Abstract
Mycoplasma gallisepticum is a bacterial pathogen of poultry that is estimated to cause annual losses exceeding $780 million. The National Poultry Improvement Plan guidelines recommend regular surveillance and intervention strategies to contain M. gallisepticum infections and ensure mycoplasma-free avian stocks, but several factors make detection of M. gallisepticum and diagnosis of M. gallisepticum infection a major challenge. Current techniques are laborious, require special expertise, and are typically plagued by false results. In this study, we describe a novel detection strategy which uses silver nanorod array–surface-enhanced Raman spectroscopy (NA-SERS) for direct detection of avian mycoplasmas. As a proof of concept for use in avian diagnostics, we used NA-SERS to detect and differentiate multiple strains of avian mycoplasma species, including Acholeplasma laidlawii, Mycoplasma gallinarum, Mycoplasma gallinaceum, Mycoplasma synoviae, and M. gallisepticum, including vaccine strains 6/85, F, and ts-11. Chemometric multivariate analysis of spectral data was used to classify these species rapidly and accurately, with >93% sensitivity and specificity. Furthermore, NA-SERS had a lower limit of detection that was 100-fold greater than that of standard PCR and comparable to that of real-time quantitative PCR. Detection of M. gallisepticum in choanal cleft swabs from experimentally infected birds yielded good sensitivity and specificity, suggesting that NA-SERS is applicable for clinical detection.
INTRODUCTION
Mycoplasma gallisepticum causes chronic respiratory disease in chickens and sinusitis in turkeys, resulting in reduced egg production, stunted growth with high mortality rates among young birds, and increased condemnations at processing plants (19). The economic impact on the U.S. poultry industry has been estimated at $150 million annually for egg production alone, and while considered under control in the U.S. broiler/fryer industry, sporadic outbreaks can result in culling of entire flocks, a drastic and expensive strategy (9). The National Poultry Improvement Plan (http://www.aphis.usda.gov) designates M. gallisepticum a type II organism, i.e., an economic and food security risk requiring monitoring through regular testing of flocks.
Diagnosis of M. gallisepticum infection in the poultry industry is by serological screening with serum plate agglutination, hemagglutination inhibition, and enzyme-linked immunosorbent assay. A definitive diagnosis is made by isolation of the organism or detection by molecular methods. The challenge of M. gallisepticum detection is daunting due to its fastidious growth requirements, the presence of commensal mycoplasmas which can outgrow M. gallisepticum, and cross-reactivity with closely related species (18). Serum plate agglutination and immunofluorescence methods are rapid but often difficult to interpret due to variable titers, the potentially confounding impact of commensal mycoplasmas, and/or poor specificity. Nucleic acid-based methods have a low limit of detection, approaching that of single organisms, but are prone to inaccurate results due to reaction inhibitors and are impractical for analysis of individual birds in a flock due to cost and the expertise required. Real-time quantitative PCR (qPCR) is replacing standard PCR due to its superior sensitivity and speed, but various inhibitors can interfere with the reaction to produce false-negative results (5). For effective control in single-age broiler populations, the flock is treated as an individual (all in/all out), and destruction of the flock when a positive result is confirmed is an expensive and drastic strategy which may not be economically feasible for the broiler farmer. Likewise, in mixed-age populations on breeder or egg-laying farms, the large number of birds affected and the continuous introduction of naïve birds render this solution economically prohibitive (9).
Three vaccines are currently approved for mixed-age flocks: the F strain (Poulvac Myco F; Pfizer Animal Health-Global Poultry, Durham, NC), ts-11(Mycoplasma gallisepticum vaccine; Merial Animal Health, Gainesville, GA), and 6/85 (Mycovac; Merck Animal Health [Intervet-Schering Plough, Inc.], Millsboro, DE). The F strain retains some virulence but confers lifetime immunity (18, 29). Both ts-11 and 6/85 are less virulent than the F strain but also less effective (18), stimulating weaker protective immune responses than the F strain (1, 32). Thus, each vaccine has its own problems, but common to all is the need to differentiate exposure of birds in mixed-age populations to vaccine versus field strains, which complicates diagnostics and undermines the value of vaccination efforts.
