Abstract
Biogenic amines are low-molecular-weight organic bases whose presence in food can result in health problems. The biosynthesis of biogenic amines in fermented foods mostly proceeds through amino acid decarboxylation carried out by lactic acid bacteria (LAB), but not all systems leading to biogenic amine production by LAB have been thoroughly characterized. Here, putative ornithine decarboxylation pathways consisting of a putative ornithine decarboxylase and an amino acid transporter were identified in LAB by strain collection screening and database searches. The decarboxylases were produced in heterologous hosts and purified and characterized in vitro, whereas transporters were heterologously expressed in Lactococcus lactis and functionally characterized in vivo. Amino acid decarboxylation by whole cells of the original hosts was determined as well. We concluded that two distinct types of ornithine decarboxylation systems exist in LAB. One is composed of an ornithine decarboxylase coupled to an ornithine/putrescine transmembrane exchanger. Their combined activities results in the extracellular release of putrescine. This typical amino acid decarboxylation system is present in only a few LAB strains and may contribute to metabolic energy production and/or pH homeostasis. The second system is widespread among LAB. It is composed of a decarboxylase active on ornithine and l-2,4-diaminobutyric acid (DABA) and a transporter that mediates unidirectional transport of ornithine into the cytoplasm. Diamines that result from this second system are retained within the cytosol.
INTRODUCTION
Amino acid decarboxylation systems contribute to the adaptation of lactic acid bacteria (LAB) to various environments, such as fruits, vegetables, and animals, where LAB occur in the oral and genital cavities and in the digestive tract. LAB are also present in thousands of traditional fermented foods, where they contribute to the transformation of raw vegetables, milk, or meat into elaborated foods. The presence of LAB carrying amino acid decarboxylation systems may lead to the accumulation of the decarboxylation products, commonly known as biogenic amines. These compounds are occasionally regarded as beneficial, as is the case for γ-amino-butyric acid (42), but more often their presence in food is undesired and results in severe health problems following ingestion (13, 23, 43).
Along with histamine and tyramine, putrescine (1,4-diaminobutane) is one of the most abundant biogenic amines in several fermented foods, including wine (16, 26, 29), cheese (30), cider (15, 24), sausage (44), and fish and meat products (20). Putrescine itself does not seem to possess a directly harmful biologic activity; instead, it enhances the toxic effects of histamine and tyramine (9, 18).
The biosynthesis of putrescine occurs through the agmatine deiminase (AgDI) or the ornithine decarboxylase (ODC) pathways. Quite interestingly, the prevalence of either pathway in food-borne LAB strains depends upon the environment. Among cider and cheese LAB strains, AgDI has a major role (24, 25), while in wine, putrescine production mostly proceeds through ODC (36).
The AgDI system of Lactobacillus brevis IOEB 9809 was already thoroughly characterized in a previous publication (31), while little information is available about the ODC systems of LAB. ODC systems were characterized in detail for Gram-negative Enterobacteriaceae (34, 35). Similar to other bacterial amino acid decarboxylation systems, the Enterobacteriaceae systems consist of a decarboxylase and a precursor/product transmembrane exchanger (2, 39). Their combined actions results in amino acid intake, decarboxylation, and release of the corresponding amine. The pathway results in alkalinization of the cytosol and generation of a proton motive force, which can be exploited for acid stress resistance and/or the production of metabolic energy in the form of ATP. Decarboxylase and transporter genes are generally organized in clusters located on the bacterial chromosome or on plasmids. Among LAB, only two ornithine decarboxylases have been purified and characterized to date. ODC from Lactobacillus sp. 30a has been thoroughly characterized (17). A putative ornithine decarboxylation system was reported for strain Oenococcus oeni RM83 (32), where the corresponding genes were likely acquired by horizontal transfer (33). The same authors also reported the existence of a second group of genes in some Lactobacillus species that form a phylogenetically distinct group among ODCs. Biochemical proof about the functionality of these enzymes was recently provided only for the ODC of O. oeni (5).
The aim of this work was to gain insight into putrescine biosynthesis in fermented foods, a so-far poorly understood process, especially with respect to the ODC pathway. PCR screening and database searches for ODC homologues revealed several putative decarboxylase and transporter genes. Representative decarboxylases and transporters from diverse LAB were characterized, and decarboxylation studies using whole cells were performed. The overall results revealed an unexpected diversity within the ornithine decarboxylation systems of LAB.
MATERIALS AND METHODS
Bacterial strains and cultures.
The LAB strains employed in this study were either purchased from culture collections or originated from the collection of the Institut d'Œnologie de Bordeaux (IOEB; Bordeaux, France). A complete list of the strains employed is presented in Table S1 of the supplemental material. Strains were cultured in half-strength Man, Rogosa, and Sharpe medium (Beckton Dickinson, Sparks, MD) at pH 5.0. Growth temperatures ranged from 25°C to 37°C.
Determination of gene sequences.
PCR products (size, ∼900 bp) corresponding to an internal segment of putative ODC genes were obtained for some strains. Reaction conditions were described in a previous report (11). In strains Oenococcus oeni IOEB 89006 and Lactobacillus brevis IOEB 9906, the sequences of neighboring genomic regions were obtained by means of conventional molecular biology techniques. Briefly, the genomic DNA of the two strains was digested and submitted to enzymatic ligation. The ligation mixtures were then purified and employed as matrices for reverse PCRs. PCR products were finally sequenced. The sequence obtained with O. oeni consisted of a 3,750-bp product that contained putative ODC and ornithine/putrescine antiporter genes. These displayed 100% identity with ODC and PotE from O. oeni RM83 (CAG 34069 and CAM 07323, respectively). As for L. brevis, a 4,732-bp sequence was obtained.
Expression and purification of recombinant decarboxylases.
Decarboxylases were obtained as recombinant His-tagged fusion proteins within Escherichia coli. PCR products were submitted to ligase-independent directional cloning within pET100/D-TOPO vectors (Invitrogen, Carlsbad, CA). Chemically competent E. coli (BL21 Star One Shot; Invitrogen) was employed as the expression host. Cloning, transformation, and expression were performed following the protocols provided by the manufacturer. The products were verified by sequencing to ensure that all products were correctly inserted within the expression construct and that no mutations had intervened throughout the cloning procedure. Cell-free enzymatic extracts were submitted to purification by means of affinity chromatography. Briefly, cell-free enzymatic extracts were submitted to purification by means of affinity chromatography on a BioLogic DuoFlow chromatographic system (Bio-Rad, Marnes-la-Coquette, France) equipped with a HiTrap chelating HP column (Amersham Biosciences, Uppsala, Sweden). Elution was performed by means of a pH 7.5 potassium phosphate buffer supplemented with increasing amounts of imidazole. SDS-PAGE analysis allowed us to isolate fractions containing the pure recombinant enzymes.
