis of other types of genetic variation (rare variants, copy number variants) and perhaps more importantly—future collaborations to increase the available sample size (59)—will expand this list of genetic loci and improve our understanding of COPD susceptibility.
MATERIALS AND METHODS
Genotyping methods and study descriptions for three data sets: the ECLIPSE; NAS and NETT; and GenKOLS have been described previously (11,13,60–64). All cohorts were of self-described European white ancestry and genotyped on Illumina platforms (Human Hap550 or Quad610). Quality-control procedures included tests for subject missingness, discordances, relatedness and sex; and marker missingness, discordances, singletons and Hardy-Weinberg equilibrium.
The study protocol for COPDGene (NCT00608764) has been described previously (65). Briefly, COPDGene is a multi-center genetic and epidemiologic investigation to study COPD and other smoking-related lung diseases. Participants completed a detailed protocol, including questionnaires, pre-and post-bronchodilator spirometry, high-resolution CT scanning of the chest and blood samples for genotyping. Samples from self-described European whites were genotyped at the Center for Inherited Disease Research (CIDR) at Johns Hopkins University using the Illumina Omni1 Quad platform. COPDGene genotyping underwent a similar quality-control procedure: subjects were screened for missing call rates > 1%, relatedness by estimated identity-by-descent > 0.125, sex discrepancies, and inbreeding coefficients > 0.2. Markers were screened for missingness > 2%, minor allele frequency < 1%, and deviation of Hardy–Weinberg at P < 1 × 10−5. Out of an initial set of 1006 COPDGene subjects, only one was unable to be genotyped successfully; none of the others failed on quality-control criteria. Five additional subjects were excluded based on the presence of lung parenchymal abnormalities other than emphysema after chest CT scan review. The average per-subject genotype missing rate was 0.05%, with a maximum missing rate of 0.96%. Details on the COPDGene subjects, as well as the subjects from the other three cohorts passing initial quality-control before removal of principal component outliers are shown in Table 1.
Of the 986 763 autosomal markers in the COPDGene study successfully genotyped, 797 983 markers remained after quality-control, with >95% of these exclusions on the basis of low minor allele frequency (<1%). Combining these markers with those from the other three cohorts (ECLIPSE, NETT/NAS and GenKOLS), a total of 296 201 markers were shared among all cohorts. After imputation from the 1000 Genomes study, this number increased to ∼6.1 million SNPs. Analysis for and control of population stratification were also performed via principal components using EIGENSOFT2.0 (66) as previously described (13) for the ECLIPSE, NETT, NAS and GenKOLS cohorts. For COPDGene, pruned markers with a LD cutoff of 0.12 were chosen from those with a MAF > 0.05, Illumina GC scores of >0.8 and present in HapMap CEU subjects. Outliers were removed over five iterations with deviations beyond 6 standard deviations, along the top 10 principal components. Significant principal components were defined using the Tracy–Widom statistic (67) as previously described (13). After removal of additional subjects with cryptic relatedness between cohorts and outliers by genetic ancestry based on principal components, 3456 cases and 1908 controls (99% of subjects) remained in the analysis. Ten ECLIPSE subjects were found to have borderline FEV1/FVC ratios but were retained in the analysis.
Genotype imputation within each study was performed using MaCH 1.0.16 (68) using 100 rounds of iterations to estimate model parameters and CEU samples from HapMap2 (69) and the 1000 Genomes Project (70) (phased CEU data, March 2010) as reference populations. Markers with an imputation r2 ≤ 0.3 were dropped from further analysis. Association analysis of SNPs with case–control status was performed in each cohort using logistic regression, adjusting for age, pack-years of cigarette smoking and genetic ancestry as summarized in the principal components. Imputed genotypes were analyzed in a similar manner, using SNP dosage data in PLINK 1.07 (71). Results were analyzed among the four cohorts using fixed-effect meta-analyses (72) using METAL (73) and in R 2.12 (www.r-project.org) with the meta-package. Genomic inflation factors (74) were calculated using GenABEL (75). I2 and Cochran's Q were calculated to assess for heterogeneity; a secondary analysis using a modified random-effects model was also performed (22).
Replication genotyping was performed using the SEQUENOM MassARRAY MALDI-TOF mass spectrometer (Sequenom, San Diego, CA, USA). Details of the ICGN cohort used for replication have been previously described (13,58). Association analysis in ICGN was performed using PBAT 3.61 (76) using one-sided P-values for the same risk allele, adjusting for age and pack-years of smoking in the COPD affection status analysis, and age, pack-years, sex and height in the analysis of FEV1.
Analyses conditional on other SNPs were performed by extracting genotype dosage data (for imputed SNPs) or actual genotypes; logistic regression adjusting for significantly associated SNPs was performed in R 2.12. Calculation of the contribution of loci to sibling relative risk was obtained using overall odds ratios and minor allele frequencies (77,78). Estimation of the fraction of variance explained was calculated using logistic regression and Nagelkerke's pseudo-R2 (79) as implemented in R Design package, and alternatively using the estimates based on a liability threshold model by So et al. (80) using a prevalence of COPD in smokers of ∼10% (81).
LD was calculated for HapMap2 and 1000 Genomes CEU data using SNAP (82). Regional association plots were created using LocusZoom (83). All positions are given in reference to the Human March 2006 (NCBI36/hg18) assembly.
AUTHOR CONTRIBUTIONS
Study design: E.K.S., M.H.C., A.A.L., D.S., S.G.P., X.K., W.H.A., R.T-S., D.A.L., P.B., A.G., T.H.B., E.A.R., B.J.M., J.E.H., J.D.C., H.O.C., L.D.E., W.M., J.V., J.C.Y., A.A., P.M.A.C., B.C., C.C., S.R., E.W.
Sample collection and phenotyping: A.A.L., D.S., S.G.P., X.K., W.H.A., R.T-S., D.A.L., P.B., A.G., E.A.R., B.J.M., J.D.C., E.K.S., C.P.H., D.L.D., H.O.C., L.D.E., W.M., J.V., J.C.Y., A.A., P.M.A.C., B.C., C.C., S.R., E.W.