In this study, we developed and evaluated a diagnostic platform that uses a silver nanorod array (NA) for surface-enhanced Raman spectroscopy (SERS) detection and identification of multiple strains of avian mycoplasma species in poultry. The silver nanorods can enhance the light-analyte interaction when a biomolecule absorbed on the Ag nanorod surface is excited by a laser beam. The vibrations of the chemical bonds in the biomolecules are amplified due to an enhanced local electromagnetic field (20). Such strong vibrations will transfer the small vibrational energy from the laser beam so that after the interaction, the wavelength of the scattered radiation is red shifted (Stokes shift), having an energy signature less than that of the incident energy. The Stokes shift due to bacteria tends to be highly specific and characteristic of biological samples (7, 26, 27). Our NA platform allows collection and measurement of a consistent and highly reproducible Raman spectral signature enhanced on the order of 108 (6) and detects and differentiates with 93 to 100% sensitivity and specificity three strains of the related human pathogen Mycoplasma pneumoniae, including 97% sensitivity and specificity with detection in true clinical throat swab specimens (15). Here, we assessed the capacity of NA-SERS to discriminate laboratory cultures of five avian mycoplasma species, including field and vaccine M. gallisepticum strains, as well as a direct comparison of the lower limits of detection with standard PCR and qPCR. We differentiated M. gallisepticum field and vaccine strains, Mycoplasma synoviae, and common commensal mycoplasmas with 98% accuracy, discriminating each with high confidence. Not surprisingly, classification in the biochemically complex background of choanal cleft swabs was promising but somewhat less robust than that obtained with pure laboratory samples.
MATERIALS AND METHODS
Mycoplasma strains, culture conditions, and quantification.
The M. gallisepticum strains examined here were as follows: Rlow (passages 14 to 20) (29), A5969 (passage 347) (17), S6 (passage 238) (34), vaccine strain F (passage 204) (21), vaccine strain ts-11 (passage 93) (32), and vaccine strain 6/85 (passage 199) (10) (Intervet, Inc., Millsboro, DE). Each was cultured by incubating a 25-μl inoculum at 37°C in 25 ml of Frey's medium (pH 7.8) containing 0.3% glucose, 1.9 mM thallium acetate, 1,000,000 units/liter penicillin G, and 12% swine serum (11). Avian mycoplasmas Acholeplasma laidlawaii (passage 38) and Mycoplasma gallinaceum Tully DD (passage level unknown) were likewise grown in Frey's medium. The Mycoplasma synoviae strain WVU 1853 (passage 44) was grown in Frey's medium plus reduced NAD (0.02%). Mycoplasma gallinarum strain SrBM285 (passage level unknown) was grown in Modified ORT medium with arginine as a carbon and energy source (4).
Upon observation of a pH indicator color change signaling growth, a 200-μl aliquot was removed for PCR analysis (see below). A 100-μl aliquot was likewise removed for titration by serial dilution in Frey's medium and analysis of 8 replicates in 96-microwell trays for most-probable number according to color-changing units (CCU) (30). A portion of each dilution was also inoculated in duplicate onto Frey's agar plates to measure CFU, which exhibited good agreement with CCU determinations. The remaining cultures were chilled on ice and centrifuged (Sorvall RC5C Plus; Thermo Scientific, Asheville, NC) at 10,000 rpm at 4°C for 25 min, washed 3 times with chilled distilled H2O, and suspended in 1 ml distilled H2O. Protein content for these undiluted samples was assessed by the Pierce bicinchoninic acid (BCA) method (Thermo Scientific, Rockford, IL).
PCR analysis.
DNA was extracted by using a Qiagen QIAamp minikit (Valencia, CA) according to the manufacturer's recommendations, except that the 50-μl volume of eluate was incubated in the spin column for 5 min before the final centrifugation. PCR analysis was carried out using an Epicentre FailSafe PCR PreMix selection kit (Epicentre Biotechnologies, Madison, WI) and primers targeting the interspacer region between the 16S and the 23S rRNA genes as described previously (28) to yield an expected 812-bp product. We visualized 10-μl volumes of the PCR products on a 2% agarose gel with ethidium bromide staining. For qPCR analysis, serial 10-fold dilutions (starting at 4.67 × 105 CFU/μl) of the M. gallisepticum Rlow strain were analyzed and compared to the University of Georgia Poultry Diagnostic Research Center Mycoplasma Diagnostic Lab standards as described previously (5), except that the standards were expanded to cover a wider range of dilutions.
Clinical samples.
Choanal cleft swabs were collected from commercial broilers with sterile polyester-tipped applicators (MDCI, Ltd., West Sussex, United Kingdom) during necropsy for an unrelated vaccine study (8). Four groups of 15 birds each were assessed 10 days after inoculation by aerosol with the ts-11 vaccine strain (3.6 × 106 CCU/ml), the Rlow strain (3.62 × 108 CCU/ml), and K6216D (1.35 × 108 CCU/ml; ts-11-like). The fourth group was an unchallenged control group. Swabs were collected during necropsy and immediately frozen at −80°C. PCR analysis for the presence of M. gallisepticum was performed on tracheal samples from each bird as described previously (5).
NA-SERS and chemometric analysis.
