Abstract
Copper alloy surfaces are passive antimicrobial sanitizing agents that kill bacteria, fungi, and some viruses. Studies of the mechanism of contact killing in Escherichia coli implicate the membrane as the target, yet the specific component and underlying biochemistry remain unknown. This study explores the hypothesis that nonenzymatic peroxidation of membrane phospholipids is responsible for copper alloy-mediated surface killing. Lipid peroxidation was monitored with the thiobarbituric acid-reactive substances (TBARS) assay. Survival, TBARS levels, and DNA degradation were followed in cells exposed to copper alloy surfaces containing 60 to 99.90% copper or in medium containing CuSO4. In all cases, TBARS levels increased with copper exposure levels. Cells exposed to the highest copper content alloys, C11000 and C24000, exhibited novel characteristics. TBARS increased immediately at a very rapid rate but peaked at about 30 min. This peak was associated with the period of most rapid killing, loss in membrane integrity, and DNA degradation. DNA degradation is not the primary cause of copper-mediated surface killing. Cells exposed to the 60% copper alloy for 60 min had fully intact genomic DNA but no viable cells. In a fabR mutant strain with increased levels of unsaturated fatty acids, sensitivity to copper alloy surface-mediated killing increased, TBARS levels peaked earlier, and genomic DNA degradation occurred sooner than in the isogenic parental strain. Taken together, these results suggest that copper alloy surface-mediated killing of E. coli is triggered by nonenzymatic oxidative damage of membrane phospholipids that ultimately results in the loss of membrane integrity and cell death.
INTRODUCTION
Copper is a trace element required for several essential biological processes that exhibit remarkable structural and functional conservation from bacteria to humans. The fact that copper exists in two oxidation states, cuprous Cu(I) (reduced) and cupric Cu(II) (oxidized), contributes to its ability to serve as a catalytic cofactor in diverse biological systems. Studies of copper acquisition, distribution, and homeostasis have shown that, while at appropriate concentrations copper is an essential micronutrient, it can, however, be toxic at higher concentrations (17, 21, 24, 25, 40, 49, 52). In excess, copper affects the biochemistry of macromolecules and reportedly leads to a rapid decline in membrane integrity (3, 4, 16, 37). Thus, it is no surprise that species have evolved tightly regulated mechanisms for copper homeostasis.
The publication by Wilks et al. (58) opened a new realm of possibilities for copper and copper alloy surfaces, that of passive antimicrobial sanitizing agents. Studies from several laboratories clearly demonstrated efficient and rapid killing of bacteria, fungi, and viruses upon exposure to surfaces composed of copper or copper-containing alloys but not stainless steel (10, 18, 31–33, 35, 36, 55, 57–59). These findings were confirmed by standardized testing in an approved Good Laboratory Practices facility, and the U.S. Environmental Protection Agency registered over 350 copper alloys as having antimicrobial activities against 6 different bacteria. In view of the potential importance of copper alloy surfaces in the battle against hospital-acquired infections, it is essential to understand the mechanism of contact-mediated killing by copper.
Transition metals like iron and copper catalyze the formation of reactive oxygen species (ROS), particularly hydroxyl radicals (·OH), via the Fenton-like reaction shown below (26, 51).
Cu(I) + H2O2 → Cu(II) + ·OH + OH−
The unpaired electron of the hydroxyl radical is highly reactive and capable of causing oxidative damage to cellular macromolecules. Phospholipids, the major component of plasma membranes, consist of a polar head group (composed of glycerol 3-phosphate and ethanolamine, serine, or choline) covalently bonded to two long-chain fatty acids that typically range in length from 14 to 20 carbons and may contain one or more unsaturated double bonds as well as other modifications (61). The biochemistry of enzymatic and nonenzymatic lipid peroxidation is reviewed in references 3, 16, 22, and 46). The hydroxyl radicals (·OH) formed by the Cu(I)-dependent Fenton-like reaction described above are able to drive the nonenzymatic peroxidation of the unsaturated double bonds of unsaturated fatty acids, thereby initiating a series of reactions that result in extensive structural changes of the phospholipid bilayer and loss of membrane integrity (7).
The diversity of microorganisms sensitive to copper-mediated contact killing is surprising and perhaps provides clues to the underlying mechanism (18). We hypothesize that the cellular target of the copper contact killing is common to all of the sensitive organisms and, by all appearances, is one that is not easily altered in a manner that provides resistance. Here we explore the proposal that copper alloy surface killing is mediated by the ROS-catalyzed nonenzymatic peroxidation of membrane unsaturated fatty acids, which are essential components of the bacterial envelope, which is required for cell viability.
MATERIALS AND METHODS
Strains and growth conditions.
Unless otherwise noted, the work was carried out using Escherichia coli strain 23724 (F− supE44 lacY1 thr-1 leuB6 mcrA thi-1 rfbD1 fhuA21 λ) obtained from the American Type Culture Collection. Strain PDJ1 (recD::Tn10) and the otherwise-isogenic mutant strain MWF1 (fabR::kan recD::Tn10) were obtained from Charles O. Rock of the University of Tennessee, Memphis, and are described in reference 60.
Strain ATCC 23724 was grown and maintained on Luria-Burtani (LB) broth, and titers were determined on LB agar plates. All the other strains were grown on M9 minimal liquid or solid medium supplemented with 0.4% glucose, 0.01% methionine, 0.0005% thiamine, and the appropriate antibiotics as indicated (25 μg/ml kanamycin and/or 20 μg/ml tetracycline). Cultures were grown with aeration at 37°C to early log phase (an optical density at 600 nm [OD600] of 0.3).
Metal coupon cleaning protocol.
Metal coupons consisting of 1-in.2 sheets of a specified alloy were provided by the Copper Development Association, New York, NY. The composition of the alloys used in this study were as follows: C11000 (99.90% copper [wt/wt], less than 0.04% oxygen, balance consisting of various trace elements depending on the ore body sources and the refining process), C24000 (80% copper, 20% zinc [wt/wt]), C26000 (70% copper, 30% zinc [wt/wt]), C28000 (60% copper, 40% zinc [wt/wt]), S30400 (304 stainless steel, with 18% chromium, 8% nickel, 74% iron [wt/wt]).
The coupons were individually dipped into warm 3.5% NaOH (71°C) for 30 s to degrease the alloy surface. They were immediately rinsed in deionized water and allowed to dry. The coupons were then individually acid rinsed in 10% H2SO4 for 30 s, immediately rinsed in deionized water, and allowed to dry. Following drying, the coupons were individually flame sterilized by dipping in 95% ethanol, igniting the ethanol by passing through a flame, and allowing it to burn off. The sterilized coupons were stored in sterile petri dishes.
TBARS assay.
