Abstract
During large Q fever outbreaks in the Netherlands between 2007 and 2010, dairy goat farms were implicated as the primary source of human Q fever. The transmission of Coxiella burnetii to humans is thought to occur primarily via aerosols, although available data on C. burnetii in aerosols and other environmental matrices are limited. During the outbreak of 2009, 19 dairy goat farms and one dairy sheep farm were selected nationwide to investigate the presence of C. burnetii DNA in vaginal swabs, manure, surface area swabs, milk unit filters, and aerosols. Four of these farms had a positive status during the Coxiella burnetii bulk milk monitoring program in 2009 and additionally reported abortion waves in 2008 or 2009. Eleven farms were reported as having positive bulk milk only, and five selected (control) farms had a bulk milk-negative status in 2009 and no reported Q fever history. Screening by quantitative PCR (qPCR) revealed that on farms with a history of abortions related to C. burnetii and, to a lesser extent, on farms positive by bulk milk monitoring, generally higher proportions of positive samples and higher levels of C. burnetii DNA within positive samples were observed than on the control farms. The relatively high levels of C. burnetii DNA in surface area swabs and aerosols sampled in stables of bulk milk-positive farms, including farms with a Q fever-related abortion history, support the hypothesis that these farms can pose a risk for the transmission of C. burnetii to humans.
INTRODUCTION
Q fever, a zoonosis caused by the bacterium Coxiella burnetii, caused unprecedented epidemics in the Netherlands between 2007 and 2010. During these outbreaks, epidemiological studies implicated C. burnetii-infected dairy goats as the primary source of human Q fever (13, 16, 20). The impact of the various transmission routes of C. burnetii is not well understood. When animals are infected, the main sources of C. burnetii shedding into the environment are manure, urine, milk, and especially birth materials like amnion fluid and placenta material (2, 7). Coxiella burnetii infections in humans are thought to occur primarily via aerosols generated by infected animals or animal products (22, 24). However, available data on C. burnetii in aerosols and other environmental matrices are sparse. To our knowledge, 10 studies have investigated the presence of C. burnetii in environmental samples using PCR-based methods.
Seven studies were not related to a human Q fever outbreak, four of which were performed in areas with a (suspected) high incidence among animals (4, 5, 21, 25). The three other studies reported the presence of C. burnetii DNA in dust (17) or primarily soil samples from a geographically large area in the United States (11, 14). The latter three studies investigated the presence of C. burnetii DNA in soil samples (6) and surface swabs (1, 9) in relation to human Q fever outbreaks.
The start of the first Q fever outbreak in the Netherlands in 2007 led to source-finding investigations by several Municipal Health Services in subsequent years. Vaginal swabs obtained from goats and sheep and, on occasion, surface area swabs obtained from stables revealed that C. burnetii DNA was present on most dairy goat farms, which were suspected to be a source for human Q fever cases in their vicinity (9). However, no systematic data from random farms not suspected of being a source of human infections were available for comparison. The aim of the current study was to investigate the presence of C. burnetii DNA in various animal and environmental matrices on farms with a Q fever-related abortion history, farms with a bulk milk-positive status only, and farms with a bulk milk-negative status and no known Q fever history, to support the epidemiologically identified role of C. burnetii-contaminated matrices in its transmission to humans.
MATERIALS AND METHODS
Farm selection.
All goat farms selected for this study participated in the larger integrated human-veterinary Q-VIVE study, described previously by Schimmer et al. (19). In that study, methods regarding human and animal serology used in our study are described in more detail.
All dairy goat farms had the Dutch white dairy goat as the main breed, sometimes in combination with smaller numbers of other breeds of goat. The sheep on the dairy sheep farm are of a relatively rare breed, Dutch Friesian sheep.
Farming procedures varied little among the dairy farms selected, with at least one lambing season per year, animals kept indoors in deep-litter stables, and automated milking procedures performed daily in a separate stable compartment.
The selection of C. burnetii-positive dairy goat farms in the current study was based on the following criteria: reported abortion waves due to C. burnetii among goats or sheep in 2008 or 2009 (category A) and a positive status in the voluntary (2008) or mandatory (2009) PCR-based bulk milk (tank) screening survey (category B). Farms that tested positive by bulk milk screening and that additionally experienced C. burnetii-related abortion waves were classified as farm category AB, and farms that tested positive only by bulk milk screening were classified as farm category B.
Q fever-negative farms, classified as category N, were selected when (i) serology of sampled goats in the Q-VIVE veterinary study component (19) (if available) was negative, and/or (ii) no history of Q fever was recorded at the time of selection (no reported abortion waves since 2005 and no positive test results in the serosurvey in 2008 and/or in voluntary or mandatory bulk milk monitoring surveys in 2008 and 2009).
