Abstract
The extremely acidophilic, chemolithoautotrophic Acidithiobacillus ferrooxidans is an important bioleaching bacterium of great value in the metallurgical industry and environmental protection. In this report, a mutagenesis system based on the homing endonuclease I-SceI was developed to produce targeted, unmarked gene deletions in the strain A. ferrooxidans ATCC 23270. A targeted phosphofructokinase (PFK) gene (pfkB) mutant of A. ferrooxidans ATCC 23270 was constructed by homologous recombination and identified by PCR with specific primers as well as Southern blot analysis. This potential pfkB gene (AFE_1807) was also characterized by expression in PFK-deficient Escherichia coli cells, and heteroexpression of the PFKB protein demonstrated that it had functional PFK activity, though it was significantly lower (about 800-fold) than that of phosphofructokinase-2 (PFK-B) expressed by the pfkB gene from E. coli K-12. The function of the potential PFKB protein in A. ferrooxidans was demonstrated by comparing the properties of the pfkB mutant with those of the wild type. The pfkB mutant strain displayed a relatively reduced growth capacity in S0 medium (0.5% [wt/vol] elemental sulfur in 9K basal salts solution adjusted to pH 3.0 with H2SO4), but the mutation did not completely prevent A. ferrooxidans from assimilating exogenous glucose. The transcriptional analysis of some related genes in central carbohydrate metabolism in the wild-type and mutant strains with or without supplementation of glucose was carried out by quantitative reverse transcription-PCR. This report suggests that the markerless mutagenesis strategy could serve as a model for functional studies of other genes of interest from A. ferrooxidans and multiple mutations could be made in a single A. ferrooxidans strain.
INTRODUCTION
Acidithiobacillus ferrooxidans is a Gram-negative, extremely acidophilic, chemolithoautotrophic bacterium that obtains its energy from the oxidation of ferrous iron or reduced inorganic sulfur compounds (RISCs) for carbon dioxide fixation and biosynthesis (8, 12, 23, 40). As an important bioleaching microorganism, studies on the metabolism of A. ferrooxidans have received increasing attention, and several models based on bioinformatic predictions and genomic, transcriptomic, or proteomic analyses have been proposed (1, 3, 9, 31, 32). However, there are still many hypothetical reactions and missing steps in these models, so that verifying the validity of the proposed hypotheses and filling in the gaps have already become problems.
Gene knockout is a common molecular method for functional genetic studies and has been successfully applied in a wide variety of microorganisms (10, 16, 17, 30, 45). Using gene knockout to inactivate targeted genes thought to be involved in a specific metabolic pathway and analyzing the relevant characteristics of the mutants would be an effective approach to advance our understanding of the metabolism in A. ferrooxidans.
Until now, there has been only one report on the construction of a gene-knockout mutant in A. ferrooxidans due to huge difficulties in its genetic manipulation (24). In 2000, Liu et al. constructed a recA mutant of A. ferrooxidans ATCC 33020 by homologous recombination (24). To disrupt the recA gene, a kanamycin resistance marker was inserted into the open reading frame (ORF) of recA, so the mutant ultimately contained an antibiotic resistance gene in its genome. This marker exchange mutagenesis not only causes potential threats to biosafety but also hampers the introduction of multiple mutations in the same strain because of the limited number of selectable markers available for A. ferrooxidans in its extremely acidic environment. Thus, construction of a genetic system for markerless gene knockout in A. ferrooxidans is undoubtedly essential and important for elucidating the physiological functions of multiple candidate genes involved in the different metabolic pathways.
The I-SceI-based mutagenesis system for markerless gene deletion and replacement has been reported in several microorganisms (10, 16, 17, 30). In this system, a suicide vector which carries the recognition site of the Saccharomyces cerevisiae mitochondrial endonuclease I-SceI and the homologous fragments flanking the targeted gene is first integrated into the host genome via homologous recombination. Then, a plasmid-based I-SceI gene is subsequently expressed to introduce a unique double-strand break at the I-SceI site in the genome, and this stimulates a second homologous recombination event, resulting in either a wild-type or a mutant allele. This method has been shown to produce small, markerless deletions, insertions, point mutations, as well as large deletions (10, 30).
Phosphofructokinase (PFK) catalyzes the phosphorylation of fructose-6-phosphate to fructose-1,6-biphosphate and plays a critical role in the typical glycolytic pathway in heterotrophic bacteria (34). The PFK was reported to be deficient in some obligate chemolithoautotrophs (25), and its existence in A. ferrooxidans was also quite uncertain because of no reports on the study of the PFK in this bacterium. However, in the available genome of A. ferrooxidans strain ATCC 23270, which was sequenced and annotated in 2008, a potential pfkB gene (AFE_1807) encoding a PFKB-family 6-phosphofructokinase (40) was predicted for the first time. Therefore, experimental characterization of this putative pfkB gene and its potential function in the central carbohydrate metabolism of A. ferrooxidans is essential for understanding the chemolithoautotrophic life of this bacterium.
In this study, we first describe the enzymatic characterization of PFKB protein expressed from the pfkB gene of A. ferrooxidans in Escherichia coli. We further report the construction of a pfkB mutant of A. ferrooxidans by markerless gene deletion based on the homing endonuclease I-SceI. In addition, we also compare the properties of the A. ferrooxidans pfkB mutant and the A. ferrooxidans wild type.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli DH5αλpir was used for the cloning manipulation of plasmids based on the R6K origin of replication. Genetic manipulation of other plasmids used in this study was performed in E. coli JM109. E. coli S17-1λpir bacteria were used as donors to transfer by conjugation plasmids based on the R6K origin of replication to A. ferrooxidans. E. coli strain SM10 was used for the conjugal transfer of the other plasmids used in this study.
Table 1.
Bacteria and plasmids used in this study
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| A. ferrooxidans | ||
| ATCC 23270 | Type strain | ATCCa |
| ΔAF1807 | ATCC 23270 ΔpfkB | This study |
| E. coli | ||
| JM109 | recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 relA1 λ−lac [F′ proAB lacIqZΔM15] | 27 |
| K-12 | Type strain | This lab |
| DF1010 | Δ(pfkB)201 Δ(rha-pfkA)200 recA56 relA1 tonA22 spoT1 ϕT2r | 35 |
| DH5αλpir | F−hsdR17 thi-1 gyrA Δ(lacZYA-argF) supE44 recA1 ϕ80dΔ(lacZ)M15 relA λpir | 42 |
| S17-1λpir | Tpr SmrrecA thi pro hsdR negative hsdM positive RP4::2-Tc::Mu::Km Tn7 λpir lysogen | 19 |
| SM10 | Kmrthi-1 thr leu tonA lacY supE recA::RP4-2–Tc::Mu | 37 |
| Plasmids | ||
| pUC19 | Apr, ColE1 replicon, cloning vector | TaKaRa |
| pEXT20 | Apr, ColE1 replicon, tac promoter, lacIq | 7 |
| pEXT20-K12B | pEXT20 containing pfkB gene from E. coli K-12 | This study |
| pEXT20-BN2 | pEXT20 containing pfkB gene from A. ferrooxidans ATCC 23270 | This study |
| pSW29T | Kmr, R6K replicon, oriTRP4 | 6 |
| pKIT | Kmr, R6K replicon, oriTRP4, I-SceI site | This study |
| pKIT-HR1 | pKIT containing HR1 | This study |
| pKOT | pKIT containing homologous sequences flanking pfkB gene from A. ferrooxidans ATCC 23270 | This study |
| pUC19RP12 | pUC19 containing I-SceI gene | 30 |
| pMSD1 | Smr, pBBR1 replicon, mob positive | This lab |
| pMSD1-I-SceI | pMSD1 containing I-SceI gene | This study |
ATCC, American Type Culture Collection.
