Abstract
Herein, we report that a modified gentamicin cassette and a PCR-based method can be used for targeted mutagenesis of the oral spirochete Treponema denticola. This approach minimizes polar effects and spontaneous antibiotic resistance. Therefore, it can serve as a reliable tool for genetic manipulation of T. denticola.
TEXT
The oral spirochete Treponema denticola is a member of the “red complex” bacteria. It is an important pathogen that is associated with human periodontal disease (7, 14), a chronic infection that occurs in 80% of the adult population worldwide (5, 22, 26). However, due to the paucity of genetic tools and its fastidious growth requirements, the biology and pathogenicity of T. denticola are poorly understood (7, 10, 15). Although significant progress has been achieved during the last 2 decades, tools for genetic manipulation of T. denticola are still very limited (2, 18, 19, 25, 31). The first tool applied to genetic studies in T. denticola is an erythromycin resistance cassette [erm(F)-erm(B)] developed by Li et al. (19). It has since become a standard method for genetic manipulations of T. denticola. However, the spirochete has a high spontaneous mutation rate to the presence of erythromycin, and the insertion of the erm(F)-erm(B) cassette in a targeted gene often has a polar effect on downstream genes (13, 20). These two drawbacks have substantially hampered the use of this cassette. Two modified erm(F)-erm(B) cassettes have been developed (13, 20), but the aforementioned problems still hold. Chloramphenicol and coumermycin antibiotic resistance cassettes have been used for trans-complementation of certain T. denticola mutants (4, 25). However, these two cassettes have not yet been successfully used for the targeted mutagenesis of T. denticola, as the resistance to chloramphenicol is not stable and the coumermycin resistance is not reliable due to the pleiotropic effects of gyrase mutation (24, 27).
Constructing a mutated gentamicin resistance cassette.
A gentamicin cassette (aacC1) (6, 28–30) has been recently used as a selectable marker for transposon mutagenesis in the T. denticola ATCC 35405 (Td35405) strain (31). We reasoned that this cassette could also be used as a selectable marker for targeted mutagenesis in the Td35405 strain. Unexpectedly, all of the attempts to use it for targeted mutagenesis failed. Our recent work revealed that Td35405 carries genes encoding three type II DNA restriction endonucleases. One of these enzymes (TDE0911) recognizes and cleaves targeted DNAs containing the sequence GGNCC (2). Sequence analysis shows that the aacC1 cassette contains a cleavage site recognized by TDE0911 (G97GCCC). We hypothesized that the failure of the aacC1 cassette in Td35405 could be due to the restriction cleavage mediated by TDE0911. To test this hypothesis, the cleavage site (G97GCCC) within aacC1 was mutated to A97GCCC by site-directed mutagenesis. The wild-type and mutated (designated aacCm) cassettes were treated with the crude cell extract of Td35405. As expected, the aacC1 cassette was digested into two fragments; but the mutated cassette remained intact (Fig. 1), indicating that the mutation successfully protected aacCm from cleavage.
Fig 1.
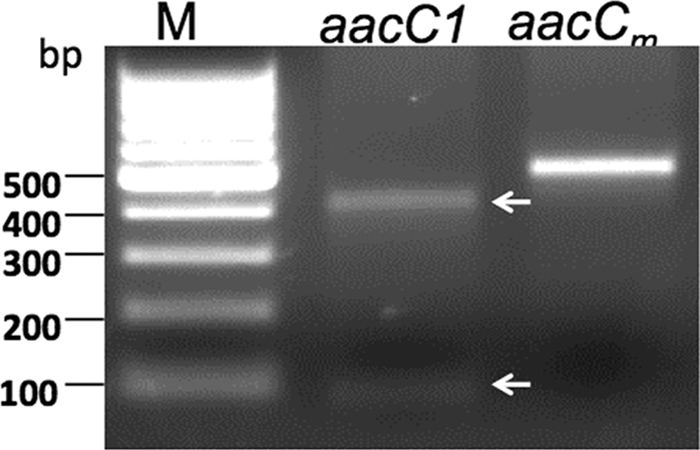
The mutated aacCm cassette is resistant to the cleavage of T. denticola. PCR amplified 534 bp of the wild-type aacC1 cassette, and mutated aacCm fragments were treated with the crude cell extract of Td35405. The samples obtained were then analyzed by 2% agarose gel electrophoresis and visualized with ethidium bromide staining. Lane M contains DNA markers. The two white arrows point toward the cleaved products.
