Abstract
Streptococcus gordonii, an important primary colonizer of dental plaque biofilm, specifically binds to salivary amylase via the surface-associated amylase-binding protein A (AbpA). We hypothesized that a function of amylase binding to S. gordonii may be to modulate the expression of chromosomal genes, which could influence bacterial survival and persistence in the oral cavity. Gene expression profiling by microarray analysis was performed to detect genes in S. gordonii strain CH1 that were differentially expressed in response to the binding of purified human salivary amylase versus exposure to purified heat-denatured amylase. Selected genes found to be differentially expressed were validated by quantitative reverse transcription-PCR (qRT-PCR). Five genes from the fatty acid synthesis (FAS) cluster were highly (10- to 35-fold) upregulated in S. gordonii CH1 cells treated with native amylase relative to those treated with denatured amylase. An abpA-deficient strain of S. gordonii exposed to amylase failed to show a response in FAS gene expression similar to that observed in the parental strain. Predicted phenotypic effects of amylase binding to S. gordonii strain CH1 (associated with increased expression of FAS genes, leading to changes in fatty acid synthesis) were noted; these included increased bacterial growth, survival at low pH, and resistance to triclosan. These changes were not observed in the amylase-exposed abpA-deficient strain, suggesting a role for AbpA in the amylase-induced phenotype. These results provide evidence that the binding of salivary amylase elicits a differential gene response in S. gordonii, resulting in a phenotypic adjustment that is potentially advantageous for bacterial survival in the oral environment.
INTRODUCTION
It is well established that tooth-borne bacterial biofilms (also known as dental plaques) are etiologic for the two most common oral diseases, dental caries and periodontal disease (29). Essential to oral biofilm development is the initial colonization of the oral cavity by the commensal (also known as “normal”) microflora. Throughout evolutionary history, humans and their commensal microbial partners have evolved together with mutual advantage. While most commensal organisms benefit human health, some can transition to serve as pathogens when ecologic factors are perturbed, as, for example, following the administration of broad-spectrum antibiotics that select for antibiotic-resistant forms, resulting in imbalances of the microflora that foster opportunistic infections. The presence of abundant commensal bacteria may prevent pathogens from increasing in numbers and thus prevent shifts in the ecological balance toward a pathogenic state. It is thus imperative to learn details of the biology of commensal bacteria, such as oral streptococci, in order to understand the ecologic factors that support a healthy flora and prevent the transition to a pathogenic flora (1).
Dental plaque formation is a complex process that involves the participation of a variety of salivary components (43). One such interaction is that between amylase and several predominant oral commensal streptococcal species (7, 10, 11, 14, 26, 36, 37, 40–42, 44, 46, 49). Amylase binds to a number of oral streptococcal species, collectively referred to as the amylase-binding streptococci (ABS) (13, 14, 22, 40). The binding of amylase to Streptococcus gordonii is saturable, irreversible, calcium independent, and inhibited by starch and maltotriose, but not by maltose, suggesting the involvement of a specific receptor on the bacterial surface (13, 15, 40). Amylase bound to streptococcal cells retains enzymatic activity to mediate the hydrolysis of starch to fermentable oligosaccharides in vitro (13, 15, 37, 41). Thus, streptococcus-bound salivary amylase hydrolyzes dietary starch that can be further metabolized for streptococcal nutrition. ABS are abundant in supragingival plaque immediately after tooth cleaning (43), highlighting their role in the initiation of plaque development. Notably, only animals with measurable salivary amylase activity have ABS in their mouths (43), suggesting that streptococcal amylase binding has evolved with the ability of mammalian hosts to produce salivary amylase. These findings offer compelling evidence that the binding of amylase to these bacteria influences their colonization of the oral cavity. Furthermore, the ABS survive and persist in this host niche for prolonged periods and facilitate colonization by other bacterial species, including commensals and pathogens, to form multispecies biofilms (23, 39).
Previous in vitro studies found that amylase-binding proteins (ABPs) play a role in bacterial adhesion and biofilm formation (30, 37). Amylase binding is mediated by a specific protein, amylase-binding protein A (AbpA), found on the surfaces of S. gordonii cells (36, 42). Interestingly, AbpA-deficient S. gordonii mutants were able to colonize rat mouths as well as the parental strains, suggesting that bacterial factors in addition to AbpA are involved in colonization and survival in vivo (46). In light of these findings, we have considered other, potentially novel functions for this interaction. Thus, the goals of this study were (i) to determine the transcriptional profile of S. gordonii in response to salivary α-amylase, (ii) to determine the role of AbpA in the amylase-induced gene response, and (iii) to examine the phenotypic changes that occur in S. gordonii in response to amylase binding.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The fully sequenced S. gordonii strain Challis CH1 (GenBank accession no. CP000725) was the primary strain for study. A kanamycin-resistant derivative of S. gordonii strain CH1 that carries a chromosomal aphIII gene replacing a phage integrase gene (SGO_2076) was constructed previously for use in animal studies (J. M. Tanzer et al., submitted for publication) and was designated strain KS1. The analysis of growth curves of S. gordonii strains showed no decline in KS1 vitality. An abpA-deficient derivative of KS1 was constructed by inserting the tet(M) gene, encoding tetracycline resistance (EMBL accession no. X56353), into the SnaB1 site of the chromosomal abpA gene of KS1, as previously described (Tanzer et al., submitted). Briefly, a 1.5-kb fragment containing abpA was PCR amplified from the CH1 chromosomal template and was cloned into pGEM-T (Promega). A 2.3-kb fragment containing the tet(M) gene and its promoter was PCR amplified from pAM620 (50) using primers with engineered 5′ HincII sites and was cloned into pGEM-T. The tet(M) fragment was released from the plasmid by digestion with HincII and was cloned into the internal SnaBI site of abpA in pGEM-T. Cloned fragments were checked for fidelity by nucleic acid sequencing. After digestion of the plasmid with BamHI, the cloned 3.8-kb abpA::tet(M) double-stranded DNA (dsDNA) fragment was used to transform competent strain KS1. Putative transformants were selected on agar medium with 10 μg tetracycline/ml, and the expected chromosomal sequence was confirmed by nucleic acid sequencing. Growth curves of strains CH1 and KS1 and of the KS1 abpA mutant were conducted in chemically defined medium. All three strains grew essentially identically to each other, as shown in Fig. S3 in the supplemental material. Statistical analysis using a one-way analysis-of-variance (ANOVA) test found no significant difference between the growth curves of these three strains (with 3 growth curve experiments conducted on separate days for each strain).
All S. gordonii strains were routinely cultured in the chemically defined medium (CDM) (20, 30). Bacteria from frozen stocks were streaked onto Todd-Hewitt (TH) (Difco, Detroit, MI) agar plates and were cultured overnight at 37°C under microaerophilic conditions in a candle jar. Tetracycline supplementation (10 μg/ml) was used only for initial culture of the KS1 abpA mutant from freezer stock; no antibiotics were added to subsequent subcultures used in experiments, and the disruption of the abpA gene was confirmed by an amylase ligand dot blot assay.
Purification of human salivary α-amylase.
Human parotid saliva from several healthy donors was duct collected, as described previously (6, 33). This saliva was extensively dialyzed against distilled water, lyophilized, resuspended in chromatography buffer (40), and subjected to Bio-Gel P60 gel filtration chromatography (6, 40). The peaks corresponding to nonglycosylated amylase were analyzed for purity by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting; selected peaks were pooled, dialyzed, lyophilized, and stored at −20°C. For the experiments in this study, nonglycosylated amylase was resuspended in simulated salivary buffer (0.021 M sodium phosphate buffer, 0.036 M NaCl, 0.96 mM CaCl2) (2, 6) and was used at 0.4 mg/ml, corresponding to its salivary concentration.
To ensure that the amylase fractions were not contaminated with salivary IgA, the chromatography peaks from saliva purifications were tested by Western blotting (8) with an anti-amylase or anti-IgA antibody. Briefly, 20 μl from the apex fraction of each peak (see Fig. S1, p6 and p7, in the supplemental material) and from the pooled peaks (see Fig. S1, A/g and A/n) was resolved on a 12% SDS-polyacrylamide gel, transferred to an Immobilon-P polyvinylidene difluoride (PVDF) membrane (Millipore, Bedford, MA), blocked with 5% milk in Tris-buffered saline containing 0.1% Tween 20 (TBST), washed in TBST, and overlaid with a rabbit anti-amylase antibody (1:1,000) (Calbiochem-Merck, Rockland, MA) in 1% milk–TBST for 30 min (10). After washing, the blot was incubated with a goat anti-rabbit IgG antibody conjugated with alkaline phosphatase (1:7,500) (Bio-Rad, Hercules, CA) in 1% milk–TBST for 30 min and was developed with SigmaFAST 5-bromo-4-chloro-3-indolylphosphate (BCIP)–nitroblue tetrazolium (NBT) (Sigma). Alternatively, the blot was directly overlaid with a goat anti-human IgA antibody conjugated with alkaline phosphatase (Sigma) and was developed with SigmaFAST BCIP-NBT.