The staggered inclusion of farms into the Q-VIVE study was due to the introduction of mandatory vaccination for small ruminants for the south of the country in 2009 and nationwide in 2010. Eight farms were located within the mandatory vaccination area of 2009 and were sampled in October and November 2009. The 12 remaining farms from the rest of the country were sampled in April to June 2010.
The characteristics of the 19 dairy goat farms and the dairy sheep farm selected for screening for the presence of C. burnetii in animal and environmental matrices are described in Table 1. Fifteen farms were positive for C. burnetii in bulk milk (farms A to O), and 5 were negative (farms P to T). On farms with an abortion history due to C. burnetii (farms A to D), abortion rates were 19% on farm A (April 2009), 33% on farm B (April 2009), 33% on farm C (April of 2008), and 7% on farm D (February 2009).
Table 1.
Characteristics of farms investigated and serology of farm animals and occupantsa
| Farm | Farm category | Lambing season(s) (wk) | Date of 1st vaccination (day-mo-yr) | Date of sampling (day-mo-yr) | Date of culling (day-mo-yr) | Distance to nearest bulk milk-positive farm (m) | Animal serology |
Human serology of farmers and family |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % positive samples | No. of positive samples/total no. of samples | F | S | C | O | |||||||
| A | AB | 1–36 | 9-7-2009 | 13-11-2009 | 8-2-2010 | 16.329 | 71.4 | 15/21 | P | P | R | |
| B | AB | 3–40 | 5-5-2009 | 12-11-2009 | 17-2-2010 | 792 | 42.9 | 9/21 | R | P | P | |
| C | AB | NA | 18-6-2009 | 22-10-2009 | 4-2-2010 | 12.470 | NA | NA | NA | |||
| D | AB | 1–27 | 12-4-2010 | 19-4-2010 | 23-1-2010 | 14.505 | 80.0 | 52/65 | R | R | ||
| E | B | 11–15 | 21-7-2009 | 13-11-2009 | 21-1-2010 | 5.575 | 42.9 | 9/21 | P | P | P | |
| F | B | 1–18 | 7-7-2009 | 20-11-2009 | 10-2-2010 | 2.613 | 71.4 | 15/21 | P | P | ||
| G | B | 1–27 and 40–52 | 19-3-2010 | 21-5-2010 | 15-2-2010 | 755 | 9.5 | 2/21 | P | R | ||
| H | B | 8–52 | 7-4-2010 | 21-5-2010 | 11-2-2010 | 1.284 | 41.9 | 13/31 | R | P | P | |
| I | B | 10–41 | 24-3-2010 | 16-4-2010 | 4-2-2010 | 3.346 | 63.6 | 56/88 | P | R | R | |
| J | B | NA | 8-7-2009 | 18-11-2009 | 8-2-2010 | 3.104 | 71.4 | 15/21 | NA | |||
| K | B | 13–21 | 28-4-2010 | 15-4-2010 | 16-2-2010 | 11.045 | 45.8 | 11/24 | P | P | R | |
| L | B | 3–21 | 26-4-2010 | 19-5-2010 | 5-2-2010 | 10.114 | 92.3 | 12/13 | P | R/P | ||
| M | B | 2–40 | 17-5-2010 | 22-4-2010 | 5-2-2010 | 5.515 | 52.4 | 11/21 | P | R | P | |
| N | B | 2–52 | 19-3-2010 | 9-4-2010 | 5-2-2010 | 9.149 | 56.5 | 26/46 | R | P | P | |
| O | B | 18–31 and 43–52 | 12-5-2010 | 3-6-2010 | 2-2-2010 | 6.690 | 66.7 | 2/3 | R | R | ||
| P | N | 4–13 and 24–32 | 27-5-2010 | 5-11-2009 | NA | 2.848 | NA | NA | P | P | P | |
| Q | N | 18–31 | 16-9-2009 | 5-11-2009 | NA | 8.395 | 0 | 0/21 | N | P | P | |
| R | N | 10–21 | 4-5-2010 | 19-4-2010 | NA | 25.026 | 0 | 0/21 | P | P | ||
| S | N | 8–10 | 16-12-2009 | 22-4-2010 | NA | 23.177 | 0 | 0/21 | N | N | ||
| T | N | 6–18 | 28-5-2010 | 19-5-2010 | NA | 10.059 | 0 | 0/22 | N | N/P | ||
Farms are categorized as farms with a C. burnetii-related abortion history (category AB), farms with a bulk milk-positive status in 2009 (category B), or control farms (category N). All farms were dairy goat farms, except for farm D, which was a dairy sheep farm. F, farmer; S, spouse/partner; C, child of farmer (can be an adult child); O, other family member or employee; P, past infection (IgG phase II level of >1:32, either with or without an IgG phase I level of >1:32, and IgM phase I and II negative); R, recent infection (IgM phase II level of >1:32, either with or without IgG phase I, and/or IgG phase II levels of >1:32); N, negative; NA, information not available (no farm questionnaire or no participation in animal or human sampling).
