Abstract
Four hyperthermophilic members of the bacterial genus Thermotoga (T. maritima, T. neapolitana, T. petrophila, and Thermotoga sp. strain RQ2) share a core genome of 1,470 open reading frames (ORFs), or about 75% of their genomes. Nonetheless, each species exhibited certain distinguishing features during growth on simple and complex carbohydrates that correlated with genomic inventories of specific ABC sugar transporters and glycoside hydrolases. These differences were consistent with transcriptomic analysis based on a multispecies cDNA microarray. Growth on a mixture of six pentoses and hexoses showed no significant utilization of galactose or mannose by any of the four species. T. maritima and T. neapolitana exhibited similar monosaccharide utilization profiles, with a strong preference for glucose and xylose over fructose and arabinose. Thermotoga sp. strain RQ2 also used glucose and xylose, but was the only species to utilize fructose to any extent, consistent with a phosphotransferase system (PTS) specific for this sugar encoded in its genome. T. petrophila used glucose to a significantly lesser extent than the other species. In fact, the XylR regulon was triggered by growth on glucose for T. petrophila, which was attributed to the absence of a glucose transporter (XylE2F2K2), otherwise present in the other Thermotoga species. This suggested that T. petrophila acquires glucose through the XylE1F1K1 transporter, which primarily serves to transport xylose in the other three Thermotoga species. The results here show that subtle differences exist among the hyperthermophilic Thermotogales with respect to carbohydrate utilization, which supports their designation as separate species.
INTRODUCTION
Metabolic engineering efforts to develop microorganisms capable of producing biofuels through the fermentation of sugars have largely focused on manipulating central and intermediary metabolism to direct electrons to the desired bioenergy product (1, 54, 55). In most cases, this presumes that the carbon source is homogenous and readily utilized by the microorganism. However, if lignocellulosic feedstocks are to be processed to biofuels efficiently, microbial systems capable of simultaneously processing mixtures of simple and complex sugars are highly desirable since such systems could circumvent or minimize biomass pretreatment requirements. The strategies used by microorganisms capable of utilizing heterogeneous mixtures of complex carbohydrates rely heavily on the regulation of glycoside hydrolase inventories and the presence of ATP-binding cassette (ABC) transporters to move sugars across cell boundaries (51, 52). Understanding these strategies can ultimately lead to metabolically engineered microorganisms that coordinate the uptake and processing of carbohydrates with the synthesis of biofuel molecules.
Members of the genus Thermotoga have been isolated from locations throughout the world, including Italy, France, The Netherlands, the Azores, Japan, and Africa (4, 12, 19, 21, 25, 28, 39, 53). These bacteria possess a large number of glycoside hydrolases and ABC transporters relative to their genome size (7, 41), which allow them to ferment a wide variety of simple and complex carbohydrates, forming hydrogen (or hydrogen sulfide), acetate, and CO2 as the primary metabolic products (10). Polysaccharides that can support growth of Thermotoga include xylan, laminarin, carboxymethyl cellulose, starch, glucomannan, galactomannan, and chitin (7, 11). However, Thermotoga species lack multifunctional enzymes with both endoglucanase and carbohydrate binding module (CBM) domains, at least partially explaining their inability to grow on crystalline cellulose (3). Nevertheless, Thermotoga strains have been extensively investigated as sources of hyperthermophilic mannanases, xylanases, and amylases (2, 40, 45). These bacteria are also noteworthy for high-yield hydrogen production (3 to 4 mol/mol glucose), which may be related to the synergistic utilization of NADH and reduced ferredoxin by a bifurcating hydrogenase (9, 46).
Regulatory networks related to carbohydrate utilization in Thermotoga have been predicted by transcriptomics and comparative genomics (11, 38) and, in some cases, validated by characterization of transcriptional regulators (D. Rodionov, personal communication). Transcription of genes in a particular regulon is generally repressed by the transcriptional regulator and induced by binding of the regulator to an effector (often a mono- or disaccharide). For example, genes in the CelR regulon are involved in catabolism and transport of β-linked glucans and glucomannan. Binding of the CelR regulator (TM1218) to the predicted effector (cellobiose) induces transcription of two endoglucanases (TM1524 and TM1525), a mannobiose transporter (TM1219 to TM1223), a transporter of oligosaccharides derived from xyloglucans (TM0300 to TM0304), and a cellobiose phosphorylase (TM1848) (33, 38). The BglR regulon is also induced by cellobiose and was found to be upregulated during growth on various β-linked glucans, including laminarin, barley glucan, and pustulan (11). This regulon's single operon includes the regulator (TM0032), a laminarinase (TM0024), a β-glucosidase (TM0025), and a cellobiose/laminaribiose transporter (TM0027 to TM0031) (33, 38).
The XylR regulon is composed of xylan utilization genes and is organized into 7 or 8 operons, depending on the Thermotoga species. Binding of the XylR repressor (TM0110) to DNA is disrupted by the inducers xylose and glucose in vitro (D. Rodionov, personal communication). This large regulon includes two xylanases (TM0061 and TM0070), a xylobiose/xylotriose transporter (TM0071 to TM0075), a transporter of unknown xylan degradation products (TM0056 to TM0060), the xylose/glucose transporter XylE1F1K1 (TM0112, TM0114, and TM0115), a β-xylosidase (TM0076), a xylose isomerase (TM1667), and a xylulokinase (TM0116) (11, 33, 38, 50).
Another regulon of interest here (GluR) consists of two transporters organized in one operon that were, until recently, uncharacterized. This operon shows variability from one species to another, as Thermotoga petrophila is lacking one of the two transporters and Thermotoga sp. strain RQ2 is missing an α-amylase. Nearly the entire operon is missing from the Thermotoga maritima strain used to generate the whole-genome sequence published in 1999 (34), but a recent study has shown that the strain of T. maritima deposited in DSMZ has all of these genes, except for the gene coding for α-amylase (5). The transporters in this operon from T. maritima were characterized; TreEFG was shown to bind trehalose, sucrose, and glucose, while XylE2F2K2 was shown to bind xylose, glucose, and fucose (5). The regulator encoded in TM1847 is predicted to bind glucose (38).
Given the wide range of geographic locations inhabited by Thermotoga species, it is interesting to consider the underlying factors that make them so adaptive as well as what, if anything, differentiates one species from another with respect to growth physiology on carbohydrates. Genome sequences are available for several hyperthermophilic Thermotoga species, including T. maritima (34), T. petrophila (56), T. neapolitana, and Thermotoga sp. strain RQ2 (GenBank accession no. AE000512, CP000702, CP000916, and CP000969, respectively). An understanding of the physiological characteristics encoded in the “core” genome, as well as the features that are associated with the “non-core” open reading frames (ORFs), could provide new insights into the intricacies of microbial carbohydrate utilization strategies. To this end, growth of four hyperthermophilic Thermotoga species on glucose, a mixture of six monosaccharides, and a polysaccharide mixture was examined to probe the relationship between genotype and phenotype. Even among these closely related hyperthermophiles, sugar utilization patterns and corresponding transcriptomes revealed distinguishing features that could be important in selecting a platform for biofuel production.