Amylase ligand dot blot.
Dot blot analysis was modified from the method of Konto-Ghiorghi et al. (24). Briefly, whole bacteria at an optical density at 600 nm (OD600) of 0.3 were pelleted by centrifugation, washed in phosphate-buffered saline (PBS), and resuspended in PBS to obtain a similar OD600 of ∼2.9. A 10-μl bacterial suspension was spotted onto a nitrocellulose membrane, dried for 20 min at room temperature, blocked in 5% milk–PBS for 30 min, incubated with amylase (0.1 mg/ml) in 1% milk–PBS for 30 min, washed in PBS, and then overlaid with a rabbit anti-amylase antibody (1:1,000) (Calbiochem) in 1% milk–PBS for 30 min. After washing, the blot was incubated with goat anti-rabbit IgG, conjugated with alkaline phosphatase (1:7,500) (Bio-Rad), in 1% milk–PBS for 30 min and was developed with SigmaFAST BCIP-NBT (Sigma).
Isolation of RNA from S. gordonii.
In order to identify amylase-regulated genes, S. gordonii CH1 was cultured statically in 40 ml CDM at 37°C in a candle jar to mid-log phase, corresponding to an optical density at 600 nm of 0.5 to 0.6. The mid-log-phase bacterial culture was then divided into two aliquots of equal volume. Bacterial cells from all aliquots were pelleted by centrifugation at 6,000 × g in a Sorvall RC6 centrifuge at 20°C and were washed once with simulated salivary buffer preconditioned to 37°C. Simulated salivary buffer containing 0.4 mg/ml purified, nonglycosylated salivary amylase (native amylase) and warmed to 37°C was added to the cells of the first aliquot. As a negative control, simulated salivary buffer containing 0.4 mg/ml of the same salivary amylase that had first been denatured by heating to 100°C for 15 min (18) and cooled to 37°C was added to the cells of the second aliquot. Each aliquot (the amylase-treated aliquot and the control aliquot) was incubated statically for 15 min at 37°C in a candle jar.
Total RNA was immediately isolated by the hot acid–phenol method as described previously (48), followed by treatment with Turbo DNase (Applied Biosystems/Ambion, Austin, TX) according to the manufacturer's protocol. The remaining contaminants were removed using the RNeasy minikit column (Qiagen, Valencia, CA) with the cleanup protocol. Total RNA was quantified using the NanoDrop 2000 spectrophotometer, and RNA integrity was determined by agarose gel electrophoresis. Total RNA was used immediately for cDNA synthesis.
Microarray probe preparation.
S. gordonii total RNA was reverse transcribed to aminoallyl-cDNA and was indirectly labeled with Cy dyes according to the standard operating protocol of the Pathogen Functional Genomics Resource Center (PFGRC) at the J. Craig Venter Institute (http://pfgrc.jcvi.org/index.php/microarray.html). Briefly, 4 μg of total RNA was reverse transcribed using SuperScript II or III (Invitrogen, Carlsbad, CA) reverse transcriptase in a deoxynucleotide mixture containing aminoallyl-dUTP (Ambion). After alkaline hydrolysis to remove the RNA template, aminoallyl-cDNA was purified with QIAquick PCR columns (Qiagen), and was dried in a SpeedVac instrument. The cDNA was labeled with the appropriate Cy3 or Cy5 N-hydroxysuccinimide (NHS) dye (GE Healthcare Bio-Sciences Corp., Piscataway, NJ), and unbound Cy dyes were removed from the probe using the QIAquick PCR column (Qiagen). The quality of the probes and their specific activity were confirmed by a spectrophotometric scan from 200 to 700 nm on a NanoDrop 2000 spectrophotometer. Only labeled cDNAs with >200 pmol of dye incorporation per sample and a ratio of <50 nucleotides/dye molecule were used for hybridizations. The Cy dye-labeled cDNA from the native-amylase-treated aliquot of the culture was mixed with Cy dye-labeled cDNA from the denatured-amylase-treated control aliquot, and this mixture was used to probe the S. gordonii microarray slides. Initially, cDNA from cells exposed to native amylase was labeled with Cy5, and cDNA from cells exposed to denatured amylase was labeled with Cy3. Each amylase exposure experiment was repeated four times. To confirm microarray results, the cDNA from each strain was labeled with the opposite Cy dye and was hybridized to similar arrays for the flip-dye comparison.
Microarray hybridization.
A collection of 2,151 70-mer oligonucleotides (Invitrogen, Carlsbad, CA) representing all loci in the S. gordonii Challis CH1 genome (48) were spotted onto Nexterion Slide E microarray glass slides (Schott North America Inc., Louisville, KY) in replicates of four for each S. gordonii microarray slide, along with the DNA microarray controls described below. The oligonucleotides were spotted onto a QArray2 printer (Genetix USA Inc., Boston, MA) located in the genomics core at Roswell Park Cancer Institute in Buffalo, NY. This number of replicates compensates for spot-to-spot variation within each slide and allows for greater overall precision of the method. Quality control experiments were rerun on each new batch of printed microarray slides. These consisted of self-self hybridizations and determination of total spot saturation using SYBR dyes (Molecular Probes, Eugene, OR) that intercalate between DNA strands to check spotting consistency.
Cy dye-labeled cDNA probes were hybridized to S. gordonii microarray slides according to the Nexterion Slide E DNA application protocol (Schott). Briefly, slides were prewashed sequentially in a 0.1% Triton X-100 solution, a 1 mM HCl solution, and a 100 mM KCl solution before blocking with Nexterion blocking solution (Schott). The Cy dye-labeled cDNAs from the two samples were mixed, dried, and resuspended in Nexterion Hyb solution. The probe mixture was heated to 95°C and was pipetted onto a dried microarray slide. A coverslip (Lifter Slip; Erie Scientific Co., Portsmouth, NH) was placed over the slide, which was then sealed in a hybridization chamber (Corning Inc., Lowell, MA) and incubated in a 42°C water bath for 16 h. After hybridization, the slides were washed sequentially with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.2% SDS, 2× SSC, and 0.2× SSC solutions, dried briefly by centrifugation, and kept in a desiccator in a light-tight container until scanning.
Microarray analysis.
The fluorescent spots on the microarray were scanned using the Axon 4000A laser scanner (Axon Instruments, Foster City, CA), and Genepix software, version 6.0, was used to extract the raw data, which were analyzed using the TM4 Microarray software suite (www.jcvi.org/cms/research/software). The Spotfinder program was used to analyze the quality and fluorescence of individual spots. Raw Spotfinder data were then normalized in MIDAS by applying the LOWESS algorithm (block mode; smooth parameter, 0.33). The MeV program was used for comparison of the results of the normalized data, providing numerical values corresponding to each specific gene expressed, and for visualization of the data. Statistical analysis was performed in MeV using the t test (alpha parameter, 0.1) and the statistical analysis of microarray (SAM) test; a change of ±1.7-fold was considered significant.
qRT-PCR.