Genotyping: B.J.K., M.H.C., S.G.P., X.K., T.H.B., T.M., J.B.H.
Informatics: M.H.C., J.S.S., J.P.Z., B.J.K., E.S.W., P.J.C., B.E.H.
Statistical analysis: M.H.C., C.L., T.H.B., T.M., J.B.H, M.S., E.K.S.
Manuscript writing: M.H.C., E.K.S.
FUNDING
This work was supported by US National Institutes of Health (NIH) grants R01 HL075478, R01 HL084323, P01 HL083069, P01 HL105339 and U01 HL089856 (E.K.S.); K12HL089990 and K08 HL097029 (M.H.C.); and U01 HL089897 (J.D.C.). M.S. is a recipient of a postdoctoral fellowship from the Niels Stensen Foundation. The National Emphysema Treatment Trial was supported by the National Heart, Lung, and Blood Institute, the Centers for Medicare and Medicaid Services and the Agency for Healthcare Research and Quality. The Normative Aging Study is supported by the Cooperative Studies Program/ERIC of the US Department of Veterans Affairs and is a component of the Massachusetts Veterans Epidemiology Research and Information Center (MAVERIC). The Norway GenKOLS study (Genetics of Chronic Obstructive Lung Disease, GSK code RES11080), the ECLIPSE study (clinicaltrials.gov identifier NCT00292552; GSK code SCO104960) and the ICGN study are funded by GlaxoSmithKline. The COPDGene® project is also supported by the COPD Foundation through contributions made to an Industry Advisory Board comprised of AstraZeneca, Boehringer Ingelheim, Novartis, Pfizer and Sunovion.
ACKNOWLEDGEMENTS
The members of the COPDGene® study group include: Ann Arbor VA: Jeffrey Curtis, MD (PI), Ella Kazerooni, MD (RAD). Baylor College of Medicine, Houston, TX: Nicola Hanania, MD, MS (PI), Philip Alapat, MD, Venkata Bandi, MD, Kalpalatha Guntupalli, MD, Elizabeth Guy, MD, Antara Mallampalli, MD, Charles Trinh, MD (RAD), Mustafa Atik, MD. Brigham and Women's Hospital, Boston, MA: Dawn DeMeo, MD, MPH (Co-PI), Craig Hersh, MD, MPH (Co-PI), George Washko, MD, Francine Jacobson, MD, MPH (RAD). Columbia University, New York, NY: R. Graham Barr, MD, DrPH (PI), Byron Thomashow, MD, John Austin, MD (RAD).
Duke University Medical Center, Durham, NC: Neil MacIntyre, Jr, MD (PI), Lacey Washington, MD (RAD), H. Page McAdams, MD (RAD). Fallon Clinic, Worcester, MA: Richard Rosiello, MD (PI), Timothy Bresnahan, MD (RAD). Health Partners Research Foundation, Minneapolis, MN: Charlene McEvoy, MD, MPH (PI), Joseph Tashjian, MD (RAD). Johns Hopkins University, Baltimore, MD: Robert Wise, MD (PI), Nadia Hansel, MD, MPH, Robert Brown, MD (RAD), Gregory Diette, MD. Los Angeles Biomedical Research Institute at Harbor UCLA Medical Center, Los Angeles, CA: Richard Casaburi, MD (PI), Janos Porszasz, MD, PhD, Hans Fischer, MD, PhD (RAD), Matt Budoff, MD.
Michael E. DeBakey VAMC, Houston, TX: Amir Sharafkhaneh, MD (PI), Charles Trinh, MD (RAD), Hirani Kamal, MD, Roham Darvishi, MD. Minneapolis VA: Dennis Niewoehner, MD (PI), Tadashi Allen, MD (RAD), Quentin Anderson, MD (RAD), Kathryn Rice, MD. Morehouse School of Medicine, Atlanta, GA: Marilyn Foreman, MD, MS (PI), Gloria Westney, MD, MS, Eugene Berkowitz, MD, PhD (RAD). National Jewish Health, Denver, CO: Russell Bowler, MD, PhD (PI), Adam Friedlander, MD, David Lynch, MB (RAD), Joyce Schroeder, MD (RAD), John Newell, Jr., MD (RAD).
Temple University, Philadelphia, PA: Gerard Criner, MD (PI), Victor Kim, MD, Nathaniel Marchetti, DO, Aditi Satti, MD, A. James Mamary, MD, Robert Steiner, MD (RAD), Chandra Dass, MD (RAD). University of Alabama, Birmingham, AL: William Bailey, MD (PI), Mark Dransfield, MD (Co-PI), Hrudaya Nath, MD (RAD). University of California, San Diego, CA: Joe Ramsdell, MD (PI), Paul Friedman, MD (RAD).
University of Iowa, Iowa City, IA: Geoffrey McLennan, MD, PhD (PI), Edwin JR van Beek, MD, PhD (RAD), Brad Thompson, MD (RAD), Dwight Look, MD. University of Michigan, Ann Arbor, MI: Fernando Martinez, MD (PI), MeiLan Han, MD, Ella Kazerooni, MD (RAD). University of Minnesota, Minneapolis, MN: Christine Wendt, MD (PI), Tadashi Allen, MD (RAD). University of Pittsburgh, Pittsburgh, PA: Frank Sciurba, MD (PI), Joel Weissfeld, MD, MPH, Carl Fuhrman, MD (RAD), Jessica Bon, MD. University of Texas Health Science Center at San Antonio, San Antonio, TX: Antonio Anzueto, MD (PI), Sandra Adams, MD, Carlos Orozco, MD, Mario Ruiz, MD (RAD).
Administrative Core: James Crapo, MD (PI), Edwin Silverman, MD, PhD (PI), Barry Make, MD, Elizabeth Regan, MD, Sarah Moyle, MS, Douglas Stinson.