Sampling of animal and environmental matrices.
For each individual farm, approximately 25 samples were obtained in the stables from several animal and environmental matrices. For each farm, we aimed to obtain 5 vaginal swabs from animals, 10 surface area swabs from horizontal (dust-accumulating) surfaces, 5 manure samples, 3 milk unit filters (if available), and 3 aerosol samples. In addition, four aerosol samples were obtained in the four wind directions at a radial distance of 500 m from the farms. All samples were taken by the same veterinarian from the Dutch Food and Consumer Product Safety Authority (nVWA).
A random sample of animals was selected on each farm, and vaginal swabs were obtained from animals by using sterile cotton swabs (VWR International, Netherlands). Surface area swabs were obtained from horizontal (dust-accumulating) surfaces, such as windowsills or low stable compartment boundary walls. Surface swabs were obtained by swabbing a sterile cotton swab (VWR International, Netherlands) in a single motion over a length of approximately 2 m.
Manure goat droppings were collected from stable floors and added to 50-ml Greiner tubes. Milk unit filters were obtained from the automated milk units. Aerosol samples were obtained by using a Sartorius MD8 Airport device (10) equipped with cellulose nitrate filters with a pore size of 8 μm. The flow rate was set to 50 liters per minute with a sampling time of 10 min, resulting in a filtered air sample of 500 liters. After aerosol collection, cellulose nitrate filters were transferred onto sterile petri dishes. All samples were transported to the laboratory and stored at −20°C.
Sample processing, DNA extraction, and qPCR.
Samples were processed and DNA was extracted by use of the NucliSens magnetic extraction kit (bioMérieux, France). Small modifications were made to the manufacturer's guidelines for DNA extraction. Vaginal and surface area swabs were added to 10 ml of NucliSens lysis buffer and vortexed for 10 s. Following an incubated period of 10 min, the swabs were removed.
Ten grams of goat manure droppings was added to phosphate-buffered saline (PBS) in 50-ml Greiner tubes (Greiner Bio-one, Netherlands), using a 1:2 ratio of manure to PBS. This mixture was homogenized for nearly 2 h on a rotating tube holder at 10 rpm. Greiner tubes were centrifuged (Varifuge 3.2RS; Heraeus) at 9,000 × g for 10 min. The supernatant was transferred into a new Greiner tube, and 1 ml of the supernatant was used for DNA extraction.
For each milk unit filter, one-third of the filter (±20 cm) was cut from the bottom of the filter and used for sample processing. This part of the milk unit filter was cut into two squared-centimeter portions, which were added to a 250-ml bottle (VWR International, Netherlands) with 50 ml of NucliSens lysis buffer. The bottles were placed onto a horizontal shaker and homogenized overnight at 50 rpm. The NucliSens lysis buffer containing the DNA extract of the filters was transferred from the bottles into 50-ml Greiner tubes.
For aerosol samples, 10 ml of NucliSens lysis buffer was added to petri dishes containing cellulose nitrate filters, and the dishes were placed onto a horizontal shaker for 6 h at 50 rpm. NucliSens lysis buffer was transferred from the petri dishes into 15-ml Greiner tubes.
As an internal control, 50 μl of a Bacillus thuringiensis spore suspension (1.2 × 105 spores) was added to each sample before extraction. All samples were placed at room temperature for 1 h to complete lysis. From this point onwards, DNA extraction procedures were carried out according to the manufacturer's protocol.
The multiplex quantitative PCR (qPCR) assay used for C. burnetii (A. de Bruin, M. Koning, L. de Heer, R. Q. J. van der Plaats, I. Janse, and B. J. van Rotterdam, submitted for publication) is an improved version of a method described previously (9). This assay was slightly modified by the removal of the icd signature sequence, as one single-copy target (com1) and one multicopy target (IS1111) were proven previously to be sufficient for screening purposes (9).
In addition, to improve the detection of fragmented DNA, oligonucleotides for the detection of com1 and IS1111 signature sequences were modified to yield shorter amplification products. This resulted in a slightly improved sensitivity for the IS1111 target (8.8 copies, compared to 10.4 copies per reaction in the previous assay).