Media and growth conditions.
The E. coli strains were grown at 37°C with shaking at 180 rpm in Luria-Bertani (LB) broth or on LB agar plates. The strains of A. ferrooxidans were cultured at 30°C with shaking at 180 rpm in Fe(II) medium (62 mM FeSO4 · 7H2O in 9K basal salts solution [28] adjusted to pH 2.0 with H2SO4) or S0 medium (0.5% [wt/vol] elemental sulfur in 9K basal salts solution adjusted to pH 3.0 with H2SO4). The compositions of solid 2:2 medium for A. ferrooxidans and mating medium were described previously (28). To test the phosphofructokinase activity, E. coli DF1010 cells were cultured in M63 mannitol medium (minimal medium 63 supplemented with 1 μg/ml thiamine hydrochloride, 0.4% [wt/vol] mannitol) (4). The following antibiotics were used for selection in E. coli: ampicillin (80 μg/ml), kanamycin (80 μg/ml), and streptomycin (80 μg/ml). In addition, kanamycin (100 μg/ml) and streptomycin (100 μg/ml) were used for selection in A. ferrooxidans on 2:2 medium plates, and kanamycin (300 μg/ml) and streptomycin (300 μg/ml) were used in 9K liquid medium.
General molecular techniques.
Isolation of genomic DNA from cells of E. coli and A. ferrooxidans was accomplished with a bacterial DNA kit obtained from Omega Bio-Tek, which was used according to the manufacturer's instructions. PCR was performed with PTC-200 DNA Engine (Bio-Rad) with TaKaRa Taq or PrimeSTAR HS DNA polymerase (TaKaRa). The PCR conditions were optimized for each primer pair (Table 2). The recovery of DNA fragments from agarose gels was performed with a gel extraction kit (Omega Bio-Tek). Plasmids were isolated with a Plasmid Mini Kit I from Omega Bio-Tek. Restriction enzymes and T4 DNA ligase were purchased from TaKaRa and used as recommended. Transformation of E. coli cells and agarose gel electrophoresis were performed as described elsewhere (36). The PCR products and recombinant clones were analyzed by restriction enzyme digestions and confirmed by sequencing. Primer synthesis and sequencing reactions were performed by Shanghai Biosune Biotechnology Company. The A. ferrooxidans mutants were identified by Southern blot analysis with Amersham Hybond-N+ positively charged nylon transfer membranes (GE Healthcare) and a DIG High Prime DNA Labeling and Detection Starter Kit I (Roche).
Table 2.
Primers used in this study
| Primer | Sequence (5′ to 3′)a |
|---|---|
| K12BF | GCCAGAATTCGCTGATTCGGTGCCAGAC |
| K12BR | GCTGAAGCTTGTTGCCGACAGGTTGGTG |
| BN2F | GCGCGAATTCTATAGGAGGAAATGATGACCGAAAACAACACC |
| BN2R | CGGCAAGCTTCTGTCTCCCATACCTCCC |
| 29KpnI | CGGCGGTACCTAGGGATAACAGGGTAATAGCGCTTTTCCGCTGCATAACCC |
| 29SalI | CTAAGTCGACTGATCAACGCGTCTCGAGGCCGGCCAGCCTCGCAGAGCAGG |
| HR1F | GACGGAATTCCCGCGGCCTGACGCTGGAAG |
| HR1R | GCCGTCTAGACGGTCATGACCGCGCCTCC |
| HR2F | GCCGTCTAGAGGGTACCCCAAAAAAGGCCC |
| HR2R | GACGGAGCTCCCATCAGTATAGAACAGTCGGGG |
| K59 | ATACCGTAAAGCACGAGG |
| K629 | CCCTGAATGAACTCCAAG |
| AF1F | TTGCCAAAGGGTTCGTGT |
| AF1R | GGGTTGAGGGTGAGGGTAG |
| AF2F | TAACGGAAGCGCGTGTACTC |
| AF2R | TGACATGGGAATTGCGCGC |
| DOF | CATGCTGCAGGGTGCTCACTTACAACTTCCG |
| DOR | CATGGAGCTCGGCCATCTGTTACTCCTCGTC |
| SMF | TGTCAGAGGCGGAGAATC |
| SMR | CAAGTCAGAGGGTCCAATC |
| cbbL1F | GGATTTCACCAAGGACGAT |
| cbbL1R | GAACTCTGCCCGCTCAT |
| cbbL2F | GGTCGCTTCCGGTGGTAT |
| cbbL2R | GCCTTGCCTTCCTTCTCG |
| cbbMF | CCGTCTATCATCAGGAATGG |
| cbbMR | TCCAGCACTCGTAAGCCT |
| pgmF | TTCGGGCGTCATTACTACAC |
| pgmR | TCGGCATAGGCAAAGTCA |
| pgiF | ATGACCAACGCCCAACAG |
| pgiR | CCCAATCCCAGAAACCGA |
| zwfF | CAGGAGTTGGATGCCTATGA |
| zwfR | CCGGGTAGGTGAGGATGTAG |
| pfkBF | GGCGTGGATGATCATTTCTA |
| pfkBR | ATTGGGTTTGATCAGGAAGG |
Artificial restriction sites are underlined.
Construction of expression plasmids for pfkB genes in E. coli.
To determine the 6-phosphofructokinase activity of the potential PFKB protein expressed from the putative pfkB gene (AFE_1807) from A. ferrooxidans ATCC 23270, the phosphofructokinase-2 (PFK-B) expressed by the pfkB gene from E. coli K-12 was used as a control. The 1,043-bp DNA fragment containing the pfkB gene from E. coli K-12 was amplified with the primers K12BF and K12BR, using the genomic DNA of E. coli K-12 as the template. After digestion with EcoRI and HindIII, the amplified DNA fragment was inserted into the corresponding cloning sites in the expression vector pEXT20, producing pEXT20-K12B. To amplify the potential pfkB gene (AFE_1807) from A. ferrooxidans ATCC 23270, primers BN2F and BN2R were used and genomic DNA from A. ferrooxidans ATCC 23270 was used as the template. The resulting 1,049-bp PCR fragment was ligated into pEXT20 after digestion with EcoRI and HindIII to generate pEXT20-BN2.
Analysis of 6-phosphofructokinase activity in E. coli DF1010.
The plasmid-based pfkB genes were expressed in E. coli DF1010 in M63 mannitol medium. Cells were grown in LB broth containing 80 μg/ml of ampicillin to an optical density of 0.8 at 600 nm. Then 3% of the cultures was inoculated to fresh M63 mannitol medium, and expression of the pfkB genes was induced with 1 mM isopropyl-β-d-thiogalactopyranoside overnight. Cells were collected by centrifugation (6,000 × g, 5 min) and washed three times with 50 mM Tris-HCl buffer (pH 7.5). The cell extracts for enzymatic assays were obtained by ultrasonication (80 Hz, 6 times with 5 s of treatment and 10-s intervals in between) on ice and subsequent centrifugation (13,000 × g, 20 min) at 4°C. The protein concentrations of the supernatants were measured with Coomassie brilliant blue G-250. The activities of 6-phosphofructokinase were determined using the coupling enzyme assay according to the method proposed by Kotlarz and Buc (21). All the coupling enzymes and chemicals required for enzymatic analysis were purchased from Sigma-Aldrich. A cell extract of E. coli K-12 was used as a positive control, while E. coli DF1010 containing plasmid pEXT20 was used as a negative control. One unit of PFK activity was defined as the amount of enzyme that catalyzed the conversion of 1 μmol of 6-phosphofructose to fructose-1,6-biphosphate per minute.