Decarboxylation assays of purified enzymes.
Enzymatic reactions were performed on 200-μl volumes. Four micrograms of enzyme and 0.4 mM pyridoxal-phosphate (PLP) were employed. Reaction mixtures were incubated for up to 1 h at 37°C. Reaction velocities were determined by sampling at regular intervals (typically 10, 30, and 60 min). Different substrate concentrations (DABA, lysine, or ornithine at 0.5 to 100 mM) were employed for the calculation of kinetic parameters. pH activity profiles were established at 10 mM substrate concentrations. Enzymatic activity was stopped by addition of 10 μl trichloroacetic acid (20%, vol/vol), and samples were stored at −20°C until analysis. Kinetic parameters were calculated with the aid of the Prism 5.04 program (Graphpad Software, La Jolla, CA). Assays were performed in 0.05 M buffers. Depending upon the pH of the reaction mixture, different buffers were prepared: citrate (3.0 to 6.0), phosphate (6.0 to 8.0), and borax (8.0 to 10.0). Changes in buffer composition had no influence on the enzymatic activity. For modeling pH dependence of enzymatic activity, a classical diprotic model was employed (37), and pH activity profiles were established with the aid of GraFit 7.0 (Erithacus Software, Horley, United Kingdom).
Decarboxylation assays on whole cells.
Strains were cultured until mid-exponential phase (0.5 to 1.5 OD units ml−1, depending upon the strain). Cells were harvested, washed twice with potassium phosphate buffer (0.1 M, pH 5.0), and finally resuspended at 1.0 OD units ml−1 in the same buffer containing either ornithine, lysine, or diaminobutyric acid at a concentration of 10 mM. Cell suspensions were incubated at 25°C (O. oeni), 30°C (L. brevis), or 37°C (L. gasseri and L. casei). Suspensions were finally centrifuged (10 min at 10,000 × g), and supernatants were directly analyzed. Pellets were washed two times with buffer and resuspended in saturated NaHCO3 solution at a final concentration of 10 OD units ml−1. Preliminary trials indicated that supplementary extraction treatments of the biomass (e.g., freeze-thaw cycles or incubation with lysozyme) did not significantly improve extraction. Derivatization and thin-layer chromatography (TLC) analysis were then performed as described below.
TLC.
An analytical method was expressly developed with the aim of detecting and quantifying all decarboxylation products. This was based upon sample derivatization with fluorescent dansyl chloride, separation by TLC, and quantification by densitometric analysis.
Fifty-microliter aliquots of each sample were supplemented with 50 μl of a saturated NaHCO3 solution and 100 μl dansyl chloride solution (5 mg ml−1 in acetone). Reactions mixtures were incubated at 55°C for 1 h to allow for the formation of fluorescent adducts. Ten-microliter aliquots of the reaction mixtures were deposed on TLC silica 60 plates (Merck, Darmstadt, Germany). Plates were developed for 7 cm in different eluent mixtures, depending upon the products analyzed. Chloroform-triethylamine (4/1) and chloroform-triethylamine-methanol (15/4/1) were used for diamine and agmatine determinations, respectively. TLC plates were illuminated at a wavelength of 312 nm, and images were captured by means of an Infinity 1000 imaging system (Vilber-Lourmat, Marne-la-Vallée, France). The images were analyzed with the aid of BIO-1D software (Vilber-Lourmat). All reagents and standards for TLC analysis were obtained from Sigma-Aldrich (Saint-Quentin-Fallavier, France). The method allowed for the quantification of amine concentrations as low as 5 μM.
Cloning and expression of transporter genes.
The gene encoding the PotE homologue of O. oeni IOEB 89006 was amplified from chromosomal DNA by PCR with primers potE-fw (5′-GCGAAACCATGGCTCAAGAGAAAAAGAAAATGGGCGT) and potE-rv (5′-GCGAAATCTAGATGTCATATCAGTTGGTAAAACAGC), introducing NcoI and XbaI restriction sites. Phusion high-fidelity DNA polymerase (Thermo Scientific) was used for amplification, following a procedure provided by the manufacturer. The PCR product was, after digestion with NcoI and XbaI (Fermentas), ligated (T4 DNA ligase; Fermentas) into the NICE system expression vector pNZ8048 (12), which was digested with the same enzymes. The ligation mixture was transformed into Lactococcus lactis NZ9000. Chloramphenicol-resistant clones were tested to confirm they harbored the correct construct, which was designated pNZpotE. The putative amino acid transporter gene of L. gasseri ATCC 33323 was cloned in two steps. The first 1,416 bp was amplified using primers lg1880-fw (5′-GCGAAACCATGGATAAACCAGATGTCTTAAATGATTTAGAACCG) and lg1800-rvm (5′-GCGAAACTGCAGAAAACGAATCCTAATCCAAAC) and cloned into pNZ8048, using NcoI and PstI restriction sites. In a second step, the complete gene was amplified using primers lg1880-fw and lg1800-rv (5′-GCGAAACTGCAGGATTTTATTGCTACAAAAGATTGG) followed by restriction with AatII (endogenous restriction site) and PstI. The fragment that included the 200-bp 3′ end of the gene was cloned into the first construct by replacement of the AatII-PstI fragment, resulting in pNZ-lg1880. For gene expression, L. lactis cells harboring the constructs were grown to mid-exponential phase (OD600, ∼0.6) in M17 supplemented with 0.5% (wt/vol) glucose, induced with 5 mg/liter of nisin, and left to grow for one more hour.
Transport assays.
L. lactis cells expressing the transporter genes and control cells harboring the empty vector pNZ8048 were washed and resuspended in 100 mM potassium phosphate buffer, pH 6, to an OD600 of 2.0. Glucose was added to 0.2% (wt/vol), and cells were kept on ice until the transport assays. Samples of 100 μl were incubated for 5 min at 30°C with constant stirring. At time zero, [14C]ornithine (PerkinElmer, Waltham, MA) or l-[14C]putrescine (Amersham, Diegem, Belgium) was added to a final concentration of 17.5 μM or 4.7 μM, respectively. In the exchange experiments, unlabeled putrescine, ornithine, or l-lysine was added to the final concentrations indicated below. In competition experiments to determine Km and Ki values, cells were incubated with 17.5 μM 14C-labeled ornithine and unlabeled substrate at concentrations ranging from 0 to 1 mM (lysine and ornithine) or 0 to 10 mM (DABA). Uptake was stopped by addition of 2 ml of ice-cold 0.1 M LiCl solution, followed by filtration through a 0.45-μM-pore-size nitrocellulose filter (BA 85; Whatman GmbH, Dassel, Germany). The filter was washed once with 2 ml of 0.1 M LiCl and submerged in Emulsifier Scintillator Plus scintillation fluid (Packard Bioscience, Meriden, CT), and radioactivity was measured in a Tri-Carb 2000CA liquid scintillation analyzer (Packard Instruments).