Silver nanorod arrays were prepared for generating enhanced Raman signals as described previously (6, 35). Individual 1-cm by 1-cm substrates or 1-in. by 3-in. substrates with 40 3-mm-diameter wells formed by polydimethylsiloxane (2) were used for NA-SERS characterization. Bacterial suspensions in H2O were diluted 1:10 in high-performance liquid chromatography (HPLC)-grade methanol and dispersed by syringe passage 10 times (22-gauge needle) before 1-μl volumes were loaded onto NA substrates and allowed to air dry overnight. Choanal swabs were eluted in 500 μl sterile distilled H2O, dispersed by syringe passage (22-gauge needle), and fixed by a 1:10 dilution in HPLC-grade methanol before they were loaded onto NA substrates. SERS spectra were acquired using a Renishaw inVia Reflex multiwavelength confocal imaging microscope (Hoffman Estates, IL). A Leica apochromatic 5× objective (numerical aperture = 0.12) allowed spatial averaging of the samples, illuminating a 1,265-μm2 area on the substrate. Spectral signals were collected in 10-s acquisitions with a 785-nm near-infrared diode laser source (Renishaw) operating at 5% power capacity (15 mW). Sample imaging was accomplished by line-focus optics (diffraction grating of 1,200 lines/mm). Spectra from sample-free substrates and methanol-only controls were collected from 10 random substrate locations each. Mycoplasma samples were analyzed on 3 independent substrates, with spectra from 5 random locations on each examined to assess spectral variation within and among substrates. Alternatively, spectra were collected with Bruker optics on a Senterra dispersive Raman microscope (Billerica, MA) using the 785-nm laser at a 25-mW power level, a 10× objective, and a 10-s acquisition with a resolution of 3 to 5 wave numbers, using OPUS 6.5 software.
Spectra were first averaged using the GRAMS32/A1 spectral software package (Galactic Industries, Nashua, NH) to assess signal-to-noise spectral quality and baseline corrected using a concave rubber band algorithm (OPUS, Bruker Optics, Inc., Billerica, MA) which performed 10 iterations on 64 points to aid in a preliminary evaluation/comparison and peak assignment. Spectra were processed for chemometric analysis as described previously (15). Briefly, raw spectra were preprocessed using the first derivative of each and a 15-point, 2nd-order polynomial Savitsky-Golay algorithm, followed by normalization to unit vector length with Matlab R2009A software (Natick, MA) supplemented with PLS toolbox 5.8 and 6.0 (Eigenvector, Wenatchee, WA). Unit vector normalization allows direct intensity comparison between individual samples and between variables within a sample. The unsupervised methods of principal component analysis (PCA) and hierarchical cluster analysis (HCA) were used to explore clustering of similar spectra using Matlab software (version 2009A). The diagonal matrix (y block) for partial least-squares discriminatory analysis (PLS-DA) was generated in Excel and imported into PLS toolbox 5.8 and 6.0 (Eigenvector, Wenatchee, WA) operating in Matlab 2009A to perform PLS-DA. Both the x and the y block data were mean centered prior to PLS-DA (3). Model robustness was tested by cross-validation with Venetian Blinds, which withheld 10% of the data during model calibration and then applied the model to these sample “unknowns” to assess its predictive power.
RESULTS
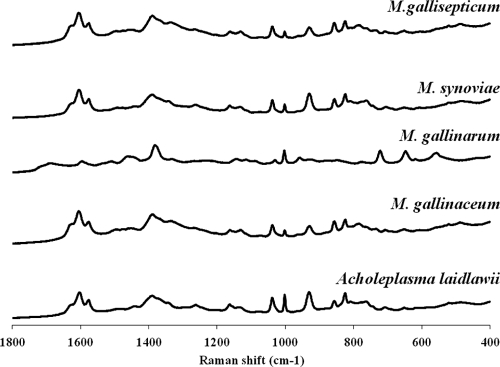
We evaluated NA-SERS for the ability to detect and discriminate 5 different avian mycoplasma species, including 3 strains of the pathogen M. gallisepticum (A5969, S6, and Rlow, treated initially as a single class), the pathogen M. synoviae, and the commensal species Acholeplasma laidlawii, M. gallinarum, and M. gallinaceum. We used methanol to fix avian specimens because formalin used for NA-SERS analysis of the human pathogen M. pneumoniae (15) caused clumping here, quenching the enhancement; it is not clear why M. gallisepticum but not M. pneumoniae exhibited clumping with formalin fixation. Multiple spectra were collected for each analyte from different, randomly selected locations on the NA substrates to demonstrate reproducibility (data not shown). The averaged spectral signatures (n = 15) for each strain were very similar (Fig. 1), demonstrating a need for chemometric processing for multivariate analysis to facilitate interpretation. PLS-DA is a supervised method for spectral analysis that utilizes prior knowledge of the classes (in this case the mycoplasma species) as a training set, preserving spectral variation within classes while emphasizing variation among classes (3). PLS-DA classified the spectra for all species with 93 to 100% sensitivity and likewise with similar specificity for all species except M. synoviae, which was distinguishable with only 80% specificity (Table 1). Cross-validated (CV) errors for this initial analysis were as low as 3%. Finding the hyperplane/vector in multivariate space to maximize variance is more difficult with six classes than with two, and follow-up analysis of the same data set with classification as positive or negative for M. synoviae yielded clear differentiation from the other species, with 97% sensitivity and 93% specificity (Table 2). Thus, the information necessary to distinguish M. synoviae was present in the spectra and accessible with the appropriate chemometric parameters.