Construction of pfkB gene-knockout mutant.
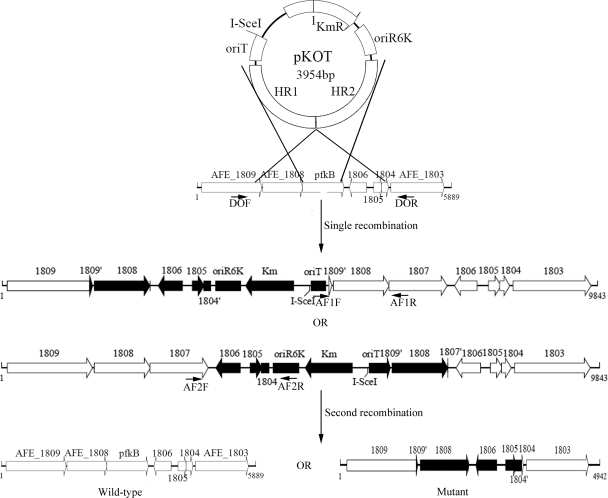
A schematic diagram for the construction of the pfkB-knockout mutant of A. ferrooxidans ATCC 23270 is shown in Fig. 1. The plasmids involved are listed in Table 1.
Fig 1.
Schematic diagram for construction of the pfkB-knockout mutant. The suicide plasmid pKOT was introduced into A. ferrooxidans ATCC 23270 and integrated into the genome, resulting in either one of the two possible single recombinants. Then plasmid pMSD1-I-SceI, which expresses the I-SceI endonuclease, was introduced into the single recombinant, producing a double-stranded break at the I-SceI site and stimulating the host recombinational repair machinery. This second recombination event generated a mutant or resulted in reversion to wild type. The integrated kanamycin resistance gene was eliminated in any of the types of second recombinants. The annealing sites for the primers used to identify various types of recombinants are indicated.
(i) Construction of pKIT.
Primers 29KpnI and 29SalI were used to insert an 18-bp I-SceI site as well as BclI, MluI, and XhoI sites into pSW29T. PCR was performed using the two primers mentioned above and plasmid DNA of pSW29T as the template. After digestion with KpnI and SalI, the amplified 306-bp DNA fragment was used to replace the corresponding KpnI-SalI fragment in pSW29T. The resulting plasmid, pKIT, was used as the backbone of the suicide plasmid pKOT.
(ii) Construction of suicide plasmid pKOT.
To inactivate the pfkB gene of A. ferrooxidans ATCC 23270, two pairs of primers (primers HR1F and HR1R and primers HR2F and HR2R) were used to amplify the homologous region upstream (HR1) and downstream (HR2) of the pfkB gene from the genome of A. ferrooxidans ATCC 23270. The amplified HR1 sequence (positions 1572178 → 1571179) was digested with EcoRI and XbaI, and the amplified HR2 sequence (positions 1570227 → 1569224) was digested with XbaI and SacI. Then, the digested HR1 and HR2 sequences were inserted in tandem into the corresponding cloning site in pKIT, producing the suicide plasmid pKOT that was used for pfkB gene deletion.
(iii) Conjugative transfer of pKOT into A. ferrooxidans ATCC 23270.
The conjugation experiments were modified based on the method of Peng et al. (28). E. coli S17-1λpir cells containing pKOT were cultured in LB broth in the presence of kanamycin until the exponential growth phase. The A. ferrooxidans ATCC 23270 recipient cells were grown in S0 medium to the stationary phase. The donor and recipient cells were harvested by centrifugation (8,000 × g, 5 min), washed three times with 2:2 basal salt buffer, and then mixed in an approximate 1:2 ratio. A small quantity of this cell suspension (100 μl, equivalent to approximately 108 cells) was spotted onto a piece of filter paper (0.45-μm pore size, 25-mm diameter) that was placed upon the mating medium. After 5 to 7 days of incubation at 30°C, the cells on the filter were resuspended with 2:2 basal salt buffer, diluted, and plated on 2:2 medium plates with or without kanamycin. The plates were incubated at 30°C for 15 days. The apparent recombination frequency was calculated on the basis of the number of single-recombinant colonies grown on 2:2 kanamycin medium divided by the number of recipient colonies grown on nonselective 2:2 medium.
(iv) Selection and verification of single recombination events.
The colonies grown on 2:2 medium plates containing kanamycin were picked and cultured in 10 ml of Fe(II) medium at 30°C for 5 days for recovery growth. Then they were transferred to S0 medium containing kanamycin. After 5 to 7 days of incubation at 30°C, cells were harvested for isolation of genomic DNA, which was used as the template for PCR verification of single recombination events. Primers K59 and K629 were selected to amplify the internal fragment of the kanamycin resistance aph gene on pKOT. Primer pairs AF1F-AF1R and AF2F-AF2R were used to verify the two types of single recombination events.
(v) Introduction of pMSD1-I-SceI into single-recombinant cells of A. ferrooxidans ATCC 23270.
Plasmid pMSD1 was modified from the broad-host-range vector pBBR1MCS-2 by replacing the kanamycin resistance marker with a streptomycin resistance gene. An 804-bp XbaI-PstI fragment from plasmid pUC19RP12 containing the I-SceI gene was ligated into the corresponding cloning sites in pMSD1 to generate pMSD1-I-SceI. Plasmid pMSD1-I-SceI was transformed into E. coli SM10. E. coli SM10 cells containing pMSD1-I-SceI were used as donors for conjugation. A. ferrooxidans ATCC 23270 cells that had a single recombination event were used as recipients. The mobilizable pMSD1-I-SceI was transferred from donor cells to recipient cells by the same conjugation procedure mentioned above, except for 48 h of incubation at 30°C. To select for transconjugants, the cells were plated on 2:2 medium plates containing streptomycin, as well as solid 2:2 medium without streptomycin. The plates were incubated at 30°C for 15 days. The apparent plasmid transfer frequency was calculated as the number of colonies that grew on 2:2 streptomycin medium per recipient colonies that grew on nonselective 2:2 medium.
(vi) Screening of double recombination events.
Colonies from 2:2 streptomycin medium plates were picked and cultured in 10 ml of Fe(II) medium at 30°C for 5 days for recovery of growth and then subcultured twice in S0 medium containing streptomycin for 5 to 7 days at 30°C. The cells were harvested for isolation of genomic DNA, which was used as the template for PCR verification of double recombination mutants. Primers DOF and DOR were used to screen and verify the double recombinants of A. ferrooxidans ATCC 23270.
(vii) Elimination of plasmid pMSD1-I-SceI from double recombination mutants.
The targeted mutants were subcultured 3 to 5 times in S0 medium without streptomycin and plated on solid 2:2 medium without streptomycin. The single colonies were grown again in liquid S0 medium, and the cultures were plated on 2:2 medium with and without streptomycin to screen for the spontaneous loss of plasmid pMSD1-I-SceI. The cells were also harvested for PCR verification. Primers SMF and SMR were used to amplify the streptomycin resistance gene fragment in pMSD1-I-SceI to verify the elimination of pMSD1-I-SceI.