Constructing a vector for deletion of the prcA gene.
To test whether the aacCm cassette is functional in T. denticola, we decided to choose the prcA gene for targeted mutagenesis. The prcA gene (TDE0761) encodes a lipoprotein that is a component of the outer membrane protease complex (dentilisin). The function of PrcA in the dentilisin complex has been extensively studied (11–13). In addition, prcA is located within the prcB-prcA-prtP operon. Previous studies report that the insertion of either erm(F)-erm(B) or erm(B) in prcA could decrease the expression level of the gene downstream, prtP (12, 13). Thus, targeting prcA could help us to determine whether the insertion of aacCm cassette in a targeted gene has any polar effect on its downstream genes. A PCR-based method, which was recently used in the Lyme disease spirochete, Borrelia burgdorferi (21), was modified to construct a vector for the targeted mutagenesis of prcA. The scheme that is illustrated in Fig. 2 aims to replace the entire open reading frame of prcA (1,920 bp) in frame with the promoterless aacCm cassette (the coding sequence from the start codon ATG to the TAA stop codon). The vector was constructed via three PCR amplification steps. In the first step, the upstream prcB gene (primers P1/P2), the downstream prtP gene (primers P5/P6), and the aacCm cassette (primers P3/P4) were PCR amplified. In the second step, the prcB and aacCm fragments were fused via PCR amplification with primers P1/P4. In the third step, the prcB-aacCm and prtP fragments were linked via PCR amplification with primers P1/P6. The final fragment prcB-aacCm-prtP[r] (2,132 bp) was cloned into pGEM-T Easy vector (Promega, Madison, WI). The final vector was named PrcA::aacCm and confirmed by DNA sequencing analysis (Roswell Park Cancer Institute DNA Sequencing Laboratory, Buffalo, NY). The primer sequences are listed in Table 1.
Fig 2.

Schematic illustration of constructing the PrcA::aacCm vector for the deletion of prcA gene. The vector was constructed via three PCR amplification steps. Arrows indicate the relative positions of PCR primers for constructing the vector. The sequences of these primers are listed in Table 1.
Table 1.
Oligonucleotide primers used in this study
| Primer | Sequencea | Note | Directionb |
|---|---|---|---|
| P1 | CCGAATCTCGGCACCTGT | 5′-Flanking region of prcA | F |
| P2 | GCGTAACATACTCGTCCTCCTATAAAT | 5′-Flanking region of prcA | R |
| P3 | GGAGGACGAGTATGTTACGCAGCAGCAACG | aacCm cassette | F |
| P4 | GACCTCCTTAGGTGGCGGTACTTGGG | aacCm cassette | R |
| P5 | GCCACCTAAGGAGGTCTTTAAGATGAAG | 3′-Flanking region of prcA | F |
| P6 | GATAGTGCCCGAACAGTG | 3′-Flanking region of prcA | R |
| P7 | ATGTTACGCAGCAGCAACG | aacCm | F |
| P8 | TTAGGTGGCGGTACTTGGG | aacCm | R |
| P9 | GCTGCGGCGTTATGGTGT | Detected prcA | F |
| P10 | CCGTCGTCGAGGGGAAAG | Detected prcA | R |
| P11 | GCAGCGGGTACAAAGATAC | 5′-Flanking region of prcB | F |
The underlined sequences are engineered overlapping sequences for PCR amplifications.
F, forward; R, reverse.
Isolation of gentamicin-resistant prcA mutants.