To confirm the transcription of selected differentially expressed genes identified by microarray analysis, S. gordonii strain CH1 cells were exposed to native or denatured amylase for 15 min at 37°C in a candle jar in the same manner as for microarray experiments. Total RNA was extracted as described previously and was reverse transcribed to cDNA with SuperScript II (Invitrogen, Carlsbad, CA) reverse transcriptase using the manufacturer's protocol. Prior to quantitative reverse transcription-PCR (qRT-PCR), the cDNA in each sample was normalized to the same concentration (2 ng/ml) using a NanoDrop 2000 spectrophotometer. The gene-specific primers designed using Primer BLAST (National Center for Biotechnology Information [www.ncbi.nlm.nih.gov/]) and synthesized by Invitrogen are listed in Table 1. To ensure that the efficiency of the assay was between 90 and 100%, qRT-PCR was performed using serial dilutions of the cDNA template with gyrA primers. Serial 10-fold dilutions of primers were tested with cDNA as a template to optimize each reaction. Each reaction mixture (total volume, 25 μl) contained 5 μl of the template, 3.5 μl of diethyl pyrocarbonate (DEPC)-treated water, 12.5 μl of Power SYBR green PCR master mixture (Applied Biosystems, Foster City, CA), and 2 μl each of the forward and reverse primers (100 nM). Relative qRT-PCR was performed on the ABI 7500 thermal cycler (Applied Biosystems), using uniform cycling conditions (95°C for 10 min, followed by 40 cycles of 30 s at 95°C and 1 min at 56°C) for Power SYBR green (Applied Biosystems). Melt (dissociation) curve analysis was done at the conclusion of the initial run of each set of primers to verify the amplification of a single product. Assay controls for each run included (i) amplification with a primer set for gyrase A (gyrA) as the endogenous control and (ii) amplification with a reaction mixture without a template. Results for samples exposed to native amylase were normalized to those for samples exposed to denatured amylase. All reactions were run in triplicate and were repeated with four independent biological replicates. Statistical analysis of qRT-PCR data was performed on the relative quantity (RQ) values or log10 RQ values by using the Student paired t test to compare the expression of each specific gene in native-amylase-treated cells with the expression of the same gene in denatured-amylase-treated cells. Intrareplica comparisons between the microarray and qRT-PCR data for the selected upregulated and downregulated genes showed Pearson correlation coefficients (r) of 0.99 for SGO_1697, 0.93 for SGO_1693, 0.92 for SGO_1698, 0.97 for SGO_1686, 0.8 for SGO_1688, 0.88 for SGO_1890, and 0.83 for SGO_0091. For the scope of genes, the correlation resulted in a Pearson coefficient of 0.89.
Table 1.
Primers used in qRT-PCR
| Primer name | Primer sequence |
|---|---|
| SGO_0091 IMP-F | TCAGTCGTGGCTGGGGTGTT |
| SGO_0091 IMP-R | TCCCTGCACTAGAGGCCAGC |
| SGO_1834-F | AGTTGCTCAAGCAGCTCACGAA |
| SGO_1834-R | AGAAGCCACCGTCAACAGCC |
| SGO_0671 murD-F | GCAGGTGGCTTGGACCGAGG |
| SGO_0671 murD-R | GCTGGCGTTGGCAGGACTGA |
| SGO_1541 atpC-F | AGCTGAGTCCGCCACAATCG |
| SGO_1541 atpC-R | AGGTCAAGGTACGCCGGGTC |
| SGO_1546 atpF-F | CTGCCACATCGCCCTTGATTGA |
| SGO_1546 atpF-R | GAAGCACTGGCAGGAAGCCG |
| SGO_1599 sodA-F | TGGTGGACACCTTAACCACGCT |
| SGO_1599 sodA-R | ACCAAGCCCAGCCTGATCCA |
| SGO_1890 PTS-F | TCAACCGCTTTGTCGCCCAG |
| SGO_1890 PTS-R | CGTGCAAATGGTGCTGACGGA |
| SGO_1833 lip-F | AGCAGGCATTATTCTAGCGTGT |
| SGO_1833 lip-R | CCTGCCCCTAGAGTAGCGCAAA |
| SGO_1697 acpP-F | GAGTCAACTTTCGATGATCTTGAAGCA |
| SGO_1697 acpP-R | ACCTTCCTCAGTTTCGATTTGGATGT |
| SGO_1688 accD-F | TGGAGGGGCACGTATGCAGG |
| SGO_1688 accD-R | ACTCGACGACCAGCGAATCCA |
| SGO_1686-F | TGCCTTGGGGGACTGTAAACCT |
| SGO_1686-R | AGCCACAACTCCCAGAGCCA |
| SGO_0832-F | CACTTTCAGCCGCTGGTCCG |
| SGO_0832-R | GCCTGCTTCATAGCATCACGTCC |
| SGO_1693 fabG-F | TACGAGCCGCAACCTCACGA |
| SGO_1693 fabG-R | TGACTAAGGCGCGTCAGGGAG |
| SGO_1698 fabH-F | ATGCCATCAACAGCGGCTCG |
| SGO_1698 fabH-R | AGACCGCAGTGCCTCGATCA |
| SGO_1495-F | AGCATGCCGGGAAACCTGGA |
| SGO_1495-R | TTGACTCGCTTTCAGCAGCGG |
| SGO_0107-F | AGGCTGAGGCTCGTCCAAGAC |
| SGO_0107-R | AAGGGCGAGCACCTGATCCAA |
| SGO_1273 pol-rpoD-F | GCACGGGTAATGGCCTGACG |
| SGO_1273 pol-rpoD-R | GAGCAGGGCGATACTGAGGCT |
| GyrA-F | GCGGATTGTTGTAACCGAGT |
| GyrA-R | ACGGACACCCTCACGATTAG |
Phenotypic characterization of S. gordonii following amylase binding. (i) Effect of salivary amylase on bacterial growth.
S. gordonii strains were cultured statically at 37°C in a candle jar to mid-log phase and were then exposed to native or denatured salivary amylase. The growth of the bacterial cultures was monitored by measuring the OD600 of the vortexed culture at every hour for an additional 6 h after the addition of amylase. The difference between the OD of native-amylase-treated cells and the OD of denatured-amylase-treated cells at each time point was calculated for S. gordonii strain CH1. In the same manner, the difference between the ODs of cells treated with native versus denatured amylase at each time point was calculated for the KS1 AbpA− mutant. Similarly, CFU were determined after incubation for 3 and 4 h at 37°C in a candle jar by plating serial dilutions on TH plates, and the CFU counts were used to compare native-amylase-treated with denatured-amylase-treated parts of the same S. gordonii strain CH1 or KS1 abpA mutant culture at the 3- and 4-h time points. Statistical analysis was performed using Student's t test, where the means of the differences for strain CH1 were compared to the means of the differences for the mutant. In addition, to rule out effects of amylase binding on chain length, mean bacterial chain lengths were determined for each condition microscopically by counting 10 random fields per slide and 10 random chains per field.
(ii) pH sensitivity.
Wild-type (WT) S. gordonii CH1 was tested for resistance to low pH. Bacteria from frozen stocks were streaked onto a TH plate and were cultured overnight at 37°C in a candle jar. The KS1 abpA mutant was cultured under the same conditions on a TH plate supplemented with tetracycline (10 μg/ml). Several colonies from strain CH1 were inoculated into 10 ml of CDM and were cultured statically to late-log phase (OD600, 0.7 to 0.8). The culture was then divided equally (5 ml each) and was pelleted at 6,000 × g for 5 min. To the bacterial pellet, 0.5 ml of salivary buffer containing either native or denatured salivary amylase (0.4 mg/ml in each case) was added, resuspended, and incubated for 15 min at 37°C. After incubation, bacteria were vortexed, and 10 μl was transferred into 1 ml of simulated salivary buffer previously adjusted with 2 N HCl to a pH of either 3, 5, 7, or 9 and was incubated for 1 h at room temperature. Following this, serial dilutions were plated on TH plates and were cultured for 24 h at 37°C in a candle jar in order to enumerate the remaining CFU. In parallel, the same experiment was conducted with the AbpA− mutant. The numbers of CFU were compared using paired two-tailed Student t tests, where the amylase-treated cells were paired with cells derived from the same culture exposed to denatured amylase.
(iii) Triclosan resistance.
The bacteria were tested for sensitivity to triclosan, an antibacterial agent known to affect fatty acid synthesis (FAS) (32). Wild-type S. gordonii CH1 and the KS1 abpA mutant were cultured to mid-log phase and were exposed to native or heat-treated amylase as described above. After 15 min of incubation with salivary amylase, triclosan (Irgasan; Sigma, St. Louis, MO) was added to a final concentration of 0.01%. The OD600 was measured every 30 min for 5 h.
(iv) Resistance to detergent (bile salts).
S. gordonii strain CH1 and the KS1 abpA mutant were tested for resistance to deoxycholate, a bile salt that emulsifies lipids and disrupts bacterial lipid membranes, causing cell lysis. Bacteria were cultured to mid-log phase as described above. After exposure to either native or denatured salivary amylase, sodium deoxycholate (Calbiochem) was added to a final concentration of 100 μg/ml, and the OD600 was measured every hour for 3 h (31).
Microarray data accession number.
Microarray data were deposited in NCBI's Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession no. GSE31830.
RESULTS
Transcriptome profiling of S. gordonii in response to amylase binding.