Southern blot analysis.
To confirm the genotype of the pfkB mutant, Southern blot analysis was performed using EcoRV and ApaI digests of genomic DNA. The 1,010-bp EcoRI-XbaI HR1 fragment digested from pKIT-HR1 was isolated and used as the probe for Southern blot analysis.
Growth properties of wild-type versus pfkB mutant of A. ferrooxidans ATCC 23270.
To measure the growth of the pfkB mutant, cells from both the mutant and the wild type were initially cultured in S0 medium until early stationary phase, and then they were collected by centrifugation (6,000 × g, 5 min). The concentrated cells were resuspended with 9K basal salt solution, and 106 cells were inoculated into fresh S0 medium with or without 1 g/liter of glucose. Growth was monitored by measuring the change in optical density at 600 nm. The glucose levels in the cultures were determined by the glucose oxidase-peroxidase system using a commercial enzymatic assay kit purchased from Shanghai Rongsheng Biotechnology Company. The amount of sulfate in the cultures was measured using the turbidimetric method (20). The depletion of glucose or production of sulfate was calculated by the difference between the initial and final concentrations in the culture media. To determine the dry weight, cells were filtered through qualitative filter paper before they were collected by centrifugation (6,000 × g, 5 min). The concentration of ferrous iron in the liquid medium was determined by the modified o-phenanthroline method (15). Liquid medium without any bacteria was used as a negative control.
RNA extraction and qRT-PCR.
The wild-type and pfkB mutant cells of A. ferrooxidans were grown in S0 medium (with and without 1 g/liter of glucose) until mid-log phase and collected by centrifugation. Total RNA extraction was performed using the RNAlater solution and PureLink RNA minikit from Ambion according to the manufacturer's instructions. DNA was eliminated from the samples by using RNase-free DNase I (Fermentas), and the absence of contaminating DNA was examined by PCR. RNA quality and concentration were tested by agarose gel electrophoresis and a NanoDrop-1000 spectrophotometer (NanoDrop Technologies). For quantitative reverse transcription-PCR (qRT-PCR), cDNA was generated from RNA samples by a RevertAid first-strand cDNA synthesis kit (Fermentas). The qRT-PCR was performed with a LightCycler 480 apparatus (Roche) and the SYBR Premix Ex Taq from TaKaRa. The thermal cycling conditions were 95°C for 2 min and 40 cycles (95°C for 20 s, 54°C for 20 s, and 72°C for 15 s), followed by a final extension at 72°C for 2 min and subsequent melting curve analysis. The qRT-PCRs were performed in triplicate with two biological replicates. Primers used for qRT-PCR are shown in Table 2, and three genes (alaS, map, and rpoC) were used for normalization (26). The relative gene expression values were calculated by using the method of Pfaffl (29).
RESULTS
Functional expression of pfkB genes in E. coli DF1010 and analysis of 6-phosphofructokinase activity.
The ORF of the annotated pfkB gene (AFE_1807) in A. ferrooxidans ATCC 23270 contains 960 bp of nucleotides and encodes a polypeptide of 319 amino acids with a calculated molecular mass of 33.7 kDa. The potential 6-phosphofructokinase encoded by pfkB (AFE_1807) in A. ferrooxidans ATCC 23270 and the minor ATP-PFK (PFK-B or PFK2) encoded by pfkB (eco:b1723) in E. coli K-12 were annotated as members of the PFK-B sugar kinase family. The other PFK (PFK-A or PFK1) from E. coli K-12 belongs to the PFK-A family and displays different kinetic properties. The function of the putative PFKB in A. ferrooxidans ATCC 23270 was analyzed by heteroexpression of the pfkB gene (AFE_1807) and subsequent enzymatic analysis in E. coli DF1010, along with the expression of the pfkB gene from E. coli K-12 as a control.
E. coli DF1010 cannot grow on M63 mannitol solid medium because of the lack of phosphofructokinase (4). When the cells contain plasmid pEXT20-BN2 or pEXT20-K12B, then E. coli DF1010 was able to grow on M63 mannitol plates. These results suggested that the pfkB gene in A. ferrooxidans ATCC 23270 encodes an active phosphofructokinase that was expressed in E. coli DF1010. However, the cells of E. coli DF1010(pEXT20-BN2) grew poorly. The cell density and the size of colonies of E. coli DF1010(pEXT20-BN2) were smaller than those of DF1010(pEXT20-K12B). The PFKB protein encoded by the pfkB gene from A. ferrooxidans ATCC 23270 accounted for about 28.8% of the total proteins in the E. coli DF1010 cell extract, while the PFK-B protein encoded by the pfkB gene from E. coli K-12 was about 44.8% of the total proteins in the E. coli DF1010 cell extract (SDS-PAGE results are not shown). The specific activities of the PFK proteins in their cell extracts were also determined and compared (Table 3).
Table 3.
Specific activities of PFK proteins in their cell extracts
| Cell-free extract | Sp act of PFK (U/mg protein) |
|---|---|
| E. coli DF1010 | 0.0 |
| E. coli K-12 | 0.329 ± 0.007 |
| E. coli DF1010(pEXT20) | 0.0 |
| E. coli DF1010(pEXT20-K12B) | 270.247 ± 4.289 |
| E. coli DF1010(pEXT20-BN2) | 0.328 ± 0.005 |
Construction of pfkB mutant of A. ferrooxidans ATCC 23270.
An essential criterion for construction of a markerless gene-knockout system based on the I-SceI homing endonuclease is a targeted genome lacking the I-SceI recognition site. We first determined that the sequenced genome of A. ferrooxidans type strain ATCC 23270 lacks the 18-bp I-SceI recognition sequence. This was confirmed when no degradation of the genomic DNA was detected after treatment with the I-SceI nuclease.
To create a knockout mutant of A. ferrooxidans ATCC 23270, an initial plasmid, pKIT, was constructed as described above (see Materials and Methods) and could be further used as a platform for construction of all other mutants of this strain. The plasmid pKIT was modified from pSW29T and contained a kanamycin resistance gene, an oriTRP4 for mobilization by the RP4 conjugative machinery, an 18-bp I-SceI recognition site, a multiple-cloning site (MCS) of more than 10 unique cloning sites, and the pir-dependent origin of replication from the R6K replicon (Fig. 1). To knock out the pfkB gene of A. ferrooxidans ATCC 23270, the suicide plasmid pKOT was constructed by inserting two homologous fragments flanking the pfkB gene (HR1 and HR2) into the MCS of pKIT. Once transferred into A. ferrooxidans by conjugation, pKOT cannot replicate in recipient cells of A. ferrooxidans and was integrated in the genome by homologous recombination via the plasmid-containing homologous sequences under the pressure of kanamycin selection.
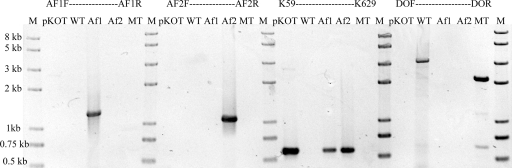
Of the colonies that grew on kanamycin selective plates, 15 were randomly picked and cultured for the identification of single recombination. A 571-bp fragment was amplified from the internal sequence of the kanamycin resistance aph gene by PCR using primers K59 and K629 in all 15 colonies (Fig. 2). A 1,259-bp PCR product was detected in seven of them with primer pair AF1F-AF1R, and a 1,087-bp PCR fragment was detected in the other eight colonies with primers AF2F and AF2R. Of the 15 colonies, 7 represented one type of single recombinant and 8 represented the other type, suggesting that a ratio of 1:1 was obtained. The apparent single recombination frequency was about 5.11 ± 2.66 × 10−7.