Fig 1.
NA-SERS spectra of selected avian mycoplasma species. Fifteen spectra from random spots were averaged for each species. The M. gallisepticum strain is Rlow.
Table 1.
Specificity and sensitivity of NA-SERS discrimination of avian mycoplasma species by PLS-DA
| Modeled classa | Sensitivity (CVb) | Specificity (CV) | Class error (CV) | RMSECVc |
|---|---|---|---|---|
| M. gallisepticum | 0.944 | 0.951 | 0.053 | 0.246 |
| M. gallinaceum | 0.933 | 0.992 | 0.038 | 0.201 |
| A. laidlawii | 0.933 | 0.950 | 0.058 | 0.236 |
| M. synoviae | 0.933 | 0.800 | 0.133 | 0.287 |
| M. gallinarum | 1.000 | 1.000 | 0 | 0.033 |
| Methanol | 1.000 | 0.930 | 0.035 | 0.238 |
Six latent variables (n = 135), accounting for 77% of the captured x variance, were used to generate the model.
CV, cross validation by Venetian Blinds with 10% of the data.
RMSECV, root mean squared error in cross validation.
Table 2.
PLS-DA classification of the original species data set as positive or negative for M. synoviaea
| Modeled classb | Sensitivity (CV) | Specificity (CV) | Class error (CV) | RMSECV |
|---|---|---|---|---|
| M. synoviae positive | 0.978 | 0.939 | 0.042 | 0.200 |
| M. synoviae negative | 0.939 | 0.978 | 0.042 | 0.200 |
The abbreviations are the same as those in Table 1.
Six latent variables (n = 135), accounting for 88% of the captured x variance, were used to generate the model.
We next examined the related M. gallisepticum strains Rlow, S6, and A5969. Rlow was originally a clinical isolate but has not been observed subsequently in natural infections and is now primarily used as a challenge organism for laboratory studies and trials. S6 is a virulent strain originally isolated from the brain of an infected turkey (34), while A5969 is a high-passage strain which is avirulent (33). Analysis of their NA-SERS spectra by PLS-DA classified these strains with 96 to 100% sensitivity and specificity (Table 3). When the vaccine strains 6/85, F, and ts-11 were included in the model, PLS-DA no longer distinguished each with statistical significance (sensitivity and specificity as low as 70%) (data not shown). However, examination of the same data set with classification according to virulence correctly differentiated all members of each class with >96% accuracy (Table 4), again reflecting the challenge of maximizing variance among a larger number of classes.
Table 3.
PLS-DA classification of NA-SERS spectra of three M. gallisepticum strainsa
| Modeled classb | Sensitivity (CV) | Specificity (CV) | Class error (CV) | RMSECV |
|---|---|---|---|---|
| Rlow | 0.981 | 0.969 | 0.036 | 0.144 |
| S6 | 0.981 | 0.967 | 0.036 | 0.100 |
| A5969 | 1.000 | 1.000 | 0.000 | 0.117 |
The abbreviations are the same as those in Table 1.
Model generated with 7 latent variables (n = 82), capturing 87% of x variance and 93% of y variance.
Table 4.
PLS-DA classification of M. gallisepticum strains according to virulence
| Modeled classa | Sensitivity (CV) | Specificity (CV) | Class error (CV) | RMSECV |
|---|---|---|---|---|
| Virulent MG (Rlow and S6) | 0.969 | 0.986 | 0.023 | 0.226 |
| Avirulent MG (A5969 and vaccine strains) | 0.986 | 0.969 | 0.023 | 0.226 |
Model generated with 5 latent variables (n = 101), capturing 70% of the x variance and 87% of the y variance.