In order to determine the potential role of host salivary amylase as an environmental signal to S. gordonii, we used microarray analysis to test the effect of S. gordonii exposure to salivary amylase on its gene expression. CDM was used to culture the bacteria in all experiments, in order to eliminate the possible variable effects of complex carbohydrates or proteins found in a compound medium. As a control for protein exposure, we exposed the bacteria to heat-denatured salivary amylase. It was found previously that treating amylase with heat eliminates its ability to bind to S. gordonii and abolishes its enzymatic activity (4). Bacterial cultures were cultured to mid-log phase, when AbpA is maximally present on the surfaces of bacteria (13). The culture was divided into two aliquots; one was treated with purified native salivary amylase and the other with denatured salivary amylase. Nonglycosylated amylase was used in order to avoid possible interactions of the bacteria with glycosyl moieties of amylase. This amylase fraction was tested by Western blotting (see Fig. S1 in the supplemental material) and was found to be free of IgA, which could possibly bind to the bacteria to elicit a response. Based on preliminary experiments, S. gordonii gene expression was found to be optimal after a 15-min exposure to salivary amylase.
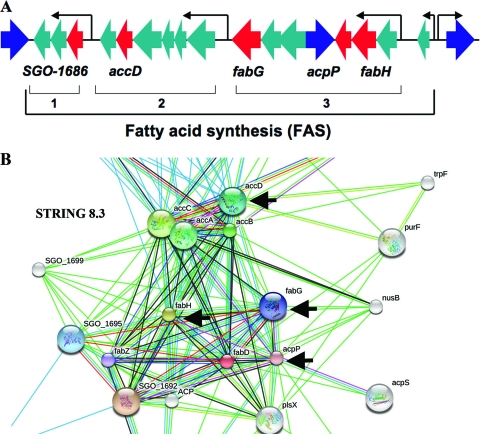
Microarray analysis identified 17 genes upregulated 2-fold or more in response to a 15-min amylase exposure, while 16 genes were downregulated (Table 2). Several genes belonging to the fatty acid synthesis (FAS) locus (Fig. 1A and B) were upregulated by as much as 4-fold in response to the binding of native amylase compared to exposure to denatured amylase. These genes, which are involved in the initiation of fatty acid synthesis and product elongation, include acpP, encoding acyl carrier protein (ACP) (SGO_1697); accD, encoding acetyl coenzyme A (acetyl-CoA) carboxylase subunit beta (SGO_1688); fabG, encoding 3-ketoacyl-(acyl-carrier-protein) reductase (SGO_1693); fabH, encoding 3-oxoacyl-(acyl carrier protein) synthase III (SGO_1698); and one of the genes from the same chromosomal location, SGO_1686, whose product has been designated a hypothetical protein. These genes were selected for confirmation by qRT-PCR.
Table 2.
S. gordonii CH1 genes upregulated or downregulated upon exposure to amylase as determined by microarray analysis
| Protein encoded | NCBI locus tag | Data analysis |
|||
|---|---|---|---|---|---|
| High stringency |
Low stringency |
||||
| Log 2 (ratio) | Fold | Log 2 (ratio) | Fold | ||
| Upregulated genes | |||||
| Acyl carrier protein | SGO_1697 | 1.57 | 2.98 | 2.09 | 4.28 |
| Acetyl-CoA carboxylase, carboxyl transferase, beta subunit | SGO_1688 | 2.12 | 4.34 | 2.13 | 4.39 |
| Hypothetical protein | SGO_0832 | 1.92 | 3.79 | 1.69 | 3.22 |
| Imidazoleglycerol-phosphate dehydratase | SGO_1407 | 1.22 | 2.34 | 1.18 | 2.27 |
| Hypothetical protein | SGO_1686 | 1.89 | 3.71 | 2.11 | 4.32 |
| Phosphoribosyl-AMP cyclohydrolase | SGO_1403 | 1.05 | 2.07 | 0.92 | 1.89 |
| Putative transcriptional regulator | SGO_1495 | 1.08 | 2.11 | 1.29 | 2.44 |
| 3-Oxoacyl-(acyl-carrier-protein) reductase | SGO_1693 | 1.01 | 2.01 | 1.10 | 2.14 |
| Conserved domain protein | SGO_2052 | 1.31 | 2.49 | 0.94 | 1.91 |
| LPXTG cell wall surface protein | SGO_0107 | 0.92 | 1.89 | 1.10 | 2.15 |
| 3-Oxoacyl-[acyl-carrier-protein] synthase III (beta-ketoacyl-ACP synthase III) (KAS III) | SGO_1698 | 0.87 | 1.83 | 1.01 | 2.02 |
| 4-Hydroxybenzoate octaprenyltransferase-related protein | SGO_0939 | 1.00 | 2.00 | 0.80 | 1.74 |
| Trehalose PTS enzyme II | SGO_1653 | 0.72 | 1.65 | 0.77 | 1.71 |
| Transcriptional regulator | SGO_1792 | 0.78 | 1.71 | 0.65 | 1.57 |
| ATP synthase (F/14-kDa) subunit | SGO_0133 | 0.86 | 1.82 | 0.82 | 1.77 |
| RNA polymerase sigma factor (rpoD) | SGO_1273 | 0.78 | 1.72 | 0.83 | 1.78 |
| Conserved domain protein | SGO_0018 | 0.63 | 1.55 | 0.98 | 1.97 |
| Downregulated genes | |||||
| Integral membrane protein | SGO_0091 | −1.59 | 3 | −1.69 | 3.33 |
| Conserved hypothetical protein | SGO_1834 | −1.76 | 3.4 | −1.73 | 3.33 |
| PTS system, fructose (mannose)-specific IID | SGO_1890 | −1.65 | 3.2 | −1.83 | 3.57 |
| UDP-N-acetylmuramoylalanine-d-glutamate ligase | SGO_0671 | −1.34 | 2.5 | −1.38 | 2.6 |
| Ribosomal protein L7A family | SGO_0545 | −1.89 | 3.8 | −1.86 | 3.7 |
| ABC transporter, ATP-binding protein SP1553 | SGO_1118 | −1.23 | 2.3 | −1.27 | 2.4 |
| ATP synthase Fo, B subunit | SGO_1546 | −1.01 | 2.0 | −0.77 | 1.7 |
| Pyruvate oxidase | SGO_0292 | −1.04 | 2.0 | −0.63 | 1.56 |
| Manganese-dependent superoxide dismutase | SGO_1599 | −0.86 | 1.8 | −0.82 | 1.78 |
| Lipoprotein, putative | SGO_1833 | −0.96 | 1.9 | −0.86 | 1.84 |
| Glycosyl hydrolase, family 38 | SGO_1768 | −0.89 | 1.8 | −0.98 | 2.00 |
| Voltage-gated chloride channel family protein | SGO_1166 | −0.80 | 1.7 | −0.89 | 1.75 |
| ATP synthase F1, epsilon subunit | SGO_1541 | −0.81 | 1.78 | −0.88 | 1.85 |
| Phosphate import ATP-binding protein PstB 1 (ABC phosphate transporter 1) | SGO_1059 | −0.81 | 1.78 | −0.90 | 1.88 |
| SUF system FeS assembly protein, NifU family | SGO_1719 | −0.88 | 1.85 | −0.87 | 1.84 |
| Conserved hypothetical protein | SGO_0010 | −0.79 | 1.75 | −0.81 | 1.78 |
Fig 1.
Fatty acid synthesis (FAS) genes. (A) FAS gene loci (green). Genes that are upregulated in amylase-treated bacteria compared to denatured-amylase-treated bacteria (red) positioned in the three distinct polycistronic regions (1, 2, and 3) and clustered in the chromosome among other genes transcribed in the opposite direction (blue). (B) Predicted functional relationship of FAS genes plotted by STRING 8.3. Black arrows indicate genes upregulated upon exposure to native amylase.
Among the downregulated genes, a large effect was noted for a gene encoding an integral membrane protein (IMP), SGO_0091. This gene was analyzed using STRING 8.3 software (http://string-db.org) and was found to be a predicted functional partner of SGO_0090, encoding a transcriptional regulator of the TetR family (21, 48). Other genes significantly downregulated in response to salivary amylase included a gene encoding phosphotransferase transport system (PTS) fructose (mannose)-specific subunit IID (SGO_1890); murD, encoding UDP-N-acetylmuramoyl-l-alanyl-d-glutamate synthetase (SGO_0671); two genes involved in oxidation phosphorylation, atpF, encoding ATP synthase subunit B (SGO_1546), and atpC, encoding ATP synthase F1, epsilon subunit (SGO_1541); and a gene encoding a putative lipoprotein (SGO_1833) (Table 2).
qRT-PCR analysis.