Genetic Analysis Core: Terri Beaty, PhD, Barbara Klanderman, PhD, Nan Laird, PhD, Christoph Lange, PhD, Michael Cho, MD, Stephanie Santorico, PhD, John Hokanson, MPH, PhD, Dawn DeMeo, MD, MPH, Nadia Hansel, MD, MPH, Craig Hersh, MD, MPH, Jacqueline Hetmanski, MS, Tanda Murray.
Imaging Core: David Lynch, MB, Joyce Schroeder, MD, John Newell, Jr, MD, John Reilly, MD, Harvey Coxson, PhD, Philip Judy, PhD, Eric Hoffman, PhD, George Washko, MD, Raul San Jose Estepar, PhD, James Ross, MSc, Rebecca Leek, Jordan Zach, Alex Kluiber, Jered Sieren, Heather Baumhauer, Verity McArthur, Dzimitry Kazlouski, Andrew Allen, Tanya Mann, Anastasia Rodionova.
PFT QA Core, LDS Hospital, Salt Lake City, UT: Robert Jensen, PhD.
Biological Repository, Johns Hopkins University, Baltimore, MD: Homayoon Farzadegan, PhD, Stacey Meyerer, Shivam Chandan, Samantha Bragan.
Data Coordinating Center and Biostatistics, National Jewish Health, Denver, CO: James Murphy, PhD (deceased), Douglas Everett, PhD, Carla Wilson, MS, Ruthie Knowles, Amber Powell, Joe Piccoli, Maura Robinson, Margaret Forbes, Martina Wamboldt.
Epidemiology Core, University of Colorado School of Public Health, Denver, CO: John Hokanson, MPH, PhD, Marci Sontag, PhD, Jennifer Black-Shinn, MPH, Gregory Kinney, MPH.
Principal investigators and centers participating in ECLIPSE include: Bulgaria: Y. Ivanov, Pleven; K. Kostov, Sofia. Canada: J. Bourbeau, Montreal; M. Fitzgerald, Vancouver; P. Hernández, Halifax; K. Killian, Hamilton; R. Levy, Vancouver; F. Maltais, Montreal; D. O'Donnell, Kingston. Czech Republic: J. Krepelka, Praha.
Denmark: J. Vestbo, Hvidovre. The Netherlands: E. Wouters, Horn. New Zealand: D. Quinn, Wellington. Norway: P. Bakke, Bergen, Slovenia: M. Kosnik, Golnik. Spain: A. Agusti, Jaume Sauleda, Palma de Mallorca. Ukraine: Y. Feschenko, Kiev; V. Gavrisyuk, Kiev; L. Yashina, Kiev. UK: L. Yashina, W. MacNee, Edinburgh; D. Singh, Manchester; J. Wedzicha, London. USA: A. Anzueto, San Antonio, TX; S. Braman, Providence. RI; R. Casaburi, Torrance CA; B. Celli, Boston, MA; G. Giessel, Richmond, VA; M. Gotfried, Phoenix, AZ; G. Greenwald, Rancho Mirage, CA; N. Hanania, Houston, TX; D. Mahler, Lebanon, NH; B. Make, Denver, CO; S. Rennard, Omaha, NE; C. Rochester, New Haven, CT; P. Scanlon, Rochester, MN; D. Schuller, Omaha, NE; F. Sciurba, Pittsburgh, PA; A. Sharafkhaneh, Houston, TX; T. Siler, St Charles, MO; E. Silverman, Boston, MA; A. Wanner, Miami, FL; R. Wise, Baltimore, MD; R. ZuWallack, Hartford, CT.
Steering Committee: H. Coxson (Canada), C. Crim (GlaxoSmithKline, USA), L. Edwards (GlaxoSmithKline, USA), D. Lomas (UK), W. MacNee (UK), E. Silverman (USA), R. Tal Singer (Co-chair, GlaxoSmithKline, USA), J. Vestbo (Co-chair, Denmark), J. Yates (GlaxoSmithKline, USA).
Scientific Committee: A. Agusti (Spain), P. Calverley (UK), B. Celli (USA), C. Crim (GlaxoSmithKline, USA), B. Miller (GlaxoSmithKline, USA), W. MacNee (Chair, UK), S. Rennard (USA), R. Tal-Singer (GlaxoSmithKline, USA), E. Wouters (The Netherlands), J. Yates (GlaxoSmithKline, USA).
The National Emphysema Treatment Trial was supported by the National Heart, Lung, and Blood Institute contracts N01HR76101, N01HR76102, N01HR76103, N01HR76104, N01HR76105, N01HR76106, N01HR76107, N01HR76108, N01HR76109, N01HR76110, N01HR76111, N01HR76112, N01HR76113, N01HR76114, N01HR76115, N01HR76116, N01HR76118 and N01HR76119.
Co-investigators in the NETT Genetics Ancillary Study also include J. Benditt, G. Criner, M. DeCamp, P. Diaz, M. Ginsburg, L. Kaiser, M. Katz, M. Krasna, N. MacIntyre, R. McKenna, F. Martinez, Z. Mosenifar, J. Reilly, A. Ries, P. Scanlon, F. Sciurba and J. Utz.
International COPD Genetics Network (ICGN) investigators: Edwin K. Silverman, Brigham & Women's Hospital, Boston, MA, USA; David A. Lomas, Cambridge Institute for Medical Research, University of Cambridge, Cambridge, UK; Barry J. Make, National Jewish Medical and Research Center, Denver, CO, USA; Alvar Agusti and Jaume Sauleda, Hospital Universitari Son Dureta, Fundación Caubet-Cimera and Ciber Enfermedades Respiratorias, Spain; Peter M.A. Calverley, University of Liverpool, UK; Claudio F. Donner, Division of Pulmonary Disease, S. Maugeri Foundation, Veruno (NO), Italy; Robert D. Levy, University of British Columbia, Vancouver, Canada; Peter D. Paré, University of British Columbia, Vancouver, Canada; Stephen Rennard, Section of Pulmonary & Critical Care, University of Nebraska Medical Center, Omaha, NE, USA; Jørgen Vestbo, Department of Cardiology and Respiratory Medicine, Hvidovre Hospital, Copenhagen, Denmark; Emiel F.M. Wouters, University Hospital Maastricht, The Netherlands.