The TBARS (thiobarbituric acid-reactive substances) assay kit was obtained from ZeptoMetrix Corporation, and the TBARS assay was performed as described by the manufacturer. E. coli strains were grown to early log phase (OD600 of about 0.3) in LB or M9 medium, harvested from 100 ml of culture medium, washed once with 0.85% NaCl, and resuspended in 0.85% NaCl to give a final volume of 200 or 500 μl, as indicated. For cells exposed to CuSO4 in liquid cultures, 100-μl samples of concentrated cells were used for the TBARS assay. Exposure to the metal coupon surface was carried out as follows. Samples (100 μl) of concentrated cells were spread on the coupon surface in a sterile petri dish and allowed to air dry. Drying was complete in about 15 min. Cells were collected from the coupon surface by repeated washing and scraping with the micropipette tip into 100 μl of sterile 0.85% NaCl and harvested from the coupon wash by centrifugation. The entire sample was used for the TBARS assay. To summarize, the harvested cells were resuspended in 100 μl of SDS by gentle swirling, 2.5 ml of TBA buffer reagent was added, and the covered tubes were incubated in a water bath at 95°C for 60 min. After the tubes were allowed to cool to room temperature, they were put into an ice bath for 10 min, and 2 ml of the reaction mixture was transferred to microcentrifuge tubes for debris removal by centrifugation at 3,000 rpm for 15 min. The absorbance at 532 nm of the supernatant was determined using a Shimadzu Biospec-mini spectrophotometer. The absorbance obtained for the experimental sample was compared to a standard curve obtained using the malondialdehyde (MDA) provided with the TBARS assay kit. The results are reported as nmoles MDA equivalents per 109 cells. Each experiment was repeated with three independent cultures.
Microscopic cell viability assay.
The LIVE/DEAD BacLight bacterial viability kit (Invitrogen) for microscopy and quantitative assays was used to visually monitor cell viability. The kit contains a mixture of two stains, SYTO9 and propidium iodide, which differentially enter bacterial cells and monitor bacterial cell viability as a function of membrane integrity. SYTO9, a green fluorescent nucleic acid stain, is capable of passing the intact membrane of both viable and dead or dying bacterial cells. Propidium iodide, a red fluorescent nucleic acid stain, only enters bacterial cells with intact but compromised membranes that are either dead or dying. When both dyes are present, the propidium iodide causes a reduction in the SYTO9 fluorescence. Thus, cells with a compromised membrane will stain red, whereas cells with an intact membrane will stain green. Cells were stained as described in the manufacturer's protocol and observed using a Zeiss fluorescence Axioscope microscope (fluorescein isothiocyanate [FITC] and rhodamine filters) and an AxoCam ICm1 camera. Total magnification was 1,000× with oil immersion.
RESULTS
Exposure of Escherichia coli to increasing concentrations of CuSO4 correlates with decreased survival and increased TBARS production.
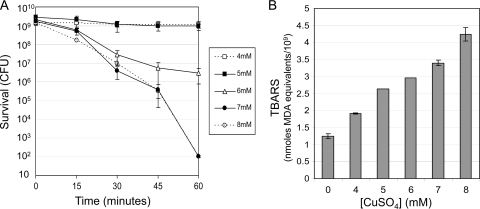
It is well established that exposure to copper alloy surfaces or to copper ions in solution is toxic to bacterial species (reviewed in references 17, 18, 28, 34, 38, 44, 53, and 56. Nonetheless, questions have arisen as to whether the same mechanisms are operating in these different exposure environments (10, 12, 34). The presence of copper ions in the medium has been shown to inhibit proliferation of wild-type strains of E. coli at concentrations as low as 0.2 to 2.5 mM (17, 38) but may not be killing the cells. To investigate this, we explored the relationship between exposure to copper ions in liquid medium, phospholipid oxidation (as measured by the TBARS assay), and bacterial cell death. CuSO4 was added to a log-phase Escherichia coli (ATCC 23724) culture to a final concentration of 4 to 8 mM, and survival was followed for 60 min. The results are shown in Fig. 1A. Little to no killing was seen at the 4 and 5 mM CuSO4 concentrations, although no significant increase in the number of CFU was observed either. It appears that at these low concentrations CuSO4 copper ions are bacteriostatic but not bactericidal. Toxic effects of copper ions on cellular metabolism are well known (18, 25, 41, 51), and the inhibition of hydratases via the binding of Cu(I) to the Fe-S clusters in these proteins has been demonstrated (27). It appears that E. coli is able to tolerate these low Cu2+ ion concentrations (18, 25, 40, 51). This is not the case at higher concentrations. At 6 mM CuSO4, a low rate of killing was observed, about 3 logs of killing over the 60-min time course. At 7 to 8 mM this rate appeared to reach a maximum of about 7 to 8 logs of killing, leaving few to no survivors after 1 h. Overall, the rate of killing correlated with the CuSO4 concentration.
Fig 1.
E. coli survival and lipid peroxidation following exposure to different CuSO4 concentrations in liquid medium. E. coli strain ATCC 23724 was grown at 37°C in LB medium to mid-log phase (OD600, 0.3). At time zero, CuSO4 was added to the culture to the indicated concentration, and incubation continued for the course of the experiment. (A) At the indicated time, cells were harvested by centrifugation from 100 ml of culture and resuspended in 200 μl of 0.85% NaCl. Survival, reported as the number of CFU, was determined by diluting culture samples in 0.85% NaCl and plating on LB agar. The error bars indicate standard deviations from three independent cultures for which titers were determined in duplicate. (B) CuSO4 was added to the cultures to the indicated final concentration and harvested 60 min after the addition of CuSO4, as described for panel A. The control (0) is the sample taken at time zero, prior to the addition of CuSO4. Membrane lipid peroxidation products were determined as TBARS and are reported as nmoles MDA equivalents/109 cells (see Materials and Methods). The error bars indicate standard deviations from three independent cultures assayed in duplicate.
A variety of methods are available for quantification of long-chain unsaturated fatty acid peroxidation (reviewed in references 3, 8, 13, 30, and 45). We used the TBARS method to monitor lipid peroxidation, a well-established straightforward colorimetric assay that is widely accepted as a valid measure of lipid oxidation but not one without some controversy, which is reviewed at length in the Discussion, below, and references 23 and 39.
TBARS production in E. coli exposed to CuSO4 in the growth medium is shown in Fig. 1B. At time zero and after 60 min of growth in medium containing the indicated CuSO4 concentration, approximately 109 to 1010 cells were harvested, and TBARS levels were assayed. Growth in the presence of CuSO4 caused an increase in the level of TBARS that correlated directly with the concentration of CuSO4 in the growth medium. It should be noted that TBARS levels increased even at the lowest concentrations of 4 to 5 mM, which did not appear to kill cells. At these low concentrations, no increase or decrease in colony count was observed, suggesting that 4 to 5 mM CuSO4 is bacteriostatic but not bactericidal.
Exposure to copper alloy surfaces leads to cell death, TBARS production, and DNA degradation.