Genes encoding proteins of the FAS pathway, including accD (encoding acetyl-CoA carboxylase subunit beta), acpP (encoding acyl carrier protein), fabG [encoding 3-ketoacyl-(acyl-carrier-protein) reductase], and fabH [encoding 3-oxoacyl-(acyl carrier protein) synthase III], were confirmed by qRT-PCR to be significantly (P < 0.001) upregulated (Table 3) in native-amylase-treated bacteria compared to bacteria treated with denatured amylase. Two proteins of unknown function—hypothetical proteins SGO_0832 and SGO_1686—were also significantly upregulated in amylase-treated bacteria (Table 3). Six genes confirmed by qRT-PCR to be downregulated following exposure to salivary amylase compared to exposure to denatured amylase included genes encoding an integral membrane protein (SGO_0091), PTS fructose (mannose)-specific IID, ATP synthase subunit B (atpF), a putative lipoprotein (SGO_1833), a conserved hypothetical protein (SGO_1834), and manganese-dependent superoxide dismutase (sodA) (SGO_1599) (Table 3). Among the downregulated genes, atpC, encoding ATP synthase F1, epsilon subunit (SGO_1541), and murD, encoding UDP-N-acetylmuramoyl-l-alanyl-d-glutamate synthetase (SGO_0671), showed statistically significant (P < 0.05) 1.54- and 1.55-fold downregulation, respectively. These data are consistent with the microarray results.
Table 3.
S. gordonii CH1 genes upregulated or downregulated upon exposure to amylase as determined by qRT-PCR
| NCBI locus tag | Gene description by NCBI and STRING 8.3 | Mean fold differencea ± SE | P |
|---|---|---|---|
| Upregulated genes | |||
| SGO_0107b | LPXTG cell wall surface protein | 0.72 ± 0.04 | <0.001 |
| SGO_0832 | Hypothetical protein | 33.95 ± 14.33 | <0.05 |
| SGO_1495b | Putative transcriptional regulator | 1.33 ± 0.17 | <0.001 |
| SGO_1686 | Hypothetical protein | 10.42 ± 0.71 | <0.001 |
| SGO_1273b | RNA polymerase sigma factor (rpoD) | 0.99 ± 0.08 | 0.4655 |
| SGO_1688 | Acetyl-CoA carboxylase subunit beta (accD) | 18.28 ± 2.42 | <0.001 |
| SGO_1697 | Acyl carrier protein (acpP) | 20.89 ± 2.71 | <0.001 |
| SGO_1693 | 3-Ketoacyl-(acyl-carrier-protein) reductase (fabG) | 35.51 ± 3.41 | <0.001 |
| SGO_1698 | 3-Oxoacyl-(acyl carrier protein) synthase III (fabH) | 18.38 ± 1.90 | <0.001 |
| Downregulated genes | |||
| SGO_0091 | Integral membrane protein | 201.56 ± 73.22 | <0.05 |
| SGO_1890 | PTS system fructose (mannose)-specific IID | 39.58 ± 25.02 | <0.1 |
| SGO_1546 | ATP synthase subunit B (atpF) | 2.49 ± 0.58 | <0.05 |
| SGO_1833 | Lipoprotein, putative | 2.84 ± 0.66 | <0.05 |
| SGO_1834 | Conserved hypothetical protein | 2.81 ± 0.99 | <0.1 |
| SGO_1599 | Manganese-dependent superoxide dismutase (sodA) | 2.09 ± 0.22 | <0.01 |
| SGO_1541 | ATP synthase F1, epsilon subunit (atpC) | 1.54 ± 0.10 | <0.01 |
| SGO_0671 | UDP-N-acetylmuramoyl-l-alanyl-d-glutamate synthetase (murD) | 1.55 ± 0.18 | <0.05 |
| SGO_1786 | Glycosyl hydrolase, family 38 | 1.46 ± 0.72 | 0.2724 |
From four independent assays.
Gene upregulated by microarray analysis that did not prove to be upregulated by qRT-PCR.
Two transcription regulators, SGO_1273 and SGO_1495, and the “LPXTG cell wall surface protein” (SGO_0107), which were upregulated by microarray analysis, were also tested by qRT-PCR. The expression of these three genes was not significantly changed by the addition of amylase in the qRT-PCR experiments, which used the same RNA as that used in the microarray experiments. Overall, the qRT-PCR results verified the microarray data for 12 genes showing significant changes in expression in S. gordonii in response to a 15-min exposure to purified human parotid nonglycosylated amylase.
Role of AbpA in amylase-induced gene response.
The KS1 abpA mutant was tested to determine whether mutation of the abpA gene affects the response of bacteria to amylase exposure. Amylase ligand dot blot analysis of whole cells demonstrated that inactivation of abpA eliminated amylase binding to the surface of the S. gordonii KS1 abpA mutant (Fig. 2). We tested the expression of selected genes shown to be upregulated in the S. gordonii strains CH1 and KS1 after amylase exposure compared to the KS1 AbpA− mutant. As expected, these genes were upregulated in both CH1 and KS1. In contrast, qRT-PCR analysis of the FAS genes in the AbpA− mutant did not show significant differences between cells treated with native versus denatured amylase (Fig. 3). Thus, mutation of abpA eliminated the amylase-dependent gene response, suggesting that the gene response observed following amylase binding involves AbpA.
Fig 2.

A dot blot amylase overlay assay was used to demonstrate amylase binding to bacterial cells. Whole bacterial cells were pelleted from broth culture, washed, and standardized to an OD600 of 2.9 in PBS. Each sample (10 μl) was spotted onto a nitrocellulose membrane, blocked with skim milk, overlaid with salivary amylase (0.1 mg/ml) followed by a rabbit anti-amylase antibody, and developed with alkaline phosphatase-conjugated goat anti-rabbit IgG and SigmaFAST BCIP-NBT (Sigma). CH1, S. gordonii Challis CH1; AbpA−, the KS1 amylase-binding protein A mutant; AbpB−, the KS1 amylase-binding protein B mutant.
Fig 3.
Expression of fatty acid synthesis genes after a 15-min exposure of bacteria to salivary amylase compared to the control culture exposed to denatured salivary amylase, as determined by qRT-PCR. Filled bars, WT S. gordonii; striped bars, KS1 AbpA− mutant. gyrA was used as the endogenous control. Each bar represents the log10 of the mean fold difference in expression (native amylase − denatured amylase) ± the standard deviation from four independent experiments. Significance levels: ⁎, P < 0.01; ⁎⁎, P < 0.001. Results similar to those seen with CH1 were obtained with KS1.
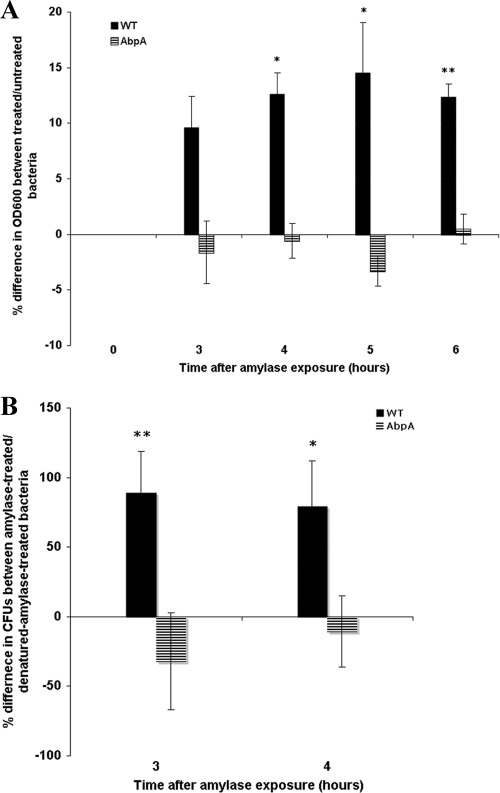
Effect of amylase on bacterial growth.
One of the genes of the fab cluster, fabG, which was highly upregulated in response to salivary amylase in S. gordonii strain CH1, has been reported previously to be an essential gene for bacterial growth in Escherichia coli (51). The fact that fabG was induced by amylase exposure led us to test the effect of amylase exposure on bacterial growth. After incubation with salivary amylase, S. gordonii strain CH1 increased in optical density faster, by 12% on average, than did cells from the same culture exposed to denatured amylase (Fig. 4A). In contrast, no difference in growth was noted between native-amylase-treated and denatured-amylase-treated AbpA− mutant cells (Fig. 4A). A change in bacterial growth was also verified by plating the amylase-treated culture at 3 and 4 h and comparing the CFU of native-amylase-treated bacteria with those of denatured-amylase-treated bacteria (Fig. 4B). These data suggest that the growth of S. gordonii may be enhanced upon exposure to salivary amylase. The fact that there was no increase in growth in the AbpA− mutant exposed to native amylase compared to denatured amylase further suggests that AbpA plays a role in the amylase response by S. gordonii. No significant change in bacterial chain length between treated and control bacteria was observed (data not shown), suggesting that the observed increase in CFU following exposure to salivary amylase is not due to a reduction in chain length.
