Abstract
Hypersaline close-to-saturation environments harbor an extremely high concentration of virus-like particles, but the number of haloviruses isolated so far is still very low. Haloviruses can be directly studied from natural samples by using different culture-independent techniques that include transmission electron microscopy, pulsed-field gel electrophoresis, and different metagenomic approaches. Here, we review the findings of these studies, with a main focus on the metagenomic approaches. The analysis of bulk viral nucleic acids directly retrieved from the environment allows estimations of viral diversity, activity, and dynamics and tentative host assignment. Results point to a diverse and active viral community in constant interplay with its hosts and to a “hypersalineness” quality common to viral assemblages present in hypersaline environments that are thousands of kilometers away from each other.
INTRODUCTION: HYPERSALINE ENVIRONMENTS AND THEIR VIRUSES
Hypersaline environments harbor the highest viral densities reported so far for aquatic systems (Table 1), with concentrations up to more than 109 virus-like particles (VLP) per milliliter. The reason for this abundance is intriguing, although there are basically two hypotheses for explaining this phenomenon: these high numbers are due either to the physical stability of viruses in high-salt systems or to the high viral production reached in these systems (12, 57). When analyzed along a salinity gradient (from marine or freshwater to extremely hypersaline, salt-saturated environments), the number of viruses, which is normally correlated to the number of cells, increases with salt (see Fig. 1 for an example). In the transition to the most concentrated systems, over 25% total salts, a sharper increase in virus concentration can be observed, as reported for Mediterranean coastal multipond solar salterns (26) and a series of natural systems in Senegal covering salinities from brackish to near salt saturation (12). This increase may be due to the lack of bacterivory or very low abundances (2 × 105 to 3 × 105 per liter) of nanoflagellate predators reported for these two analyzed systems. In these cases, the abundance of viruses may thus be favored by other biological forces, e.g., the lack of predators in the environment. However, this cannot be generalized to all hypersaline systems, since high numbers (7 × 106 to 28 × 106 per liter) of heterotrophic and extremely halophilic grazers actively ingesting prokaryotes have been described in 31% salt waters in a solar saltern located in western South Korea (40). In addition, viral persistence in water is positively correlated with salinity, with rates of persistence above 97% in close-to-saturation waters (12).
Table 1.
Viral surveys based on culture-independent techniques carried out in hypersaline environments
| Environment (% salinity) | VLP/ml (× 108) | VLP/cell ratio | Technique(s)a | Reference(s) or source |
|---|---|---|---|---|
| La Trinitat salterns, northeast Spain (3.7–37) | 0.4–15 | 5–10 | TEM, in situ measurements of viral activity | 26 |
| Santa Pola salterns, southeast Spain (4–38) | 0.1–10 | 5–10 | TEM, PFGE, metagenomics, metatranscriptomics | 20, 26, 54,56 |
| Crystallizer CR-30 (34) | 15.2 | 44 | ||
| Dead Sea, Israel (34) | 0.08–0.73 | 0.9–42 | TEM | 38 |
| Mono Lake, California (7–8.5) | 0.5–10 | 5.4–142 | TEM, PFGE, metagenomics | 15, 28, 51 |
| Great Salt Lake, Utah (24–30) | 30–62 | 100 | TEM | 10 |
| San Diego salterns, California (6–30) | 1.53–28.7 | 10.9–42.8 | metagenomics | 21, 47 |
| Kaolak solar salterns, Senegal (14–24) | Up to 0.4 | 10 | TEM, in situ measurements of viral activity | 11 |
| Lake Retba, Senegal (29–36) | 5.8–6.9 | 2 | TEM, metagenomics, in situ measurements of viral activity | 11, 58 |
| Sfax salterns, Tunisia (13.8–36) | 1.92–134 | 1.7–50 | TEM, PFGE, metagenomics | Boujelben et al.b |
TEM, transmission electron microscopy; PFGE, pulsed-field gel electrophoresis.
I. Boujelben, P. Yarza, C. Almansa, J. Villamor, S. Maalej, J. Antón, and F. Santos, unpublished data.
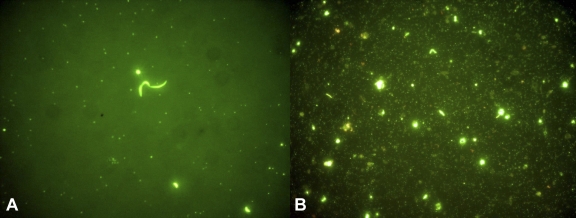
Fig 1.