E. coli cells were exposed to metal surfaces by using 1-in.2 sheets, referred to as coupons, of the following compositions (details in Materials and Methods): 304 stainless steel (S30400); 99.90% copper (C11000); and copper-zinc alloys C24000, C26000, and C28000, containing 80%, 70%, and 60% copper, respectively, with the remainder zinc. At the indicated time of exposure, cell survival, TBARS formation, and genomic DNA integrity were followed. To obtain accurate data, it was necessary to apply the cell sample to the coupon surface and to reproducibly recover a sufficient sample of exposed cells to carry out the desired assay. The TBARS method requires a minimum sample size of 109 to 1010 E. coli cells for each time point in order to obtain colorimetric readings in the mid-range of the MDA standard curve. The methods used in published studies, while adequate for determining cell survival, were not adequate for the TBARS assay, because only very small numbers of cells were applied to the coupon surface. To address this technical problem, we tried a number of different approaches, including increasing the size of the coupon, but none was satisfactory or provided sufficiently reproducible recovery of cells following copper exposure. Reluctantly, we were forced to settle on the moist/dry “hybrid” exposure scheme described below.
A 100-μl sample of a cell suspension containing approximately 109 to 1010 cells of E. coli strain ATCC 23724 was evenly spread over the surface of the metal coupons and allowed to air dry. During the 15-min drying period, the cells were exposed to the copper surface under moist rather than dry conditions. Based on previous studies (reviewed in reference 18), this initial moist exposure would likely delay the onset of rapid contact killing. This caveat should be kept in mind when analyzing our results. At the indicated times, the bacterial cells were recovered from the metal coupons by vigorous washing, as described in Materials and Methods, and the recovered cell sample was used to determine cell survival and to assay TBARS production. The results are shown in Fig. 2.
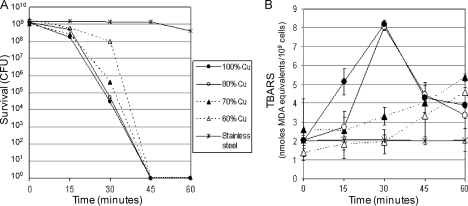
Fig 2.
E. coli survival and lipid peroxidation on copper-zinc alloy surfaces containing different copper concentrations. E. coli strain ATCC 23724 was grown in LB medium to mid-log phase (OD600, 0.3), harvested by centrifugation from 100 ml of culture, and resuspended in 0.85% NaCl to a final volume of 500 μl. Samples (100 μl) of concentrated cells were spread over the surface of metal coupons of 304 stainless steel (S30400), 99.90% copper (C11000), and copper-zinc alloys C24000, C26000, and C28000, containing 80%, 70%, 60% copper, respectively (with the remainder zinc). (A) Following the indicated time of exposure, the cells were washed from the coupon surface with 100 μl of 0.85% NaCl, and samples were taken to determine survival titers, as described for Fig. 1A. (B) Results of the TBARS assay on cells exposed and recovered from the different alloys at the indicated times, as described for panel A. The results represent at least two independent trials. The legend applies to both panels A and B.
As anticipated, bacterial killing on the copper-containing alloys was biphasic: slow for the first 15 min and, depending on the alloy, significantly more rapid after 15 min of exposure, the point at which the samples had dried (Fig. 2A). E. coli cells exposed to the coupons containing the highest concentrations of copper, C11000 and C24000, appeared to be killed rapidly, and killing initiated soon after the samples dried on the surface of the coupon. Alloy C26000, containing 70% copper–30% zinc showed evidence of a slight delay in the onset of rapid killing while alloy C28000, containing the lowest concentration of copper, 60% copper–40% zinc, exhibited a delay of almost 30 min before the onset of rapid killing. Despite the delays on the C26000 and C28000 coupons, little or no survival was observed after 45 min of exposure to the copper-containing alloys. In contrast, less than 1 log of killing was observed during the 60-min exposure to the 304 stainless steel coupons. Thus, there is a direct correlation between the copper content of the alloy and the exposure time needed to initiate rapid killing.
The time course of TBARS production in E. coli cells exposed to these same copper alloy coupons is shown in Fig. 2B. TBARS levels increased above background levels in E. coli cells exposed to each of the copper alloy surfaces, but the kinetics differed significantly and in a manner that correlated well with the killing curve shown in Fig. 2A. In cells exposed to the 60% copper alloy (C28000), TBARS production appeared to show an initial lag of about 30 min, at which point levels began to increase but at a low rate. In cells exposed to the 70% copper alloy (C26000), there was a shorter delay, perhaps 15 min, followed by a similar low rate of increase in TBARS levels. Cells exposed to the 80% copper alloy (C24000) appeared to exhibit a very short delay, but this was followed by an extremely rapid rise in TBARS levels. Surprisingly, the level of TBARS peaked at 30 min and then decreased precipitously at 45 min, but they did not return to initial levels. No delay in the onset of TBARS production was observed in E. coli cells exposed to the 99.90% copper alloy (C11000). TBARS production began immediately at a very high rate, peaked at 30 min, and then dropped to a lower level but which was still significantly higher than at time zero. The basis for this precipitous decrease is not clear, but one likely explanation is that the reactive compounds being monitored by the TBARS assay are themselves subject to chemical breakdown to compounds that do not react with TBA. A similar peak of TBARS levels was observed in E. coli cells exposed to the oxidizing effects of TiO2 and UV light (29). Finally, no increase in TBARS above initial levels was observed in cells exposed to 304 stainless steel.
Warnes et al. (53, 54) reported that exposure of strains of Enterococcus faecalis to copper surfaces caused random DNA fragmentation, and they postulated that DNA degradation occurs early in the cell death process and plays an active role. We investigated whether this was also the case for E. coli strain ATCC 23724 and whether differences in the copper content of the alloy surface impacted the timing or extent of the degradation. Total genomic DNA was isolated from E. coli cells exposed to and recovered from coupons of 304 stainless steel (S30400), 99.90% copper (C11000), and copper-zinc alloys C24000, C26000, and C28000 and size separated by PAGE gel analysis (Fig. 3). The large-sized fragments produced by the DNA isolation procedure ran near the top of the gel. Small differences in the amount of this total genomic DNA from lane to lane of each panel are due to variations in cell recovery from the coupon. To control for this variation, each exposure series was performed in triplicate. Representative gels are shown.
Fig 3.