Fig 2.
PCR analyses of the single and double recombinants. The numbers on the left indicate the sizes of some fragments based on the molecular size marker (lanes M). The primer pairs and templates for each reaction are indicated above the corresponding lane. Lanes WT and MT, wild type and pfkB mutant of A. ferrooxidans ATCC 23270, respectively; lanes Af1 and Af2, the two types of single recombinants in Fig. 1, respectively.
To promote a second recombination event, a broad-host-range plasmid containing the I-SceI gene, pMSD1-I-SceI, was constructed and transferred by conjugation from E. coli SM10 into one of the single recombinants of A. ferrooxidans ATCC 23270. The apparent plasmid transfer frequency was approximately 2.22 ± 0.67 × 10−4. After it was transferred to a single recombinant of A. ferrooxidans, plasmid pMSD1-I-SceI could replicate and was stably maintained in the transconjugant under selecting pressure. Plasmid pMSD1-I-SceI carries an I-SceI gene whose product specifically recognizes and cuts the 18-bp I-SceI site, stimulating homologous recombination in the host cells. Of the colonies that grew on streptomycin selective plates, 98 were randomly picked and screened by PCR (see Materials and Methods). A 2,129-bp fragment is expected when the genomic DNA from a double-recombinant mutant is amplified with primers DOF and DOR. Under these conditions, 94 remained single crossovers and only 4 converted to double crossovers. Among the four double-crossover recombinants, half of them reverted to wild type and the other half were considered to be potential mutants (Fig. 2), which is consistent with the theoretical ratio of 1:1 (Fig. 1).
Thus, the ratio of double-recombinant mutants was about 2.0% in all of the Smr transconjugants, which was much higher than the rate of single recombinants (5.11 ± 2.66 × 10−7). However, selection for the double-crossover mutants was more difficult because of the absence of a selectable marker, and the purity of the initially obtained mutants was unknown. Therefore, the subsequent multiple subcultures of the potential mutants alternately in liquid and solid medium without streptomycin were necessary not only for the spontaneous loss of plasmid pMSD1-I-SceI but also for the purification of the mutants. During this period, both identification of the double-recombinant mutants and the absence of plasmid pMSD1-I-SceI should be performed at intervals.
Genetic analysis of pfkB mutant of A. ferrooxidans ATCC 23270.
To identify the mutation in the potential pfkB mutant, genomic DNA from mutant and wild-type A. ferrooxidans ATCC 23270 was purified, and both PCR analysis and Southern blot hybridization were performed. Using primers DOF and DOR, a 2,129-bp fragment was amplified with the genomic DNA from the mutant and a larger fragment (3,096-bp) was obtained from the wild type, as predicted (Fig. 2). The amplified DNA fragment was also verified by sequencing.
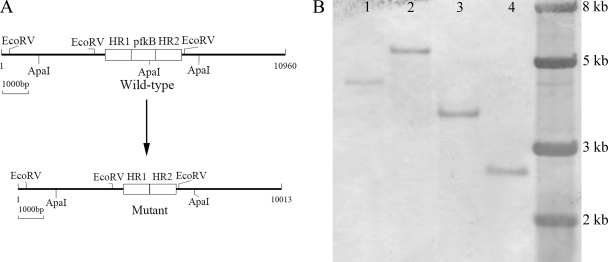
After labeling with digoxigenin, the 1,010-bp HR1 was used to probe the chromosomal DNA from the mutant and the wild type, which had been digested with EcoRV or ApaI. As expected, a 4.3-kb ApaI band and a 3.5-kb EcoRV band were obtained with the wild type, whereas a 5.2-kb ApaI band and a 2.5-kb EcoRV band were obtained with the mutant (Fig. 3B). The Southern blot hybridization results suggest that the pfkB gene was knocked out of the chromosomal DNA in the mutant of A. ferrooxidans ATCC 23270, as shown in Fig. 3A.
Fig 3.
Identification of a pfkB gene-knockout mutant of A. ferrooxidans ATCC 23270. (A) Schematic representation of the genomic region encoding the pfkB gene and the same region after mutation. (B) Southern blot analysis of ApaI-digested genomic DNA from the wild type (lane 1) and the pfkB mutant (lane 2) of A. ferrooxidans ATCC 23270 and EcoRV-digested genomic DNA from the wild type (lane 3) and the pfkB mutant (lane 4) of A. ferrooxidans ATCC 23270. The molecular size marker was loaded on the right lane, and the sizes of its fragments are indicated.
Growth properties of A. ferrooxidans ATCC 23270 wild type and its pfkB mutant.
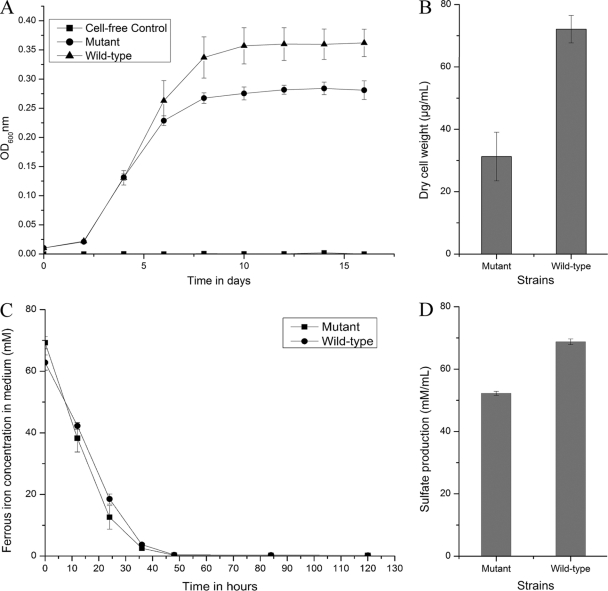
When sulfur powder was used as the sole energy source, the wild-type cells grew well and the pfkB mutant strain displayed a relatively reduced growth capacity (Fig. 4A). The final cell dry weight and sulfate production were also decreased for the pfkB mutant strain compared with the wild-type strain, as shown in Fig. 4B and D. However, the mutant strain did not have any perceptible change in ferrous oxidation when cultured in Fe(II) medium (Fig. 4C).
Fig 4.
Comparison of the wild type and the pfkB mutant of A. ferrooxidans ATCC 23270. (A) Growth of the wild type and the pfkB mutant of A. ferrooxidans ATCC 23270 in S0 medium (OD600, optical density at 600 nm); (B) final cell dry weight of the wild type and the pfkB mutant of A. ferrooxidans ATCC 23270 in S0 medium; (C) ferrous oxidation by the wild type and the pfkB mutant of A. ferrooxidans ATCC 23270 in Fe(II) medium; (D) sulfate production by the wild type and the pfkB mutant of A. ferrooxidans ATCC 23270 in S0 medium. Experiments were performed in triplicate. Each data point is given as the arithmetic mean value. The error bars indicate the standard deviations.
Glucose assimilation of A. ferrooxidans ATCC 23270 wild type and its pfkB mutant.