Nucleotide sequence accession number.
The L. brevis sequence contained a putative ODC gene and an ornithine/putrescine antiporter gene and was deposited in GenBank under accession number JN120479.
RESULTS
Collection screening and database searches disclosed several uncharacterized putative ODC systems among LAB.
In previous work (11), a collection of 275 cider and wine LAB strains was screened for the presence of genes coding for ODCs by using primers based upon the gene encoding the well-characterized ODC of Lactobacillus sp. 30a (17). The same approach was adopted for the screening of 40 more LAB strains originating from diverse food-related environments (see Table S1 in the supplemental material). Overall, four strains scored positive: the Lactobacillus sp. 30a control strain, two O. oeni strains (namely, IOEB 89006 and IOEB 9915), and Lactobacillus brevis strain IOEB 9906.
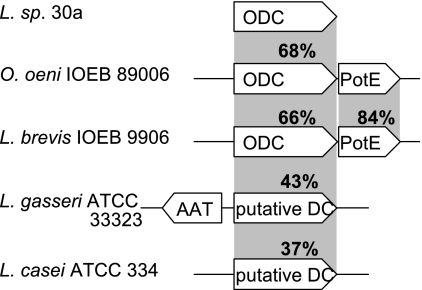
The ODC from Lactobacillus sp. 30a was previously characterized (17). The other ODC-positive strains, namely, O. oeni IOEB 89006, O. oeni IOEB 9915, and Lactobacillus brevis IOEB 9906, were investigated in further detail. The complete gene sequences of the putative ODCs and the adjacent amino acid transporters that supposedly are associated with these ODCs (ornithine/putrescine exchangers PotE) were determined. The sequences of the putative ODCs and transporters from the two O. oeni strains were identical, so the study of strain IOEB 9915 was abandoned. The O. oeni and L. brevis ODCs shared 68% and 66% amino acid sequence identities with the Lactobacillus sp. 30a enzyme, respectively. The two transporter proteins shared 84% sequence identity (Fig. 1).
Fig 1.
Schematic representation of different decarboxylation systems. Percent identities with amino acid sequences of decarboxylase from Lactobacillus sp. 30a and the transporter from O. oeni are reported. PotE, ornithine/putrescine antiporter; AAT, putative amino acid transporter.
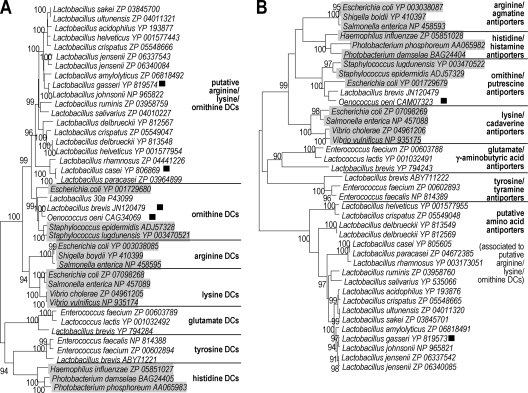
Database searches using BlastP (1) and the O. oeni, L. brevis, and Lactobacillus sp. 30a ODCs as queries revealed no closely related homologues in LAB but disclosed a large family of more distantly related proteins annotated as putative arginine/lysine/ornithine decarboxylases exclusively found in Lactobacillus species. These enzymes shared up to 43% amino acid sequence identity with the Lactobacillus sp. 30a ODC (Fig. 1). To better understand their phylogenetic relationships, all protein sequences were compared in a neighbor-joining tree with the aid of MEGA4 software (47). The tree contained representative amino acid decarboxylases of the aspartate aminotransferase family, including all known substrate specificities (Fig. 2A). Decarboxylases indentified in LAB or closely related species included ornithine decarboxylases, glutamate decarboxylases, and tyrosine decarboxylases, as well as putative arginine/lysine/ornithine decarboxylases. Decarboxylases not described in LAB to date include lysine decarboxylases, arginine decarboxylases, and histidine decarboxylases, which are frequently observed in Gram-negative bacteria. Histidine decarboxylation is a trait of LAB as well, but the decarboxylase is from a different family, the pyruvoyl-dependent decarboxylases (14). Phylogenetic groups accurately reflected the different substrate specificities, as already reported (7). All basic amino acid decarboxylases (i.e., arginine, lysine, and ornithine decarboxylases) form separate subgroups. ODCs of the LAB Lactobacillus sp. 30a, O. oeni, and L. brevis are in the same cluster as ODCs found in Gram-negative bacteria. The putative arginine/lysine/ornithine decarboxylases form a separate cluster that is closely related but distinct from the ODC cluster. Interestingly, in most species the putative decarboxylase genes were adjacent to a putative amino acid transporter gene, even though gene disposition differed from what was observed for O. oeni and L. brevis (Fig. 1). The neighbor-joining tree constructed on the basis of the alignment of the transporter amino acid sequences showed the same clusters, which is in line with the functional relation between the decarboxylase and corresponding transporter in one species (Fig. 2B). The most striking difference between the two trees was the phylogenetic relation of the putative arginine/lysine/ornithine decarboxylases and corresponding transporters. While the decarboxylases were close to the ODCs of Gram-positive and Gram-negative species, the transporters seemed to form a separate clade, distant from the ornithine/putrescine exchangers PotE. According to the transporter classification system (41), both transporter types are members of the APC superfamily of transporters but belong to distinct families (19). The putative amino acid transporters from Lactobacillus spp. associated with the putative l-arginine/ l-lysine/ l-ornithine decarboxylases are closest to the GGA (glutamate:GABA antiporter; TC 2.A.3.7) family, whereas PotE homologues are members of the APA (basic amino acid/polyamine antiporter; TC 2.A.3.2) family. The difference between the decarboxylase/transporter pairs is emphasized by the organization on the chromosome. ODCs and PotE are organized in an operon in which the potE gene is downstream of the odc gene. In contrast, the amino acid transporter gene of L. gasseri is located upstream of the decarboxylase and the genes are transcribed divergently (Fig. 1).