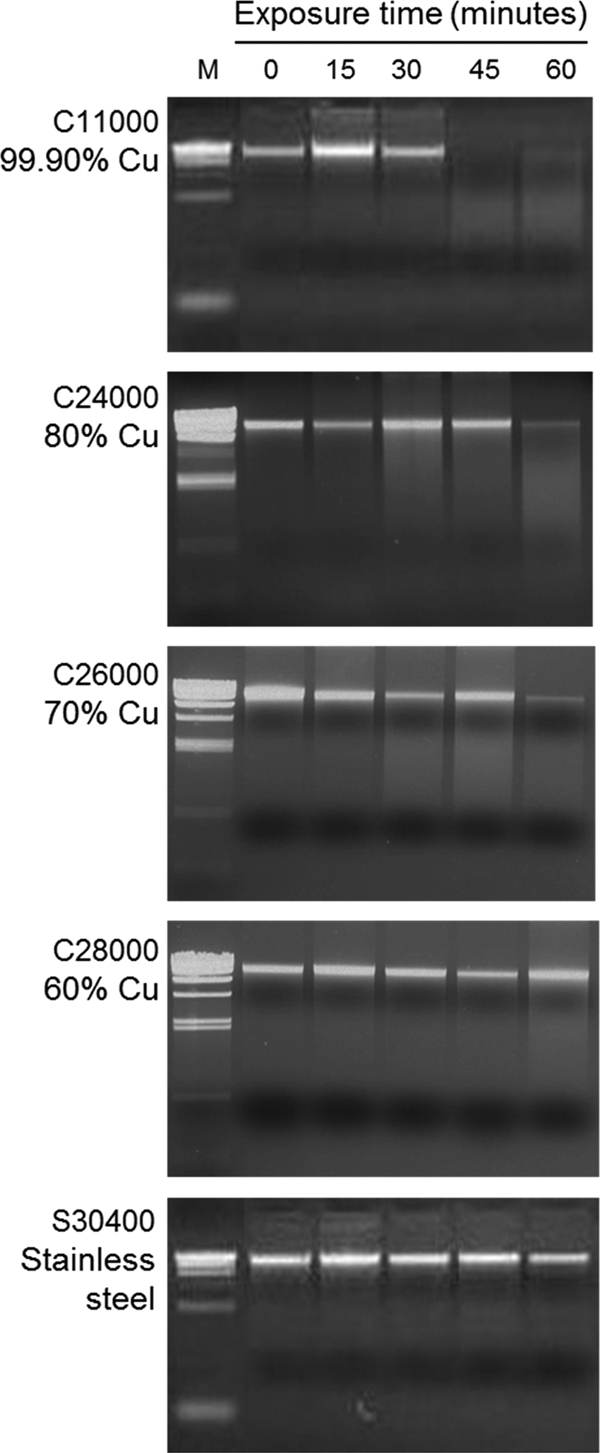
DNA degradation following copper surface exposure. E. coli strain ATCC 23724 was grown, harvested, and resuspended in 0.85% NaCl and exposed to coupon surfaces of the indicated alloy as described for Fig. 2. At the indicated times, the cells were washed from the coupon surface with 100 μl of 0.85% NaCl, total DNA was isolated using the Promega Wizard SV genomic DNA purification system, and the DNA was size separated in a 1% agarose gel. The size markers (M) are HindIII-digested λ phage DNA.
Exposure to 99.90% copper alloy (C11000) coupons (top panel) caused rapid and extensive degradation of genomic DNA after the 30-min time point. DNA degradation was complete by 45 min and, reproducibly, no genomic DNA was found in the 45- and 60-min samples. It should also be noted that no shorter DNA fragments could be observed in the gels, suggesting that degradation produced random-sized fragments with no preferred target sites. Exposure to 80% (C24000) and 70% (C26000) alloy also led to rapid DNA degradation, but the onset of the degradation was delayed until after 45 min of exposure, and small but reproducible amounts of genomic DNA fragments were still present at the 60-min time point. No loss in genomic DNA was observed in alloy C28000-exposed (60% copper) cells or in the 304 stainless steel-exposed cells over the course of the 60 min of the experiment. Two important conclusions emerge from this result. First, the exposure time required to activate DNA degradation is inversely correlated with the copper content of the alloy, that is, the higher the copper content, the sooner the onset of DNA degradation. Second, cell death precedes DNA degradation in cells exposed to alloys containing 60 to 80% copper. This was most clear for alloy C28000 (60% copper), for which few if any survivors were present at the 45- and 60-min exposure times (Fig. 2A), with no evidence of DNA degradation (Fig. 3).
The results shown in Fig. 2B and 3 suggest that the 30-min time point represents a critical turning point for cells exposed to 99.90% copper (C11000). At about 30 min, we found approximately 50% survival, TBARS levels reached a maximum level and peaked, and DNA degradation was initiated. We used Live/Dead staining to follow E. coli cell viability over this same time course. E. coli strain ATCC 23724 was grown, harvested, and exposed to the 99.90% copper (C11000) coupon surface, as described for Fig. 2. The cells were then washed from the coupon surface, and the recovered cells were stained as described by the LIVE/DEAD kit manufacturer for observation by phase and fluorescence microscopy. This test kit monitors cell death indirectly by measuring membrane integrity. The presence of intracellular propidium iodide, which enters cells when the membrane ceases to function as a semipermeable membrane, is used as an indicator of cell viability. The results in Fig. 4 correlate with the survival curve shown in Fig. 2B. The percentage of dead cells (or cells with incompetent membranes) was approximately 50% at 30 min and increased to more than 90% at 45 min. By 60 min, few if any live cells were observed either by this assay or by the cell titer assay. No evidence of cell death or loss of membrane integrity was observed in E. coli cells exposed to 304 stainless steel (data not shown).
Fig 4.
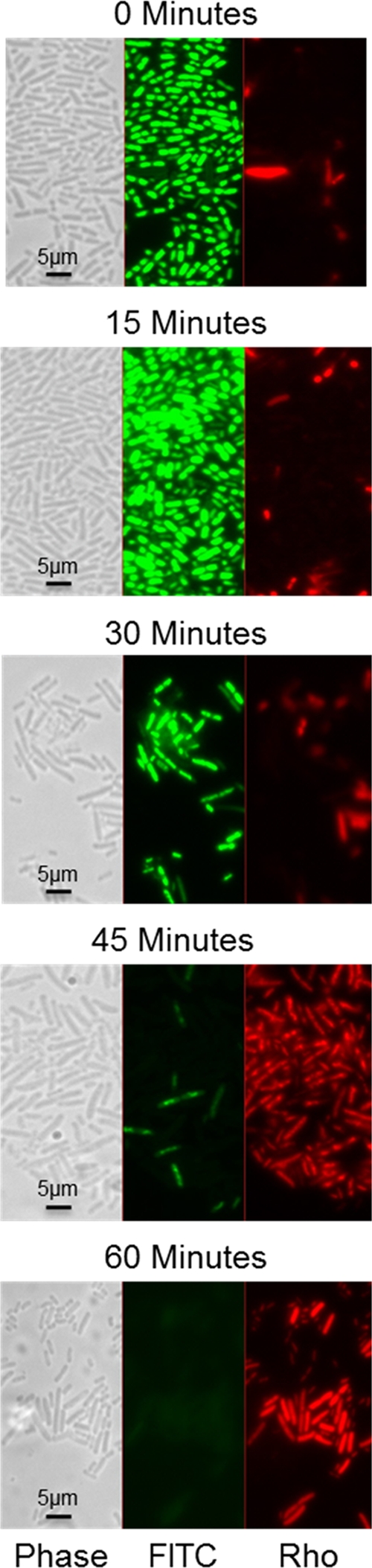
Microscopic assay of membrane integrity following exposure to a 99.90% copper surface. E. coli strain ATCC 23724 was grown, harvested, and exposed to 99.90% copper (C11000) coupons, as described for Fig. 2. At the indicated times of exposure, the cells were washed from the coupon surface with 100 μl of 0.85% NaCl and prepared for the Live/Dead BacLight assay as described by the manufacturer (Invitrogen). Cells were observed using a Zeiss fluorescece Axioscope and an AxoCam ICm1 camera. The panels, from left to right, were viewed with phase-contrast illumination, a FITC filter (green), and a rhodamine filter (Rho; red). Total magnification used was 1,000× with oil immersion.