Conflict of Interest statement. R.T.-S., X.K., L.D.E, J.Y. and C.C. are current employees of GlaxoSmithKline. E.K.S. has received grant support from GlaxoSmithKline for studies of COPD genetics and honoraria and consulting fees from AstraZeneca. D.A.L. has received educational grants, fees for speaking and acts as a consultant for GlaxoSmithKline. H.O.C. has received grant support, honorarium, travel expenses, and has contract service agreements with GlaxoSmithKline or GlaxoSmithKline Canada; has a service agreement with Spiration Inc., has received grant support from a Canadian Institutes of Health—Industry (Wyeth) partnership grant, and has received a fee for speaking at a conference and related travel expenses from AstraZeneca (Australia). S.J.R. consulted or participated in advisory boards for: ABIM, Able Associates, Adelphi Research, Almirall, APT, Aradigm, Argenta, AstraZeneca, BI (ACCP), Biostrategies, BoomCom, Britnall and Nicolini, Capital Research, Chiesi, Clinical Advisors, CommonHealth, Complete Medical Group, Consult Complete, COPDForum, DataMonitor, Decision Resources, Defined Health, Dey, Dunn Group, Easton Associates, Enterprise Analysis, Equinox, Forest, Fulcrum, Gerson Lehman, GSK, Guidepoint, Hoffman LaRoche, IMS, Informed, Inspire, Insyght, KOL Connection, Leerink Swan, M. Pankove, MDRx Financial, MedaCorp, Medimmune, Mpex, Novartis, Nycomed, Oriel, Otsuka, Pearl, Pennside Partners, Pfizer, Pharma Ventures, Pharmaxis, Pick Research, Prescott, Price Waterhouse, Propagate, Pulmatrix, Pulmonary Reviews, Quadrant, Reckner Associates, Recruiting Resource, Reviews and Trends in COPD/Convergent Health Solutions, Roche, Sacoor, Schering, Schlesinger Medical, Scimed, Smith Research, Sudler and Hennessey, Talecris, Theravance, UBC, Uptake Medical, Vantage Point; has received lecture fees from AAAAI, Am Col Osteopathic Physicians, Asan Medical Center, ATS, AstraZeneca, California Soc Allergy, Convergent Health Solutions for Reviews and Trends in COPD, COPDFoundation, Creative Educational Concepts, Dey, Duke, France Foundation, Information TV, University of California-Los Angeles, Network for Continuing Education, Novartis, Nycomed, Otsuka, Pfizer, Sarasota Mem Hospital, Spanish Thoracic Society, University of Washington, University of Alabama-Birmingham, University of Pittsburgh, University of British Columbia, University of California-Davis, VA Sioux Falls; and has received industry-sponsored grants from AstraZeneca, Biomarck, Centocor, GlaxoSmithKline, Mpex, Nabi, Novartis, Otsuka, Pfizer.
REFERENCES
- 1.Rabe K.F., Hurd S., Anzueto A., Barnes P.J., Buist S.A., Calverley P., Fukuchi Y., Jenkins C., Rodriguez-Roisin R., van Weel C., et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am. J. Respir. Crit. Care Med. 2007;176:532–555. doi: 10.1164/rccm.200703-456SO. doi:10.1164/rccm.200703-456SO. [DOI] [PubMed] [Google Scholar]
- 2.Miniño A.M., Xu J., Kochanek K.D. National Vital Statistics Reports: Deaths: Preliminary Data for 2008. Vol. 59. Hyattsville, Maryland: National Center for Health Statistics; 2010. [PubMed] [Google Scholar]
- 3.Lokke A., Lange P., Scharling H., Fabricius P., Vestbo J. Developing COPD: a 25 year follow up study of the general population. Thorax. 2006;61:935–939. doi: 10.1136/thx.2006.062802. doi:10.1136/thx.2006.062802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burrows B., Knudson R.J., Cline M.G., Lebowitz M.D. Quantitative relationships between cigarette smoking and ventilatory function. Am. Rev. Respir. Dis. 1977;115:195–205. doi: 10.1164/arrd.1977.115.2.195. [DOI] [PubMed] [Google Scholar]
- 5.Silverman E.K., Chapman H.A., Drazen J.M., Weiss S.T., Rosner B., Campbell E.J., O'Donnell W.J., Reilly J.J., Ginns L., Mentzer S., et al. Genetic epidemiology of severe, early-onset chronic obstructive pulmonary disease. Risk to relatives for airflow obstruction and chronic bronchitis. Am. J. Respir. Crit. Care Med. 1998;157:1770–1778. doi: 10.1164/ajrccm.157.6.9706014. [DOI] [PubMed] [Google Scholar]
- 6.McCloskey S.C., Patel B.D., Hinchliffe S.J., Reid E.D., Wareham N.J., Lomas D.A. Siblings of patients with severe chronic obstructive pulmonary disease have a significant risk of airflow obstruction. Am. J. Respir. Crit. Care Med. 2001;164:1419–1424. doi: 10.1164/ajrccm.164.8.2105002. [DOI] [PubMed] [Google Scholar]
- 7.Palmer L.J., Celedon J.C., Chapman H.A., Speizer F.E., Weiss S.T., Silverman E.K. Genome-wide linkage analysis of bronchodilator responsiveness and post-bronchodilator spirometric phenotypes in chronic obstructive pulmonary disease. Hum. Mol. Genet. 2003;12:1199–1210. doi: 10.1093/hmg/ddg125. doi:10.1093/hmg/ddg125. [DOI] [PubMed] [Google Scholar]
- 8.Silverman E.K., Mosley J.D., Palmer L.J., Barth M., Senter J.M., Brown A., Drazen J.M., Kwiatkowski D.J., Chapman H.A., Campbell E.J., et al. Genome-wide linkage analysis of severe, early-onset chronic obstructive pulmonary disease: airflow obstruction and chronic bronchitis phenotypes. Hum. Mol. Genet. 2002;11:623–632. doi: 10.1093/hmg/11.6.623. doi:10.1093/hmg/11.6.623. [DOI] [PubMed] [Google Scholar]