Our measurements showed quantification cycle (Cq) values for IS1111 that were always lower than those for com1, as was expected due to multiple copies within the C. burnetii genome (15).
For each qPCR, 3 μl of DNA extract, obtained from animal and environmental matrices, was tested in triplicate undiluted and in 10-fold dilutions.
Coxiella burnetii-positive samples were categorized into two classes with increasing C. burnetii DNA contents: (i) IS1111 positive and (ii) IS1111 and com1 positive.
The proportions of positive samples per matrix were compared among the three farm categories (category AB versus category B, category AB versus category N, and category B versus category N) by using Fisher exact tests. For comparisons of the Cq values obtained by qPCR among the three farm categories, a Wilcoxon rank sum test was used.
RESULTS
Serology of ruminants and farm family members at participating farms.
Based on serology, the percentages of positive small ruminants were on average 64.8% for the farms with a C. burnetii-related abortion history (category AB), 55.9% among the bulk milk-positive farms (category B), and, by preselection, 0% among the control farms (category N). There was a significant difference in the percentages of positive small ruminants between farms with an abortion history and bulk milk-positive farms (P < 0.05 by Fisher exact test).
The risk of transmission to humans, measured as the proportion of farms with at least one household member or worker with a recent infection according to their serological profile, was found to be related to the Q fever status of the farm. Recent infections were found at all 3 farms (100%) with a C. burnetii-related abortion history (one category AB farm did not participate in the human component of the study), in comparison to 8 out of 11 (73%) bulk milk-positive farms (category B) and none (0%) of the 5 control farms (category N). In more detail, of the eight sera taken at farms in category AB, 50% had a profile matching a recent infection, and 50% had a profile matching a past infection. For the 27 sera taken from farms in category B, these rates were 37% and 63%, respectively. For the 13 sera from farms in category N, these figures were 0% and 62%, respectively, with the remaining 38% testing negative.
Presence of Coxiella burnetii DNA in animal and environmental matrices.
The presence of C. burnetii DNA in animal and environmental samples obtained from the 19 dairy goat farms and 1 dairy sheep farm is summarized in Table 2.
Table 2.
Screening for C. burnetii DNA in animal and environmental matrices obtained from 19 dairy goat farms and 1 dairy sheep farma
| Matrix | Farm category | Total no. of samples | No. of IS1111-positive samples | No. of IS1111- + com1-positive samples | % positive samples | Mean Cq (SD) |
Categories compared |
P value |
||
|---|---|---|---|---|---|---|---|---|---|---|
| com1 | IS1111 | Fisher exact testb | Wilcoxon rank sum testc | |||||||
| Vaginal swabs | AB | 45 | 18 | 27 | 100 | 37.1 (1.0) | 33.2 (1.6) | AB and B | <0.05 | <0.01 |
| B | 55 | 30 | 15 | 81.8 | 37.3 (1.5) | 34.6 (1.8) | AB and N | <0.01 | <0.01 | |
| N | 25 | 6 | 0 | 24 | ND | 36.5 (0.8) | B and N | <0.01 | 0.02 | |
| Manure | AB | 19 | 10 | 1 | 57.9 | 37.0 (2.2) | 36.1 (1.4) | AB and B | 0.59 | 0.70 |
| B | 50 | 19 | 5 | 48 | 36.0 (3.0) | 35.7 (2.7) | AB and N | <0.01 | NA | |
| N | 25 | 0 | 0 | 0 | ND | ND | B and N | <0.01 | NA | |
| Milk unit filters | AB | 6 | 0 | 6 | 100 | 33.1 (2.2) | 27.3 (2.0) | AB and B | 1.00 | 0.02 |
| B | 33 | 9 | 23 | 97 | 34.7 (1.4) | 31.4 (3.3) | AB and N | <0.01 | 0.04 | |
| N | 13 | 2 | 0 | 15,4 | ND | 35.4 (0.9) | B and N | <0.01 | 0.09 | |
| Surface area swabs | AB | 40 | 0 | 40 | 100 | 34.1 (2.5) | 28.3 (2.4) | AB and B | NA | <0.01 |
| B | 110 | 13 | 97 | 100 | 36.0 (1.8) | 30.9 (3.1) | AB and N | <0.01 | <0.01 | |
| N | 50 | 17 | 9 | 52 | 35.1 (1.6) | 34.9 (1.5) | B and N | <0.01 | <0.01 | |
| Aerosols (stables) | AB | 15 | 1 | 14 | 100 | 36.1 (1.8) | 31.0 (1.7) | AB and B | 1.00 | 0.02 |
| B | 33 | 14 | 18 | 97 | 36.7 (1.7) | 33.7 (3.0) | AB and N | NA | <0.01 | |
| N | 15 | 13 | 2 | 100 | 35.2 (0.3) | 35.7 (1.6) | B and N | 1.00 | 0.02 | |
| Aerosols (500 m) | AB | 16 | 12 | 3 | 93.8 | 39.2 (2.3) | 36.8 (1.9) | AB and B | 0.26 | 0.14 |
| B | 42 | 30 | 3 | 78.6 | 39.0 (0.4) | 37.6 (2.0) | AB and N | 0.09 | 0.33 | |
| N | 18 | 10 | 2 | 66.7 | 37.4 (0.7) | 37.2 (1.9) | B and N | 0.35 | 0.94 | |
Farms are categorized into three farm categories (categories AB, B, and N), and for each animal or environmental matrix, the proportions of positive samples and C. burnetii DNA contents expressed by averaged Cq values are shown for C. burnetii target combinations of com1 and IS1111. Category AB, abortion wave in 2008 or 2009 (and bulk milk positive); category B, bulk milk positive only; category N, bulk milk negative and no known Q fever history. NA, not applicable; ND, not detected.