In summary, the results presented above demonstrate that, in E. coli cells exposed to metallic copper alloy surfaces, there is a direct correlation between the copper content of the alloy and (i) the time of onset of killing, (ii) the rate of cell death, (iii) the kinetics of TBARS production, and (iv) the induction of genomic DNA degradation. Moreover, in cells exposed to 99.90% copper, the time period in which rapid cell death was observed and TBARS levels peaked coincided with the time period of rapid loss of membrane integrity. In cells exposed to the 60% copper alloy, DNA degradation was clearly shown to occur post-cell death.
An increased ratio of unsaturated to saturated fatty acids in E. coli results in increased sensitivity to a 99.90% copper surface.
The targets of the ROS formed by the Fenton-like reaction of Cu(I)-to-Cu(II) reduction have been proposed to be membrane proteins and lipids, specifically, unsaturated fatty acids of plasma membrane phospholipids. If peroxidation of plasma membrane unsaturated fatty acids is a significant factor contributing to copper alloy-mediated contact killing, membranes containing increased levels of unsaturated fatty acids should exhibit increased sensitivity to copper killing.
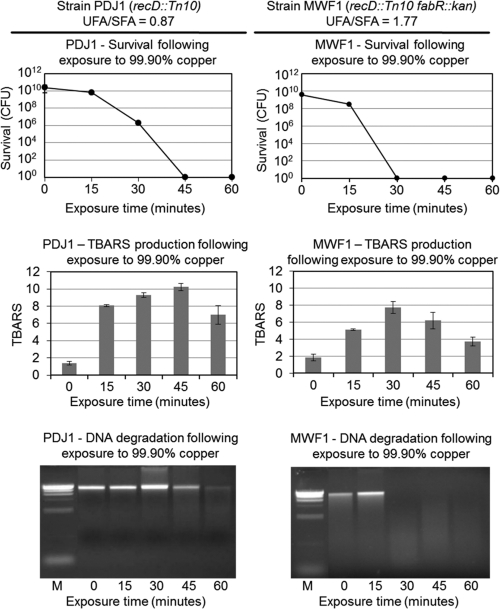
To test our hypothesis, we investigated copper alloy surface killing in an E. coli mutant strain carrying an alteration in a gene responsible for the regulation of unsaturated fatty acid biosynthesis and that modulates the relative levels of unsaturated versus saturated fatty acids (UFA:SFA) in the plasma membrane (reviewed in references 15, 60, and 61. The fabA and fabB genes encode essential enzymes for the biosynthesis of unsaturated fatty acids in E. coli, and a repressor encoded by fabR regulates the expression of these genes. Loss of FabR repression increases the rate of synthesis of unsaturated fatty acids and thus increases the relative level of unsaturated fatty acids in the cell membranes. The parental control strain PDJ1 (fabR) and the otherwise-isogenic strain MWF1 (fabR::kan) carrying a transposon insertion in fabR were isolated and characterized by Zhang et al. (60), who reported that the UFA:SFA ratio in strain PDJ1 is 0.87 and in strain MWF1 it is 1.77, a nearly 2-fold increase.
Figure 5 compares strains PDJ1 and MWF1 with regard to their sensitivities to killing by copper alloy surfaces, TBARS production, and total genomic DNA degradation. The strains were grown on minimal M9 medium to mid-log phase, but in other regards were exposed to 99.90% copper coupons (C11000) as described for Fig. 2. Similar to E. coli strain ATCC 23724, the onset of the rapid phase of cell death in the parental strain PDJ1 occurred at about 15 min, the time when the sample had fully dried on the coupon surface, and no survivors were found at 45 min. In contrast, while rapid killing also initiated at 15 min in MWF1 cells, no survivors were present at 30 min in MWF1 cells, indicating that the rate of killing was approximately 2-fold faster than that seen in the fabR::kan mutant strain. While TBARS production in MWF1 was not faster than that observed in PDJ1, it peaked earlier, by about 30 min, and the peak coincided with the total degradation of genomic DNA. DNA degradation was first observed in the PDJ1 cells at 45 min, with some residual genomic DNA present even at 60 min. In the MWF1 cells, genomic DNA degradation was complete by 30 min.
Fig 5.
Increased relative unsaturated fatty acid levels correlate with increased E. coli killing, lipid peroxidation, and DNA degradation following exposure to a 99.90% copper surface. E. coli strains PDJ1 (recD::Tn10) and MWF1 (fabR::kan recD::Tn10) were grown to mid-log phase (OD600, 0.3) in M9 minimal medium supplemented with 0.4% glucose, 0.01% methionine, 0.0005% thiamine, and 25 μg/ml kanamycin or 20 μg/ml tetracycline, respectively. Cells were exposed to 99.90% copper (C11000) coupons, as described for Fig. 2. UFA:SFA is the ratio of unsaturated to saturated fatty acids as reported by Zhang et al. (60). Survival, TBARS production, and DNA degradation were assayed as described for Fig. 2 and 4.
In summary, the results in Fig. 5 indicate that increased relative levels of unsaturated fatty acids cause increased sensitivity to copper alloy surface killing and, as shown for E. coli strain ATCC 23724, cell death coincides with the peak of TBARS levels and the time of total genomic DNA degradation.
DISCUSSION
Organisms defend themselves from copper toxicity via a variety of mechanisms that tightly control intracellular copper ion levels to an acceptable range and localize copper ions to the appropriate subcellular compartment (18, 25, 40, 51). These include copper-binding proteins in the periplasm and cytoplasm, copper chaperones, and membrane transporters for the export of free intracellular copper ions. Recent studies have presented clear evidence that surface-released free copper ions are the causative agent in copper alloy-mediated contact killing and that intracellular copper is detected soon after exposure to the copper alloy surface (9–12, 34). Consistent with this, studies found that strains carrying null mutations in the genes encoding components of the plasma membrane P-type ATPase copper efflux transporter CopA, the tripartite outer-inner membrane-spanning copper efflux system encoded by CusCFBA, and the periplasmic multicopper oxidase CueO exhibit increased sensitivity to growth in medium containing copper ions (17, 28, 38). Additionally, increased copper tolerance has been identified in bacteria isolated from a variety of environments, such as mining effluents, manure from animal farms, and the surface of copper coins (12). Where the causative genes have been identified, they were found to encode components of copper efflux systems overexpressed from plasmids (1, 5, 19). These environmentally selected mutations are not truly resistant to copper alloy surface killing; killing is simply delayed rather than prevented (10, 12). Therefore, while the toxicity of intracellular copper appears to play a role in copper alloy-mediated contact killing, it does not appear to be the key contributing factor.