- 9.Castaldi P.J., Cho M.H., Cohn M., Langerman F., Moran S., Tarragona N., Moukhachen H., Venugopal R., Hasimja D., Kao E., et al. The COPD genetic association compendium: a comprehensive online database of COPD genetic associations. Hum. Mol. Genet. 2010;19:526–534. doi: 10.1093/hmg/ddp519. doi:10.1093/hmg/ddp519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hersh C.P., Demeo D.L., Lange C., Litonjua A.A., Reilly J.J., Kwiatkowski D., Laird N., Sylvia J.S., Sparrow D., Speizer F.E., et al. Attempted replication of reported chronic obstructive pulmonary disease candidate gene associations. Am. J. Respir. Cell Mol. Biol. 2005;33:71–78. doi: 10.1165/rcmb.2005-0073OC. doi:10.1165/rcmb.2005-0073OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pillai S.G., Ge D., Zhu G., Kong X., Shianna K.V., Need A.C., Feng S., Hersh C.P., Bakke P., Gulsvik A., et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5:e1000421. doi: 10.1371/journal.pgen.1000421. doi:10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilk J.B., Chen T.H., Gottlieb D.J., Walter R.E., Nagle M.W., Brandler B.J., Myers R.H., Borecki I.B., Silverman E.K., Weiss S.T., et al. A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet. 2009;5:e1000429. doi: 10.1371/journal.pgen.1000429. doi:10.1371/journal.pgen.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho M.H., Boutaoui N., Klanderman B.J., Sylvia J.S., Ziniti J.P., Hersh C.P., DeMeo D.L., Hunninghake G.M., Litonjua A.A., Sparrow D., et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat. Genet. 2010;42:200–202. doi: 10.1038/ng.535. doi:10.1038/ng.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hancock D.B., Eijgelsheim M., Wilk J.B., Gharib S.A., Loehr L.R., Marciante K.D., Franceschini N., van Durme Y.M., Chen T.H., Barr R.G., et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat. Genet. 2010;42:45–52. doi: 10.1038/ng.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Repapi E., Sayers I., Wain L.V., Burton P.R., Johnson T., Obeidat M., Zhao J.H., Ramasamy A., Zhai G., Vitart V., et al. Genome-wide association study identifies five loci associated with lung function. Nat. Genet. 2010;42:36–44. doi: 10.1038/ng.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lambrechts D., Buysschaert I., Zanen P., Coolen J., Lays N., Cuppens H., Groen H.J., Dewever W., van Klaveren R.J., Verschakelen J., et al. The 15q24/25 susceptibility variant for lung cancer and chronic obstructive pulmonary disease is associated with emphysema. Am. J. Respir. Crit. Care Med. 2010;181:486–493. doi: 10.1164/rccm.200909-1364OC. doi:10.1164/rccm.200909-1364OC. [DOI] [PubMed] [Google Scholar]
- 17.Young R.P., Hopkins R.J., Whittington C.F., Hay B.A., Epton M.J., Gamble G.D. Individual and cumulative effects of GWAS susceptibility loci in lung cancer: associations after sub-phenotyping for COPD. PLoS ONE. 2011;6:e16476. doi: 10.1371/journal.pone.0016476. doi:10.1371/journal.pone.0016476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Durme Y.M., Eijgelsheim M., Joos G.F., Hofman A., Uitterlinden A.G., Brusselle G.G., Stricker B.H. Hedgehog-interacting protein is a COPD susceptibility gene: the Rotterdam Study. Eur. Respir. J. 2010;36:89–95. doi: 10.1183/09031936.00129509. doi:10.1183/09031936.00129509. [DOI] [PubMed] [Google Scholar]
- 19.Young R.P., Hopkins R.J., Hay B.A., Epton M.J., Black P.N., Gamble G.D. Lung cancer gene associated with COPD: triple whammy or possible confounding effect? Eur. Respir. J. 2008;32:1158–1164. doi: 10.1183/09031936.00093908. doi:10.1183/09031936.00093908. [DOI] [PubMed] [Google Scholar]
- 20.Young R.P., Whittington C.F., Hopkins R.J., Hay B.A., Epton M.J., Black P.N., Gamble G.D. Chromosome 4q31 locus in COPD is also associated with lung cancer. Eur. Respir. J. 2010;36:1375–1382. doi: 10.1183/09031936.00033310. doi:10.1183/09031936.00033310. [DOI] [PubMed] [Google Scholar]
- 21.Chappell S.L., Daly L., Lotya J., Alsaegh A., Guetta-Baranes T., Roca J., Rabinovich R., Morgan K., Millar A.B., Donnelly S.C., et al. The role of IREB2 and transforming growth factor beta-1 genetic variants in COPD: a replication case-control study. BMC Med. Genet. 2011;12:24. doi: 10.1186/1471-2350-12-24. doi:10.1186/1471-2350-12-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han B., Eskin E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am. J. Hum. Genet. 2011;88:586–598. doi: 10.1016/j.ajhg.2011.04.014. doi:10.1016/j.ajhg.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thorgeirsson T.E., Gudbjartsson D.F., Surakka I., Vink J.M., Amin N., Geller F., Sulem P., Rafnar T., Esko T., Walter S., et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat. Genet. 2010;42:448–453. doi: 10.1038/ng.573. doi:10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tobacco and Genetics (TAG) Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat. Genet. 2010;42:441–447. doi: 10.1038/ng.571. doi:10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saccone N.L., Culverhouse R.C., Schwantes-An T.H., Cannon D.S., Chen X., Cichon S., Giegling I., Han S., Han Y., Keskitalo-Vuokko K., et al. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS Genet. 2010;6:pii: e1001053.. doi: 10.1371/journal.pgen.1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siedlinski M., Cho M.H., Bakke P., Gulsvik A., Lomas D.A., Anderson W., Kong X., Rennard S.I., Beaty T.H., Hokanson J.E., et al. Genome-wide association study of smoking behaviours in patients with COPD. Thorax. 2011;66:894–902. doi: 10.1136/thoraxjnl-2011-200154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larson R.K., Barman M.L. The familial occurrence of chronic obstructive pulmonary disease. Ann. Intern. Med. 1965;63:1001–1008. doi: 10.7326/0003-4819-63-6-1001. [DOI] [PubMed] [Google Scholar]