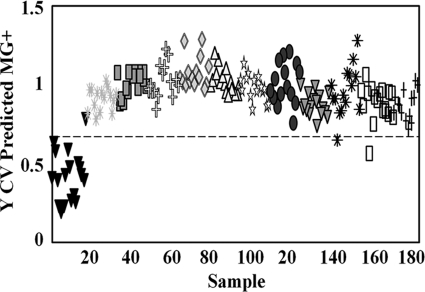
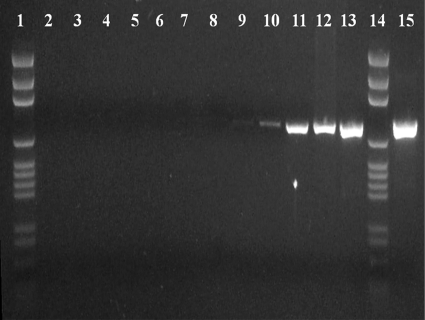
Limits of detection were assessed on 10 serial 10-fold dilutions of M. gallisepticum Rlow with a starting concentration of 4.67 × 105 CFU/μl. PLS-DA correctly classified all dilutions relative to the methanol background with 94 to 98% accuracy (Fig. 2 and data not shown). This reflected a lower limit of detection of <1 CFU, consistent with that observed for M. pneumoniae (15) but difficult to extrapolate to the single-cell level due to the tendency for mycoplasmas to clump and their poorly defined plating efficiency. Starting protein content in the undiluted sample was 0.5 μg/μl, indicating detection of protein at femtomolar quantities, consistent with previously reported SERS sensitivity (13, 14). Linear regression analysis failed to demonstrate a strong correlation between intensity and concentration (data not shown). We also compared NA-SERS directly to standard PCR and qPCR. We observed the expected 812-bp product by standard PCR only in dilutions of 100 to 10−6 (Fig. 3, lanes 15, 13, 12, 11, 10, 9, and 8, respectively), albeit very weakly in the 10−6 dilution (lane 8). This corresponds to a lower detection limit of 10 CFU/PCR loaded on the gel. In contrast, qPCR, like NA-SERS, detected M. gallisepticum in all dilutions, and a linear correlation was observed through the 10−8 dilution (Table 5).
Fig 2.
PLS-DA output of serial dilutions of Rlow. Samples were classified as positive or negative for M. gallisepticum, with the horizontal dotted line indicating the threshold for a 50% probability that the samples are being misclassified. The initial concentration was 4.7 × 105 CFU/μl (gray asterisks). Solid inverted triangles, methanol control. The remaining symbols denote increasing serial 10-fold dilutions (100 to 10−10) from left to right. Results are shown for the samples (n = 183), with 3 latent variables capturing 26% of the x variance and 81% of the y variance. Cross-validated sensitivity, 98%; cross-validated specificity, 94%.
Fig 3.
PCR results for M. gallisepticum Rlow serial dilutions. Lanes 1 and 14, DNA ladder; lane 2, negative control for PCR (no template); lane 3, negative control for DNA extraction (no template); lanes 4 to 13 and 15, dilutions of 10−10 to 100, respectively.
Table 5.
Summary of qPCR analysis of M. gallisepticum dilution series
| Dilution | Avg crossing threshold valuea |
|---|---|
| 100 | 11.18 ± 0.14 |
| 10−1 | 14.72 ± 0.16 |
| 10−2 | 17.44 ± 0.25 |
| 10−3 | 20.88 ± 0.08 |
| 10−4 | 24.85 ± 0.04 |
| 10−5 | 27.00 ± 0.10 |
| 10−6 | 30.47 ± 0.38 |
| 10−7 | 31.56 ± 0.32 |
| 10−8 | 32.33 ± 0.17 |
| 10−9 | 31.96 ± 0.12 |
| 10−10 | 31.67 ± 0.01 |
Regression analysis of the starting concentrations versus the threshold cycle gave an R2 value of 99% for the first eight dilutions (100 to 10−8). The 10−9 and 10−10 dilutions were recognized by the qPCR (mglp) primers but did not exhibit a linear relationship between threshold number and concentration.
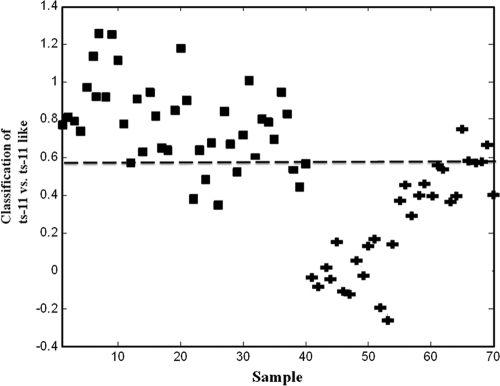
To determine sensitivity and specificity in a biochemically complex clinical background, we analyzed by NA-SERS choanal samples collected as part of a vaccine challenge study involving ts-11. Samples from birds infected with Rlow and strain K6216D (ts-11-like field isolate) were classified as M. gallisepticum positive based on serology and standard PCR, while samples from birds infected with ts-11 or uninfected controls were classified as M. gallisepticum negative, also based on serology and standard PCR. PLS-DA accurately distinguished samples from the positive field isolate and negative background/vaccine strain classes with 88% CV sensitivity and 85% CV specificity (data not shown). Whereas qPCR results could not distinguish the ts-11 vaccine strain from the ts-11-like field isolate K6216D (8), NA-SERS discriminated the two strains with 90% sensitivity and 82% specificity with a 14% error rate (Fig. 4).
Fig 4.