Previous studies by Espirito Santo et al. (10) and others (reviewed in reference 18) concluded that an as-yet-unidentified component of the bacterial cell membrane is the primary target in cells exposed to dry copper surfaces. This investigation explores the possibility that oxidation of membrane phospholipids is the primary mediator of copper alloy surface killing. Wilks et al. (58) showed that survival of the pathogen Escherichia coli O157 varied inversely with the copper content of the alloy tested. We made use of this finding to tease apart the physiological events resulting from exposure to copper alloy surfaces. The kinetics of cell death, DNA degradation, and lipid oxidation (as measured by TBARS production) were compared in cells exposed to metallic copper surfaces composed of alloys containing different amounts of copper ranging from 60% to 99.90% and were shown to directly correlate with the copper content of the alloy. Additionally, using a genetic approach, it was shown that a mutation (fabR::kan) that increases the relative level of unsaturated fatty acids in an otherwise-isogenic E. coli strain causes increased sensitivity to copper alloy surface exposure. Taken together, these results implicate the copper-dependent oxidation of unsaturated fatty acids in the E. coli membrane as the major cause of the rapid, efficient cell death observed in cells exposed to dry metallic copper alloy surfaces.
Nonenzymatic lipid peroxidation is a three-step process: initiation, propagation, and termination (3, 8, 13, 16, 20, 22). The hydroxyl radical (·OH) produced by Cu(I) via the Fenton-like reaction initiates membrane lipid peroxidation by removing a hydrogen from a —CH2 group close to the carbon-carbon double bond of an unsaturated fatty acid (reaction 1). This is followed by reaction with molecular oxygen and molecular rearrangement to produce a lipid peroxyl radical (reaction 2), which, in turn, is capable of removing another hydrogen from a site near a different unsaturated double bond (reaction 3). Reaction 3, the propagation step, occurs predominantly in polyunsaturated fatty acids (PUFAs) due to the proximity of unsaturated double bonds, thereby producing a chain reaction that amplifies the amount of lipid peroxidation. No evidence in the literature precludes the possibility of a propagation event via reactions between different fatty acid molecules but is reported to be unlikely.
Reaction 1: LH + OH· → L· + HOH
Reaction 2: L· + O2 → LOO· (lipid peroxyl radical)
Reaction 3: LH + LOO· → L· + LOOH
Propagation can be terminated by a “chain-breaking” reaction such as (i) the breakdown of unstable LOO· to various products, including aldehydes, (ii) the reaction of LOO· with an antioxidant to form LOOH, or (iii) the reaction of LOO· with another radical.
Approximately 50% of the E. coli membrane phospholipids are composed of the monounsaturated fatty acids, cis-vaccenic acid (18:1Δ11) and palmitoleate (C16:1Δ9), with no significant levels of PUFA (50, 60). While the propagation step of lipid peroxidation may not occur in E. coli membranes, evidence suggests that E. coli membrane lipids can undergo oxidation. E. coli exposed to oxidizing agents, including hydrogen peroxide and titanium oxide, exhibits increased TBARS levels that are proposed to result from the production of reactive aldehydes and other compounds formed by the breakdown of oxidized lipids (29, 42, 43). This conclusion is based on the finding that expression of enzymes that protect cells from oxidation is also protective against oxidizing agents in E. coli. Overexpression of the E. coli yqhD gene encoding an aldehyde reductase protects against treatment with compounds that generate ROS. Conversely, deletion of yqhD causes increased sensitivity to these compounds and increases TBARS production. Taken together, these studies (described in references 29, 42, and 43) support the use of the TBARS assay as a valid measure of membrane lipid oxidation in E. coli. Thus, the increased TBARS production observed here in E. coli strains exposed to copper alloy surfaces or to copper ions in solution is an indicator of copper-induced membrane lipid oxidation.
Further evidence that unsaturated fatty acids are the target of copper contact killing in the E. coli membrane comes from studies of an E. coli strain (MWF1), with a null mutation in fabR, the gene encoding the FabR repressor, which expresses increased relative levels of unsaturated versus saturated fatty acids. By all measures, strain MWF1, which has a 2-fold-higher level of unsaturated fatty acids than the otherwise-isogenic parental strain PDJ1, exhibited a significant increase in sensitivity to copper alloy contact killing. The time of onset of the rapid phase of contact killing was foreshortened, and the rate of killing was approximately 2-fold faster. Additionally, the time point at which nearly 100% killing was reached was significantly earlier than in PDJ1 and coincided with the peak in TBARS levels and DNA degradation, which was about 15 min earlier than in PDJ1. We propose that the increase in the relative level of unsaturated fatty acids provides a higher concentration of potential reaction sites for copper-induced nonenzymatic lipid oxidation and greater opportunity for membrane structural changes and loss of membrane integrity, which we believe serve as initiators of copper alloy contact killing. Oxidized fatty acids fragment to shorter species, form lipid-lipid and lipid-protein cross-links, and cause bends or even circularization of the chain of carbons. Such structural changes distort the phospholipid bilayer and disrupt the biophysical characteristics of the membrane, resulting in a concomitant loss of membrane integrity (7). Thus, the role of the plasma membrane as a selectively permeable barrier that separates the cytoplasm from the environment and regulates entry into and exit from the cell is seriously impaired and quickly results in cell death. We cannot rule out the possibility that increased membrane fluidity due to the elevated levels of unsaturated fatty acids has an impact on the activity of membrane-associated enzymes, such as the CusCFBA copper efflux system.
Warnes et al. (54) suggested that degradation of genomic DNA is the primary cause of cell death in bacteria exposed to high copper content surfaces. The results illustrated here in Fig. 2A and 3 do not support this conclusion. Rather, they demonstrate that DNA degradation is a secondary effect of copper surface exposure. No survivors were found following 45 min of exposure to the C28000 copper-zinc alloy containing 60% copper, the alloy with the lowest copper content tested. Nevertheless, genomic DNA degradation was not observed at 45 min or even after 60 min of exposure. This finding clearly separates cell death and DNA degradation and demonstrates that, under certain exposure conditions, E. coli cells die without degrading their DNA. Oxidizing radicals and copper ions have been reported to attack DNA directly, causing degradation, and is a likely cause of the observed random fragmentation of genomic DNA (2, 14, 48). Nevertheless, in view of reports of E. coli signaling pathways that monitor the integrity of the cell envelope, it is tempting to propose that these may also respond to the types of membrane damage described here (reviewed in reference 47).
The results reported here suggest that oxidation of unsaturated fatty acids of the E. coli membrane is the initiating event of copper surface killing. Unsaturated fatty acids are essential and irreplaceable components of biological membranes (7, 15, 60, 61). Changes in fatty acid composition alter the physical properties of the lipid bilayer, affecting membrane fluidity and thus indirectly regulating the activity of integral membrane proteins, which in the case of prokaryotes includes the cytochromes and the enzymes for phospholipid biosynthesis (15, 61). Minimal estimates of 15 to 20% unsaturated fatty acids have been reported for E. coli, and genetic alterations that eliminate unsaturated fatty acid synthesis or decrease expression to levels below this threshold are lethal (6). Consistent with published findings (58), resistance to copper surface-mediated contact killing by genetic mutations is exceedingly rare and may not be possible to achieve by one or a few simple genetic modifications. Given this, the introduction of copper alloy surfaces into the hospital room environment is an attractive weapon in the war on nosocomial infections.