Comparison based on the proportion of positive samples, using nonparametric Fisher exact tests.
Comparison based on Cq values, using nonparametric Wilcoxon rank sum tests.
A comparison per matrix among the three farm categories was based on the number of positive samples, which can be divided into two categories: IS1111 positive or IS1111 and com1 positive. Due to the presence of multiple copies of the IS1111 target within the C. burnetii genome (15), the amplification of this target is expected to occur before the amplification of the single-copy target com1. This was reflected in our data, where for samples showing positive results for both targets com1 and IS1111, Cq values of IS1111 were consistently lower than those of com1. For matrices derived from animal samples such as vaginal swabs, manure droppings, and milk unit filters, the percentage of positive samples was highest for the category AB farms with a C. burnetii-related abortion history, followed by bulk tank milk-positive farms (category B), and was lowest for the control farms (category N). Significant differences in the numbers of positive samples were found primarily between category AB farms and the control (category N) farms and between category B farms and the control (category N) farms. A similar tendency was observed for aerosol samples taken at 500 m in all four wind directions of the farm, although these values were not statistically significant. For aerosol samples taken from the stables, the percentage of positive samples was (almost) 100% for all three farm categories. In addition, the C. burnetii DNA contents in the animal and environmental matrices were compared between farm categories based on averaged Cq values for the targets com1 and IS1111. Significant differences between farm categories were found for both animal and environmental matrices. For vaginal swabs, surface area swabs, and aerosols obtained from the stables, Cq values for the target IS1111 were significantly lower (indicating a high C. burnetii DNA content) on farms with a C. burnetii-related abortion history (category AB) than on farms with a positive status by bulk milk screening only (category B) and on the control farms (category N). No significant differences were observed between farm categories for the manure samples (compared only between category AB and B farms, as category N farms had no positive manure samples), for a single-farm category comparison (farms B to N) for the milk unit filters, and for aerosol samples obtained from a radial distance of 500 m from the farms. Significant differences were found in C. burnetii DNA contents among the three farm groups (categories AB, B, and N) for aerosols obtained in the stables, while the proportions of positive stable aerosol samples were not significant between these farm groups. Finally, based on Cq values for both targets com1 and IS1111, aerosol samples obtained from the stables contained higher levels of C. burnetii DNA than did the aerosol samples obtained at a 500-m distance from the farms (P < 0.01 by Kruskal-Wallis test).
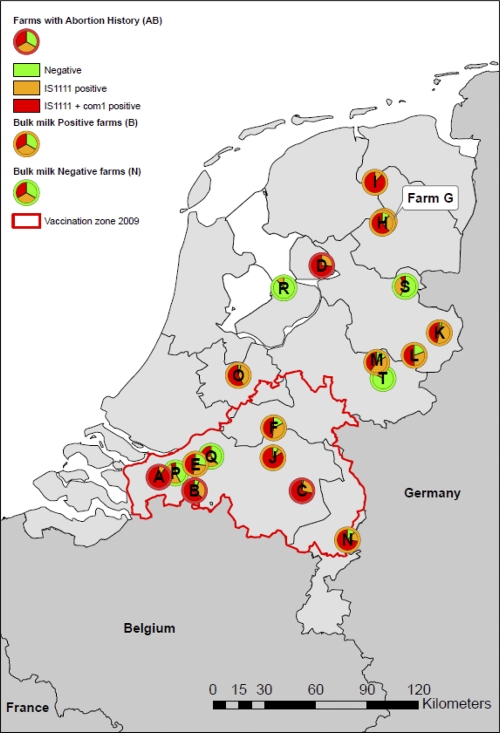
In Fig. 1, the locations of the farms, the farm categories, and the proportions of negative, IS1111-positive, and IS1111- and com1-positive samples per farm are indicated (in green, orange, and red, respectively). Ten dairy goat farms and the dairy sheep farm were located outside the mandatory vaccination area of 2009, and nine dairy goat farms were located within this area.