PLS-DA output from the analysis of choanal swabs from birds infected with the ts-11 vaccine strain (squares) or the ts-11-like field isolate K6216D (plus signs). Results are shown for the samples (n = 70) (samples 1 to 40 from ts-11-infected birds and samples 41 to 70 from K6216D-infected birds) and 2 latent variables, capturing 19% of the x variance and 79% of the y variance. CV sensitivity, 90%; CV specificity, 82%; CV error rate, 14%; root mean squared error in cross validation (RMSECV), 34%.
DISCUSSION
We demonstrated the capacity of NA-SERS to detect and differentiate avian mycoplasma species by multivariate chemometric analysis and the potential for diagnostic application in poultry. M. gallisepticum was clearly distinguished from three common avian flora mycoplasma species which can interfere with serologic detection. The capacity of NA-SERS to differentiate the three vaccine strains from virulent M. gallisepticum strains could have practical epidemiological applications where multiple vaccination protocols to contain M. gallisepticum in layer flocks are utilized, and thus more than one vaccine strain may be present (9). When the three vaccine strains were classified individually in a separate model, the 6/85 and ts-11were accurately discriminated (CV error rate < 0.05), while the F strain had an 11% CV error rate, with an overall sensitivity of 86% and an overall specificity of 90% (data not shown). As our protocol is further refined by modification in sample handling and chemometric analysis to reduce nonrelevant noise, this CV error rate is expected to decrease.
Our initial focus was mycoplasma detection with minimal sample processing, and we have only begun to explore what structural features define the mycoplasma spectral fingerprint. The vibrational energies from specific molecules have previously been characterized based upon the relative movements of atoms within the molecule (16, 22). The spectral patterns exhibit strong protein components, evidenced by vibrational bands at 1,602 cm−1 (phenylalanine), 1,440 to 60 cm−1 (C-H2 deformation), 1,372 cm−1 (C-H bend protein), 1,261 cm−1 (amide III), 1,001 cm−1 (phenylalanine), 1,132 cm−1 (C-O-C protein stretch), 960 cm−1 (C-N stretch), 876 cm−1 (tryptophan), and 829 cm−1 (exposed tyrosine) (26, 31). Obvious targets are surface-exposed mycoplasma adhesin proteins, which are likely to be in close proximity to the NA substrate. M. gallisepticum proteins GapA (MGA_0934), PlpA (MGA_1199), and CrmA (MGA_0928) are logical starting points for addressing this question, as studies comparing Rlow and Rhigh protein profiles demonstrate a correlation between their presence and virulence (24). Preliminary studies with M. pneumoniae strains with altered derivatives of the P30 adhesin protein suggest that this strategy will be informative (unpublished).
PLS-DA multivariate analysis failed to discriminate M. synoviae from M. gallisepticum at a high confidence interval when these species were evaluated with several other avian species collectively and attempts were made to classify each individually. Perhaps significantly, both M. gallisepticum and M. synoviae express members of the VlhA family of lipoproteins on their surfaces (23, 25), where they are likely positioned in close proximity to the silver of the NA and might be expected to contribute significantly to the spectral signature of each. We speculate that shared features of the VlhA proteins may contribute to the reduced capacity to distinguish these species when considered individually among a large group of mycoplasmas. It is noteworthy that PLS-DA with the same data set successfully distinguished M. synoviae from all other species when the number of classes was reduced (i.e., samples were classified as either positive or negative for M. synoviae). Thus, a stepwise classification approach might be more effective than modeling multiple classes at once when NA-SERS and PLS-DA are used to distinguish more than two species. It might be possible to use support vector algorithms to automatically perform this sequential pattern recognition bifurcation (one versus all) to define the class margins in a manner very different from that of PLS-DA.
Since NA-SERS detected the mycoplasma spectral signature in samples below the CFU level, it will be important to further define a detection threshold for diagnostics. Like PCR assays, our platform does not require intact organisms, and thus debris from lysed cells likely contributes to the spectral signature and the ability to detect mycoplasmas with high sensitivity. Our ability to detect femtomolar-level protein in the highest dilution is consistent with values reported previously for NA-SERS (13). Future studies will require that we correlate limits of detection with genome number, for example, as an alternative metric for comparison with competing techniques.