ACKNOWLEDGMENTS
This research was supported by student research training grants to Queensborough Community College—CUNY from the NSF STEP (DUE-0652963) and NSF REU (DUE-0754673) programs, a Professional Staff Congress CUNY Award from the City University of New York (to N.G.), and funding from the Copper Development Association, NY (to N.G.). Equipment for the project was purchased from the New York State Department of Education Perkins Grant program (to N.G.). Additional support came from an NIH Bridges to the Baccalaureate grant (GM50070 to SUNY—Stony Brook with a subcontract to Queensborough Community College).
We thank Charles O. Rock of the University of Tennessee, Memphis, for providing strains PDJ1 and MWF1, Rachel Hammer for her experimental contributions at the start of this project, Susan A. Rotenberg and Robert Bittman of Queens College—CUNY for valuable discussions, and Teresa R. Salas, Laura Rachiele, and Annette Lopez for their technical assistance, which greatly facilitated the progress of this research project.
Footnotes
Published ahead of print 13 January 2012
REFERENCES
- 1. Brown NL, Barrett SR, Camakaris J, Lee BT, Rouch DA. 1995. Molecular genetics and transport analysis of the copper-resistance determinant (pco) from Escherichia coli plasmid pRJ1004. Mol. Microbiol. 17:1153–1166 [DOI] [PubMed] [Google Scholar]
- 2. Cadet J, Douki T, Ravanat JL. 2008. Oxidatively generated damage to the guanine moiety of DNA: mechanistic aspects and formation in cells. ACC Chem. Res. 41:1075–1083 [DOI] [PubMed] [Google Scholar]
- 3. Catala A. 2006. An overview of lipid peroxidation with emphasis in outer segments of photoreceptors and the chemiluminescence assay. Int. J. Biochem. Cell Biol. 38:1482–1495 [DOI] [PubMed] [Google Scholar]
- 4. Cervantes C, Gutierrez-Corona F. 1994. Copper resistance mechanisms in bacteria and fungi. FEMS Microbiol. Rev. 14:121–137 [DOI] [PubMed] [Google Scholar]
- 5. Cooksey DA. 1994. Molecular mechanisms of copper resistance and accumulation in bacteria. FEMS Microbiol. Rev. 14:381–386 [DOI] [PubMed] [Google Scholar]
- 6. Cronan JE, Jr, Gelmann EP. 1973. An estimate of the minimum amount of unsaturated fatty acid required for growth of Escherichia coli. J. Biol. Chem. 248:1188–1195 [PubMed] [Google Scholar]
- 7. Cronan JE, Jr, Gelmann EP. 1975. Physical properties of membrane lipids: biological relevance and regulation. Bacteriol. Rev. 39:232–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Devasagayam TPA, Boloor KK, Ramasarma T. 2003. Methods for estimating lipid peroxidation: an analysis of merits and demerits. Indian J. Exp. Biol. 40:300–308 [PubMed] [Google Scholar]
- 9. Elguindi J, et al. 2011. Metallic copper corrosion rates, moisture content, and growth medium influence survival of copper ion-resistant bacteria. Appl. Microbiol. Biotechnol. 89:1963–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Espirito Santo C, et al. 2011. Bacterial killing by dry metallic copper surfaces. Appl. Environ. Microbiol. 77:794–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Espirito Santo C, Taudte N, Nies DH, Grass G. 2008. Contribution of copper ion resistance to survival of Escherichia coli on metallic copper surfaces. Appl. Environ. Microbiol. 74:977–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Espirito Santo CE, Morais PV, Grass G. 2010. Isolation and characterization of bacteria resistant to metallic copper surfaces. Appl. Environ. Microbiol. 76:1341–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fernandez J, Perez-Alvarez JA, Fernandez-Lopez JA. 1997. Thiobarbituric acid test for monitoring lipid oxidation in meat. Food Chem. 59:345–353 [Google Scholar]
- 14. Frelon S, Douki T, Favier A, Cadet J. 2003. Hydroxyl radical is not the main reactive species involved in the degradation of DNA bases by copper in the presence of hydrogen peroxide. Chem. Res. Toxicol. 16:191–197 [DOI] [PubMed] [Google Scholar]
- 15. Fujita Y, Matsuoka H, Hirooka K. 2007. Regulation of fatty acid metabolism in bacteria. Mol. Microbiol. 66:829–839 [DOI] [PubMed] [Google Scholar]
- 16. Girotti AW. 1998. Lipid hydroperoxide generation, turnover, and effector action in biological systems. J. Lipid Res. 39:1529–1542 [PubMed] [Google Scholar]
- 17. Grass G, Rensing C. 2001. Genes involved in copper homeostasis in Escherichia coli. J. Bacteriol. 183:2145–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grass G, Rensing C, Solioz M. 2011. Metallic copper as an antimicrobial surface. Appl. Environ. Microbiol. 77:1541–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hasman H, Aarestrup FM. 2002. tcrB, a gene conferring transferable copper resistance in Enterococcus faecium: occurrence, transferability, and linkage to macrolide and glycopeptide resistance. Antimicrob. Agents Chemother. 46:1410–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hermes-Lima M. (ed.) 2005. Oxygen in biology and biochemistry: role of free radicals. John Wiley & Sons, Inc., Hoboken, NJ [Google Scholar]
- 21. Kim BE, Nevitt T, Thiele DJ. 2008. Mechanisms for copper acquisition, distribution and regulation. Nat. Chem. Biol. 4:176–185 [DOI] [PubMed] [Google Scholar]
- 22. Kohen R, Nyska A. 2002. Oxidation of biological systems: oxidative stress phenomena, antioxidants, redox reactions, and methods for their quantification. Toxicol. Pathol. 30:620–650 [DOI] [PubMed] [Google Scholar]
- 23. Kosugi H, Kikugawa K. 1989. Potential thiobarbituric acid-reactive substances in peroxidized lipids. Free Radic. Biol. Med. 7:205–207 [DOI] [PubMed] [Google Scholar]
- 24. Laliberte J, Labbe S. 2008. The molecular bases for copper uptake and distribution: lessons from yeast. Med. Sci. (Paris) 24:277–283 [DOI] [PubMed] [Google Scholar]
- 25. Linder MC, Hazegh-Azam M. 1996. Copper biochemistry and molecular biology. Am. J. Clin. Nutr. 63:797S–811S [DOI] [PubMed] [Google Scholar]
- 26. Liochev SI, Fridovich I. 2002. The Haber-Weiss cycle, 70 years later: an alternative view. Redox Rep. 7:55–57 [DOI] [PubMed] [Google Scholar]
- 27. Macomber L, Imlay JA. 2009. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. U. S. A. 106:8344–8349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Macomber L, Rensing C, Imlay JA. 2007. Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. J. Bacteriol. 189:1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maness PC, et al. 1999. Bactericidal activity of photocatalytic TiO2 reaction: toward an understanding of its killing mechanism. Appl. Environ. Microbiol. 65:4094–4098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meagher EA, FitzGerald GA. 2000. Indices of lipid peroxidation in vivo: strengths and limitations. Free Radic. Biol. Med. 28:1745–1750 [DOI] [PubMed] [Google Scholar]