Fig 2.
Neighbor-joining trees based upon the alignment of PLP-dependent amino acid decarboxylases (A) and amino acid transporters (B). Sequences were aligned with the aid of MEGA4 software (44) by employing ClustalW as the algorithm. Percent values from 1,000 bootstrap replicates are indicated; values less than 90% are not shown. Representative sequences are presented for each substrate specificity. Non-LAB sequences are shaded. The proteins that were characterized in the present study are marked with a black square.
Enzymes from O. oeni and L. brevis are typical ODCs, in contrast to L. gasseri and L. casei decarboxylases.
To better understand the role of the putative decarboxylation systems detected in LAB, the putative ODCs from L. brevis IOEB 9906 and O. oeni IOEB 89006 and the putative arginine/lysine/ornithine decarboxylases from Lactobacillus gasseri ATCC 33323 and Lactobacillus casei ATCC 334 were selected for further study as representatives of the two groups of decarboxylases. The four genes were cloned in Escherichia coli, in frame with an upstream sequence encoding a His tag. This allowed for the production of soluble recombinant proteins that were purified by affinity chromatography. For each protein, a pure product was obtained with the expected molecular mass, ranging from 79 to 87 kDa (verified by SDS-PAGE) (results not shown). Decarboxylation activities of the four recombinant proteins with the four basic amino acids ornithine, arginine, lysine, and DABA as substrates were measured by determining the respective decarboxylation products (i.e., putrescine, agmatine, cadaverine, and diaminopropane) using a method based upon dansylation and thin-layer chromatography (see Materials and Methods). The pH of the reaction medium was 5.5 for the enzymes from O. oeni and L. brevis and 7.5 for those from L. gasseri and L. casei. These pH values were chosen on the basis of preliminary trials that allowed us to establish an approximate range of optimal activity for each recombinant decarboxylase (results not shown). The substrates were added at various concentrations (0.5 to 100 mM), and initial decarboxylation rates were estimated.
Steady-state kinetic parameters were estimated whenever reliable initial rates could be determined (Table 1). The ODCs from O. oeni and L. brevis showed a clear preference for ornithine as the substrate. The affinities for ornithine were 0.7 and 1 mM, respectively, while the affinities for DABA and lysine were 5 to 10 times lower. However, the most prominent difference was observed in the maximal rates resulting in a catalytic efficiency, expressed as the kcat/Km ratio, of the enzyme from O. oeni that was 500-fold and 1,100-fold higher with ornithine than with lysine and DABA, respectively. The decarboxylase from L. brevis was somewhat less strict, with catalytic efficiencies for lysine and DABA decarboxylation that were 116- and 60-fold lower than for ornithine, respectively. The putative arginine/lysine/ornithine decarboxylases from L. gasseri and L. casei displayed a substrate preference for DABA. The affinities were in a range between 4 and 12 mM, similar to that observed for the two ODCs for this substrate. A higher maximal rate was observed for the L. gasseri enzyme with DABA as substrate, with a catalytic efficiency of 1.8 × 103 M−1 min−1 that, nevertheless, was 2 orders of magnitude lower than that observed for the ODCs with ornithine as the substrate. The L. casei enzyme appeared to have a somewhat stronger preference for DABA over ornithine than the L. gasseri enzyme. The rates of lysine decarboxylation by the L. gasseri enzyme and lysine and ornithine decarboxylation by the L. casei enzyme were too low to allow for an accurate estimation of kinetic parameters (less than 0.1% conversion with 20 to 80 mM substrate after 1 h). In such cases, the catalytic efficiency would be at least 10-fold lower than that obtained for the preferred substrate. In summary, ODCs are ornithine decarboxylases that show low activities with DABA and lysine as substrates. The enzymes annotated as arginine/lysine/ornithine decarboxylases have a lower specificity, decarboxylating ornithine and DABA, but most of all they have a much lower efficiency than the ODCs. Arginine decarboxylation was never observed.
Table 1.
Steady-state kinetic parameters and substrate specificities of the four decarboxylases
| Microorganism | Reaction pH | Ornithine |
DABA |
l-Lysine |
l-Arginine |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| kcat (min−1)a | Km (mM)a | kcat/Km (M−1 min−1) | kcat (min−1)a | Km (mM)a | kcat/Km (M−1 min−1) | kcat (min−1)a | Km (mM)a | kcat/Km (M−1 min−1) | kcat (min−1) | Km (mM) | ||
| O. oeni | 5.5 | 416 ± 18 | 0.7 ± 0.1 | 5.5 × 105 | 2.7 ± 0.2 | 4.9 ± 1.6 | 0.5 × 103 | 5.6 ± 0.6 | 5.0 ± 1.6 | 1.1 × 103 | NAb | NA |
| L. brevis | 5.5 | 290 ± 13 | 1.0 ± 0.1 | 2.8 × 105 | 27 ± 4 | 11.2 ± 3.6 | 2.4 × 103 | 34 ± 4 | 7.2 ± 2.9 | 4.6 × 103 | NA | NA |
| L. gasseri | 7.5 | 8.3 ± 0.5 | 7.2 ± 1.4 | 1.2 × 103 | 21 ± 2 | 12.1 ± 2.2 | 1.8 × 103 | NDc | ND | ND | NA | NA |
| L. casei | 7.5 | ND | ND | ND | 3 ± 0.1 | 3.9 ± 0.7 | 0.7 × 103 | ND | ND | ND | NA | NA |
Values are means and standard deviations of triplicates.
NA, no detectable activity.
ND, low activity did not allow estimation of kinetic parameters.
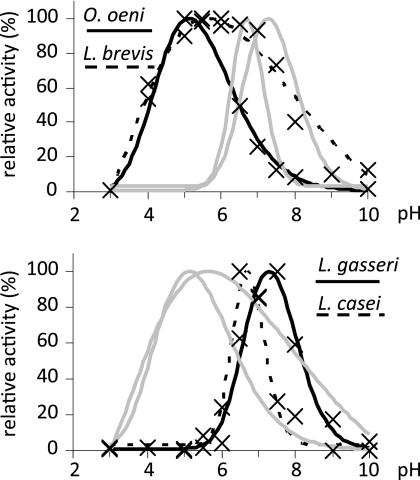
The purified enzymes from the two groups of decarboxylases showed different pH profiles. Figure 3 shows the profiles for the preferential substrates (i.e., ornithine for O. oeni and L. brevis and DABA for L. gasseri and L. casei). The pH activity optima of decarboxylases from O. oeni and L. brevis were significantly lower than those of L. gasseri and L. casei. The enzyme from L. brevis showed a broad profile with an optimum at pH 5.6, which is similar to that of the ODC of Lactobacillus sp. 30a reported before (17). The O. oeni decarboxylase reached its peak of activity at pH 5.1 and was significantly less active at pH values above 6.0. The pH profiles of the decarboxylases from L. casei and L. gasseri were relatively narrow and displayed activity optima at pH 6.7 and 7.3, respectively.