Sybr gold-stained water samples from two ponds of different salinities from the same Mediterranean coastal saltern. Pictures show the virus-like particles and cells present in a volume of 70 pl of water. (A) Medium (13.8%)-salinity pond; (B) high (36%)-salinity pond.
Another remarkable feature is the high bacterium-to-virus ratio that has been found in many hypersaline environments. As shown in Table 1, this ratio can be above 100 free VLP/cell, which is well above the average ratio among viruses and their hosts in aquatic systems, which normally ranges from 5 to 10 free VLP/cell (62). Although this high ratio seems to be a common trend in hypersaline waters, there are also cases in which this value is lower than in, for instance, marine waters. Such is the case of Lake Retba, with only 2 VLP/cell (58). In any case, as stated by Baxter et al. (10), these values cannot reveal the actual ratio between a specific host-virus pair. For instance, in the work by Guixa-Boixareu et al. (26), the burst size (i.e., maximum number of viruses per cell) of square cells was in the range of 100 to 380 VLP/cell, while the rest of the infected cells had between 6 and 35 VLP/cell. This uneven distribution has to be taken into account when analyzing the free virus assemblage in the environment.
Since the isolation of the first halophilic virus in 1974 and until the very recent work by Atanasova et al. (7), a total of around 26 haloviruses (that is, less than one virus per year, which contrasts with their high abundance in nature) had been isolated from cultures of halophilic Archaea and none of them from extremely halophilic Eukarya or Bacteria, with the exception of two viruses infecting the extremely halophilic proteobacterium Salicola marasensis, reported as unpublished results in reference 30. These studies, reviewed on Mike Dyall-Smith's webpage, indicated that many haloviruses show the typical head and tail morphology, with double-stranded DNA (dsDNA) genomes ranging from 7 to 230 kbp. Very recently (7), Atanasova et al. have isolated 49 new viruses infecting different extremely halophilic hosts (4 Bacteria and 45 Archaea) isolated from nine spatially distant hypersaline environments. Most of these new viruses had head and tail shapes, although icosahedral and pleomorphic viruses were also observed. In addition, haloviruses with other morphologies and genomes have also been isolated (see below). As will be discussed in this minireview, the most common traits (shape and genome size) found in haloviruses in their environments are not mirrored by the general characteristics displayed by haloviruses isolated using pure cultures. As pointed out by Baxter et al. (10), “a true picture of (viral) diversity will come from coupling methods and from exploring new hypersaline environments.” In agreement with Roine and Oksanen (50), these methods should go from the measurement of the bulk biological processes and physicochemical conditions to transmission electron microscopy (TEM), metagenome analysis, and, very importantly, isolation of new viruses infecting ecologically relevant extremely halophilic organisms, including Archaea, Bacteria, and Eukarya. It is worth keeping in mind that, although some of the most abundant extremely halophilic Archaea and Bacteria (Haloquadratum and Salinibacter, respectively) have now been cultivated (6, 14, 16), no viruses have been isolated that infect them.
We will review here the information on haloviruses directly retrieved from the environment, using different kinds of culture-independent approaches. We will not focus on the techniques (caveats are discussed in references 10 and 50) but rather on the results and the information they provide on the ecology of viruses infecting extremely halophilic microorganisms.
TRANSMISSION ELECTRON MICROSCOPY
A direct approach to study viral diversity in nature is the observation of negatively stained samples using TEM, which was first used to characterize marine viruses (reference 59 and references therein). This approach not only allows for the morphological characterization of the viral assemblage but also provides quantitative data on virus abundance (although viral concentrations can be routinely measured by light microscopy in an easier and faster way [36]). The first observation by TEM of an extremely halophilic microorganism infected by viruses was done by Martin Kessel in 1983 (29). This author showed pictures of a “square-shaped halobacterium” (now known as Haloquadratum sp.) infected by what resembled head-tail viruses (definitely not lemon shaped). The first systematic TEM characterization of viruses inhabiting hypersaline waters was that of Guixa-Boixareu et al. (26), who studied the microbial communities of two coastal multipond solar salterns in Spain. Soon later, the viruses in the Dead Sea (38) were also observed under TEM. Later, more salterns (20, 54) and hypersaline lakes, such as the Retba Lake in Senegal, Great Salt Lake in Utah, and Mono Lake in California, were studied (10, 15, 58).