MATERIALS AND METHODS
Design of the multispecies cDNA microarray.
A Thermotoga multispecies cDNA microarray was developed by using a combination of previously constructed cDNA probes from T. maritima (7, 23), along with new probes representing ORFs specific to three other species (T. neapolitana, Thermotoga sp. strain RQ2, and T. petrophila). At the time that the multispecies array was constructed, a complete genome sequence was available for T. petrophila, a draft genome sequence was available for T. neapolitana, and suppressive subtractive hydbridization had been used to compare T. maritima and Thermotoga sp. strain RQ2 (36). Due to the incomplete genome information that was available for Thermotoga sp. strain RQ2 at that time, in some cases, there are multiple probes for the same gene. Note that the Thermotoga sp. strain RQ2 genome is now available (GenBank accession no. CP000969).
The genomes of T. neapolitana and T. petrophila were compared to that of T. maritima using GenomeBlast (27). ORFs with amino acid sequences that were not 70% identical with 80% coverage compared to the other three species were considered to be unique to that bacterium. Vector NTI Advance 10 (Invitrogen, Carlsbad, CA) was used to design probes for these unique ORFs. Probes were produced by PCR using genomic DNA as the template and primers obtained from Integrated DNA Technologies (Coralville, IA). PCR products were spotted onto UltraGAPS slides (Corning), as previously described (7). A few of the smaller probes were ordered directly as oligonucleotides from Integrated DNA Technologies (Coralville, IA).
Growth of Thermotoga species on simple and complex carbohydrates.
Cell densities were determined by epifluorescence microscopy using acridine orange stain, as described previously (48). For monosaccharide utilization experiments, the four species were grown on modified sea salts medium to minimize the effect on the refractive index baseline during high-performance liquid chromatography (HPLC) analysis (with the concentration of sea salts lowered to 10 g/liter and resazurin omitted) (17). The monosaccharide mixture (containing glucose, xylose, arabinose, mannose, galactose, and fructose) was prepared in solution, sterilized by filtration, and added to autoclaved medium at a final concentration of 0.25 g/liter of each sugar. Samples taken were passed through a 0.22-μm-pore filter. Monosaccharide concentrations were determined by HPLC with a Shodex SP0810 column at 80°C (52). Water was used as the mobile phase at a flow rate of 0.2 ml/min.
For transcriptional analysis experiments, four Thermotoga species (T. maritima MSB8, T. neapolitana DSM 4359, T. petrophila RKU-1, and Thermotoga sp. strain RQ2) were each grown in pure culture at 80°C on artificial seawater (ASW) medium (6), supplemented with either glucose (0.25% [wt/vol]) (Sigma-Aldrich, St. Louis, MO) or a polysaccharide mixture (0.083% [wt/vol]) consisting of equal parts by mass of lichenan (>85% purity) (Megazyme, Wicklow, Ireland), konjac glucomannan (90% purity) (Jarrow Formulas, Los Angeles, CA), pectin (>85% purity) (Sigma-Aldrich), galactomannan (95% purity) (Sigma-Aldrich), carboxymethyl cellulose (∼99% purity) (Sigma-Aldrich), xylan from oat spelts (90% purity) (Sigma-Aldrich), and xylan from birchwood (90% purity) (Sigma-Aldrich). Polysaccharides were added to autoclaved medium without sterilization. Prior to extracting total RNA, cultures were passed at least six times on either glucose or the polysaccharide mixture, using a 1% (vol/vol) inoculum. Polysaccharide- or glucose-grown cultures were harvested during mid-exponential phase (3 × 107 to 7 × 107 cells/ml) and quickly cooled in a dry ice-ethanol bath, and cells were collected by centrifugation. Cultures grown on polysaccharides were passed through a coffee filter prior to centrifugation. RNA extraction, reverse transcription, labeling, hybridization, and slide scanning were performed with modifications of previously described protocols (7, 23). Loess normalization, analysis of variance (ANOVA) normalization, and ANOVA row-by-row modeling were performed using JMP Genomics (SAS Institute, Cary, NC). Fold changes at or above 2-fold with significant values at or above the Bonferroni correction (typically equivalent to a P value of approximately 10−5) were considered to be significant differential transcription.
Microarray data accession numbers.
Full results, additional experimental details, and more information about the multispecies array have been deposited in NCBI's Gene Expression Omnibus (13) and are accessible through GEO platform accession no. GPL13655 and GEO series accession no. GSE29557.
RESULTS AND DISCUSSION
Genomic comparison of Thermotoga species.
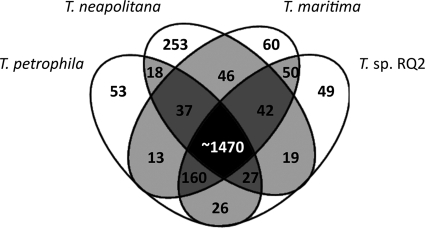
The 10 Thermotoga species currently identified can clearly be divided into one group with optimum growth temperatures of 65 to 70°C and another group with optimum growth temperatures of 75 to 80°C (Table 1). 16S rRNA sequences indicate that the isolates with higher optimum growth temperatures are more closely related (≥99% identical) to each other than to the less thermophilic Thermotoga strains (15). Complete genome sequences are available for seven Thermotoga species, and all of these, except for T. lettingae and T. thermarum, belong to the group of higher-temperature species (Table 1). Only the 16S rRNA sequence was available for T. naphthophila when this study was initiated, so it was not included; note that T. petrophila, which is closely related to T. naphthophila, was included (35). To probe the significance of differences among these species, the genomes of T. maritima, T. neapolitana, Thermotoga sp. strain RQ2, and T. petrophila were compared on an ORF-by-ORF basis. Based on 70% amino acid sequence identity over 80% of the ORF, a core genome of approximately 1,470 ORFs (Fig. 1) was defined for the four hyperthermophilic Thermotoga species. In general, genes involved in central metabolism are highly conserved. All four species have complete glycolytic, pentose phosphate, and Entner-Doudoroff pathways. T. maritima and T. neapolitana have an extra transketolase that is not present in the other two species, but the physiological role and phenotypic implications of this enzyme are unknown.
Table 1.
Genomic characteristics of Thermotoga speciesa
| Species or strain | Isolation site | Location | Topt (°C) | % 16S rRNA identity to T. maritima | No. of protein-coding ORFs |
|---|---|---|---|---|---|
| T. maritima | Geothermally heated sea floor | Vulcano, Italy | 80 | 100 | 1,858 |
| Thermotoga sp. strain RQ2 | Geothermally heated sea floor | São Miguel, Azores | 80 | 99.7 | 1,819 |
| T. neapolitana | Submarine thermal vent | Naples, Italy | 77 | 99.4 | 1,905 |
| T. petrophila | Oil reservoir production fluid | Niigata, Japan | 80 | 99.2 | 1,785 |
| T. naphthophila | Oil reservoir production fluid | Niigata, Japan | 80 | 99.0 | 1,768 |
| T. thermarum | Continental solfataric spring | Djibouti, Africa | 70 | 91.8 | 1,945 |
| T. lettingae | Sulfate-reducing bioreactor | Wageningen, Netherlands | 65 | 90.8 | 2,040 |
| T. elfii | Oil-producing well | Africa | 66 | 90.7 | |
| T. subterranea | Deep continental oil reservoir | East Paris Basin, France | 70 | 90.6 | |
| T. hypogea | Oil-producing well | Cameroon, Africa | 70 | 90.2 |
Shading indicates species with an optimum growth temperature (Topt) of 75 to 80°C.