- 31. Mehtar S, Wiid I, Todorov SD. 2008. The antimicrobial activity of copper and copper alloys against nosocomial pathogens and Mycobacterium tuberculosis isolated from healthcare facilities in the Western Cape: an in-vitro study. J. Hosp. Infect. 68:45–51 [DOI] [PubMed] [Google Scholar]
- 32. Michels HT, Noyce JO, Keevil CW. 2009. Effects of temperature and humidity on the efficacy of methicillin-resistant Staphylococcus aureus challenged antimicrobial materials containing silver and copper. Lett. Appl. Microbiol. 49:191–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mikolay A, et al. 2010. Survival of bacteria on metallic copper surfaces in a hospital trial. Appl. Microbiol. Biotechnol. 87:1875–1879 [DOI] [PubMed] [Google Scholar]
- 34. Molteni C, Abicht HK, Solioz M. 2010. Killing of bacteria by copper surfaces involves dissolved copper. Appl. Environ. Microbiol. 76:4099–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Noyce JO, Michels H, Keevil CW. 2007. Inactivation of influenza A virus on copper versus stainless steel surfaces. Appl. Environ. Microbiol. 73:2748–2750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Noyce JO, Michels H, Keevil CW. 2006. Use of copper cast alloys to control Escherichia coli O157 cross-contamination during food processing. Appl. Environ. Microbiol. 72:4239–4244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ohsumi Y, Kitamoto K, Anraku Y. 1988. Changes induced in the permeability barrier of the yeast plasma membrane by cupric ion. J. Bacteriol. 170:2676–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Outten FW, Huffman DL, Hale JA, O'Halloran TV. 2001. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J. Biol. Chem. 276:30670–30677 [DOI] [PubMed] [Google Scholar]
- 39. Pegg RB. 2001. Spectrophotometric measurement of secondary lipid oxidation products. Curr. Protoc. Food Anal. Chem. Suppl. 1:D2.4.1–D2.4.18 [Google Scholar]
- 40. Pena MM, Lee J, Thiele DJ. 1999. A delicate balance: homeostatic control of copper uptake and distribution. J. Nutr. 129:1251–1260 [DOI] [PubMed] [Google Scholar]
- 41. Pena MM, Puig S, Thiele DJ. 2000. Characterization of the Saccharomyces cerevisiae high affinity copper transporter Ctr3. J. Biol. Chem. 275:33244–33251 [DOI] [PubMed] [Google Scholar]
- 42. Perez JM, Arenas FA, Pradenas GA, Sandoval JM, Vasquez CC. 2008. Escherichia coli yqhD exhibits aldehyde reductase activity and protects from the harmful effect of lipid peroxidation-derived aldehydes. J. Biol. Chem. 283:7346–7353 [DOI] [PubMed] [Google Scholar]
- 43. Perez JM, et al. 2007. Bacterial toxicity of potassium tellurite: unveiling an ancient enigma. PLoS One 2:e211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Quaranta D, et al. 2011. Mechanisms of contact-mediated killing of yeast cells on dry metallic copper surfaces. Appl. Environ. Microbiol. 77:416–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rael LT, et al. 2004. Lipid peroxidation and the thiobarbituric acid assay: standardization of the assay when using saturated and unsaturated fatty acids. J. Biochem. Mol. Biol. 37:749–752 [DOI] [PubMed] [Google Scholar]
- 46. Roberts LJ, II, Fessel JP, Davies SS. 2005. The biochemistry of the isoprostane, neuroprostane, and isofuran pathways of lipid peroxidation. Brain Pathol. 15:143–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ruiz N, Silhavy TJ. 2005. Sensing external stress: watchdogs of the Escherichia coli cell envelope. Curr. Opin. Microbiol. 8:122–126 [DOI] [PubMed] [Google Scholar]
- 48. Schweigert N, et al. 2000. DNA degradation by the mixture of copper and catechol is caused by DNA-copper-hydroperoxo complexes, probably DNA-Cu(I)OOH. Environ. Mol. Mutagen. 36:5–12 [DOI] [PubMed] [Google Scholar]
- 49. Turski ML, Thiele DJ. 2007. Drosophila Ctr1A functions as a copper transporter essential for development. J. Biol. Chem. 282:24017–24026 [DOI] [PubMed] [Google Scholar]
- 50. Ulrich AK, de Mendoza D, Garwin JL, Cronan JE., Jr 1983. Genetic and biochemical analyses of Escherichia coli mutants altered in the temperature-dependent regulation of membrane lipid composition. J. Bacteriol. 154:221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Valko M, Morris H, Cronin MT. 2005. Metals, toxicity and oxidative stress. Curr. Med. Chem. 12:1161–1208 [DOI] [PubMed] [Google Scholar]
- 52. Vonk WI, Wijmenga C, van de Sluis B. 2008. Relevance of animal models for understanding mammalian copper homeostasis. Am. J. Clin. Nutr. 88:840S–845S [DOI] [PubMed] [Google Scholar]
- 53. Warnes SL, Green SM, Michels HT, Keevil CW. 2010. Biocidal efficacy of copper alloys against pathogenic enterococci involves degradation of genomic and plasmid DNAs. Appl. Environ. Microbiol. 76:5390–5401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Warnes SL, Keevil CW. 2011. Mechanism of copper surface toxicity in vancomycin-resistant enterococci following wet or dry surface contact. Appl. Environ. Microbiol. 77:6049–6059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Weaver L, Michels HT, Keevil CW. 2008. Survival of Clostridium difficile on copper and steel: futuristic options for hospital hygiene. J. Hosp. Infect. 68:145–151 [DOI] [PubMed] [Google Scholar]
- 56. Weaver L, Noyce JO, Michels HT, Keevil CW. 2010. Potential action of copper surfaces on methicillin-resistant Staphylococcus aureus. J. Appl. Microbiol. 109:2200–2205 [DOI] [PubMed] [Google Scholar]
- 57. Wheeldon LJ, et al. 2008. Antimicrobial efficacy of copper surfaces against spores and vegetative cells of Clostridium difficile: the germination theory. J. Antimicrob. Chemother. 62:522–525 [DOI] [PubMed] [Google Scholar]
- 58. Wilks SA, Michels H, Keevil CW. 2005. The survival of Escherichia coli O157 on a range of metal surfaces. Int. J. Food Microbiol. 105:445–454 [DOI] [PubMed] [Google Scholar]
- 59. Wilks SA, Michels HT, Keevil CW. 2006. Survival of Listeria monocytogenes Scott A on metal surfaces: implications for cross-contamination. Int. J. Food Microbiol. 111:93–98 [DOI] [PubMed] [Google Scholar]
- 60. Zhang YM, Marrakchi H, Rock CO. 2002. The FabR (YijC) transcription factor regulates unsaturated fatty acid biosynthesis in Escherichia coli. J. Biol. Chem. 277:15558–15565 [DOI] [PubMed] [Google Scholar]
- 61. Zhang YM, Rock CO. 2008. Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 6:222–233 [DOI] [PubMed] [Google Scholar]