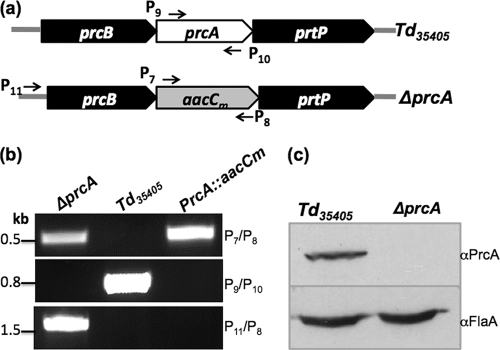
The PrcA::aacCm plasmid was purified from an Escherichia coli dam dcm mutant strain (New England BioLabs, Ipswich, MA), as previously described (2). The obtained unmethylated vector was linearized and transformed into Td35405 competent cells. The antibiotic-resistant clones were selected on TYGVS (1) semisolid agar plates containing gentamicin (20 μg/ml) (31). After a 14-day incubation, 18 antibiotic-resistant colonies appeared on the plates. Eight colonies were selected and analyzed by PCR for the presence of the aacCm cassette (primers P7/P8) and absence of the prcA gene (primers P9/P10). The results showed that all of the colonies examined contained the targeted mutation (data not shown). One clone (ΔprcA mutant clone) was further analyzed by PCR with different pairs of primers (Fig. 3a) and by Western blotting using a PrcA-specific antibody. The results showed that the entire prcA gene was replaced by aacCm as expected (Fig. 3b). Furthermore, the cognate gene product was completely abolished in the mutant (Fig. 3c), indicating that aacCm can be used for the targeted mutagenesis of T. denticola.
Fig 3.
Isolation and characterization of ΔprcA mutant. (a) Illustration of the prcA locus in Td35405 (wild type) and the aacCm cassette in the ΔprcA mutant. (b) PCR analysis showing that the prcA gene was deleted and replaced by aacCm in the ΔprcA mutant. The primers used for PCR analysis are labeled in panel a. (c) Western blot analysis of the ΔprcA mutant. Similar amounts of whole-cell lysates from the wild type and the mutant were analyzed by SDS-PAGE and then probed with a specific PrcA antibody (αPrcA, anti-PrcA antibody). Immunoblots were developed using horseradish peroxidase secondary antibody with an enhanced chemiluminescence (ECL) luminol assay as previously described (2, 3).
The insertion of aacCm in prcA has no polar effect on prtP.
Insertion of an antibiotic resistance cassette in a targeted gene can interfere with expression of downstream genes. This effect has compromised the interpretation of results and needs to be ruled out via genetic complementation or by other studies. Given that there is no shuttle vector available for the Td35405 strain, an antibiotic resistance marker that avoids polar effects and a reliable method of targeting it to a desired locus would be very useful. In the ΔprcA mutant, the entire prcA gene was deleted and replaced in frame by the promoterless aacCm cassette. The aacCm cassette is transcribed from the promoter upstream of prcA. To test whether there are polar effects on the downstream gene, the level of PrtP in the ΔprcA mutant and two previously constructed prcA mutants (13, 16) was measured by Western blotting using a specific antibody against PrtP. A 72-kDa PrtP protein was detected in the wild type and the ΔprcA mutant (Fig. 4). However, PrtP was not detected in the PNE mutant [in which prcA was inactivated by an erm(F)-erm(B) cassette] (16) or the CF547 mutant [in which prcA was disrupted by an erm(B) cassette] (13), two mutants that were constructed using different approaches. Taken together, these observations suggest that the approach reported here can avoid the potential polar effect in the targeted mutagenesis of T. denticola that has been observed previously.
Fig 4.
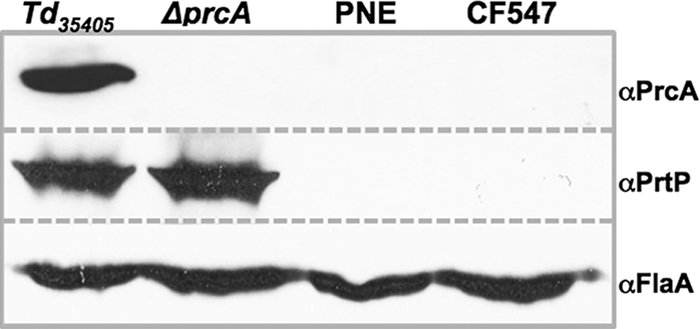
Western immunoblots of Td35405, ΔprcA mutant, and two previously constructed prcA mutants (PNE and CF547) (13, 16). The blots were probed with antibodies against T. denticola PrcA, PrtP, or FlaA. FlaA expression was used as a loading control (3, 13).