Fig 1.
Venn diagram summarizing the shared ORFs of the selected hyperthermophilic Thermotoga species, based on 70% identity and 80% coverage at the amino acid level over the entire ORF. Numbers indicate the ORFs shared by subsets of species.
These four species have notable genomic differences with respect to carbohydrate utilization and transport (Table 2). Non-core genes include an arabinose utilization locus, present only in T. petrophila and Thermotoga sp. strain RQ2 (TRQ2_0657 to TRQ2_0667), which consists of a predicted arabinose transporter (Table 2) and four enzymes that are apparently targeted to the secretome. Mannan-derived oligosaccharides are likely transported into the cell by the products of TM1746 to TM1750, before being further hydrolyzed by two intracellular glucomannanases (TM1751 and TM1752) (8), which are only present in T. maritima and Thermotoga sp. strain RQ2. A recently characterized glucose/xylose transporter (5) is missing from T. petrophila (Table 2), while a nearby α-amylase (CTN_0781) is missing from T. maritima and Thermotoga sp. strain RQ2. There are two relatively large loci which are not well characterized and are unique to T. neapolitana. The first (CTN_0355 to CTN_0373) includes a glycoside hydrolase family 31 (GH31) enzyme (CTN_0355) and two putative monosaccharide transporters (Table 2), while the second (CTN_1540 to CTN_1555) includes another GH31 enzyme and an uncharacterized transporter (Table 2). The question arises as to whether these variations in glycoside hydrolases and ABC transporters relate to carbohydrate utilization patterns.
Table 2.
Carbohydrate transporter binding proteins identified in genomes of hyperthermophilic Thermotoga species
| Predicted substrate(s) | Gene of species or strain: |
|||
|---|---|---|---|---|
| T. maritima | T. neapolitana | T. petrophila | Thermotoga sp. strain RQ2 | |
| α-Arabinosides | TM0277 | CTN_0408 | Tpet_0646-7 | TRQ2_0671 |
| Cellobiose, laminaribiose | TM0031 | CTN_0664 | Tpet_0892 | TRQ2_0914 |
| Chitobiose | TM0810 | CTN_1767 | Tpet_0118 | TRQ2_0116 |
| Maltose | TM1204 | CTN_1367 | Tpet_1552 | TRQ2_1614 |
| Maltose | TM1839 | CTN_0765 | Tpet_0966 | TRQ2_0984 |
| Mannobiose | TM1223 | CTN_1348 | Tpet_1545 | TRQ2_1595 |
| Rhamnose related | TM1067 | CTN_1502-3 | Tpet_1677, Tpet_1687 | TRQ2_0510 |
| Trehalose, maltose | treE | CTN_0780 | Tpet_0954 | TRQ2_0970 |
| Xylan related | TM0056 | CTN_0638 | Tpet_0868 | TRQ2_0890 |
| Xylobiose, xylotriose | TM0071 | CTN_0622 | Tpet_0853 | TRQ2_0876 |
| Xylose, glucose | TM0114 | CTN_0576 | Tpet_0810 | TRQ2_0833 |
| Unknown carbohydrate | TM0309 | CTN_0378-9 | Tpet_0608 | TRQ2_0622 |
| Unknown carbohydrate | TM1235 | CTN_1337 | Tpet_1534 | TRQ2_1583 |
| Unknown monosaccharide | TM0595 | CTN_0067 | Tpet_0322 | TRQ2_0340 |
| Galactoside | TM1199 | CTN_1375 | TRQ2_1619 | |
| Glucose, xylose | xylE2 | CTN_0777 | TRQ2_0973 | |
| Mannose-glucuronic acid related | TM1855 | CTN_0790 | Tpet_0942 | |
| Myo-inositol | TM0418 | CTN_0252 | Tpet_0503 | |
| Pectin/galacturonate-related | TM0432 | Tpet_1679 | TRQ2_0512 | |
| Ribose | TM0958 | CTN_1618 | Tpet_1794 | |
| Arabinose | Tpet_0636 | TRQ2_0661 | ||
| Mannan related | TM1746 | TRQ2_1079 | ||
| Mannooligosaccharides | TM1226 | TRQ2_1592 | ||
| Xyloglucooligosaccharides | TM0300 | TRQ2_0631 | ||
| β-Glucoside | CTN_0660 | |||
| Fructose (PTS) | TRQ2_0639 | |||
| Galactoside | CTN_1372 | |||
| Pectin/galacturonate related | Tpet_0485 | |||
| Pectin/galacturonate related | CTN_0240 | |||
| Unknown carbohydrate | CTN_1541 | |||
| Unknown monosaccharides | CTN_0358 | |||
| Unknown monosaccharides | CTN_0364 | |||
Utilization of a mixture of monosaccharides.
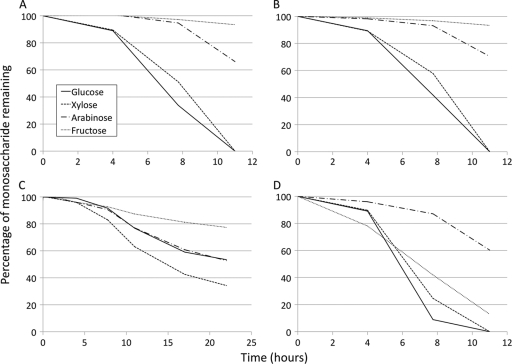
The existence of monosaccharide transporters that are not conserved among all four species (Table 2) suggests that these species may show different sugar preferences. To test this, all four species were grown on equal amounts of glucose, galactose, mannose, fructose, arabinose, and xylose. Consumption of the individual monosaccharides was monitored by HPLC. T. maritima and T. neapolitana have very similar monosaccharide utilization patterns (Fig. 2A and B), and there are no well-annotated monosaccharide transporters that differ between them (Table 2). Both species clearly coutilize glucose and xylose, while utilizing arabinose to a lesser extent. Thermotoga sp. strain RQ2 (Fig. 2D) has a similar pattern, except that fructose is consumed almost as quickly as glucose and xylose. This is most certainly due to the fructose-specific phosphotransferase system (PTS), which is present in Thermotoga sp. strain RQ2 but missing from the other three species (Table 2). All six genes of the PTS locus were significantly upregulated on fructose compared to glucose in Thermotoga sp. strain RQ2 (A. D. Frock and R. M. Kelly, unpublished results), supporting the annotation of this PTS as fructose specific. Similar results were seen for a homologous PTS in Caldicellulosiruptor saccharolyticus (52). Although T. petrophila was initially reported to be unable to grow on xylose (49), in this experiment, xylose was depleted faster than the other monosaccharides (Fig. 2C). Comparison of T. petrophila's utilization pattern to the other species reveals that T. petrophila shows less of a preference for glucose, which can be traced to the absence of a transporter that will be discussed in further detail below.