TEM studies indicate that there are basically four kinds of morphologies: the well-known fusiforms (spindle or lemon shaped), head-tail VLP, spherical VLP, and filamentous VLP, the last of which was recently proposed as a new category (10). Finally, unusual morphologies have been also observed in hypersaline settings, such as the six-point stars found in the Dead Sea (38). In their characterization of the viral assemblage in the hypersaline Lake Retba, Senegal, Sime-Ngando et al. (58) provide pictures of extraordinary quality showing not only the spindle-shaped viruses found in high-salt concentrations but also a panoply of other morphologies, resembling hairpins, rods, chains of small globules, hooks, and tadpoles, among others. One of these unusually shaped virus types was also found in solar salterns in Alicante, Spain, as shown in Fig. 2D.
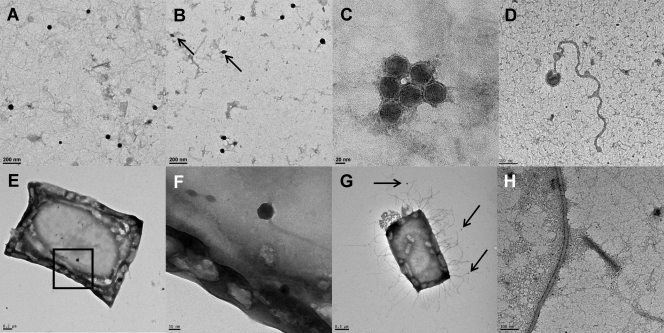
Fig 2.
Transmission electron micrographs of negatively stained viruses and cells from Mediterranean solar salterns. (A) Spherical and tailed haloviruses from a medium-salinity pond (13.8%); (B) tailed and lemon-shaped viruses (indicated by arrows) from a high-salinity pond (36%); (C) detail of an aggregate of spherical haloviruses; (D) detail of particles with unusual shape; (E) Haloquadratum sp.-infected cells (the framed area is enlarged in panel F); (F) lemon-shaped and tailed haloviruses found in association with Haloquadratum sp.; (G) haloviruses (indicated by arrows) apparently attached to the extracellular fibrils of a square cell; (H) detail of a filamentous halovirus.
The distributions of VLP morphologies change with the salinity gradient, with an increase of spindle-shaped viruses that can reach very high concentrations, e.g., 25% in crystallizer ponds from Mediterranean coastal solar salterns, as described in reference 26. These spindle-shaped viruses can even dominate the community, as in Lake Retba (58), where they account for 46% of the total particles, and only less than 1% had a head-tail morphology typical of bacterial DNA viruses (and also of many haloarchaeal viral isolates). In Great Salt Lake, lemon-shaped viruses that appear to have a membrane enclosure and a small tail at one end are very abundant but not dominant. Viruses with similar morphologies are known to infect different kinds of Archaea, including the haloarchaeon Haloarcula hispanica (9). TEM pictures in reference 26 indicate that the square archaeon Haloquadratum walsbyi (one of the most abundant prokaryotes in many hypersaline waters) is infected also, but not only by viruses of these morphologies.
Although in electron microscopy it is sometimes difficult to distinguish artifacts from real traits, Fig. 2G appears to show a square archaeon with lemon-shaped viruses attached to the extracellular net of fibrils. Another square cell has lemon-shaped viruses inside (there is also a head-tail virus, but it seems to be outside the cell and not in the cytoplasm) (Fig. 2F). Therefore, the increase of fusiform viruses is most likely due to the increase of square archaea very frequently observed along salinity gradients (11, 26).
In their study of Mono Lake (a highly alkaline, moderately hypersaline environment, with salinity ranging from 7 to 8.5% total salts) (15), lemon-shaped and filamentous viruses were not observed, which could indicate that these morphologies correspond to viruses infecting extremely halophilic organisms, most likely haloarchaea. However, these differences could also be due to the high alkalinity of the lake. In any case, as discussed above, haloarchaea are infected also by viruses of different morphologies.
PULSED-FIELD GEL ELECTROPHORESIS
Pulsed-field gel electrophoresis (PFGE) is a culture-independent approach that can be used to examine the range of genome sizes of the dominant members of virioplankton. The viral fraction of a sample is collected, embedded in agarose, and treated with proteinase and detergents, and the remaining nucleic acids are separated electrophoretically. The resulting fingerprint is a quick way of characterizing a viral community and comparing different samples. However, this method has a relatively low resolution, since different virus genomes can have the same size and therefore would appear as the same band in a gel, although the potential diversity within any particular band can be further analyzed by cloning and sequencing or by combining this technique with hybridization, as described below.