- 28.Laurell C.-B., Eriksson A. The electrophoretic alpha-1 globulin pattern of serum in alpha 1-antitrypsin deficiency. Scand. J. Clin. Lab. Invest. 1963;15:132–140. [Google Scholar]
- 29.Wang J., Spitz M.R., Amos C.I., Wilkinson A.V., Wu X., Shete S. Mediating effects of smoking and chronic obstructive pulmonary disease on the relation between the CHRNA5-A3 genetic locus and lung cancer risk. Cancer. 2010;116:3458–3462. doi: 10.1002/cncr.25085. doi:10.1002/cncr.25085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeMeo D.L., Mariani T., Bhattacharya S., Srisuma S., Lange C., Litonjua A., Bueno R., Pillai S.G., Lomas D.A., Sparrow D., et al. Integration of genomic and genetic approaches implicates IREB2 as a COPD susceptibility gene. Am. J. Hum. Genet. 2009;85:493–502. doi: 10.1016/j.ajhg.2009.09.004. doi:10.1016/j.ajhg.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Otulakowski G., Duan W., O'Brodovich H. Global and gene-specific translational regulation in rat lung development. Am. J. Respir. Cell Mol. Biol. 2009;40:555–567. doi: 10.1165/rcmb.2008-0284OC. doi:10.1165/rcmb.2008-0284OC. [DOI] [PubMed] [Google Scholar]
- 32.Lin S., Ikegami M., Xu Y., Bosserhoff A.K., Malkinson A.M., Shannon J.M. Misexpression of MIA disrupts lung morphogenesis and causes neonatal death. Dev. Biol. 2008;316:441–455. doi: 10.1016/j.ydbio.2008.02.003. doi:10.1016/j.ydbio.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Groenman F., Rutter M., Caniggia I., Tibboel D., Post M. Hypoxia-inducible factors in the first trimester human lung. J. Histochem. Cytochem. 2007;55:355–363. doi: 10.1369/jhc.6A7129.2006. doi:10.1369/jhc.6A7129.2006. [DOI] [PubMed] [Google Scholar]
- 34.Hersh C.P., Demeo D.L., Lazarus R., Celedon J.C., Raby B.A., Benditt J.O., Criner G., Make B., Martinez F.J., Scanlon P.D., et al. Genetic association analysis of functional impairment in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2006;173:977–984. doi: 10.1164/rccm.200509-1452OC. doi:10.1164/rccm.200509-1452OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sterner-Kock A., Thorey I.S., Koli K., Wempe F., Otte J., Bangsow T., Kuhlmeier K., Kirchner T., Jin S., Keski-Oja J., et al. Disruption of the gene encoding the latent transforming growth factor-beta binding protein 4 (LTBP-4) causes abnormal lung development, cardiomyopathy, and colorectal cancer. Genes Dev. 2002;16:2264–2273. doi: 10.1101/gad.229102. doi:10.1101/gad.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Urban Z., Hucthagowder V., Schurmann N., Todorovic V., Zilberberg L., Choi J., Sens C., Brown C.W., Clark R.D., Holland K.E., et al. Mutations in LTBP4 cause a syndrome of impaired pulmonary, gastrointestinal, genitourinary, musculoskeletal, and dermal development. Am. J. Hum. Genet. 2009;85:593–605. doi: 10.1016/j.ajhg.2009.09.013. doi:10.1016/j.ajhg.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.London S.J., Idle J.R., Daly A.K., Coetzee G.A. Genetic variation of CYP2A6, smoking, and risk of cancer. Lancet. 1999;353:898–899. doi: 10.1016/S0140-6736(98)04984-8. doi:10.1016/S0140-6736(98)04984-8. [DOI] [PubMed] [Google Scholar]
- 38.Nakajima M., Yamamoto T., Nunoya K., Yokoi T., Nagashima K., Inoue K., Funae Y., Shimada N., Kamataki T., Kuroiwa Y. Characterization of CYP2A6 involved in 3′-hydroxylation of cotinine in human liver microsomes. J. Pharmacol. Exp. Ther. 1996;277:1010–1015. [PubMed] [Google Scholar]
- 39.Hukkanen J., Jacob P., 3rd, Benowitz N.L. Metabolism and disposition kinetics of nicotine. Pharmacol. Rev. 2005;57:79–115. doi: 10.1124/pr.57.1.3. doi:10.1124/pr.57.1.3. [DOI] [PubMed] [Google Scholar]
- 40.Al Koudsi N., Ahluwalia J.S., Lin S.K., Sellers E.M., Tyndale R.F. A novel CYP2A6 allele (CYP2A6*35) resulting in an amino-acid substitution (Asn438Tyr) is associated with lower CYP2A6 activity in vivo. Pharmacogenomics J. 2009;9:274–282. doi: 10.1038/tpj.2009.11. doi:10.1038/tpj.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mwenifumbo J.C., Al Koudsi N., Ho M.K., Zhou Q., Hoffmann E.B., Sellers E.M., Tyndale R.F. Novel and established CYP2A6 alleles impair in vivo nicotine metabolism in a population of Black African descent. Hum. Mutat. 2008;29:679–688. doi: 10.1002/humu.20698. doi:10.1002/humu.20698. [DOI] [PubMed] [Google Scholar]
- 42.Bloom J., Hinrichs A.L., Wang J.C., von Weymarn L.B., Kharasch E.D., Bierut L.J., Goate A., Murphy S.E. The contribution of common CYP2A6 alleles to variation in nicotine metabolism among European-Americans. Pharmacogenet. Genomics. 2011;21:403–416. doi: 10.1097/FPC.0b013e328346e8c0. doi:10.1097/FPC.0b013e328346e8c0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rao Y., Hoffmann E., Zia M., Bodin L., Zeman M., Sellers E.M., Tyndale R.F. Duplications and defects in the CYP2A6 gene: identification, genotyping, and in vivo effects on smoking. Mol. Pharmacol. 2000;58:747–755. doi: 10.1124/mol.58.4.747. [DOI] [PubMed] [Google Scholar]