Fig 2.
Utilization of a mixture of six monosaccharides by four Thermotoga species. Mannose and galactose were not significantly utilized by any of the four species, so these two sugars were omitted from the figures for clarity. (A) T. maritima; (B) T. neapolitana; (C) T. petrophila; (D) Thermotoga sp. strain RQ2.
Design of the Thermotoga multispecies cDNA microarray.
Transcriptomic data for the four Thermotoga species were obtained to provide further insights into metabolic differences arising from carbohydrate processing. Previously, a whole-genome cDNA microarray for T. maritima was used to investigate a range of physiological and ecological issues pertaining to the growth of this bacterium (7, 10, 11, 22–24, 29, 30, 42, 43, 47). The high level of sequence identity between these four species suggested that common microarray probes could be used, expanding the transcriptomics analysis to several Thermotoga species. To include those non-core genes absent from the original T. maritima array, probes were added based on sequences from T. neapolitana (304 probes), Thermotoga sp. strain RQ2 (284 probes), and T. petrophila (60 probes).
Monosaccharide and polysaccharide transcriptomes for Thermotoga species.
To compare the genes expressed for carbohydrate utilization in these species, transcriptomes for each species during growth on a mixture of polysaccharides (including glucan, mannan, xylan, and pectin) were compared to that during growth on a single monosaccharide (glucose). As discussed above, these species possess a significant number of carbohydrate active enzymes and transporters that are not conserved among all four species. However, few of these genes were differentially transcribed on the polysaccharide mix compared to glucose. The most significant differential transcription of non-core genes involved a mannan utilization locus (TM1746 to TM1752) (8) and a mannooligosaccharide binding protein (Table 2) (33), which are present only in T. maritima and Thermotoga sp. strain RQ2. For these two species, these genes were upregulated on the polysaccharide mix compared to glucose (Table 3). It is not known how the absence of these genes affects mannan utilization by T. neapolitana and T. petrophila. All four species do have an extracellular endo-1,4-β-mannanase (TM1227) and a mannobiose transporter (TM1219 to TM1223) (33), which may be sufficient for mannan consumption.
Table 3.
ORFs responding in hyperthermophilic Thermotoga species during growth on polysaccharide mixture compared to growth on glucose
| Probe | Product | Fold change in expression ina: |
|||
|---|---|---|---|---|---|
| T. maritima | T. neapolitana | T. petrophila | Thermotoga sp. strain RQ2 | ||
| Mannan utilization | |||||
| TM1746 | Mannan ABC transporter, periplasmic oligopeptide-binding protein | 4.6 | Absent | Absent | 3.7 |
| TM1747 | Mannan ABC transporter, permease protein | 3.7 | Absent | Absent | 8.3 |
| TM1748 | Mannan ABC transporter, permease protein | Absent | Absent | ||
| TM1749 | Mannan ABC transporter, ATP-binding protein | 6.4 | Absent | Absent | 4.3 |
| TM1750 | Mannan ABC transporter, ATP-binding protein | 5.5 | Absent | Absent | 6.7 |
| TM1751 | Endoglucanase | 5.9 | Absent | Absent | 2.5 |
| ManR operon | |||||
| TM1224 | ManR transcriptional regulator, ROK family | 3.3 | 2.4 | 8.7 | |
| TM1226 | Mannooligosaccharide ABC transporter, sugar-binding protein | 5.4 | Absent | Absent | 18.3 |
| TM1227 | Endo-1,4-β-mannosidase | 24.1 | 12.7 | 3.9 | 15.3 |
| CelR regulon | |||||
| TM0312 | Predicted dehydrogenase | 2.1 | 2.1 | ||
| TM0313 | Predicted aldo/keto reductase | 2.0 | 2.6 | ||
| TM1218 | CelR transcriptional regulator, LacI family | 3.8 | 3.9 | ||
| TM1219 | Mannobiose ABC transporter, ATP-binding protein | 2.6 | 2.5 | ||
| TM1223 | Mannobiose ABC transporter, sugar-binding protein | 10.0 | 25.4 | 4.9 | 18.8 |
| TM1524 | Endoglucanase | 7.7 | 6.8 | 3.9 | |
| TM1525 | Endoglucanase | 9.5 | 7.3 | ||
| TM1848 | Cellobiose phosphorylase | 10.2 | 4.7 | 19.7 | 9.4 |
| TRAP transporter | |||||
| TM0322 | TRAP transporter, periplasmic substrate-binding protein, putative | 8.1 | 14.7 | ||
| TM0323 | TRAP transporter, small transmembrane component | 7.0 | |||
| TM0324 | TRAP transporter, large transmembrane component | 3.9 | |||
| TM0325 | Predicted sugar dehydrogenase | 2.8 | 12.0 | ||
| TM0326 | Transcriptional regulator, RpiR family | −3.6 | 2.6 | ||
| TM0327 | Phosphoglycerate dehydrogenase, putative | 6.2 | |||
| Processing of α-glucans | |||||
| TM0752 | α-Glucosidase, putative | 5.3 | 2.2 | ||
| TM0767 | Maltodextrin glycosyltransferase | −2.7 | −6.1 | ||
| TM1834 | α-Glucosidase | −3.6 | |||
| TM1839 | Maltose ABC transporter, periplasmic maltose-binding protein | −3.8 | −2.3 | −6.6 | |
| TM1841 | Hypothetical protein | −2.5 | |||
| TM1842 | Hypothetical protein | −2.6 | −3.2 | −5.9 | |
| TM1843 | Hypothetical protein | −3.2 | |||
| TM1844 | Hypothetical protein | −5.1 | |||
| TM1845 | Pullulanase | −5.1 | |||
| CTN_1407 | α-Glucan phosphorylase | −2.3 | |||
| TRQ2_1647a | α-Glucan phosphorylase | −2.2 | −2.5 | ||
| BglR operon | |||||
| TM0023 | Methyl-accepting chemotaxis protein | −7.8 | −2.4 | ||
| TM0024 | Laminarinase | −2.6 | −11.8 | ||
| TM0025 | β-Glucosidase | −2.1 | 2.1 | −6.7 | |
| TM0026 | Hypothetical protein | −3.1 | |||
| TM0027 | Cellobiose/laminaribiose ABC transporter, ATP-binding protein | −2.0 | −4.7 | −2.6 | |
| TM0028 | Cellobiose/laminaribiose ABC transporter, ATP-binding protein | −2.0 | −8.0 | −2.4 | |
| TM0029 | Cellobiose/laminaribiose ABC transporter, permease protein | −5.0 | −2.6 | ||
| TM0031 | Cellobiose/laminaribiose ABC transporter, sugar-binding protein | −2.7 | −11.6 | −2.7 | |
| TM0032 | BglR transcriptional regulator, ROK family | −4.2 | |||
Shaded values, significantly upregulated on the polysaccharide mix; unshaded values, significantly upregulated on glucose; blank spaces, no statistically significant change; Absent, absent from the indicated genome. The designation “TRQ2_1647a” indicates that there is more than one microarray probe for TRQ2_1647.