On the basis of genomic analysis of A. ferrooxidans ATCC 23270, the genes encoding carbohydrate transporters and a glucokinase were annotated and A. ferrooxidans ATCC 23270 was predicted to possess the genes essential for glucose glycolytic metabolism (40). So, we tested whether the growth of A. ferrooxidans ATCC 23270 on S0 medium could be stimulated by glucose and whether this stimulation could be affected by mutation of the pfkB gene.
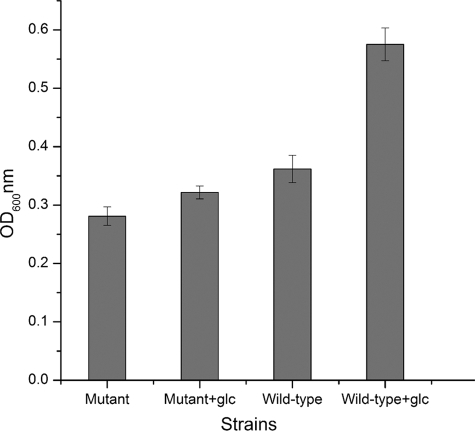
As shown in Fig. 5, 1 g/liter of glucose stimulated the growth of both the wild-type strain and the pfkB mutant, on the basis of the final cell density. The increase for the wild type was about 58% and that for the pfkB mutant was about 14%. However, the glucose levels in both culture media were just slightly decreased (∼0.1 g/liter), which was consistent with the characteristics of obligate chemolithotrophs that incorporate only limited amounts of organic compounds for their biosynthesis (25).
Fig 5.
Final cell density of the wild type and the pfkB mutant of A. ferrooxidans ATCC 23270 in S0 medium with or without 1 g/liter of glucose.
Transcriptional analysis of A. ferrooxidans ATCC 23270 and its pfkB mutant.
To investigate the transcriptional response to pfkB gene knockout of A. ferrooxidans ATCC 23270, the mRNA abundances of selected genes involved in central carbohydrate metabolism were determined by qRT-PCR assays (Table 4). The mRNA of pfkB was not detected in the pfkB mutant, which is in accordance with the genetic analysis. In the pfkB mutant, the mRNA levels of both cbbL2 and cbbM, which were involved in fixation of CO2, and pgi for glucose-6-phosphate isomerase were decreased, while the mRNA levels of cbbL1 and zwf were increased. Gene pgm was not differentially transcribed in the mutant compared to the wild-type strain (Table 4).
Table 4.
Subset of genes involved in central carbohydrate metabolism in A. ferrooxidans ATCC 23270 and changes of expression in response to pfkB mutation
| Gene | Locus | Gene description | Fold change (SD) |
|---|---|---|---|
| cbbL1 | AFE_1691 | Ribulose bisphosphate carboxylase large chain | 1.47 (0.17) |
| cbbL2 | AFE_3051 | Ribulose bisphosphate carboxylase large chain | 0.34 (0.18) |
| cbbM | AFE_2155 | Ribulose bisphosphate carboxylase large chain, form II | 0.59 (0.03) |
| pgm | AFE_2324 | Phosphoglucomutase | 0.88 (0.18) |
| pgi | AFE_2924 | Glucose-6-phosphate isomerase | 0.24 (0.09) |
| zwf | AFE_2025 | Glucose-6-phosphate 1-dehydrogenase | 1.51 (0.28) |
| pfkB | AFE_1807 | Carbohydrate kinase, PFKB family | —a |
—, pfkB mRNA was not detected in the mutant.
The relative mRNA levels of these selected genes of the wild-type and pfkB mutant strains in response to glucose supplementation were also investigated (Table 5). The qRT-PCR analyses revealed that cbbL1 was downregulated in both strains in the presence of glucose. The cbbL2, cbbM, pgi, and zwf genes were more actively transcribed for the pfkB mutant when cells were grown in the presence of glucose. The addition of glucose increased the pfkB mRNA level in the wild-type strain.
Table 5.
Changes of gene expression in wild-type A. ferrooxidans ATCC 23270 and its pfkB mutant in response to glucose supplementation
| Gene | Fold change (SD) |
|
|---|---|---|
| Wild type | pfkB mutant | |
| cbbL1 | 0.25 (0.07) | 0.24 (0.04) |
| cbbL2 | 0.86 (0.34) | 1.79 (0.58) |
| cbbM | 0.80 (0.09) | 1.34 (0.16) |
| pgm | 1.24 (0.24) | 0.99 (0.12) |
| pgi | 0.74 (0.23) | 2.18 (0.39) |
| zwf | 1.27 (0.14) | 1.92 (0.57) |
| pfkB | 1.47 (0.06) | |
DISCUSSION
This is the first report of construction of a markerless gene mutation in the extremely acidophilic organism A. ferrooxidans. Besides the allelic exchange, other strategies to generate marker-free mutations in A. ferrooxidans had also been considered, such as bacteriophage λ-Red and Cre-loxP recombination. Due to the low efficiency and poor reproducibility of plasmid transfer by electrotransformation (22) and the obligate chemolithoautotrophic characteristics of A. ferrooxidans, we eventually focused on the I-SceI-based homologous recombination system. Transfers of both the suicide plasmid and the helper plasmid containing the I-SceI gene from E. coli to A. ferrooxidans ATCC 23270 were performed using the well-established method of conjugation (28). Clean deletion of the potential pfkB gene can be used as a model for the application of this system in A. ferrooxidans.
With our conjugation procedure for A. ferrooxidans ATCC 23270, the apparent transfer frequency of mobilizable broad-host-range plasmids was about 10−4 transconjugants per recipient. The apparent recombination frequency of the mobilizable suicide plasmid pKOT was approximately 10−7; that is, single-recombination events occurred at a rate of 10−7 single recombinants per recipient or 10−3 per transconjugant. This is consistent with the results in E. coli that homologous recombination occurs at a frequency of about 10−3 to 10−4 less than the plasmid transfer frequency (13, 41). Although the initial crossover event occurred at a relatively low efficiency, potential single recombinants were generated and screened with little effort because of the availability of the kanamycin-selectable marker that was integrated into the chromosome of the recipient.
Once pMSD1-I-SceI was introduced into the single-recombinant cells, the I-SceI endonuclease was expressed and stimulated a second recombination. In E. coli cells, the frequency of spontaneous intramolecular recombination was increased by 2 to 3 orders of magnitude by I-SceI endonuclease (30). Apparently, this effect depended on the concentration of I-SceI endonuclease in the cell. Thus, it might be more efficient to screen for the double-recombination mutants after several passages in the presence of pMSD1-I-SceI. In addition, the second recombination event could occur at any time during cell growth. If it takes place during an early stage of growth, most of the cells in this colony would be composed of double recombinants, but if it occurs later, the percentage of double recombinants would be relatively low. Therefore, the initially identified double-recombinant mutants need further purification. A very faint positive PCR band was detected in some of the 94 cultures that remained single recombinants, and a very faint kanamycin resistance PCR band was also detected in one primary colony of a potential mutant. Having no way to concentrate the mutant with another round of selection, it is advisable to select targeted colonies with clearly positive PCR bands to decrease the labor needed to obtain a pure mutant.