Fig 4.
(A) Comparison of culture growth between bacteria treated with native amylase and those treated with denatured amylase. Filled bars, WT S. gordonii; striped bars, KS1 AbpA− mutant. Bacteria were cultured in chemically defined medium to mid-log phase, when salivary amylase was added to a final concentration of 0.4 mg/ml (0 h). (A) The OD600 was measured after 3 h and was monitored every hour thereafter for the next 3 h. (B) The culture was plated so as to determine CFU. Each bar represents the mean difference in culture growth between native-amylase-treated and denatured-amylase-treated bacteria of the same culture (n = 3). ⁎, P < 0.05; ⁎⁎, P < 0.01.
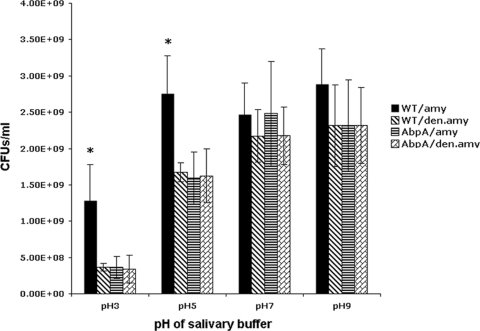
Amylase-induced resistance to low pH.
We hypothesized that the observed increased expression of genes responsible for the initiation of fatty acid synthesis may have an effect on membrane-dependent pH resistance. The presence of host amylase in the bacterial milieu could be indicative of the availability of additional utilizable carbohydrates, thus priming the bacterium for carbohydrate uptake. Hydrolysis of starch by amylase would supply maltose, which, when metabolized by bacteria, would lower the culture pH. Exposure of S. gordonii to salivary amylase could increase its resistance to low pH. Indeed, the survival of S. gordonii CH1 was significantly increased in a buffer at pH 3 and pH 5 after amylase treatment. As expected, there was no difference in survival between native-amylase-treated and denatured-amylase-treated bacteria at pH 7 or pH 9 (Fig. 5). The survival of the AbpA− mutant under acidic or basic conditions was not affected by amylase treatment, again suggesting a role for AbpA in survival in an acidic environment after amylase exposure.
Fig 5.
Effects of salivary amylase on S. gordonii survival under various pH conditions. WT S. gordonii was first cultured in chemically defined medium to mid-log phase, then exposed either to native salivary amylase (WT/amy) or to salivary amylase denatured by boiling (WT/den.amy), and finally incubated further in a salivary buffer adjusted to various pHs. The KS1 AbpA− mutant was also similarly exposed to native (AbpA/amy) or denatured (AbpA/den.amy) amylase and was further incubated in a salivary buffer adjusted to various pHs. ⁎, P < 0.05.
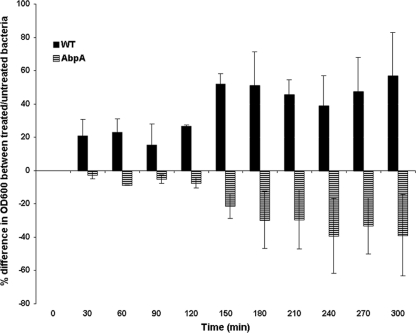
Amylase exposure increases resistance to triclosan.
To further support the finding that amylase binding affects the expression of fatty acid synthesis genes, resulting in a change in phenotype, we compared the triclosan resistance of S. gordonii cells exposed to native or denatured amylase. Triclosan is a bacteriostatic antiseptic that inhibits enoyl-ACP reductases encoded by the fab gene cluster (9). S. gordonii cells were found to be more resistant to triclosan following amylase binding than control cells exposed to denatured salivary amylase (Fig. 6). The effect on the AbpA− mutant was reversed, i.e., cells exposed to denatured salivary amylase were more resistant to triclosan than cells exposed to native salivary amylase (Fig. 6). The amylase-induced expression of FAS genes allows S. gordonii strain CH1 to overcome the growth suppression caused by triclosan, while the AbpA− mutant abolishes this effect, and growth is suppressed. These results suggest that the expression of FAS enzymes is increased following exposure to salivary amylase and that this increase is sufficient to overcome the suppressing effect of triclosan.
Fig 6.
Resistance of S. gordonii to triclosan after exposure to salivary amylase. Filled bars, WT S. gordonii; striped bars, KS1 AbpA− mutant. Each bar represents the mean difference in growth (as indicated by the OD600) between bacteria treated with native versus denatured amylase after the addition of triclosan to a final concentration of 0.1 mg/ml at 0 min and upon monitoring every 30 min. Data represent three independent experiments (P < 0.001).
Resistance to detergent (bile salts).
We considered the possibility that transcriptional regulation of fab cluster genes could alter the fatty acid composition of the membrane and increase its sensitivity to lysis by the detergent deoxycholate, as described for Streptococcus pneumoniae (31). After exposure of native-amylase-treated or denatured-amylase-treated S. gordonii CH1 to deoxycholate, no change in sensitivity to bile salts was observed. Similarly, there was no difference in lysis between KS1 AbpA− mutant cells treated with native versus denatured amylase following exposure to deoxycholate. These results suggest that increased expression of fatty acid synthesis genes in amylase-treated S. gordonii strain CH1 likely does not cause a change in the composition of the membranes; however, increased expression of FAS genes may modulate overall bacterial growth and membrane turnover.
DISCUSSION
S. gordonii expresses endogenous intracellular amylase, which is not secreted, and is thus unable to metabolize starch as a primary nutritional source unless it binds host salivary amylase (13, 17, 37, 41, 45). α-Amylase hydrolyzes starch at α-1,4 glucosidic linkages to form various maltooligosaccharides and eventually maltose. Once bound to the streptococcal surface, host amylase retains its enzymatic activity, allowing the organism to metabolize dietary starch efficiently (13, 37, 41). The presence of both starch and amylase in the oral cavity likely allows S. gordonii to persist and proliferate. Previous studies identified genes in S. gordonii that were differentially expressed in response to exposure to human whole saliva (16). Considering that amylase is one of the most abundant components of human saliva, this interaction could mediate differential gene expression and could serve as an important factor for bacterial host adaptation.
The results of this study provide evidence that the binding of amylase to the surface of S. gordonii elicits differential gene expression in the bacterial cell. This appears to result in an alteration of the bacterial phenotype that may be advantageous for bacterial survival in the oral environment. Here we show evidence that after exposure to salivary amylase, S. gordonii is better able to tolerate an acidic environment. The bacterium also appears to grow better in the presence of bound amylase. Salivary amylase upregulates genes involved in the synthesis of fatty acids, which are key components of bacterial membranes. Considering the fact that in all the experiments we performed, the presence of complex carbohydrate as a source of nutrition in the medium was avoided through the use of a chemically defined medium, it is clear that S. gordonii not only binds host amylase but also responds to its binding by differential regulation of gene expression and phenotype adjustment.
The amylase-induced upregulation of FAS genes could be a mechanism for the “fine tuning” of bacterial metabolism to redirect acetyl-CoA from the citric acid cycle to fatty acid synthesis for membrane building and bacterial proliferation. In the first step of fatty acid synthesis, acetyl-CoA from the citric acid cycle receives a CO2 group and forms malonyl-CoA; this involves the action of acetyl-CoA carboxylase (accD) (9). Next, 3-oxoacyl-(acyl carrier protein) synthase III (fabH) initiates the condensation of acetyl-CoA with malonyl-CoA and attaches it to acyl carrier protein (acpP), creating acetoacetyl-ACP (9). Then 3-ketoacyl-(acyl-carrier-protein) reductase (fabG) reduces the 3-carbon ketone of acetoacetyl-ACP to hydroxyl and converts it to hydroxybutanoyl-ACP. These steps are repeated to elongate the fatty acid chain. The acyl carrier protein is essential for the cycle, since it carries the synthesizing fatty acid chain. In fact, acpP is an essential gene in E. coli that possesses several promoters, ensuring that mutation of one of the promoters does not prevent the transcription of fully functional protein (51). Another essential gene, accD, previously reported in E. coli temperature-sensitive mutations, blocked fatty acid synthesis (9, 27). KAS III-3-oxoacyl-(acyl carrier protein) synthase III, encoded by fabH, produces only C4 through C6 compounds and cannot make long-chain fatty acids (47), suggesting that this enzyme initiates fatty acid synthesis (9, 19, 35). fabG, encoding 3-ketoacyl-(acyl-carrier-protein) reductase, is also an essential gene; its transcriptional termination in Salmonella enterica serovar Typhimurium blocked cell growth (51). Considering the fact that several amylase-induced genes affect bacterial growth and proliferation, the findings of the present study suggest that upregulated fatty acid synthesis genes in amylase-exposed S. gordonii modulate the initiation of fatty acid synthesis and stimulate proliferation. Thus, the effect of upregulation of FAS genes may be to increase the overall amount of fatty acids, but it does not change the proportion of fatty acids in the membrane (see Table S1 in the supplemental material).