- 44.Le Marchand L., Derby K.S., Murphy S.E., Hecht S.S., Hatsukami D., Carmella S.G., Tiirikainen M., Wang H. Smokers with the CHRNA lung cancer-associated variants are exposed to higher levels of nicotine equivalents and a carcinogenic tobacco-specific nitrosamine. Cancer Res. 2008;68:9137–9140. doi: 10.1158/0008-5472.CAN-08-2271. doi:10.1158/0008-5472.CAN-08-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Timofeeva M.N., McKay J.D., Smith G.D., Johansson M., Byrnes G.B., Chabrier A., Relton C., Ueland P.M., Vollset S.E., Midttun O., et al. Genetic polymorphisms in 15q25 and 19q13 loci, cotinine levels, and risk of lung cancer in EPIC. Cancer Epidemiol. Biomarkers Prev. 2011;20:2250–2261. doi: 10.1158/1055-9965.EPI-11-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bernaards C.M., Twisk J.W., Snel J., Van Mechelen W., Kemper H.C. Is calculating pack-years retrospectively a valid method to estimate life-time tobacco smoking? A comparison between prospectively calculated pack-years and retrospectively calculated pack-years. Addiction. 2001;96:1653–1661. doi: 10.1046/j.1360-0443.2001.9611165311.x. doi:10.1046/j.1360-0443.2001.9611165311.x. [DOI] [PubMed] [Google Scholar]
- 47.Njolstad I., Arnesen E., Lund-Larsen P.G. Smoking, serum lipids, blood pressure, and sex differences in myocardial infarction. A 12-year follow-up of the Finnmark Study. Circulation. 1996;93:450–456. doi: 10.1161/01.cir.93.3.450. [DOI] [PubMed] [Google Scholar]
- 48.Schunkert H., Konig I.R., Kathiresan S., Reilly M.P., Assimes T.L., Holm H., Preuss M., Stewart A.F., Barbalic M., Gieger C., et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 2011;43:333–338. doi: 10.1038/ng.784. doi:10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kohansal R., Martinez-Camblor P., Agusti A., Buist A.S., Mannino D.M., Soriano J.B. The natural history of chronic airflow obstruction revisited: an analysis of the Framingham offspring cohort. Am. J. Respir. Crit. Care Med. 2009;180:3–10. doi: 10.1164/rccm.200901-0047OC. doi:10.1164/rccm.200901-0047OC. [DOI] [PubMed] [Google Scholar]
- 50.Kanner R.E., Connett J.E., Williams D.E., Buist A.S. Effects of randomized assignment to a smoking cessation intervention and changes in smoking habits on respiratory symptoms in smokers with early chronic obstructive pulmonary disease: the Lung Health Study. Am. J. Med. 1999;106:410–416. doi: 10.1016/s0002-9343(99)00056-x. doi:10.1016/S0002-9343(99)00056-X. [DOI] [PubMed] [Google Scholar]
- 51.Anthonisen N.R., Skeans M.A., Wise R.A., Manfreda J., Kanner R.E., Connett J.E. The effects of a smoking cessation intervention on 14.5-year mortality: a randomized clinical trial. Ann. Intern. Med. 2005;142:233–239. doi: 10.7326/0003-4819-142-4-200502150-00005. [DOI] [PubMed] [Google Scholar]
- 52.Anthonisen N.R., Connett J.E., Kiley J.P., Altose M.D., Bailey W.C., Buist A.S., Conway W.A., Jr, Enright P.L., Kanner R.E., O'Hara P., et al. Effects of smoking intervention and the use of an inhaled anticholinergic bronchodilator on the rate of decline of FEV1. The Lung Health Study. JAMA. 1994;272:1497–1505. doi:10.1001/jama.272.19.1497. [PubMed] [Google Scholar]
- 53.Carpenter M.J., Strange C., Jones Y., Dickson M.R., Carter C., Moseley M.A., Gilbert G.E. Does genetic testing result in behavioral health change? Changes in smoking behavior following testing for alpha-1 antitrypsin deficiency. Ann. Behav. Med. 2007;33:22–28. doi: 10.1207/s15324796abm3301_3. doi:10.1207/s15324796abm3301_3. [DOI] [PubMed] [Google Scholar]
- 54.Audrain J., Boyd N.R., Roth J., Main D., Caporaso N.F., Lerman C. Genetic susceptibility testing in smoking-cessation treatment: one-year outcomes of a randomized trial. Addict. Behav. 1997;22:741–751. doi: 10.1016/s0306-4603(97)00060-9. doi:10.1016/S0306-4603(97)00060-9. [DOI] [PubMed] [Google Scholar]
- 55.Risch N., Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. doi:10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 56.Evangelou E., Trikalinos T.A., Salanti G., Ioannidis J.P. Family-based versus unrelated case-control designs for genetic associations. PLoS Genet. 2006;2:e123. doi: 10.1371/journal.pgen.0020123. doi:10.1371/journal.pgen.0020123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kelleher C.M., Silverman E.K., Broekelmann T., Litonjua A.A., Hernandez M., Sylvia J.S., Stoler J., Reilly J.J., Chapman H.A., Speizer F.E., et al. A functional mutation in the terminal exon of elastin in severe, early-onset chronic obstructive pulmonary disease. Am. J. Respir. Cell Mol. Biol. 2005;33:355–362. doi: 10.1165/rcmb.2005-0206OC. doi:10.1165/rcmb.2005-0206OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Patel B.D., Coxson H.O., Pillai S.G., Agusti A.G., Calverley P.M., Donner C.F., Make B.J., Muller N.L., Rennard S.I., Vestbo J., et al. Airway wall thickening and emphysema show independent familial aggregation in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2008;178:500–505. doi: 10.1164/rccm.200801-059OC. doi:10.1164/rccm.200801-059OC. [DOI] [PubMed] [Google Scholar]