Fig 1.
Screening for C. burnetii DNA in animal and environmental matrices on 19 dairy goat farms and 1 dairy sheep farm in the Netherlands. Farm categories (categories AB, B, and N) are indicated by red, orange, and green pie chart borders, respectively. The proportions of negative, IS1111-positive, and IS1111- and com1-positive samples are indicated in green, orange, and red, respectively.
DISCUSSION
In our study, generally both the proportions of C. burnetii-positive animal and environmental samples and the C. burnetii DNA contents within positive samples were highest on farms with a C. burnetii-related abortion history (category AB), followed by the farms with positive bulk milk (category B), and were lowest on the control farms (category N). The higher proportions of positive samples and higher levels of C. burnetii DNA content on the category AB and category B farms suggest that these farms pose an increased transmission risk for humans. This was supported by the serological results obtained from the farm occupants. The only exception to this general rule was found for the aerosol samples. It seems that within stables, C. burnetii is almost always present in aerosols, but levels vary with the Q fever status of the farm. In contrast, aerosols obtained from a radial distance of 500 m from category AB and category B farms were more often found to be positive, but the less common positive samples around negative farms seemed to contain similar C. burnetii DNA levels.
Naturally, it should be noted that especially the aerosol samples obtained outside the stables are greatly influenced by the sampling conditions, such as wind direction, wind speed, humidity, and other sources contaminated with C. burnetii. As samples were taken on a random day, the results might not represent the occurrence over a longer time interval. The results for the C. burnetii DNA contents in aerosols in the vicinity of small-ruminant farms should therefore be interpreted with caution and do not allow firm conclusions.
Although clear differences in C. burnetii DNA contents were observed among the different farm categories in our study, these differences are thought to be underestimated for several reasons. First, the excretion of C. burnetii in vaginal mucus and in milk is intermittent and is known to decrease over time after lambing (7).
The sampling period for farms within the mandatory vaccination area occurred longer after the lambing period than for the farms outside this vaccination area. As these first farms included three of the four farms with abortion histories, the C. burnetii DNA contents might have been at a relatively low level at that time of the year for these farms. Second, the reported dates of the actual C. burnetii-related abortions at these farms preceded the sampling date by several months to over a year. It is known that levels of C. burnetii excretion decrease over time following a Q fever outbreak in animals, again resulting in lower levels on the sampling date (3, 8, 18). Finally, at one of the category AB farms (farm D), the pregnant sheep were already culled before sampling took place, again reducing the number of excreting animals within this farm category. Consequently, true differences are expected to be even greater than those observed in our study.
The various animal and environmental matrices studied represent the presence of C. burnetii DNA on and around small-ruminant farms for different time intervals and with different numbers of ruminants on the farm. Surface area swabs give insight into the presence of C. burnetii DNA on a farm over a longer period of time, because (contaminated) dust particles, generated by multiple small ruminants, accumulate on surface areas. Milk unit filters represent a shorter time interval but again represent many goats, as during the passage of the milk, residues accumulate on the milk unit filters. This may explain the relatively high levels of C. burnetii DNA in these two matrices.
In contrast, levels of C. burnetii DNA in vaginal swabs from individual animals were relatively low. This may be explained by the fact that vaginal swabs provide information on the shedding of C. burnetii by individual animals at the moment of sampling only. Also, low levels of C. burnetii DNA were found in aerosols, especially in samples taken outside the stables. Suspended dust particles, obtained by aerosol sampling within stables, contained fewer contaminated dust particles than did surface area swabs from those same stables. The level of C. burnetii DNA in aerosols obtained from stables represents captured contaminated aerosols at the moment of sampling only, which is influenced especially by dust-producing activities in the stable. Because of these legitimate differences between the matrices, a quantitative comparison of C. burnetii DNA contents between matrices within and between farms is difficult. Standardized quantitative sampling of accumulated dust on surface areas (surface area swabs) and dust suspended in air (aerosols) is very difficult. In addition, animal and environmental matrices are known to inhibit the qPCR assay, which complicates the accurate quantitative assessment of C. burnetii DNA in these types of matrices.