T. denticola has a low spontaneous mutation rate to the presence of gentamicin.
Thus far, the erm(F)-erm(B) cassette and its erm(B) derivative have provided the only selectable marker for targeted mutagenesis of T. denticola (13, 19). However, T. denticola often develops spontaneous resistance to erythromycin. Thus, additional efforts are required to distinguish targeted mutant clones from spontaneous mutants. To determine whether a similar scenario exists with gentamicin, the spontaneous resistance to the antibiotic was measured. For this assay, Td35405 was continuously cultivated and passed in the liquid TYGVS medium for up to 28 generations. The cells (5 × 109) were collected every 4 generations, plated on semisolid TYGVS plates containing a MIC of erythromycin (40 μg/ml) (8) or gentamicin (20 μg/ml) (31), and incubated in an anaerobic chamber for 2 weeks. The frequencies of spontaneous resistance to the antibiotics were expressed as the number of colonies divided by the number of cells plated (17, 23). The development of gentamicin resistance in T. denticola is extremely rare (spontaneous mutation frequency of <1.5 × 10−10) and is substantially lower than that of erythromycin (approximate frequency of 5 × 10−8) during the period of 28-generation cultivations, indicating that aacCm can serve as a reliable antibiotic selectable marker for constructing T. denticola mutants.
Summary.
The main objective of this study was to develop an alternative and more reliable antibiotic marker for the genetic studies of T. denticola. Compared to the erm(F)-erm(B) cassette (2,145 bp) (9) and its derivative, erm(B) (741 bp) (13), the newly constructed aacCm cassette (534 bp) is much shorter, which will be more convenient for genetic manipulations. In addition, the spontaneously occurring gentamicin resistance in T. denticola is substantially less than that of erythromycin. Moreover, the application of aacCm in combination with the PCR-based method described herein can avoid potential polar effects in targeted mutagenesis. The aacCm cassette and the methodology developed in this report will facilitate genetic studies of T. denticola (e.g., providing an additional antibiotic marker for construction of double mutations, for complementation of a mutant, and for increasing the targeted mutagenesis efficiency in T. denticola).
ACKNOWLEDGMENTS
We thank P. Rosa for providing the aacC1 cassette and R. Limberger for providing the sequence of erythromycin cassette.
This research was supported by Public Health Service grants DE018829 and DE019667 to C. Li and DE018221 to J. C. Fenno.
Footnotes
Published ahead of print 13 January 2012
REFERENCES
- 1. Arakawa S, Kuramitsu HK. 1994. Cloning and sequence analysis of a chymotrypsinlike protease from Treponema denticola. Infect. Immun. 62:3424–3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bian J, Li C. 2011. Disruption of a type II endonuclease (TDE0911) enables Treponema denticola ATCC 35405 to accept an unmethylated shuttle vector. Appl. Environ. Microbiol. 77:4573–4578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bian J, Shen H, Tu Y, Yu A, Li C. 2011. The riboswitch regulates a thiamine pyrophosphate ABC transporter of the oral spirochete Treponema denticola. J. Bacteriol. 193:3912–3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chi B, Limberger RJ, Kuramitsu HK. 2002. Complementation of a Treponema denticola flgE mutant with a novel coumermycin A1-resistant T. denticola shuttle vector system. Infect. Immun. 70:2233–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Darveau RP. 2010. Periodontitis: a polymicrobial disruption of host homeostasis. Nat. Rev. Microbiol. 8:481–490 [DOI] [PubMed] [Google Scholar]
- 6. Elias AF, et al. 2003. New antibiotic resistance cassettes suitable for genetic studies in Borrelia burgdorferi. J. Mol. Microbiol. Biotechnol. 6:29–40 [DOI] [PubMed] [Google Scholar]
- 7. Ellen RP, Galimanas VB. 2005. Spirochetes at the forefront of periodontal infections. Periodontol. 2000 38:13–32 [DOI] [PubMed] [Google Scholar]
- 8. Fenno JC, Wong GW, Hannam PM, McBride BC. 1998. Mutagenesis of outer membrane virulence determinants of the oral spirochete Treponema denticola. FEMS Microbiol. Lett. 163:209–215 [DOI] [PubMed] [Google Scholar]