Results from analysis of choanal cleft samples were promising and establish the potential for NA-SERS detection of avian mycoplasmas in biochemically complex backgrounds, as was observed recently for M. pneumoniae in human throat swab samples (15). Handheld Raman spectral equipment is commercially available, raising the possibility of on-site testing and a web-based interface, e.g., for chemometric analysis, perhaps coupled with existing testing strategies for immediate confirmation and more-complete screening of flocks. Challenges that remain include independently validating models for general application in field settings. For example, how many and which field strains will adequately represent population diversity in a rigorous model without introducing nonrelevant information to the spectral data? How might sample handling be modified to optimize performance and streamline testing? Examination of strain and species performance by PCR assays reveals a range of sensitivities and cross-reactivities (12). When protocols are executed on extracted DNA from laboratory strains, the sensitivity is superior compared to that obtained using isolation from clinical swabs. The time of swab collection affects the performance of the PCR assay. Longer incubation times allow organism numbers to increase and improve the limits of detection. NA-SERS is similarly affected by the clinical background, although not for the same reason, as the signal from low numbers of organisms against a clinical background matrix is more difficult to extract for classification. Finally, while the vaccine strains can all be detected by PCR-based methods using various primer sets, detection of the 6/85 vaccine strain, which has reduced colonization and transmissibility, is difficult. In this respect, NA-SERS outperformed the alternative, as demonstrated by our detection of 6/85 with a CV error of <0.05. Furthermore, the ability to discriminate the ts-11 vaccine strains from a ts-11-like field isolate or potential virulent revertants of ts-11 is important on farms employing that vaccine to protect their flocks, and our SERS data demonstrate superior performance compared to other molecular strategies at this time (8). Finally, we anticipate the applicability of this platform to the detection of other mycoplasmas, including species such as Mycoplasma iowae and Mycoplasma meleagridis, which are pathogens in turkeys.
ACKNOWLEDGMENTS
We thank Vivien Hsiao Yun Chu, Justin Abell, and Yu Zhu for providing substrates for this work and Victoria Laibinis for her expertise in handling avian mycoplasmas.
This work was supported by funds from the University of Georgia Veterinary Medical Experiment Station.
Footnotes
Published ahead of print 30 December 2011
REFERENCES
- 1. Abd-el-Motelib TY, Kleven SH. 1993. A comparative study of Mycoplasma gallisepticum vaccines in young chickens. Avian Dis. 37:981–987 [PubMed] [Google Scholar]
- 2. Abell JL, Driskell JD, Dluhy RA, Tripp RA, Zhao Y-P. 2009. Fabrication and characterization of a multiwell array SERS chip with biological applications. Biosens. Bioelectron. 24:3663–3670 [DOI] [PubMed] [Google Scholar]
- 3. Barker M, Rayens W. 2003. Partial least squares for discrimination. J. Chemom. 17:166–173 [Google Scholar]
- 4. Bradbury JM. 1998. Recovery of mycoplasmas from birds, p 45–52 In Miles R, Nicholas R. (ed), Mycoplasma protocols, vol 104 Humana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 5. Callison SA, et al. 2006. Development and validation of a real-time Taqman polymerase chain reaction assay for the detection of Mycoplasma gallisepticum in naturally infected birds. Avian Dis. 50:537–544 [DOI] [PubMed] [Google Scholar]
- 6. Chaney SB, Shanmukh S, Dluhy RA, Zhao Y-P. 2005. Aligned silver nanorod arrays produce high sensitivity surface-enhanced Raman spectroscopy substrates. Appl. Phys. Lett. 87:31908–31910 [Google Scholar]
- 7. Chu H, Huang Y, Zhao Y-P. 2008. Silver nanorod arrays as a surface-enhanced Raman scattering substrate for foodborne pathogenic acteria detection. Appl. Spectrosc. 62:922–931 [DOI] [PubMed] [Google Scholar]
- 8. El Gazzar M, Laibinis VA, Ferguson-Noel N. 2011. Characterization of a ts-11-like Mycoplasma gallisepticum isolate from commercial broiler chickens. Avian Dis. 55:569–574 [DOI] [PubMed] [Google Scholar]
- 9. Evans JD, et al. 2005. Mycoplasma gallisepticum: current and developing means to control the avian pathogen. J. Appl. Poult. Res. 14:757–763 [Google Scholar]
- 10. Evans RD, Hafez YS. 1992. Evaluation of a Mycoplasma gallisepticum strain exhibiting reduced virulence for prevention and control of poultry mycoplasmosis. Avian Dis. 36:197–201 [PubMed] [Google Scholar]