Our study had several limitations regarding the sampling strategy. First, ideally, all farms should have been sampled at the same time and before vaccination took place to eliminate the effect of vaccination on the study results. Originally, the entire study was planned to start in the fall of 2009, but in March 2009, it was decided suddenly by the Dutch government that the vaccination of small ruminants was to be mandatory in the south of the country. All efforts were then placed on the immediate preparation of the recruitment of farms and retrieval of sera from a sample of the goats and sheep of the small-ruminant dairy farms in the mandatory vaccination area that were willing to participate. Vaccination probably did not affect the results for the farms with abortions to a large extent, since it is known that the vaccine is not effective on ruminants that are already infected (12). However, excretion by goats and sheep at farms with relatively few affected animals, as might have been the situation for farms in category B, was possibly kept at low levels because the vaccine drastically decreased the amount of new infections, and for the few animals infected despite vaccination, excretion levels would have been reduced. Nevertheless, as results for the bulk milk-positive farms more resembled the results for the farms with abortion histories than results for the negative farms, the effect of vaccination on the outcome of our study seems to have been limited.
Second, the detection of C. burnetii in our study was based on the amplification of specific targets within the C. burnetii genome by qPCR. The viability of the C. burnetii organisms within these matrices can therefore not be assessed. The cultivation of C. burnetii from complex matrices, such as surface area swabs, manure, and milk unit filters, was not successful. In addition, the cultivation of a large number of samples was not feasible, due to the requirement for biosafety level 3 (BSL3) facilities, and was attempted only for highly positive samples. To date, the cultivation of C. burnetii in our laboratory has been successful only with highly positive placenta materials obtained from goats and sheep. However, these materials could not be obtained from the farms included in this study.
From the qPCR results obtained from the various matrices and the circumstantial evidence of the human serology of farm occupants, we conclude that levels of C. burnetii DNA in both animal and environmental matrices are high on C. burnetii-positive farms and most pronounced on farms with a C. burnetii-related abortion wave, which poses an increased risk of infection of humans living on or near these farms. Nevertheless, it should be noted that in addition to the presence of C. burnetii in aerosols and other matrices at the farms, local geographical conditions also influence the actual risk of infection for humans living in the vicinity of the farm, as was recently demonstrated (23). This complex interaction between the farm and environment and the risk to inhabitants in the vicinity are currently the subject of more in-depth investigations.
In conclusion, this study supports epidemiological findings suggesting that contaminated dust and aerosols derived from contaminated animal matrices from C. burnetii-positive dairy goat farms played a predominant role in the transmission of C. burnetii to humans during the outbreaks in the Netherlands (13, 23).
ACKNOWLEDGMENTS
This study was financially supported by the Netherlands Organization for Health Research and Development (ZonMw); the Ministry of Health, Welfare, and Sport (VWS); the Ministry of Economic Affairs, Agriculture, and Innovation (EL&I); and the Food and Consumer Product Safety Authority (nVWA).
We thank Jan van den Bergh for organizing the permission for the veterinarian of the nVWA to perform nationwide sampling as well as for providing background data on the farms. In addition, we are very grateful to all farmers for their cooperation in this study.