We did not attempt to increase the ratio of double-recombinant events among the Smr transconjugants. In E. coli, all cells were shown to be double recombinants after 60 generations (30). As an obligate chemolithoautotroph, A. ferrooxidans grows slowly and its generation time is long (about 9 h at 30°C). So, the cells for screening were merely collected by at least two sequential serial passages in S0 medium containing streptomycin. In our experiments, the apparent ratio of double-recombinant mutants seemed high (about 2.0%), but this screening procedure was considered to be the most laborious process among the whole construction of the pfkB gene-knockout mutant. Since the colonies of A. ferrooxidans ATCC 23270 cannot be replica plated on plates with and without antibiotics like E. coli because of the tiny colonies and poor growth in solid medium, an improved method for screening for double-recombinant mutants is being investigated by our laboratory. Anyway, the markerless gene replacement system is really reliable and effective since we have recently obtained a second knockout mutant of A. ferrooxidans by using the same method. Moreover, there are some other factors that also affect the efficiency of this markerless gene mutation method. Two examples are the length of homologous fragments flanking the targeted gene and the copy number of the mobilizable, replicable plasmid containing the I-SceI endonuclease. These conditions should be taken into account when generating other mutations in A. ferrooxidans in the future.
E. coli K-12 containing both pfkA and pfkB is wild type in sugar metabolism (2). About 90% of the activity present in wild-type E. coli K-12 is PFK-A. PFK-B seemed to be dispensable and permitted only slow growth on sugar in a pfkA mutant, but PFK-B could support substantial glycolytic flux when present in sufficient amounts in a pfkA mutant (5). In A. ferrooxidans ATCC 23270, only a potential pfkB gene (AFE_1807) was predicted. The expression of the potential pfkB gene in E. coli DF1010 demonstrated the functional activity of PFK, though it was significantly lower (about 800-fold) than that of the PFK-B of E. coli K-12 expressed in DF1010 (Table 3). The possible relationship between the low level of activity of this key enzyme and the obligate autotrophic life of A. ferrooxidans is still not well understood.
A. ferrooxidans was characterized as a chemolithoautotrophic bacterium, and many organic compounds were reported to inhibit its autotrophic growth at certain concentrations (25). However, there have been some reports of heterotrophic growth on glucose (38, 39). Although some occurrences of heterotrophic growth were due to the contamination of acidophilic heterotrophs (14), it is true that some strains of A. ferrooxidans exhibited stimulated growth on glucose and the different characteristics of glucose metabolism were shown to be strain dependent (11, 38). Other chemolithoautotrophs have also been reported to assimilate limited amounts of exogenous organic nutrients as a supplementary carbon source (18, 33). Recently, the growth of Acidithiobacillus caldus SM-1, which displays a pattern of central carbohydrate metabolism similar to that of A. ferrooxidans ATCC 23270, was reported to be stimulated by glucose (10 g/liter) (44). In our experiment, the growth of A. ferrooxidans ATCC 23270 was stimulated by 1 g/liter of glucose, and the stimulation was not completely abolished in the pfkB mutant, which indicated that other glucose metabolic pathways might exist in this strain. When cultured in Fe(II) medium, the indistinctive change might be attributed to the poor growth on this energy source.
A. ferrooxidans ATCC 23270 encodes two copies of the form I Rubisco complex and a copy of the form II Rubisco complex in its Calvin-Benson cycle to fix carbon dioxide from the air (8). The pfkB mutant displayed relatively lower levels of expression of the second copy of form I Rubisco (cbbL2) and form II Rubisco (cbbM), which might imply that the lower carbon dioxide fixation rate results in a reduced growth rate. Although the CO2-responsive expression of three Rubisco complexes in Hydrogenovibrio marinus (43) has been reported, the regulation and control mechanism of different copies of Rubisco complexes in vivo are not clear for A. ferrooxidans, so the opposite expression pattern displayed by the first copy of form I Rubisco (cbbL1) in response to pfkB mutation is not well understood.
So far, limited information for central carbohydrate metabolism in A. ferrooxidans is available. According to the annotated genes, A. ferrooxidans could produce endogenous glucose from degradation of glycogen and catabolize the glucose by the pentose phosphate pathway (HMP) and Embden-Meyerhof pathway (EMP) (40). In response to glucose supplementation, the enhanced mRNA levels of pfkB and zwf in the wild-type strain indicated that both pathways might be involved in exogenous glucose metabolism. The more active expressions of zwf and pgi in the pfkB mutant verified the involvement of HMP in glucose metabolism. Anyway, the whole metabolic network is too big a story for this slow-growing chemolithoautotrophic bacterium, and more work is needed to illustrate the complete carbohydrate metabolic pathway and its regulation under different substrates, such as ferrous iron and reduced inorganic sulfur compounds.
In conclusion, the construction of a markerless gene mutation system in A. ferrooxidans will undoubtedly provide a convenient way to study this organism. The regulation of specific carbon metabolism in this chemolithoautotrophic bacterium is complicated, and further investigation is still necessary. Thus, our future work will focus on a comprehensive analysis of central carbohydrate metabolism in A. ferrooxidans.
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation (30970025), the National Basic Research Program (2010CB630902), the Natural Science Foundation (ZR2010CM004), and the Key Scientific and Technological Project (2010GSF10626) of Shandong Province, People's Republic of China.
We specially thank Thomas Linn for plasmid pEXT20, Didier Mazel for plasmid pSW29T, and Gyorgy Posfai for plasmid pUC19RP12. We also thank Roberta Greenwood for proofreading the manuscript.