- 11. Frey MC, Hanson RP, Anderson DP. 1968. A medium for the isolation of avian mycoplasmas. Am. J. Vet. Res. 29:2163–2171 [PubMed] [Google Scholar]
- 12. García M, Ikuta N, Levisohn S, Kleven SH. 2005. Evaluation and comparison of various PCR methods for detection of Mycoplasma gallisepticum infection in chickens. Avian Dis. 49:125–132 [DOI] [PubMed] [Google Scholar]
- 13. Grubisha DS, Lipert RJ, Park H-Y, Driskell J, Porter MD. 2003. Femtomolar detection of prostate-specific antigen: an immunoassay based on surface-enhanced Raman scattering and immunogold labels. Anal. Chem. 75:5936–5943 [DOI] [PubMed] [Google Scholar]
- 14. Harz M, Rösch P, Popp J. 2009. Vibrational spectroscopy: a powerful tool for the rapid identification of microbial cells at the single-cell level. Cytometry A 75:104–113 [DOI] [PubMed] [Google Scholar]
- 15. Hennigan SL, et al. 2010. Detection of Mycoplasma pneumoniae in simulated and true clinical throat swab specimens by nanorod array-surface-enhanced Raman spectroscopy. PLoS One 5:e13633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jarvis RM, Goodacre R. 2004. Discrimination of bacteria using surface-enhanced Raman spectroscopy. Anal. Chem. 76:40–47 [DOI] [PubMed] [Google Scholar]
- 17. Jungherr EL, Lunginbuhl RE, Tourtelotte M, Burr WE. 1955. Significance of serological testing for chronic respiratory disease, p 315–321 In Proceedings of the 91st Meeting of the American Veterinary Medical Association, Minneapolis, MN [Google Scholar]
- 18. Kleven SH. 2008. Control of avian mycoplasma infections in commercial poultry. Avian Dis. 52:367–374 [DOI] [PubMed] [Google Scholar]
- 19. Levisohn S, Kleven SH. 2000. Avian mycoplasmosis (Mycoplasma gallisepticum). Rev. Sci. Tech. 19:425–442 [PubMed] [Google Scholar]
- 20. Liu Y-J, Zhang Z-Y, Zhao Q, Dluhy RA, Zhao Y-P. 2009. Surface enhanced Raman scattering from an Ag nanorod array substrate: the site dependent enhancement and layer absorbance effect. J. Phys. Chem. C 113:9664–9669 [Google Scholar]
- 21. Luginbuhl RE, Tourtellotte ME, Frazier MN. 1967. Mycoplasma gallisepticum—control by immunization. Ann. N. Y. Acad. Sci. 143:234–238 [DOI] [PubMed] [Google Scholar]
- 22. Maquelin K, et al. 2002. Identification of medically relevant microorganisms by vibrational spectroscopy. J. Microbiol. Methods 51:255–271 [DOI] [PubMed] [Google Scholar]
- 23. Mardassi B, Mohammed R, Gueriri I, Boughattas S, Mlik B. 2005. Duplex PCR to differentiate between Mycoplasma synoviae and Mycoplasma gallisepticum on the basis of conserved species-specific sequences of their hemagglutinin genes. J. Clin. Microbiol. 43:948–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. May M, Papazisi L, Gorton TT, Geary SJ. 2006. Identification of fibronectin-binding proteins in Mycoplasma gallisepticum strain R. Infect. Immun. 74:1777–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Noormohammadi AH. 2007. Role of phenotypic diversity in pathogenesis of avian mycoplasmosis. Avian Pathol. 36:439–444 [DOI] [PubMed] [Google Scholar]
- 26. Premasiri WR, et al. 2005. Characterization of the surface enhanced Raman scattering (SERS) of bacteria. J. Phys. Chem. B 109:312–320 [DOI] [PubMed] [Google Scholar]
- 27. Premasiri WR, Gebregziabher Y, Ziegler LD. 2011. On the difference between surface-enhanced Raman scattering (SERS) spectra of cell growth media and whole bacterial cells. Appl. Spectrosc. 65:493–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Raviv Z, et al. 2007. The Mycoplasma gallisepticum 16S-23S rRNA intergenic spacer region sequence as a novel tool for epizootiological studies. Avian Dis. 51:555–560 [DOI] [PubMed] [Google Scholar]
- 29. Rodriguez R, Kleven SH. 1980. Evaluation of a vaccine against Mycoplasma gallisepticum in commercial broilers. Avian Dis. 24:879–889 [PubMed] [Google Scholar]
- 30. Stemke GW, Robertson JA. 1982. Comparison of two methods for enumeration of mycoplasmas. J. Clin. Microbiol. 16:959–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stewart S, Fredericks PM. 1999. Surface-enhanced Raman spectroscopy of amino acids adsorbed on an electrochemically prepared silver surface. Spectrochim. Acta A Biomol. Spectrosc. 55:1641–1660 [Google Scholar]
- 32. Whithear KG. 1996. Control of avian mycoplasmoses by vaccination. Rev. Sci. Tech. 15:1527–1553 [DOI] [PubMed] [Google Scholar]
- 33. Yoder HW., Jr 1986. A historical account of the diagnosis and characterization of strains of Mycoplasma gallisepticum of low virulence. Avian Dis. 30:510–518 [PubMed] [Google Scholar]
- 34. Zander DV. 1961. Origin of S6 strain Mycoplasma. Avian Dis. 5:154–156 [Google Scholar]
- 35. Zhao Y-P, Chaney SB, Shanmukh S, Dluhy RA. 2006. Polarized surface enhanced Raman and absorbance spectra of aligned silver nanorod arrays. J. Phys. Chem. B 110:3153–3157 [DOI] [PubMed] [Google Scholar]