Fig 3.
pH activity profiles of decarboxylases from O. oeni/L. brevis (top panel) and L. gasseri/L. casei (bottom panel). In each graph the profiles of the enzymes from the opposite group are shown in shaded format. Each enzyme was tested against a 10 mM concentration of the preferential substrate. Experimental points represent mean results of triple replicates.
PotE of O. oeni catalyzes ornithine/putrescine exchange.
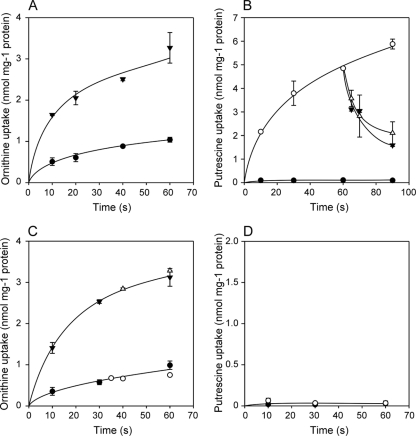
The gene encoding the PotE homologue of O. oeni, located downstream of the odc gene (Fig. 1), was expressed in L. lactis NZ9000 by using the NICE system (12). At a concentration of 17.5 μM, the initial rate of 14C-labeled l-ornithine uptake by L. lactis cells harboring the empty vector pNZ8048 was 3.0 nmol min−1 mg−1 (Fig. 4A). The background ornithine uptake in control cells is likely mediated by the arginine/ornithine exchangers ArcD1 and/or ArcD2 (6, 38) or other endogenous transporters of L. lactis. The uptake rate of ornithine increased to 9.6 nmol min−1 mg−1 in cells expressing potE, demonstrating functional expression of the transporter of O. oeni in L. lactis. Similar transport assays with [14C]putrescine as substrate showed an even more distinct effect. When present at 4.7 μM, putrescine was taken up at an initial rate of approximately 12 nmol min−1 mg−1 in cells expressing potE, whereas control cells did not show any significant uptake. Convincing evidence for the function of PotE as an ornithine/putrescine exchanger was obtained when an excess of unlabeled ornithine was added to cells that were allowed to take up putrescine for 1 min. A rapid release of putrescine from the cells was observed (Fig. 4B). Similarly, addition of an excess of unlabeled putrescine led to the release of labeled putrescine, showing the putrescine/putrescine exchange.
Fig 4.
Ornithine (A and C) and putrescine (B and D) uptake by resting cells of L. lactis NZ9000 expressing potE of O. oeni (▼ in panel A and ○ in panel B) or lgas_1880 of L. gasseri (▼ in panel C and ○ in panel D) or harboring the empty vector pNZ8048 (●). 14C-labeled ornithine and putrescine were added to 17.5 μM and 4.7 μM final concentrations, respectively. Unlabeled ornithine and putrescine at a concentration of 1 mM were added after 60 s to cells expressing potE (B; △ and ▼, respectively). Unlabeled putrescine at a concentration of 1 mM was added after 30 s to cells expressing lgas_1880 (△) and control cells harboring pNZ8048 (○) (C). Experimental points represent mean results of triple replicates.
The affinity of PotE of O. oeni for ornithine, putrescine, DABA, and lysine was determined from the inhibition of 14C-labeled ornithine uptake by different concentrations of the unlabeled substrates. The Km for ornithine was estimated at 50 to 75 μM; Ki values for lysine and DABA were estimated at less than 100 μM and between 1 and 10 mM, respectively (data not shown). Taken together, the results showed that PotE of O. oeni is an ornithine/putrescine exchanger, with affinity for lysine and cadaverine but not for DABA.
L. gasseri transporter catalyzes unidirectional ornithine transport.
The putative amino acid transporter gene located adjacent to, but divergently transcribed from, the putative arginine/lysine ornithine decarboxylase of L. gasseri was cloned and expressed in L. lactis NZ9000 as described above. A clear increase in the initial ornithine uptake rate from 2.4 nmol min−1 mg−1 in control cells to 8.4 nmol min−1 mg−1 in cells expressing the transporter was observed (Fig. 4C), demonstrating transport activity of the cloned gene product with ornithine as the substrate. In contrast, no 14C-labeled putrescine uptake could be detected (Fig. 4D), and addition of an excess of unlabeled putrescine to cells that were allowed to take up [14C]ornithine to a steady-state level did not result in efflux of accumulated ornithine (Fig. 4C). This showed that the encoded transporter is not an ornithine/putrescine exchanger but most likely a unidirectional ornithine transporter.
The affinity of the transporter for ornithine was determined by measuring the inhibition of 14C-labeled ornithine uptake at various concentrations of unlabeled ornithine. The Km was estimated to be 74 μM (data not shown).
Following the observation of DABA decarboxylation activity of the putative arginine/lysine/ornithine of L. gasseri, affinity of the transporter for DABA was tested by competition experiments as described above, using 14C-labeled ornithine uptake at a concentration of 17.5 μM and a range of concentrations of unlabeled DABA or lysine. A Ki was estimated at 12.4 mM, indicating very low affinity of the transporter for DABA. A higher affinity for lysine was observed: [14C]ornithine uptake was inhibited by unlabeled lysine, with an estimated Ki of 1.0 mM.
Decarboxylation products are released by O. oeni and L. brevis, but not by L. gasseri and L. casei.