Footnotes
Published ahead of print 30 December 2011
REFERENCES
- 1. Appia-Ayme C, et al. 2006. Microarray and bioinformatic analyses suggest models for carbon metabolism in the autotroph Acidithiobacillus ferrooxidans. Hydrometallurgy 83:273–280 [Google Scholar]
- 2. Babul J. 1978. Phosphofructokinases from Escherichia coli. Purification and characterization of the nonallosteric isozyme. J. Biol. Chem. 253:4350–4355 [PubMed] [Google Scholar]
- 3. Chi A, et al. 2007. Periplasmic proteins of the extremophile Acidithiobacillus ferrooxidans. Mol. Cell. Proteomics 6:2239–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Daldal F. 1983. Molecular cloning of the gene for phosphofructokinase-2 of Escherichia coli and the nature of a mutation, pfkB1, causing a high level of the enzyme. J. Mol. Biol. 168:285–305 [DOI] [PubMed] [Google Scholar]
- 5. Daldal F. 1984. Nucleotide sequence of gene pfkB encoding the minor phosphofructokinase of Escherichia coli K-12. Gene 28:337–342 [DOI] [PubMed] [Google Scholar]
- 6. Demarre G, et al. 2005. A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPα) conjugative machineries and their cognate Escherichia coli host strains. Res. Microbiol. 156:245–255 [DOI] [PubMed] [Google Scholar]
- 7. Dykxhoorn DM, Pierre RS, Linn T. 1996. A set of compatible tac promoter expression vectors. Gene 177:133–136 [DOI] [PubMed] [Google Scholar]
- 8. Esparza M, Cárdenas JP, Bowien B, Jedlicki E, Holmes DS. 2010. Genes and pathways for CO2 fixation in the obligate, chemolithoautotrophic acidophile, Acidithiobacillus ferrooxidans, carbon fixation in A. ferrooxidans. BMC Microbiol. 10:229–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Felício AP, et al. 2011. Differential proteomic analysis of Acidithiobacillus ferrooxidans cells maintained in contact with bornite or chalcopyrite: proteins involved with the early bacterial response. Process Biochem. 46:770–776 [Google Scholar]
- 10. Flannagan RS, Linn T, Valvano MA. 2008. A system for the construction of targeted unmarked gene deletions in the genus Burkholderia. Environ. Microbiol. 10:1652–1660 [DOI] [PubMed] [Google Scholar]
- 11. Frattini CJ, Leduc LG, Ferroni GD. 2000. Strain variability and the effects of organic compounds on the growth of the chemolithotrophic bacterium Thiobacillus ferrooxidans. Antonie Van Leeuwenhoek 77:57–64 [DOI] [PubMed] [Google Scholar]
- 12. Gale NL, Beck JV. 1967. Evidence for the Calvin cycle and hexose monophosphate pathway in Thiobacillus ferrooxidans. J. Bacteriol. 94:1052–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hamilton CM, Aldea M, Washburn BK, Babitzke P, Kushner SR. 1989. New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol. 171:4617–4622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harrison AP, Jr, Jarvis BW, Johnson JL. 1980. Heterotrophic bacteria from cultures of autotrophic Thiobacillus ferrooxidans: relationships as studied by means of deoxyribonucleic acid homology. J. Bacteriol. 143:448–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Herrera L, Ruiz P, Aguillon JC, Fehrmann A. 1989. A new spectrophotometric method for the determination of ferrous iron in the presence of ferric iron. J. Chem. Technol. Biotechnol. 44:171–181 [Google Scholar]
- 16. Horzempa J, et al. 2010. Utilization of an unstable plasmid and the I-SceI endonuclease to generate routine markerless deletion mutants in Francisella tularensis. J. Microbiol. Methods 80:106–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Janes BK, Stibitz S. 2006. Routine markerless gene replacement in Bacillus anthracis. Infect. Immun. 74:1949–1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kelly DP, Wood AP. 2006. The chemolithotrophic prokaryotes, p 441–456 In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E. (ed), The prokaryotes. Springer, New York, NY [Google Scholar]
- 19. Kienesberger S, et al. 2007. Development of experimental genetic tools for Campylobacter fetus. Appl. Environ. Microbiol. 73:4619–4630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kolmert A, Wikström P, Hallberg KB. 2000. A fast and simple turbidimetric method for the determination of sulfate in sulfate-reducing bacterial cultures. J. Microbiol. Methods 41:179–184 [DOI] [PubMed] [Google Scholar]
- 21. Kotlarz D, Buc H. 1982. Phosphofructokinases from Escherichia coli. Methods Enzymol. 90(Pt E):60–70 [DOI] [PubMed] [Google Scholar]
- 22. Kusano T, et al. 1992. Electrotransformation of Thiobacillus ferrooxidans with plasmids containing a mer determinant. J. Bacteriol. 174:6617–6623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Levicán G, Ugalde JA, Ehrenfeld N, Maass A, Parada P. 2008. Comparative genomic analysis of carbon and nitrogen assimilation mechanisms in three indigenous bioleaching bacteria: predictions and validations. BMC Genomics 9:581–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu Z, et al. 2000. Construction and characterization of a recA mutant of Thiobacillus ferrooxidans by marker exchange mutagenesis. J. Bacteriol. 182:2269–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Matin A. 1978. Organic nutrition of chemolithotrophic bacteria. Annu. Rev. Microbiol. 32:433–468 [DOI] [PubMed] [Google Scholar]
- 26. Nieto P, Covarrubias P, Jedlicki E, Holmes D, Quatrini R. 2009. Selection and evaluation of reference genes for improved interrogation of microbial transcriptomes: case study with the extremophile Acidithiobacillus ferrooxidans. BMC Mol. Biol. 10:63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nomura CT, et al. 2005. Expression of 3-ketoacyl-acyl carrier protein reductase (fabG) genes enhances production of polyhydroxyalkanoate copolymer from glucose in recombinant Escherichia coli JM109. Appl. Environ. Microbiol. 71:4297–4306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Peng JB, Yan WM, Bao XZ. 1994. Plasmid and transposon transfer to Thiobacillus ferrooxidans. J. Bacteriol. 176:2892–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pfaffl M. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pósfai G, Kolisnychenko V, Bereczki Z, Blattner FR. 1999. Markerless gene replacement in Escherichia coli stimulated by a double-strand break in the chromosome. Nucleic Acids Res. 27:4409–4415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Quatrini R, et al. 2009. Extending the models for iron and sulfur oxidation in the extreme acidophile Acidithiobacillus ferrooxidans. BMC Genomics 10:394–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ramírez P, Guiliani N, Valenzuela L, Beard S, Jerez CA. 2004. Differential protein expression during growth of Acidithiobacillus ferrooxidans on ferrous iron, sulfur compounds, or metal sulfides. Appl. Environ. Microbiol. 70:4491–4498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Robertson LA, Kuenen JG. 2006. The colorless sulfur bacteria, p 985–1011 In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E. (ed), The prokaryotes. Springer, New York, NY [Google Scholar]
- 34. Ronimus RS, Morgan HW. 2001. The biochemical properties and phylogenies of phosphofructokinases from extremophiles. Extremophiles 5:357–373 [DOI] [PubMed] [Google Scholar]
- 35. Rosey E, Oskouian B, Stewart GC. 1991. Lactose metabolism by Staphylococcus aureus: characterization of lacABCD, the structural genes of the tagatose 6-phosphate pathway. J. Bacteriol. 173:5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 37. Simon R, Priefer U, Puhler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat. Biotechnol. 1:784–791 [Google Scholar]
- 38. Sugio T, Kudo S, Tano T, Imai K. 1982. Glucose transport system in a facultative iron-oxidizing bacterium, Thiobacillus ferrooxidans. J. Bacteriol. 150:1109–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tabita R, Lundgren DG. 1971. Utilization of glucose and the effect of organic compounds on the chemolithotroph Thiobacillus ferrooxidans. J. Bacteriol. 108:328–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Valdés J, et al. 2008. Acidithiobacillus ferrooxidans metabolism: from genome sequence to industrial applications. BMC Genomics 9:597–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van Zyl LJ, Jolanda van Munster M, Rawlings DE. 2008. Construction of arsB and tetH mutants of the sulfur-oxidizing bacterium Acidithiobacillus caldus by marker exchange. Appl. Environ. Microbiol. 74:5686–5694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yin X, et al. 2009. Contributions of O island 48 to adherence of enterohemorrhagic Escherichia coli O157:H7 to epithelial cells in vitro and in ligated pig ileal loops. Appl. Environ. Microbiol. 75:5779–5786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yoshizawa Y, Toyoda K, Arai H, Ishii M, Igarashi Y. 2004. CO2-responsive expression and gene organization of three ribulose-1,5-bisphosphate carboxylase/oxygenase enzymes and carboxysomes in Hydrogenovibrio marinus strain MH-110. J. Bacteriol. 186:5685–5691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. You X-Y, et al. 2011. Unraveling the Acidithiobacillus caldus complete genome and its central metabolisms for carbon assimilation. J. Genet. Genomics 38:243–252 [DOI] [PubMed] [Google Scholar]
- 45. Zarschler K, Janesch B, Zayni S, Schäffer C, Messner P. 2009. Construction of a gene knockout system for application in Paenibacillus alvei CCM 2051T, exemplified by the S-layer glycan biosynthesis initiation enzyme WsfP. Appl. Environ. Microbiol. 75:3077–3085 [DOI] [PMC free article] [PubMed] [Google Scholar]