- 59.Silverman E.K., Vestbo J., Agusti A., Anderson W., Bakke P.S., Barnes K.C., Graham Barr R., Bleecker E.R., Boezen H.M., Burkart K.M., et al. Opportunities and challenges in the genetics of COPD 2010: an international COPD genetics conference report. COPD. 2011;8:121–135. doi: 10.3109/15412555.2011.558864. doi:10.3109/15412555.2011.558864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sorheim I.C., Johannessen A., Grydeland T.B., Omenaas E.R., Gulsvik A., Bakke P.S. Case-control studies on risk factors for chronic obstructive pulmonary disease: how does the sampling of the cases and controls affect the results. Clin. Respir. J. 2010;4:89–96. doi: 10.1111/j.1752-699X.2009.00154.x. doi:10.1111/j.1752-699X.2009.00154.x. [DOI] [PubMed] [Google Scholar]
- 61.Vestbo J., Anderson W., Coxson H.O., Crim C., Dawber F., Edwards L., Hagan G., Knobil K., Lomas D.A., MacNee W., et al. Evaluation of COPD longitudinally to identify predictive surrogate end-points (ECLIPSE) Eur. Respir. J. 2008;31:869–873. doi: 10.1183/09031936.00111707. doi:10.1183/09031936.00111707. [DOI] [PubMed] [Google Scholar]
- 62.Bell B., Rose C., Damon H. The Normative Aging Study: an interdisciplinary and longitudinal study of health and aging. Aging Hum. Dev. 1972;3:5–17. doi:10.2190/GGVP-XLB5-PC3N-EF0G. [Google Scholar]
- 63.Fishman A., Martinez F., Naunheim K., Piantadosi S., Wise R., Ries A., Weinmann G., Wood D.E. A randomized trial comparing lung-volume-reduction surgery with medical therapy for severe emphysema. N. Engl. J. Med. 2003;348:2059–2073. doi: 10.1056/NEJMoa030287. doi:10.1056/NEJMoa030287. [DOI] [PubMed] [Google Scholar]
- 64.Zhu G., Warren L., Aponte J., Gulsvik A., Bakke P., Anderson W.H., Lomas D.A., Silverman E.K., Pillai S.G. The SERPINE2 gene is associated with chronic obstructive pulmonary disease in two large populations. Am. J. Respir. Crit. Care Med. 2007;176:167–173. doi: 10.1164/rccm.200611-1723OC. doi:10.1164/rccm.200611-1723OC. [DOI] [PubMed] [Google Scholar]
- 65.Regan E.A., Hokanson J.E., Murphy J.R., Make B., Lynch D.A., Beaty T.H., Curran-Everett D., Silverman E.K., Crapo J.D. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7:32–43. doi: 10.3109/15412550903499522. doi:10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. doi:10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 67.Patterson N., Price A.L., Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. doi:10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. doi:10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Frazer K.A., Ballinger D.G., Cox D.R., Hinds D.A., Stuve L.L., Gibbs R.A., Belmont J.W., Boudreau A., Hardenbol P., Leal S.M., et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. doi:10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. doi:10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. doi:10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.de Bakker P.I., Ferreira M.A., Jia X., Neale B.M., Raychaudhuri S., Voight B.F. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum. Mol. Genet. 2008;17:R122–R128. doi: 10.1093/hmg/ddn288. doi:10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. doi:10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Devlin B., Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. doi:10.1111/j.0006-341X.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 75.Aulchenko Y.S., Ripke S., Isaacs A., van Duijn C.M. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. doi:10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 76.Lange C., DeMeo D., Silverman E.K., Weiss S.T., Laird N.M. PBAT: tools for family-based association studies. Am. J. Hum. Genet. 2004;74:367–369. doi: 10.1086/381563. doi:10.1086/381563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lin S., Chakravarti A., Cutler D.J. Exhaustive allelic transmission disequilibrium tests as a new approach to genome-wide association studies. Nat. Genet. 2004;36:1181–1188. doi: 10.1038/ng1457. doi:10.1038/ng1457. [DOI] [PubMed] [Google Scholar]
- 78.Voight B.F., Scott L.J., Steinthorsdottir V., Morris A.P., Dina C., Welch R.P., Zeggini E., Huth C., Aulchenko Y.S., Thorleifsson G., et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat. Genet. 2010;42:579–589. doi: 10.1038/ng.609. doi:10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Purcell S.M., Wray N.R., Stone J.L., Visscher P.M., O'Donovan M.C., Sullivan P.F., Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.So H.C., Gui A.H., Cherny S.S., Sham P.C. Evaluating the heritability explained by known susceptibility variants: a survey of ten complex diseases. Genet. Epidemiol. 2011;35:310–317. doi: 10.1002/gepi.20579. doi:10.1002/gepi.20579. [DOI] [PubMed] [Google Scholar]
- 81.Buist A.S., McBurnie M.A., Vollmer W.M., Gillespie S., Burney P., Mannino D.M., Menezes A.M., Sullivan S.D., Lee T.A., Weiss K.B., et al. International variation in the prevalence of COPD (the BOLD Study): a population-based prevalence study. Lancet. 2007;370:741–750. doi: 10.1016/S0140-6736(07)61377-4. doi:10.1016/S0140-6736(07)61377-4. [DOI] [PubMed] [Google Scholar]
- 82.Johnson A.D., Handsaker R.E., Pulit S.L., Nizzari M.M., O'Donnell C.J., de Bakker P.I. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. doi:10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. doi:10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]