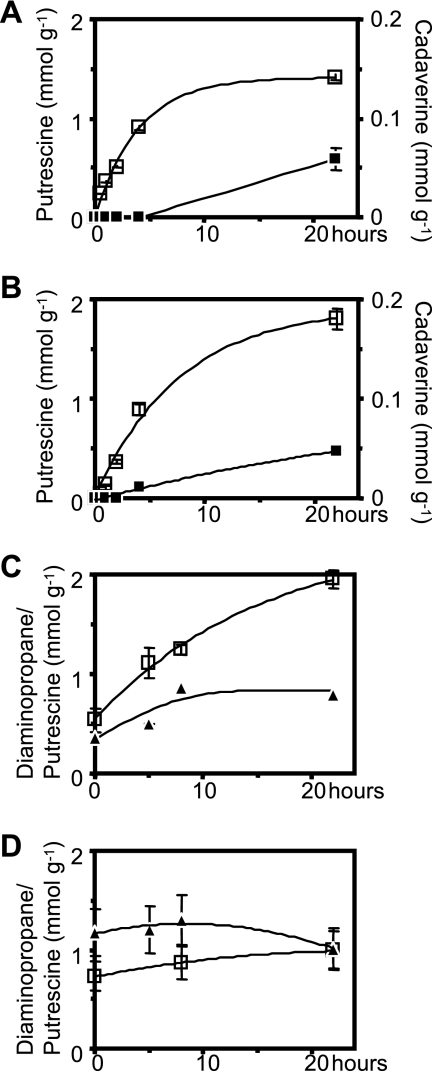
Amino acid decarboxylation activities were determined for resting cells of the four strains. Cell suspensions were incubated in the presence of 10 mM ornithine, lysine, or DABA. Samples were taken at regular intervals, and the decarboxylation products in culture supernatants and in the cytoplasm were determined separately. Analysis of supernatants revealed that O. oeni and L. brevis were able to release decarboxylation products into the medium (Fig. 5A and B). In agreement with the kinetic characteristics of the decarboxylases and transporters, the two species released at least 20-fold more putrescine than cadaverine, while diaminopropane was not detected. Analysis of the cell content revealed that in O. oeni and L. brevis relatively small amounts of putrescine (approximately 1,000-fold less than outside the cell) could be detected in the cytoplasm when ornithine was added.
Fig 5.
(A and B) Extracellular release of putrescine (□) and cadaverine (■) by O. oeni (A) and L. brevis (B). (C and D) Intracellular contents of putrescine (□) and diaminopropane (▲) in L. gasseri (C) and L. casei (D). Values are expressed in mmol or μmol per gram of wet cells. Experimental points represent mean results of triple replicates. Error bars represent standard errors.
In contrast to O. oeni and L. brevis, L. gasseri and L. casei did not release decarboxylation products into the culture medium. Relatively small amounts (0.3 to 1.2 μmol per g of wet cells) of putrescine and diaminopropane could be detected within the cytoplasm of both strains, even when no amino acid precursor was added to the medium. When 10 mM DABA was added to L. gasseri cell suspensions, intracellular concentrations of diaminopropane increased by 0.4 μmol g−1 and, most importantly, ornithine addition resulted into an almost-4-fold increase (from 0.5 to 1.9 μmol g−1) (Fig. 5C) in intracellular putrescine. In L. casei both putrescine and diaminopropane could be detected, but these levels did not vary significantly throughout the whole incubation time, regardless of the presence of candidate precursors (Fig. 5D). Cadaverine was not detected inside the cells in either the absence or presence of lysine.
DISCUSSION
The first evidence for LAB ornithine decarboxylases was obtained from strain Lactobacillus sp. 30a (40). A putative ODC system was later detected in O. oeni RM83 (32, 33). Finally, during the drafting of the manuscript, a partial functional characterization of a recombinant ODC from O. oeni was published (5). Overall, the knowledge of this subject remains sparsely distributed.
This work represents the first systematic survey on ornithine decarboxylation by LAB. Our experimentation disclosed two candidate ODC systems in strains O. oeni IOEB 89006 and L. brevis IOEB 9906, consisting of both a putative decarboxylase gene and a putative transporter gene. The genes found in O. oeni IOEB 89006 showed 100% identity to homologues described in O. oeni RM83 (32, 33). In the present work, thorough functional characterizations of the decarboxylase and the ornithine/putrescine transporter were compared with the results obtained with whole cells of the same strain. Like its orthologue from Lactobacillus sp. 30a, the enzyme accepted ornithine as the preferred substrate with relatively high specificity and reached its optimum activity at fairly acidic pH (5.1 compared to 5.8 for Lactobacillus sp. 30a). These results are substantially in agreement with those presented in the literature (5), but in addition to the preference for ornithine with respect to other substrates, they were accurately expressed in terms of ratios between the respective catalytic efficiencies. The transporter mainly catalyzed ornithine/putrescine exchange but also had affinity for lysine and its decarboxylation product cadaverine. The functionality of the ODC system was finally corroborated by the results with whole cells, which confirmed ornithine decarboxylation and putrescine release and again, to a lesser extent, lysine decarboxylation and cadaverine production. The ODC from L. brevis IOEB 9906 was also characterized. The overall results suggested that relatively strict specificity for ornithine and fairly low pH activity optima are common features of ODC systems from LAB. The product encoded by PotE from L. brevis was not characterized, but the high identity similarity with its homologue from O. oeni (84%) and the results obtained with whole cells (again showing putrescine and cadaverine release) leave little doubt as to whether this gene actually encodes an ornithine/putrescine antiporter.
This work also demonstrates the existence of a second, functionally distinct group of ornithine decarboxylase systems in LAB. The enzymes from L. gasseri and L. casei were proven to be active on DABA and ornithine. Compared to the ODCs from O. oeni and L. brevis, they showed relatively low catalytic efficiencies and relatively high pH activity optima. The decarboxylase from L. gasseri displayed a unique dual specificity for DABA and ornithine, with a slight preference for the former, a novel functionality which has not been observed for ornithine decarboxylases before. In contrast, dual specificity of amino acid decarboxylases for lysine and ornithine have been reported (27, 46). Further experimentation demonstrated that the putative amino acid transporter from L. gasseri functions as a unidirectional basic amino acid transporter, with ornithine as the preferred substrate. Surprisingly, its affinity for DABA was low. Whether this is physiologically relevant with respect to precursor supply for the decarboxylase, or whether other mechanisms for DABA supply are present, remains unclear. Nevertheless, the ornithine transport activity of the transporter, the ornithine decarboxylation activity of the decarboxylase, and the adjacent location of the respective genes strongly suggest that they are part of the same functional pathway. Experiments with whole cells showed that L. gasseri was able to accumulate putrescine within the cytoplasm when the medium was supplemented with ornithine. This supports the functionality of the decarboxylase/transporter system from L. gasseri. As far as L. casei is concerned, intracellular accumulation of diamines was not observed when either ornithine or DABA was supplied. This is not surprising, given the fact that in this strain the gene encoding the decarboxylase doesn't have a neighboring putative transporter gene. It is therefore possible that in L. casei amino acid decarboxylation is not supported by direct precursor supply from outside the cell; the reaction could instead be part of a more complex, multireaction pathway.