- 9. Fletcher HM, et al. 1995. Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect. Immun. 63:1521–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Frederick JR, Sarkar J, McDowell JV, Marconi RT. 2011. Molecular signaling mechanisms of the periopathogen, Treponema denticola. J. Dent. Res. 90:1155–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Godovikova V, Goetting-Minesky MP, Fenno JC. 2011. Composition and localization of Treponema denticola outer membrane complexes. Infect. Immun. 79:4868–4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Godovikova V, et al. 2010. Treponema denticola PrcB is required for expression and activity of the PrcA-PrtP (dentilisin) complex. J. Bacteriol. 192:3337–3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goetting-Minesky MP, Fenno JC. 2010. A simplified erythromycin resistance cassette for Treponema denticola mutagenesis. J. Microbiol. Methods 83:66–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Holt SC, Ebersole JL. 2005. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex”, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol. 2000 38:72–122 [DOI] [PubMed] [Google Scholar]
- 15. Kuramitsu HK. 2003. Molecular genetic analysis of the virulence of oral bacterial pathogens: an historical perspective. Crit. Rev. Oral Biol. Med. 14:331–344 [DOI] [PubMed] [Google Scholar]
- 16. Lee SY, et al. 2002. Cleavage of Treponema denticola PrcA polypeptide to yield protease complex-associated proteins PrcA1 and PrcA2 is dependent on PrtP. J. Bacteriol. 184:3864–3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Levin BR, Perrot V, Walker N. 2000. Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria. Genetics 154:985–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li H, Kuramitsu HK. 1996. Development of a gene transfer system in Treponema denticola by electroporation. Oral Microbiol. Immunol. 11:161–165 [DOI] [PubMed] [Google Scholar]
- 19. Li H, Ruby J, Charon N, Kuramitsu H. 1996. Gene inactivation in the oral spirochete Treponema denticola: construction of an flgE mutant. J. Bacteriol. 178:3664–3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Limberger RJ, Slivienski LL, Izard J, Samsonoff WA. 1999. Insertional inactivation of Treponema denticola tap1 results in a nonmotile mutant with elongated flagellar hooks. J. Bacteriol. 181:3743–3750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Motaleb MA, Pitzer JE, Sultan SZ, Liu J. 2011. A novel gene inactivation system reveals altered periplasmic flagellar orientation in a Borrelia burgdorferi fliL mutant. J. Bacteriol. 193:3324–3331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pihlstrom BL, Michalowicz BS, Johnson NW. 2005. Periodontal diseases. Lancet 366:1809–1820 [DOI] [PubMed] [Google Scholar]
- 23. Poggi D, Oliveira de Giuseppe P, Picardeau M. 2010. Antibiotic resistance markers for genetic manipulations of Leptospira spp. Appl. Environ. Microbiol. 76:4882–4885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Samuels DS. 2006. Antibiotic resistance in Borrelia burgdorferi: applications for genetic manipulation and implications for evolution, p 56–71 In Cabello FC, Hulinska D, Godfrey HP. (ed), Molecular biology of spirochetes. NATO Science Series. Series I. Life and behavioural sciences, vol 373 IOS Press, Amsterdam, The Netherlands [Google Scholar]
- 25. Slivienski-Gebhardt LL, Izard J, Samsonoff WA, Limberger RJ. 2004. Development of a novel chloramphenicol resistance expression plasmid used for genetic complementation of a fliG deletion mutant in Treponema denticola. Infect. Immun. 72:5493–5497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Socransky SS, Haffajee AD. 2005. Periodontal microbial ecology. Periodontol. 2000 38:135–187 [DOI] [PubMed] [Google Scholar]
- 27. Steck TR, Pruss GJ, Manes SH, Burg L, Drlica K. 1984. DNA supercoiling in gyrase mutants. J. Bacteriol. 158:397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stewart PE, Rosa PA. 2008. Transposon mutagenesis of the Lyme disease agent Borrelia burgdorferi. Methods Mol. Biol. 431:85–95 [DOI] [PubMed] [Google Scholar]
- 29. Tenover FC, et al. 1989. Development of a DNA probe from the deoxyribonucleotide sequence of a 3-N-aminoglycoside acetyltransferase [AAC(3)-I] resistance gene. Antimicrob. Agents Chemother. 33:551–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wohlleben W, et al. 1989. On the evolution of Tn21-like multiresistance transposons: sequence analysis of the gene (aacC1) for gentamicin acetyltransferase-3-I (AAC(3)-I), another member of the Tn21-based expression cassette. Mol. Gen. Genet. 217:202–208 [DOI] [PubMed] [Google Scholar]
- 31. Yang Y, Stewart PE, Shi X, Li C. 2008. Development of a transposon mutagenesis system in the oral spirochete Treponema denticola. Appl. Environ. Microbiol. 74:6461–6464 [DOI] [PMC free article] [PubMed] [Google Scholar]