Assuming that conserved genes have similar functions in all four species, these genes would be expected to exhibit similar transcriptional responses. This is true in some cases, such as the genes regulated by CelR (Table 3), which are involved in utilizing β-glucans and mannans (11). Genes in this regulon include the previously mentioned mannobiose transporter (TM1219 to TM1223), intracellular and extracellular endoglucanases (TM1524 and TM1525), and a cellobiose phosphorylase (TM1848) with a broad substrate range in vitro (44). TM1223 and TM1848 were upregulated significantly in all four species on the polysaccharide mix compared to glucose. The only genes under the control of CelR that were not upregulated on the polysaccharide mix (TM0299 to TM0307) are putatively coregulated by GloR (38), suggesting that under these conditions their expression may be repressed by this poorly studied regulator.
In other cases, transcriptional responses were less conserved among the four species. All six genes in an operon (TM0322 to TM0327) that includes a tripartite ATP-independent periplasmic (TRAP) transporter were significantly upregulated on the polysaccharide mix in Thermotoga sp. strain RQ2 (Table 3). Two of these genes were upregulated in T. neapolitana, while none were in T. petrophila or T. maritima. This locus was not observed to be responsive in T. maritima grown on 10 different carbon sources (11). TRAP transporters have primarily been shown to bind organic acids (14), so this transporter's role in polysaccharide utilization is unclear. Other genes with different responses in at least one species include those regulated by BglR and those involved in processing α-glucans (Table 3). BglR-regulated genes were generally more highly transcribed during growth on glucose, except for in T. neapolitana, where only one gene was differentially regulated (TM0025), and it was more highly transcribed on the polysaccharide mix. Genes listed in Table 3 involved in processing α-glucans were generally upregulated on glucose, except in the case of T. neapolitana, where one gene (TM0752) was upregulated on the polysaccharide mix.
Glucose transport.
Transcriptomes for growth on glucose and the polysaccharide mix facilitated the identification of a glucose-binding protein not present in the initial T. maritima genome sequence. Although Thermotoga species have been known to grow on glucose since the initial isolation of T. maritima (21), the corresponding genes responsible for glucose uptake were not identified. Glucose transport in T. neapolitana was previously shown to be dependent on ATP and sodium ions (16), and glucose binding activity was detected in cell extracts of T. maritima (31), but subsequent characterization of 15 binding proteins failed to identify one with a high specificity for glucose (33). XylE1 (the product of TM0114) is present in all four species (Table 2) and was shown to bind glucose (50), but this protein also binds xylose with high affinity (Kd [dissociation constant], ∼0.01 μM) (33). Furthermore, TM0114 was more highly transcribed in T. maritima during chemostat growth on xylose than glucose (C.-J. Chou and R. M. Kelly, unpublished results). Therefore, TM0114's involvement in in vivo glucose transport is unclear.
Here, four genes encoding sugar binding proteins were upregulated on glucose compared to the polysaccharide mix (Tables 3 and 4): TM1839, TM0031, TRQ2_0973, and TRQ2_0970 (note that TRQ2_0973 and TRQ2_0970 are homologous to CTN_0777 and CTN_0780, respectively). TM0031 was shown to bind cellobiose and laminaribiose (33), and TM1839 was shown to bind maltose, maltotriose, and trehalose (32). Both of these genes are present in all four species (Table 2). Homologs of TRQ2_0970 and TRQ2_0973 are not present in the published T. maritima genome (34), yet there was significant hybridization of T. maritima cDNA to the probes for these genes. Components of both transporters were significantly upregulated in T. maritima grown on glucose (Table 4). The microarray probes significantly upregulated in T. maritima grown on glucose correspond to a total of 2,658 bp ranging over 5,032 of the 8,862 bp that are present in Thermotoga sp. strain RQ2 but absent from the published genome for T. maritima, strongly suggesting that this entire region is actually present in the T. maritima strain used in this study. In fact, it was recently reported that the T. maritima isolate deposited in DSMZ (used in this study) contains 8,870 bp of genomic DNA that is missing from the isolate sequenced by TIGR in 1999 (5, 34). The product of the TRQ2_0970 homolog (treE) was found to bind trehalose, sucrose, and glucose, while the product of the TRQ2_0973 homolog (xylE2) was found to bind xylose, glucose, and fucose (5). Past studies that focused on T. maritima (33) failed to identify xylE2 because the sequence was missing from the sequenced strain. Noll et al. (37) predicted this genomic variation by examining gene synteny in related species and a β-glucosidase sequence in this region that was published prior to the complete genome (26). In the absence of these hints, genomic variations may not be discovered until the genome is resequenced or other analyses (e.g., RNA-seq) are used. The genomic variation was revealed by the multispecies microarray developed for this study, demonstrating the utility of microarrays in detecting mutations.
Table 4.
ORFs regulated by GluR in four hyperthermophilic Thermotoga species during growth on a polysaccharide mix compared to growth on glucose
| Probe | Product | Fold change in expression ina: |
|||
|---|---|---|---|---|---|
| T. maritima | T. neapolitana | T. petrophila | Thermotoga sp. strain RQ2 | ||
| TM1847 | GluR transcriptional regulator, ROK family | −2.8 | |||
| TRQ2_0975-0976 | GluR transcriptional regulator/glucose ABC transporter, ATP-binding protein | Absent | −2.7 | ||
| CTN_0775 | Glucose ABC transporter, ATP-binding protein | −2.4 | Absent | −4.6 | |
| TRQ2_0975a | Glucose ABC transporter, ATP-binding protein | −3.2 | Absent | −2.5 | |
| TRQ2_0975b | Glucose ABC transporter, ATP-binding protein | −3.1 | Absent | −2.2 | |
| TRQ2_0975c | Glucose ABC transporter, ATP-binding protein | −3.0 | Absent | −2.5 | |
| TRQ2_0974-0975 | Glucose ABC transporter, ATP-binding protein/permease protein | −3.0 | Absent | −3.1 | |
| TRQ2_0974 | Glucose ABC transporter, permease protein | Absent | −2.4 | ||
| CTN_0777 | Glucose ABC transporter, sugar-binding protein | −2.2 | Absent | −7.0 | |
| TRQ2_0973b | Glucose ABC transporter, sugar-binding protein | −2.9 | −2.0 | Absent | −14.4 |
| TRQ2_0973c | Glucose ABC transporter, sugar-binding protein | −3.5 | Absent | −6.3 | |
| TRQ2_0972-0973 | Glucose transporter sugar-binding protein/trehalose transporter permease protein | −2.2 | Absent | −6.4 | |
| CTN_0779b | Trehalose ABC transporter, permease protein | −2.1 | −2.4 | ||
| TRQ2_0971 | Trehalose ABC transporter, permease protein | −3.3 | −3.0 | ||
| CTN_0780 | Trehalose ABC transporter, sugar-binding protein | −2.8 | −8.4 | ||
| TRQ2_0970a | Trehalose ABC transporter, sugar-binding protein | −4.0 | −18.2 | ||
| TRQ2_0970b | Trehalose ABC transporter, sugar-binding protein | −4.2 | −13.1 | ||
The values shown represent significantly upregulated on glucose. Blank spaces, no statistically significant change; Absent, absent from the indicated genome. When probe names include two ORFs (e.g., “TRQ2_0975-0976”), the microarray probe covers parts of both ORFs (e.g., TRQ2_0975 and TRQ2_0976), as well as the intergenic region.