We established that the cell wall-attached protein AbpA plays a role in the amylase-induced gene response. Insertional inactivation of abpA drastically reduced the binding of salivary amylase to the bacterial surface and eliminated amylase-induced gene expression and phenotypic response.
Considering the fact that AbpA is a cell wall-associated protein localized at the cell surface (41), the question of its ability to convey a signal upon binding to salivary amylase arises. AbpA is maximally expressed during the mid-log phase of S. gordonii growth (13). Recent sequence analysis indicates that AbpA shares homology with several AbpA-like proteins produced by other amylase-binding oral streptococci. To date, no conserved domains have been identified (see Fig. S2 in the supplemental material). The AbpA protein is translated with a leader peptide directing its secretion outside the bacterial cell, where it attaches to the cell surface to serve as the receptor for amylase (H. Wu, personal communication). It has been suggested recently (28) that sortase B (SrtB), encoded by srtB and located immediately downstream of abpA, plays a role in the attachment of AbpA to the cell wall. AbpA does not have a classic sortase B recognition domain, such as NXZTN, as described for Listeria monocytogenes, or NPQTN, for Staphylococcus aureus (3, 5). It apparently does possess a novel, yet to be identified sortase recognition motif at the C terminus (28), which is likely responsible for the covalent binding of AbpA to the cell wall.
AbpA is present on the surface of the nascent cell wall in the annular and polar zones but is later shed into the environment (42). Its function, other than mediating the binding of amylase to streptococci, remains unknown. The fact that AbpA is present only at cell division sites and is later shed into the environment leads us to postulate that AbpA may play a role in conveying information about the environment to new daughter cells during cell division. Although AbpA is an extracellular cell wall-attached protein and lacks a membrane-spanning domain, it could potentially function as a transient coreceptor or signal transducer. Consequently, it is possible that AbpA is part of a putative signal transduction system. Analysis of the AbpA protein sequence suggests several sites for protein-protein interaction, phosphorylation, and ATP binding.
AbpA may interact with another component(s) of a putative transduction system, such as an integral membrane protein or a transport system. The latter, activated by AbpA, could carry the signal to the transcription machinery. In fact, our microarray analysis revealed two candidates for such an interaction. Two genes downregulated upon exposure to salivary amylase encoded an IMP (SGO_0091) and a PTS (SGO_1890). One or both could be involved in transmitting the signal. The IMP is a predicted functional partner of a TetR repressor family regulator (SGO_0090) and could be a member of the putative two-component signaling system (TCS) (34). Interestingly, the downregulation of this predicted TetR repressor system could be linked to upregulation of the FAS operon through the release of the suppression effect (31, 51). However, the function of the IMP as part of an amylase-triggered transduction system, as well as its role in the regulation of the FAS operon, has yet to be determined experimentally. The second candidate for such a possible regulatory role is a PTS (SGO_1890), which was downregulated upon exposure to salivary amylase. Multiple phosphotransferase systems responsible for carbohydrate transport have been shown previously to be involved in signal transduction in Gram-positive bacteria (25) and E. coli (12, 38). Thus, it is possible that AbpA could interact with the PTS on the cell surface level and initiate signaling through this carbohydrate transport system. The actual roles of these candidates in amylase-induced signal transduction have yet to be determined.
While wild-type S. gordonii showed increased resistance to triclosan after exposure to native versus denatured amylase, peculiarly, the AbpA mutant, when exposed to amylase, showed less resistance to triclosan than did the control. Since triclosan binds to and inhibits the enzymes of fatty acid synthesis, it is logical that increased expression of FAS genes in the WT after amylase exposure would cause increased levels of FAS enzymes that overcome the effect of triclosan. We can only speculate on the possible explanations for why the AbpA mutant showed the opposite effect. Perhaps the absence of AbpA protein on the surface of the bacterium results in an as yet unknown perturbation that affects the bacterial response to the presence of unbound amylase in the environment. AbpA may not be the only protein that is involved in signaling following the binding of amylase. Perhaps the deletion of AbpA from the signaling system creates a countersignal in the presence of amylase that actually reverses triclosan resistance. Studies to identify other components that may be involved in amylase-dependent signaling are in progress. We anticipate that information on the regulatory pathway will be valuable in the interpretation of this phenomenon.
Identification of the mechanisms of amylase-induced signaling in differential gene expression has implications for understanding and controlling bacterium-host interactions and also for the detection of new paradigms for the regulation of bacterial fatty acid synthesis that may stimulate the development of novel antimicrobial agents.
Now that we have demonstrated that amylase may serve as an environmental signal for the modification by S. gordonii of its gene expression and phenotype in order to adapt to a changing host environment, other questions arise. Does amylase binding provide an advantage for S. gordonii in competition with other bacterial species within the niche? Does it aid in colonization by other species? We observed that exposure to salivary amylase stimulates S. gordonii growth, as well as its survival under acidic conditions, which are often found in the oral environment. Does the interaction of S. gordonii with amylase decrease colonization by pathogenic bacteria, such as Streptococcus mutans, or does it benefit S. mutans by delivering an available source of utilizable carbohydrates? These results suggest further avenues for research to determine the fundamental role of S. gordonii in bacterium-host interaction and oral colonization.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ashu Sharma of the Department of Oral Biology, University at Buffalo, for help with qRT-PCR analysis and Jeffrey Miecznikowski, Department of Biostatistics, University at Buffalo, for assistance with statistical analysis.
This work was supported in part by the National Institute of Dental and Craniofacial Research (DE09838 and DE007034).