Footnotes
Published ahead of print 13 January 2012
REFERENCES
- 1. Amitai Z, et al. 2010. A large Q fever outbreak in an urban school in central Israel. Clin. Infect. Dis. 50:1433–1438 [DOI] [PubMed] [Google Scholar]
- 2. Arricau Bouvery N, Souriau A, Lechopier P, Rodolakis A. 2003. Experimental Coxiella burnetii infection in pregnant goats: excretion routes. Vet. Res. 34:423–433 [DOI] [PubMed] [Google Scholar]
- 3. Astobiza I, Barandika JF, Hurtado A, Juste RA, Garcia-Perez AL. 2010. Kinetics of Coxiella burnetii excretion in a commercial dairy sheep flock after treatment with oxytetracycline. Vet. J. 184:172–175 [DOI] [PubMed] [Google Scholar]
- 4. Astobiza I, et al. 2011. Coxiella burnetii shedding and environmental contamination at lambing in two highly naturally-infected dairy sheep flocks after vaccination. Res. Vet. Sci. 91:e58–e63 [DOI] [PubMed] [Google Scholar]
- 5. Astobiza I, et al. 2011. Four-year evaluation of the effect of vaccination against Coxiella burnetii on reduction of animal infection and environmental contamination in a naturally infected dairy sheep flock. Appl. Environ. Microbiol. 77:7405–7407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bamberg WM, et al. 2007. Outbreak of Q fever associated with a horse-boarding ranch, Colorado, 2005. Vector Borne Zoonotic Dis. 7:394–402 [DOI] [PubMed] [Google Scholar]
- 7. Berri M, Rousset E, Champion JL, Russo P, Rodolakis A. 2007. Goats may experience reproductive failures and shed Coxiella burnetii at two successive parturitions after a Q fever infection. Res. Vet. Sci. 83:47–52 [DOI] [PubMed] [Google Scholar]
- 8. Berri M, et al. 2001. Relationships between the shedding of Coxiella burnetii, clinical signs and serological responses of 34 sheep. Vet. Rec. 148:502–505 [DOI] [PubMed] [Google Scholar]
- 9. de Bruin A, et al. 2011. Detection of Coxiella burnetii in complex matrices by using multiplex quantitative PCR during a major Q fever outbreak in The Netherlands. Appl. Environ. Microbiol. 77:6516–6523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Engelhart S, Glasmacher A, Simon A, Exner M. 2007. Air sampling of Aspergillus fumigatus and other thermotolerant fungi: comparative performance of the Sartorius MD8 Airport and the Merck MAS-100 portable bioaerosol sampler. Int. J. Hyg. Environ. Health 210:733–739 [DOI] [PubMed] [Google Scholar]
- 11. Fitzpatrick KA, Kersh GJ, Massung RF. 2010. Practical method for extraction of PCR-quality DNA from environmental soil samples. Appl. Environ. Microbiol. 76:4571–4573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hogerwerf L, et al. 2011. Reduction of Coxiella burnetii prevalence by vaccination of goats and sheep, the Netherlands. Emerg. Infect. Dis. 17:379–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Karagiannis I, et al. 2009. Investigation of a Q fever outbreak in a rural area of The Netherlands. Epidemiol. Infect. 137:1283–1294 [DOI] [PubMed] [Google Scholar]
- 14. Kersh GJ, et al. 2010. Presence of Coxiella burnetii DNA in the environment of the United States, 2006 to 2008. Appl. Environ. Microbiol. 76:4469–4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Klee SR, et al. 2006. Highly sensitive real-time PCR for specific detection and quantification of Coxiella burnetii. BMC Microbiol. 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Koene RP, et al. 2011. A Q fever outbreak in a psychiatric care institution in The Netherlands. Epidemiol. Infect. 139:13–18 [DOI] [PubMed] [Google Scholar]
- 17. Leski TA, Malanoski AP, Gregory MJ, Lin B, Stenger DA. 2011. Application of a broad-range resequencing array for detection of pathogens in desert dust samples from Kuwait and Iraq. Appl. Environ. Microbiol. 77:4285–4292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rodolakis A, et al. 2007. Comparison of Coxiella burnetii shedding in milk of dairy bovine, caprine, and ovine herds. J. Dairy Sci. 90:5352–5360 [DOI] [PubMed] [Google Scholar]
- 19. Schimmer B, et al. 2011. Seroprevalence and risk factors of Q fever in goats on commercial dairy goat farms in the Netherlands, 2009–2010. BMC Vet. Res. 7:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schimmer B, et al. 2008. Large ongoing Q fever outbreak in the south of The Netherlands, 2008. Euro Surveill. 13(31):pii=18939. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=18939 [PubMed] [Google Scholar]
- 21. Schulz J, Runge M, Schroder C, Ganter M, Hartung J. 2005. Detection of Coxiella burnetii in the air of a sheep barn during shearing. Dtsch. Tierarztl. Wochenschr. 112:470–472 (In German.) [PubMed] [Google Scholar]
- 22. Tissot-Dupont H, Amadei MA, Nezri M, Raoult D. 2004. Wind in November, Q fever in December. Emerg. Infect. Dis. 10:1264–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van der Hoek W, Hunink J, Vellema P, Droogers P. 2011. Q fever in The Netherlands: the role of local environmental conditions. Int. J. Environ. Health Res. 21:441–451 [DOI] [PubMed] [Google Scholar]
- 24. Wallensten A, et al. 2010. Q fever outbreak in Cheltenham, United Kingdom, in 2007 and the use of dispersion modelling to investigate the possibility of airborne spread. Euro Surveill. 15(12):pii=19521. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19521 [PubMed] [Google Scholar]
- 25. Yanase T, et al. 1998. Detection of Coxiella burnetii from dust in a barn housing dairy cattle. Microbiol. Immunol. 42:51–53 [DOI] [PubMed] [Google Scholar]