Regulation of xylan utilization genes.
The most striking difference observed between these species involved genes under the control of the regulator XylR (Table 5) that are present in all four species. As described above, this regulon includes all genes necessary for hydrolyzing xylan, transporting degradation products into the cell, and assimilating the resulting sugars into the pentose phosphate pathway. One would expect that these genes would be upregulated on the polysaccharide mix, which included xylan. For three of these species, this was the case (Table 5), but in T. petrophila, these genes were surprisingly downregulated on the polysaccharide mix compared to glucose.
Table 5.
ORFs regulated by XylR in four Thermotoga species during growth on a polysaccharide mix compared to growth on glucose
| Probe | Product | Fold change in expression ina: |
|||
|---|---|---|---|---|---|
| T. maritima | T. neapolitana | T. petrophila | Thermotoga sp. strain RQ2 | ||
| TM0056 | Xylan ABC transporter, periplasmic sugar-binding protein | 6.1 | 4.8 | −5.8 | 9.7 |
| TM0057 | Xylan ABC transporter, ATP-binding protein | 5.7 | 3.6 | 6.2 | |
| TM0058 | Xylan ABC transporter, ATP-binding protein | 5.7 | −3.5 | ||
| TM0060 | Xylan ABC transporter, permease protein | 4.7 | |||
| TM0061 | Endo-1,4-β-xylanase A | 8.2 | 11.7 | −15.3 | 16.8 |
| CTN_0632 | Endo-1,4-β-xylanase A | 2.2 | 5.9 | ||
| TM0062 | Hypothetical protein | 19.4 | Absent | −6.6 | Absent |
| TM0070 | Endo-1,4-β-xylanase B | 10.6 | 2.7 | 3.1 | |
| TM0071 | Xylooligosaccharide ABC transporter, periplasmic sugar-binding protein | 13.8 | 2.6 | −7.3 | 14.4 |
| TM0072 | Xylooligosaccharide ABC transporter, permease protein | 2.1 | −4.8 | ||
| TM0073 | Xylooligosaccharide ABC transporter, permease protein | 2.1 | −3.9 | 2.9 | |
| TM0074 | Xylooligosaccharide ABC transporter, ATP-binding protein | −2.9 | |||
| TM0075 | Xylooligosaccharide ABC transporter, ATP-binding protein | −2.6 | |||
| TM0076 | Xylosidase | −2.6 | |||
| TM0077 | Acetyl xylan esterase | 2.6 | −2.2 | 2.6 | |
| TM0110 | Transcriptional regulator, XylR related | 2.5 | |||
| TM0111 | Alcohol dehydrogenase, iron-containing | −3.6 | |||
| TM0113 | xylU-related protein | −4.0 | |||
| TM0114 | Xylose/glucose ABC transporter, periplasmic sugar-binding protein | −4.6 | |||
| TM0115 | Xylose/glucose ABC transporter, ATP-binding protein | −2.7 | |||
| TM0309 | Xylan ABC transporter, periplasmic sugar-binding protein | 7.5 | 3.0 | −2.6 | 10.3 |
| TM0310 | β-d-Galactosidase | 9.2 | −2.1 | 2.8 | |
| TM1667 | Xylose isomerase | 7.2 | 2.8 | 6.3 | |
Shaded values, significantly upregulated on the polysaccharide mix; unshaded values, significantly upregulated on glucose; blank spaces, no statistically significant change; Absent, absent from the indicated genome.
One possible explanation for this unique regulation pattern is that T. petrophila is simply not using the xylan component of the polysaccharide mix. Initially, T. petrophila was reported to be unable to use xylose (49), but the cell density reached when xylose or xylan was supplemented at 2.5 g/liter was approximately 3-fold higher than that on ASW base medium (data not shown), suggesting that T. petrophila is fully capable of growing on these substrates. Also, if T. petrophila was simply not using xylan, one would expect transcript levels for these genes to be similar for both conditions, not higher on glucose. A second possible explanation for the downregulation of the XylR regulon on the polysaccharide mix in T. petrophila is that these genes are repressed by one of the other carbohydrates in the polysaccharide mix. To investigate this possibility, T. petrophila was grown with xylan as the sole carbon source, and transcript levels were compared to glucose. Again, most XylR genes were significantly downregulated on xylan compared to glucose, while no XylR genes were significantly upregulated (data not shown), suggesting that the other components of the polysaccharide mix did not repress the transcription of XylR genes.
Another possible explanation for T. petrophila's regulation of XylR genes is that these genes were actually induced by glucose. As previously mentioned, in T. maritima, Thermotoga sp. strain RQ2, and T. neapolitana, XylE2 is the only sugar binding protein whose transcript was upregulated on glucose compared to the polysaccharide mix that has also been shown to bind glucose (5). However, the transporter is missing from T. petrophila (Table 2), which causes this species to show less of a preference for glucose (Fig. 2C), as discussed above. However, T. petrophila does utilize glucose (Fig. 2C), so this species must bring glucose into the cell by some other means. The only other protein shown to bind glucose in these bacteria, XylE1 (encoded by TM0114), also binds xylose and is regulated by XylR (33, 50). The XylR transcriptional regulator (TM0110) can bind both glucose and xylose, which derepresses transcription of XylR regulon genes (D. Rodionov, personal communication). Therefore, when T. petrophila grows on glucose, it appears that glucose is brought into the cell via XylE1, and glucose binds to the XylR regulator, resulting in increased transcription of all of the XylR regulon genes. T. petrophila might be considered inefficient at utilizing glucose, because it must upregulate so many unneeded XylR genes. This is an interesting example of small differences between closely related species resulting in significantly different strategies for carbohydrate utilization and transcriptional regulation.
Summary.
T. maritima, T. neapolitana, T. petrophila, and Thermotoga sp. strain RQ2 share approximately 1,470 genes (about 75% of their genomes), yet there are differences in sugar utilization and regulatory phenotypes that relate to the genotypes of these species. The multispecies microarray revealed multiple genes that are present in the T. maritima strain from DSMZ, but missing from the published genome sequence, including a glucose transporter that had eluded previous research efforts. When given a mixture of monosaccharides, only Thermotoga sp. strain RQ2 consumed fructose, consistent with the fact that the fructose-specific phosphotransferase system is not present in the other three species. Also, the absence of xylE2 in T. petrophila impairs its ability to use glucose and necessitates the upregulation of xylE1 along with the rest of the xylan utilization regulon during growth on glucose.