Footnotes
Published ahead of print 13 January 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Avila M, Ojcius DM, Yilmaz O. 2009. The oral microbiota: living with a permanent guest. DNA Cell Biol. 28:405–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bennick A, Cannon M. 1978. Quantitative study of the interaction of salivary acidic proline-rich proteins with hydroxyapatite. Caries Res. 12:159–169 [DOI] [PubMed] [Google Scholar]
- 3. Bentley ML, Gaweska H, Kielec JM, McCafferty DG. 2007. Engineering the substrate specificity of Staphylococcus aureus sortase A. The β6/β7 loop from SrtB confers NPQTN recognition to SrtA. J. Biol. Chem. 282:6571–6581 [DOI] [PubMed] [Google Scholar]
- 4. Bergmann JE, Gulzow HJ. 1995. Detection of binding of denatured salivary α-amylase to Streptococcus sanguis. Arch. Oral Biol. 40:973–974 [DOI] [PubMed] [Google Scholar]
- 5. Bierne H, et al. 2004. Sortase B, a new class of sortase in Listeria monocytogenes. J. Bacteriol. 186:1972–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bradway SD, Bergey EJ, Jones PC, Levine MJ. 1989. Oral mucosal pellicle. Adsorption and transpeptidation of salivary components to buccal epithelial cells. Biochem. J. 261:887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brown AE, Rogers JD, Haase EM, Zelasko PM, Scannapieco FA. 1999. Prevalence of the amylase-binding protein A gene (abpA) in oral streptococci. J. Clin. Microbiol. 37:4081–4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Burnette WN. 1981. “Western blotting”: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal. Biochem. 112:195–203 [DOI] [PubMed] [Google Scholar]
- 9. Campbell JW, Cronan JE., Jr 2001. Bacterial fatty acid biosynthesis: targets for antibacterial drug discovery. Annu. Rev. Microbiol. 55:305–332 [DOI] [PubMed] [Google Scholar]
- 10. Chaudhuri B, et al. 2008. Amylase-binding protein B of Streptococcus gordonii is an extracellular dipeptidyl-peptidase. Infect. Immun. 76:4530–4537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chaudhuri B, Rojek J, Vickerman MM, Tanzer JM, Scannapieco FA. 2007. Interaction of salivary α-amylase and amylase-binding-protein A (AbpA) of Streptococcus gordonii with glucosyltransferase of S. gordonii and Streptococcus mutans. BMC Microbiol. 7:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. De Reuse H, Danchin A. 1991. Positive regulation of the pts operon of Escherichia coli: genetic evidence for a signal transduction mechanism. J. Bacteriol. 173:727–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Douglas CW. 1990. Characterization of the α-amylase receptor of Streptococcus gordonii NCTC 7868. J. Dent. Res. 69:1746–1752 [DOI] [PubMed] [Google Scholar]
- 14. Douglas CW. 1983. The binding of human salivary α-amylase by oral strains of streptococcal bacteria. Arch. Oral Biol. 28:567–573 [DOI] [PubMed] [Google Scholar]
- 15. Douglas CW, Heath J, Gwynn JP. 1992. Enzymic activity of salivary amylase when bound to the surface of oral streptococci. FEMS Microbiol. Lett. 71:193–197 [DOI] [PubMed] [Google Scholar]
- 16. Du LD, Kolenbrander PE. 2000. Identification of saliva-regulated genes of Streptococcus gordonii DL1 by differential display using random arbitrarily primed PCR. Infect. Immun. 68:4834–4837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Egland PG, Palmer RJ, Jr, Kolenbrander PE. 2004. Interspecies communication in Streptococcus gordonii-Veillonella atypica biofilms: signaling in flow conditions requires juxtaposition. Proc. Natl. Acad. Sci. U. S. A. 101:16917–16922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Heinen W, Lauwers AM. 1976. Amylase activity and stability at high and low temperature depending on calcium and other divalent cations. Experientia Suppl. 26:77–89 [DOI] [PubMed] [Google Scholar]
- 19. Jackowski S, Murphy CM, Cronan JE, Jr, Rock CO. 1989. Acetoacetyl-acyl carrier protein synthase. A target for the antibiotic thiolactomycin. J. Biol. Chem. 264:7624–7629 [PubMed] [Google Scholar]
- 20. Jenkinson HF. 1986. Cell-surface proteins of Streptococcus sanguis associated with cell hydrophobicity and coaggregation properties. J. Gen. Microbiol. 132:1575–1589 [DOI] [PubMed] [Google Scholar]
- 21. Jensen LJ, et al. 2009. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 37:D412–D416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kilian M, Nyvad B. 1990. Ability to bind salivary α-amylase discriminates certain viridans group streptococcal species. J. Clin. Microbiol. 28:2576–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kolenbrander PE. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54:413–437 [DOI] [PubMed] [Google Scholar]
- 24. Konto-Ghiorghi Y, et al. 2009. Dual role for pilus in adherence to epithelial cells and biofilm formation in Streptococcus agalactiae. PLoS Pathog. 5:e1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lengeler JW, Jahreis K. 2009. Bacterial PEP-dependent carbohydrate:phosphotransferase systems couple sensing and global control mechanisms. Contrib. Microbiol. 16:65–87 [DOI] [PubMed] [Google Scholar]
- 26. Li L, Tanzer JM, Scannapieco FA. 2002. Identification and analysis of the amylase-binding protein B (AbpB) and gene (abpB) from Streptococcus gordonii. FEMS Microbiol. Lett. 212:151–157 [DOI] [PubMed] [Google Scholar]
- 27. Li SJ, Rock CO, Cronan JE., Jr 1992. The dedB (usg) open reading frame of Escherichia coli encodes a subunit of acetyl-coenzyme A carboxylase. J. Bacteriol. 174:5755–5757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liang X, Chen YY, Wu H. 2011. Sortase B assembles amylase-binding protein A to streptococcal cell surface, abstr 20. 89th IADR, San Diego, CA. http://iadr.confex.com/iadr/2011sandiego/webprogramcd/Paper150380.html
- 29. Loesche W. 2007. Dental caries and periodontitis: contrasting two infections that have medical implications. Infect. Dis. Clin. North Am. 21:471–502 [DOI] [PubMed] [Google Scholar]
- 30. Loo CY, Corliss DA, Ganeshkumar N. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182:1374–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lu YJ, Rock CO. 2006. Transcriptional regulation of fatty acid biosynthesis in Streptococcus pneumoniae. Mol. Microbiol. 59:551–566 [DOI] [PubMed] [Google Scholar]
- 32. McMurry LM, Oethinger M, Levy SB. 1998. Triclosan targets lipid synthesis. Nature 394:531–532 [DOI] [PubMed] [Google Scholar]
- 33. Miller JL. 1960. Method of pure parotid saliva collection without cannulization. J. Dent. Res. 39:1075. [DOI] [PubMed] [Google Scholar]
- 34. Ramos JL, et al. 2005. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 69:326–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rock CO, Cronan JE. 1996. Escherichia coli as a model for the regulation of dissociable (type II) fatty acid biosynthesis. Biochim. Biophys. Acta 1302:1–16 [DOI] [PubMed] [Google Scholar]
- 36. Rogers JD, et al. 1998. Identification and analysis of a gene (abpA) encoding a major amylase-binding protein in Streptococcus gordonii. Microbiology 144(Pt 5):1223–1233 [DOI] [PubMed] [Google Scholar]
- 37. Rogers JD, Palmer RJ, Jr, Kolenbrander PE, Scannapieco FA. 2001. Role of Streptococcus gordonii amylase-binding protein A in adhesion to hydroxyapatite, starch metabolism, and biofilm formation. Infect. Immun. 69:7046–7056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rohwer JM, Postma PW, Kholodenko BN, Westerhoff HV. 1998. Implications of macromolecular crowding for signal transduction and metabolite channeling. Proc. Natl. Acad. Sci. U. S. A. 95:10547–10552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rosan B, Lamont RJ. 2000. Dental plaque formation. Microbes Infect. 2:1599–1607 [DOI] [PubMed] [Google Scholar]
- 40. Scannapieco FA, Bergey EJ, Reddy MS, Levine MJ. 1989. Characterization of salivary α-amylase binding to Streptococcus sanguis. Infect. Immun. 57:2853–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Scannapieco FA, Bhandary K, Ramasubbu N, Levine MJ. 1990. Structural relationship between the enzymatic and streptococcal binding sites of human salivary α-amylase. Biochem. Biophys. Res. Commun. 173:1109–1115 [DOI] [PubMed] [Google Scholar]
- 42. Scannapieco FA, Haraszthy GG, Cho MI, Levine MJ. 1992. Characterization of an amylase-binding component of Streptococcus gordonii G9B. Infect. Immun. 60:4726–4733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Scannapieco FA, Solomon L, Wadenya RO. 1994. Emergence in human dental plaque and host distribution of amylase-binding streptococci. J. Dent. Res. 73:1627–1635 [DOI] [PubMed] [Google Scholar]
- 44. Scannapieco FA, Torres G, Levine MJ. 1993. Salivary α-amylase: role in dental plaque and caries formation. Crit. Rev. Oral Biol. Med. 4:301–307 [DOI] [PubMed] [Google Scholar]
- 45. Shiroza T, Kuramitsu HK. 1993. Construction of a model secretion system for oral streptococci. Infect. Immun. 61:3745–3755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tanzer JM, et al. 2003. Amylase-binding proteins A (AbpA) and B (AbpB) differentially affect colonization of rats' teeth by Streptococcus gordonii. Microbiology 149:2653–2660 [DOI] [PubMed] [Google Scholar]
- 47. Tsay JT, Oh W, Larson TJ, Jackowski S, Rock CO. 1992. Isolation and characterization of the beta-ketoacyl-acyl carrier protein synthase III gene (fabH) from Escherichia coli K-12. J. Biol. Chem. 267:6807–6814 [PubMed] [Google Scholar]
- 48. Vickerman MM, Iobst S, Jesionowski AM, Gill SR. 2007. Genome-wide transcriptional changes in Streptococcus gordonii in response to competence signaling peptide. J. Bacteriol. 189:7799–7807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vorrasi J, Chaudhuri B, Haase EM, Scannapieco FA. 2010. Identification and characterization of amylase-binding protein C from Streptococcus mitis NS51. Mol. Oral Microbiol. 25:150–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yamamoto M, Jones JM, Senghas E, Gawron-Burke C, Clewell DB. 1987. Generation of Tn5 insertions in streptococcal conjugative transposon Tn916. Appl. Environ. Microbiol. 53:1069–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang Y, Cronan JE., Jr 1998. Transcriptional analysis of essential genes of the Escherichia coli fatty acid biosynthesis gene cluster by functional replacement with the analogous Salmonella typhimurium gene cluster. J. Bacteriol. 180:3295–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.