The existence of a second group of ornithine decarboxylases in LAB was first postulated by Marcobal et al. (32). To date, putative decarboxylases of this second group are considered to be typical ODCs, and the corresponding gene sequences are employed in the design of primers for the detection of putrescine producers (10). On the other hand, the reports concerning the functionality of these enzymes are rare and contradictory. Strain L. acidophilus NCFM has putative amino acid decarboxylase and transporter genes (accession numbers YP193877 and YP193876 in Fig. 2A and B, respectively) that are very close to their orthologues from L. gasseri ATCC 33323 in terms of sequence and disposition. Inactivation of the genes compromised the natural acid resistance of the strain (4). In the industrial cheese starter Lactobacillus helveticus CNRZ, a putative system of the same type was detected (accession numbers YP001577954 and YP001577955 on Fig. 2A and B, respectively), but the strain is not a putrescine producer (8). The data provided here suggest that these decarboxylases, due to their low catalytic efficiencies and the absence of a system for putrescine release, cannot account for the production of milligram amounts of putrescine.
Possible physiological functions of the newly discovered DABA/ornithine decarboxylase system may be inferred by analogy to similar systems described in Gram-negative bacteria. In E. coli, a typical ODC system coexists with a second ornithine decarboxylase, named biosynthetic (34). While the former is related to acid stress resistance, the role of the latter enzyme is to synthesize putrescine, which is retained within the cytoplasm (35). Similarly to the two types of LAB decarboxylases described in this work, the two types of enzyme display different pH optima (3). Intracellular putrescine is partly bound to nucleic acids, and its function is possibly related to the modulation of gene expression (21). Putrescine can also be addressed for the synthesis of polyamines, such as spermidine and spermine (45). In other Gram-negative bacteria species (e.g., Vibrio and Acinetobacter spp.), DABA decarboxylases have been found that also function in polyamine biosynthesis (22, 28).
Thanks to the present work, the search for putrescine producers, which so far has relied mostly upon gene annotation, is now more robustly supported by biochemical and microbiological proof. The newly characterized decarboxylase systems provide a basis for the design of more sensitive and less error-prone biomolecular tools for the detection of putrescine-producing bacteria in food and other human-related environments.
Supplementary Material
ACKNOWLEDGMENT
This work was funded by the EU commission in the framework of the BIAMFOOD project (Controlling Biogenic Amines in Traditional Food Fermentations in Regional Europe [project number 211441]).
Footnotes
Published ahead of print 13 January 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anantharam V, Allison MJ, Maloney PC. 1989. Oxalate: formate exchange: the basis for energy coupling in Oxalobacter. J. Biol. Chem. 264:7244–7250 [PubMed] [Google Scholar]
- 3. Applebaum DM, Dunlap JC, Morris DR. 1977. Comparison of biosynthetic and biodegradative decarboxylases of Escherichia coli. Biochemistry 18:1580–1584 [DOI] [PubMed] [Google Scholar]
- 4. Azcarate-Peril MA, Altermann E, Hoover-Fitzula RL, Cano RJ, Klaenhammer TR. 2004. Identification and inactivation of genetic loci involved with Lactobacillus acidophilus acid tolerance. Appl. Environ. Microbiol. 70:5315–5322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bonnin-Jusserand M, Grandvalet C, David V, Alexandre H. 2011. Molecular cloning, heterologous expression, and characterization of ornithine decarboxylase from Oenococcus oeni. J. Food Prot. 74:1309–1314 [DOI] [PubMed] [Google Scholar]
- 6. Budin-Verneuil A, Maguin E, Auffrey Y, Ehrlich DS, Pichereau V. 2006. Genetic structure and transcriptional analysis of the L-arginine deiminase (ADI) cluster in Lactococcus lactis MG 1363. Can. J. Microbiol. 52:617–622 [DOI] [PubMed] [Google Scholar]
- 7. Christen P, Mehta PK. 2001. From cofactor to enzymes. The molecular evolution of pyridoxal-5′-phosphate dependent enzymes. Chem. Rec. 1:436–447 [DOI] [PubMed] [Google Scholar]
- 8. Christiansen JK, et al. 2008. Phenotypic and genotypic analysis of amino acid auxotrophy in Lactobacillus helveticus CNRZ 32. Appl. Environ. Microbiol. 74:416–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chu CH, Bjeldanes LF. 1982. Effect of diamines, polyamines and tuna fish extracts on the binding of histamine to mucin in vitro. J. Food Sci. 47:79–80 [Google Scholar]
- 10. Coton E, et al. 2010. Origin of the putrescine-producing ability of the coagulase-negative bacterium Staphylococcus epidermidis 2015B. Appl. Environ. Microbiol. 76:5570–5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Coton M, et al. 2010. Occurrence of biogenic amine-forming lactic acid bacteria in wine and cider. Food Microbiol. 27:1078–1085 [DOI] [PubMed] [Google Scholar]
- 12. De Ruyter PG, Kuipers OP, Beerthuyzen MM, van Alen-Boerrigter I, de Vos VM. 1996. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J. Bacteriol. 178:3434–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. EFSA Panel on Biological Hazards (BIOHAZ) 2011. Scientific opinion on risk based control of biogenic amine formation in fermented foods. EFSA J. 9:2393 [Google Scholar]
- 14. Gallagher T, Snell EE, Hackert ML. 1989. Pyruvoyl-dependent histidine decarboxylase. Active site structure and mechanistic analysis. J. Biol. Chem. 264:12737–12743 [PubMed] [Google Scholar]
- 15. Garai T, Dueñas MT, Irastorza A, Martin-Alvarez PJ, Moreno-Arribas MV. 2006. Biogenic amines in natural ciders. J. Food Prot. 69:3006–3012 [DOI] [PubMed] [Google Scholar]
- 16. Glória MB, Watson BT, Simon-Sarkadi L, Daeschel MA. 1998. A survey of biogenic amines in Oregon Pinot noir and Cabernet Sauvignon wines. Am. J. Enol. Viticult. 49:279–282 [Google Scholar]