The results presented here provide further evidence that the ability of Thermotoga species to ferment individual carbohydrates in heterogeneous growth substrates is dictated by the available set of ABC transporters. The inability of some model biofuel-producing microorganisms to coferment glucose and xylose has prompted metabolic engineering efforts to knock out certain native transporters and insert heterologous systems to remedy this deficiency (18, 20). Thermotoga species are known to naturally coferment glucose and xylose and merit close examination to understand the metabolic basis for this physiological trait. It is interesting that while Thermotoga species appear to transport glucose and xylose using separate ABC transporters, no transporters have been identified that are specific for glucose or xylose. The two sugar binding proteins known to bind either sugar (XylE1 and XylE2) in fact bind both glucose and xylose. This versatility may be important in utilizing renewable heterogeneous feedstocks, which otherwise could limit biofuel production. This underscores the importance of understanding the connection between carbohydrate active enzymes, carbohydrate transporters, and their coordinated regulation.
ACKNOWLEDGMENTS
This work was supported by the U.S. Department of Energy GTL Program (DG-FG02-08ER64687). A.D.F. and S.R.G. acknowledge support from NIH Pre-doctoral Biotechnology Traineeships (NIH T32 GM008776-06).
Helpful discussions with K. Noll (University of Connecticut) and D. Rodionov (Sanford-Burnham Medical Research Institute, La Jolla, CA) are also acknowledged.
Footnotes
Published ahead of print 13 January 2012
REFERENCES
- 1. Atsumi S, Hanai T, Liao JC. 2008. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451:86–89 [DOI] [PubMed] [Google Scholar]
- 2. Ballschmiter M, Futterer O, Liebl W. 2006. Identification and characterization of a novel intracellular alkaline alpha-amylase from the hyperthermophilic bacterium Thermotoga maritima MSB8. Appl. Environ. Microbiol. 72:2206–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blumer-Schuette SE, Kataeva I, Westpheling J, Adams MW, Kelly RM. 2008. Extremely thermophilic microorganisms for biomass conversion: status and prospects. Curr. Opin. Biotechnol. 19:210–217 [DOI] [PubMed] [Google Scholar]
- 4. Bonch-Osmolovskaya EA, et al. 2003. Radioisotopic, culture-based, and oligonucleotide microchip analyses of thermophilic microbial communities in a continental high-temperature petroleum reservoir. Appl. Environ. Microbiol. 69:6143–6151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boucher N, Noll KM. 2011. Ligands of thermophilic ABC transporters encoded in a newly sequenced genomic region of Thermotoga maritima MSB8 screened by differential scanning fluorimetry. Appl. Environ. Microbiol. 77:6395–6399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chhabra SR, Kelly RM. 2002. Biochemical characterization of Thermotoga maritima endoglucanase Cel74 with and without a carbohydrate binding module (CBM). FEBS Lett. 531:375–380 [DOI] [PubMed] [Google Scholar]
- 7. Chhabra SR, et al. 2003. Carbohydrate-induced differential gene expression patterns in the hyperthermophilic bacterium Thermotoga maritima. J. Biol. Chem. 278:7540–7552 [DOI] [PubMed] [Google Scholar]
- 8. Chhabra SR, Shockley KR, Ward DE, Kelly RM. 2002. Regulation of endo-acting glycosyl hydrolases in the hyperthermophilic bacterium Thermotoga maritima grown on glucan- and mannan-based polysaccharides. Appl. Environ. Microbiol. 68:545–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chou CJ, Jenney FE, Jr, Adams MW, Kelly RM. 2008. Hydrogenesis in hyperthermophilic microorganisms: implications for biofuels. Metab. Eng. 10:394–404 [DOI] [PubMed] [Google Scholar]
- 10. Conners SB, et al. 2006. Microbial biochemistry, physiology, and biotechnology of hyperthermophilic Thermotoga species. FEMS Microbiol. Rev. 30:872–905 [DOI] [PubMed] [Google Scholar]
- 11. Conners SB, et al. 2005. An expression-driven approach to the prediction of carbohydrate transport and utilization regulons in the hyperthermophilic bacterium Thermotoga maritima. J. Bacteriol. 187:7267–7282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dahle H, Garshol F, Madsen M, Birkeland NK. 2008. Microbial community structure analysis of produced water from a high-temperature North Sea oil-field. Antonie Van Leeuwenhoek 93:37–49 [DOI] [PubMed] [Google Scholar]
- 13. Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fischer M, Zhang QY, Hubbard RE, Thomas GH. 2010. Caught in a TRAP: substrate-binding proteins in secondary transport. Trends Microbiol. 18:471–478 [DOI] [PubMed] [Google Scholar]
- 15. Frock AD, Notey JS, Kelly RM. 2010. The genus Thermotoga: recent developments. Environ. Technol. 31:1169–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Galperin MY, Noll KM, Romano AH. 1996. The glucose transport system of the hyperthermophilic anaerobic bacterium Thermotoga neapolitana. Appl. Environ. Microbiol. 62:2915–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gao J, Bauer MW, Shockley KR, Pysz MA, Kelly RM. 2003. Growth of hyperthermophilic archaeon Pyrococcus furiosus on chitin involves two family 18 chitinases. Appl. Environ. Microbiol. 69:3119–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gosset G. 2005. Improvement of Escherichia coli production strains by modification of the phosphoenolpyruvate:sugar phosphotransferase system. Microb. Cell Fact. 4:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hetzer A, Morgan HW, McDonald IR, Daughney CJ. 2007. Microbial life in Champagne Pool, a geothermal spring in Waiotapu, New Zealand. Extremophiles 11:605–614 [DOI] [PubMed] [Google Scholar]
- 20. Ho NW, Chen Z, Brainard AP. 1998. Genetically engineered Saccharomyces yeast capable of effective cofermentation of glucose and xylose. Appl. Environ. Microbiol. 64:1852–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huber R, et al. 1986. Thermotoga maritima sp-nov represents a new genus of unique extremely thermophilic eubacteria growing up to 90 degrees C. Arch. Microbiol. 144:324–333 [Google Scholar]
- 22. Johnson MR, et al. 2006. The Thermotoga maritima phenotype is impacted by syntrophic interaction with Methanococcus jannaschii in hyperthermophilic coculture. Appl. Environ. Microbiol. 72:811–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Johnson MR, et al. 2005. Population density-dependent regulation of exopolysaccharide formation in the hyperthermophilic bacterium Thermotoga maritima. Mol. Microbiol. 55:664–674 [DOI] [PubMed] [Google Scholar]
- 24. Johnson MR, et al. 2004. Functional genomics-based studies of the microbial ecology of hyperthermophilic micro-organisms. Biochem. Soc. Trans. 32:188–192 [DOI] [PubMed] [Google Scholar]
- 25. Li H, Yang SZ, Mu BZ, Rong ZF, Zhang J. 2007. Molecular phylogenetic diversity of the microbial community associated with a high-temperature petroleum reservoir at an offshore oilfield. FEMS Microbiol. Ecol. 60:74–84 [DOI] [PubMed] [Google Scholar]
- 26. Liebl W, Gabelsberger J, Schleifer KH. 1994. Comparative amino acid sequence analysis of Thermotoga maritima beta-glucosidase (BglA) deduced from the nucleotide sequence of the gene indicates distant relationship between beta-glucosidases of the BGA family and other families of beta-1,4-glycosyl hydrolases. Mol. Gen. Genet. 242:111–115 [DOI] [PubMed] [Google Scholar]
- 27. Lu G, et al. 2006. GenomeBlast: a web tool for small genome comparison. BMC Bioinformatics 7(Suppl 4):S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miroshnichenko ML, Bonch-Osmolovskaya EA. 2006. Recent developments in the thermophilic microbiology of deep-sea hydrothermal vents. Extremophiles 10:85–96 [DOI] [PubMed] [Google Scholar]
- 29. Montero CI, et al. 2007. Responses of wild-type and resistant strains of the hyperthermophilic bacterium Thermotoga maritima to chloramphenicol challenge. Appl. Environ. Microbiol. 73:5058–5065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Montero CI, et al. 2006. Colocation of genes encoding a tRNA-mRNA hybrid and a putative signaling peptide on complementary strands in the genome of the hyperthermophilic bacterium Thermotoga maritima. J. Bacteriol. 188:6802–6807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nanavati D, Noll KM, Romano AH. 2002. Periplasmic maltose- and glucose-binding protein activities in cell-free extracts of Thermotoga maritima. Microbiology 148:3531–3537 [DOI] [PubMed] [Google Scholar]
- 32. Nanavati DM, Nguyen TN, Noll KM. 2005. Substrate specificities and expression patterns reflect the evolutionary divergence of maltose ABC transporters in Thermotoga maritima. J. Bacteriol. 187:2002–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nanavati DM, Thirangoon K, Noll KM. 2006. Several archaeal homologs of putative oligopeptide-binding proteins encoded by Thermotoga maritima bind sugars. Appl. Environ. Microbiol. 72:1336–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nelson KE, et al. 1999. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature 399:323–329 [DOI] [PubMed] [Google Scholar]
- 35. Nesbo CL, Dlutek M, Doolittle WF. 2006. Recombination in Thermotoga: implications for species concepts and biogeography. Genetics 172:759–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nesbo CL, Nelson KE, Doolittle WF. 2002. Suppressive subtractive hybridization detects extensive genomic diversity in Thermotoga maritima. J. Bacteriol. 184:4475–4488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Noll KM, Lapierre P, Gogarten JP, Nanavati DM. 2008. Evolution of mal ABC transporter operons in the Thermococcales and Thermotogales. BMC Evol. Biol. 8:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Novichkov PS, et al. 2010. RegPrecise: a database of curated genomic inferences of transcriptional regulatory interactions in prokaryotes. Nucleic Acids Res. 38:D111–D118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Orphan VJ, Taylor LT, Hafenbradl D, Delong EF. 2000. Culture-dependent and culture-independent characterization of microbial assemblages associated with high-temperature petroleum reservoirs. Appl. Environ. Microbiol. 66:700–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Parker KN, et al. 2001. Galactomannanases Man2 and Man5 from Thermotoga species: growth physiology on galactomannans, gene sequence analysis, and biochemical properties of recombinant enzymes. Biotechnol. Bioeng. 75:322–333 [DOI] [PubMed] [Google Scholar]
- 41. Paulsen IT, Nguyen L, Sliwinski MK, Rabus R, Saier MH., Jr 2000. Microbial genome analyses: comparative transport capabilities in eighteen prokaryotes. J. Mol. Biol. 301:75–100 [DOI] [PubMed] [Google Scholar]
- 42. Pysz MA, et al. 2004. Transcriptional analysis of biofilm formation processes in the anaerobic, hyperthermophilic bacterium Thermotoga maritima. Appl. Environ. Microbiol. 70:6098–6112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pysz MA, et al. 2004. Transcriptional analysis of dynamic heat-shock response by the hyperthermophilic bacterium Thermotoga maritima. Extremophiles 8:209–217 [DOI] [PubMed] [Google Scholar]
- 44. Rajashekhara E, Kitaoka M, Kim YK, Hayashi K. 2002. Characterization of a cellobiose phosphorylase from a hyperthermophilic eubacterium, Thermotoga maritima MSB8. Biosci. Biotechnol. Biochem. 66:2578–2586 [DOI] [PubMed] [Google Scholar]
- 45. Saul DJ, Williams LC, Reeves RA, Gibbs MD, Bergquist PL. 1995. Sequence and expression of a xylanase gene from the hyperthermophile Thermotoga sp. strain FjSS3-B.1 and characterization of the recombinant enzyme and its activity on kraft pulp. Appl. Environ. Microbiol. 61:4110–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schut GJ, Adams MW. 2009. The iron-hydrogenase of Thermotoga maritima utilizes ferredoxin and NADH synergistically: a new perspective on anaerobic hydrogen production. J. Bacteriol. 191:4451–4457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shockley KR, et al. 2005. Genome-wide transcriptional variation within and between steady states for continuous growth of the hyperthermophile Thermotoga maritima. Appl. Environ. Microbiol. 71:5572–5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shockley KR, et al. 2003. Heat shock response by the hyperthermophilic archaeon Pyrococcus furiosus. Appl. Environ. Microbiol. 69:2365–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Takahata Y, Nishijima M, Hoaki T, Maruyama T. 2001. Thermotoga petrophila sp. nov. and Thermotoga naphthophila sp. nov., two hyperthermophilic bacteria from the Kubiki oil reservoir in Niigata, Japan. Int. J. Syst. Evol. Microbiol. 51:1901–1909 [DOI] [PubMed] [Google Scholar]
- 50. Tian Y, et al. 2007. Structure-based design of robust glucose biosensors using a Thermotoga maritima periplasmic glucose-binding protein. Protein Sci. 16:2240–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. VanFossen AL, Ozdemir I, Zelin SL, Kelly RM. 2011. Glycoside hydrolase inventory drives plant polysaccharide deconstruction by the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus. Biotechnol. Bioeng. 108:1559–1569 [DOI] [PubMed] [Google Scholar]
- 52. VanFossen AL, Verhaart MR, Kengen SM, Kelly RM. 2009. Carbohydrate utilization patterns for the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus reveal broad growth substrate preferences. Appl. Environ. Microbiol. 75:7718–7724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Windberger E, Huber R, Trincone A, Fricke H, Stetter K. 1989. Thermotoga thermarum sp-nov and Thermotoga neapolitana occurring in African continental solfataric springs. Arch. Microbiol. 151:506–512 [Google Scholar]
- 54. Yomano LP, York SW, Zhou S, Shanmugam KT, Ingram LO. 2008. Re-engineering Escherichia coli for ethanol production. Biotechnol. Lett. 30:2097–2103 [DOI] [PubMed] [Google Scholar]
- 55. Yoshida A, Nishimura T, Kawaguchi H, Inui M, Yukawa H. 2005. Enhanced hydrogen production from formic acid by formate hydrogen lyase-overexpressing Escherichia coli strains. Appl. Environ. Microbiol. 71:6762–6768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhaxybayeva O, et al. 2009. On the chimeric nature, thermophilic origin, and phylogenetic placement of the Thermotogales. Proc. Natl. Acad. Sci. U. S. A. 106:5865–5870 [DOI] [PMC free article] [PubMed] [Google Scholar]