- 17. Guirard BM, Snell EE. 1980. Purification and properties of ornithine decarboxylase from Lactobacillus sp. 30a. J. Biol. Chem. 255:5960–5964 [PubMed] [Google Scholar]
- 18. Hui JY, Taylor SL. 1985. Inhibition of in vivo histamine metabolism in rats by foodborne and pharmacologic inhibitors of diamine oxidase, histamine N-methyltransferase and monoamine oxidase. Toxicol. Appl. Pharmacol. 81:241–249 [DOI] [PubMed] [Google Scholar]
- 19. Jack DL, Paulsen IT, Saier MH. 2000. The amino acid/polyamine/organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology 146:1797–1814 [DOI] [PubMed] [Google Scholar]
- 20. Kalac P, Krausova P. 2005. A review of dietary polyamines: formation, implications for growth and health and occurrence in foods. Food Chem. 90:219–230 [Google Scholar]
- 21. Igarashi K, Kashiwagi K. 2006. Polyamine modulon in Escherichia coli: genes involved in the stimulation of cell growth by polyamines. J. Biochem. 139:11–16 [DOI] [PubMed] [Google Scholar]
- 22. Ikai H, Yamamoto S. 1997. Identification and analysis of a gene encoding L-2,4-diaminobutyrate:2-ketoglutarate 4-aminotransferase involved in the 1,3-diaminopropane production pathway in Acinetobacter baumannii. J. Bacteriol. 179:5118–5125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ladero V, Calles-Enriquez M, Fernandez M, Alvarez M. 2010. Toxicological effects of dietary biogenic amines. Curr. Nutr. Food Sci. 6:145–156 [Google Scholar]
- 24. Ladero V, et al. 2011. Biogenic amines content in Spanish and French natural ciders: application of qPCR for quantitative detection of biogenic amine-producers. Food Microbiol. 28:554–561 [DOI] [PubMed] [Google Scholar]
- 25. Ladero V, et al. 2011. Sequencing and transcriptional analysis of the biosynthesis gene cluster of putrescine-producing Lactococcus lactis. Appl. Environ. Microbiol. 77:6409–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Landete JM, Ferrer S, Polo L, Pardo I. 2005. Biogenic amines in wines from three Spanish regions. J. Agric. Food Chem. 53:1119–1124 [DOI] [PubMed] [Google Scholar]
- 27. Lee J, Michael AJ, Martynowski D, Goldsmith EJ, Phillips MA. 2007. Phylogenetic diversity and the structural basis of substrate specificity in the β/α-barrel fold basic amino acid decarboxylases. J. Biol. Chem. 282:27115–27125 [DOI] [PubMed] [Google Scholar]
- 28. Lee J, et al. 2009. An alternative polyamine biosynthetic pathway is widespread in bacteria and essential for biofilm formation in Vibrio cholerae. J. Biol. Chem. 284:9899–9907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leitão MC, Marques AP, San Romão MV. 2005. A survey of biogenic amines in commercial Portuguese wines. Food Control 16:199–204 [Google Scholar]
- 30. Linares DM, Martin MC, Ladero V, Alvarez MA, Fernandez A. 2011. Biogenic amines in dairy products. Crit. Rev. Food Sci. Nutr. 51:691–703 [DOI] [PubMed] [Google Scholar]
- 31. Lucas PM, Blancato V, Magni C, Lolkema JS, Lonvaud-Funel A. 2007. Agmatine deiminase pathway genes in Lactobacillus brevis are linked to the tyrosine decarboxylase operon in a putative acid resistance locus. Microbiology 153:2221–2230 [DOI] [PubMed] [Google Scholar]
- 32. Marcobal A, de las Rivas B, Moreno-Arribas V, Muñoz R. 2004. Identification of the ornithine decarboxylase gene in the putrescine producer Oenococcus oeni BIFI-83. FEMS Microbiol. Lett. 239:213–220 [DOI] [PubMed] [Google Scholar]
- 33. Marcobal A, de las Rivas B, Moreno-Arribas V, Muñoz R. 2006. Evidence for horizontal gene transfer as origin of putrescine production in Oenococcus oeni RM83. Appl. Environ. Microbiol. 72:7954–7958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Morris DR, Pardee AB. 1966. Multiple pathways for putrescine biosynthesis in Escherichia coli. J. Biol. Chem. 241:3129–3135 [PubMed] [Google Scholar]
- 35. Morris DR, Koffron KL. 1969. Putrescine biosynthesis in Escherichia coli. J. Biol. Chem. 244:6094–6099 [PubMed] [Google Scholar]
- 36. Nannelli F, Claisse O, Gindreau E, Lonvaud-Funel A, Lucas PM. 2008. Determination of lactic acid bacteria producing biogenic amines in wine by quantitative PCR methods. Lett. Appl. Microbiol. 47:594–599 [DOI] [PubMed] [Google Scholar]
- 37. Peller L, Alberty RA. 1959. Multiple intermediates in steady state enzyme kinetics. I. The mechanism involving a single substrate and product. J. Am. Chem. Soc. 81:5907–5914 [PubMed] [Google Scholar]
- 38. Poolman B, Driessen AJ, Konings NN. 1987. Regulation of enzyme-ornithine exchange and the L-arginine deiminase pathway in Streptococcus lactis. J. Bacteriol. 169:5597–5604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Poolman B. 1990. Precursor/product antiport in bacteria. Mol. Microbiol. 4:1629–1636 [DOI] [PubMed] [Google Scholar]
- 40. Rodwell AW. 1953. The occurrence and distribution of amino-acid decarboxylases within the genus Lactobacillus. J. Gen. Microbiol. 8:224–232 [DOI] [PubMed] [Google Scholar]
- 41. Saier MH, Tran CV, Barabote RD. 2006. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 34:D181–D186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Settanni L, Moschetti G. 2010. Non-starter lactic acid bacteria used to improve cheese quality and provide health benefits. Food Microbiol. 27:691–697 [DOI] [PubMed] [Google Scholar]
- 43. Spano G, et al. 2010. Biogenic amines in fermented foods. Eur. J. Clin. Nutr. 64:95–100 [DOI] [PubMed] [Google Scholar]
- 44. Suzzi G, Gardini F. 2003. Biogenic amines in dry fermented sausages: a review. Int. J. Food Microbiol. 88:41–54 [DOI] [PubMed] [Google Scholar]
- 45. Tabor CW, Tabor H. 1985. Polyamines in microorganisms. Microbiol. Rev. 49:81–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Takatsuka Y, Yamaguchi Y, Ono M, Kamio Y. 2000. Gene cloning and molecular characterization of lysine decarboxylase from Selenomonas ruminantium delineate its evolutionary relationship to ornithine decarboxylases from eukaryotes. J. Bacteriol. 182:6732–6741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.