PFGE has been used to study viral assemblages in coastal Mediterranean solar salterns (20, 52, 54), showing that virioplankton community structure changes along the salinity gradient. In the salterns, viral populations with genomes of 10 to 533 kb were observed, although in samples with salinities higher than 15% the diversity was lower, with viral genomes ranging from 32 to 63 kb. In very high-salt ponds (from 20% salts to saturation), only a band of around 37 kb was observed in most of the samples. When the same samples were analyzed by TEM, several morphologies were found, which most likely indicates that different viruses have genomes of similar sizes (20, 54). Furthermore, when this 37-kb band was cloned in fosmids and subsequently sequenced, an unexpectedly high diversity was found (see below), underscoring the limitations of PFGE as a diversity measurement tool.
PFGE has been also used to characterize viral diversity in the highly alkaline, moderately hypersaline Mono Lake (California) by Jiang et al. (28). These authors found a diverse viral community, with genome sizes ranging from less than 14 to more than 400 kb, with the bulk of DNA (i.e., the most abundant genomes) in the range of 30 to 60 kb, which is typical for viruses infecting prokaryotes. The presence of high-molecular-mass genomes (e.g., those over 200 kb) may suggest the presence of algal viruses, which could be an explanation of why the highest-molecular-mass bands are less abundant in the most saline ponds in solar salterns (52). However, caution must be exerted when interpreting PFGE, since, in order to get accurate size estimation, first the topology of the DNA molecules (e.g., linear or circular) has to be ascertained. In any case, the fact that in Mediterranean solar salterns VLP numbers were not correlated with chlorophyll a concentrations (26), together with the absence of the highest-molecular-mass bands in the most saline ponds (54), indicates that these high-molecular-mass bands probably correspond to the genome of algal viruses.
In the Mono Lake study (28), the contribution of a viral isolate to the total virus assemblage was monitored by hybridizing the labeled viral genome with viral DNA obtained from samples taken from different stations and at different dates within the lake. There was strong hybridization with samples taken the same day (from the same station and at similar depths), while there was no signal with samples taken at the same station 9 months earlier. This result suggests there is a turnover of viral genomes in the natural samples, as metagenomic data indicate for other hypersaline environments (see below). In a study carried out 2 years later (51) in the same environment, the authors found that viral isolates were less prevalent in the viral assemblage than uncultured viruses, reinforcing the idea that viruses readily retrieved by isolation represent only a minor fraction of the natural virus community. This may also indicate that, for haloviruses, culture-dependent and -independent techniques can be unveiling different portions of the diversity, as has happened with prokaryotic communities.
Finally, the dynamics of the viral assemblage have also been (roughly) analyzed using PFGE in solar salterns by Sandaa et al. (52), who showed by repeated sampling (6 days) that band patterns did not change with time. However, this does not necessarily mean that viral genome sequences do not change, since, as shown in other studies (54, 56), a given PFGE band can include very different viral genomes.
IN SITU INFECTION RATES
There are not many reports on the in situ viral activities in hypersaline environments apart from references 11 and 26. Schapira et al. (57) also measured in situ VLP numbers by flow cytometry, although in a moderately hypersaline environment (a natural continuous salinity gradient from 1.8 to 15.5% in a South Australian temperate coastal lagoon).
One of the issues addressed in these studies is the extent of lysogeny in the analyzed systems. In cultured haloarchaea, such as Halobacterium cutirubrum (now reclassified as Halobacterium salinarum) (reference 38 and references therein), viral lysis may be induced by lowering salinity from 30 to 17.5%, while higher salt concentrations reduce the burst size and increase the latent period of infection for the virus ΦS5100 of H. cutirubrum. Lysogeny seems prevalent in environments less favorable to bacterial hosts (33). These data support the idea (46) that a substantial part of haloviruses may be temperate instead of virulent when salinity exceeds 25%. According to reference 19, haloviruses “have probably evolved to exert minimal selective pressure on their sensitive host” by lytically infecting mainly hosts likely to be destroyed by changing environmental conditions. Finally, based on the observations that cultured haloviruses can cause chronic infections (in which viruses are released without lysis of the cell), this has been proposed as the most frequent form of virus-host interaction in high-salt systems (10), although this issue is far from being solved, as discussed below.
According to Bettarel et al. (11), “viral life strategy shows some salinity-driven dependence,” with an increase in lysogeny at higher salt concentrations, although at above 30% salt, this trend is reversed. In addition, the amount of visibly (lytically) infected cells (as observed under TEM) became undetectable beyond a salinity of 17%. However, it is difficult to reconcile this with the high diversity of host genotypes (microdiversity) found in the San Diego solar salterns explained below and with the continuous recycling of virus and their prey. This would fit better in a lytic dynamic. In addition, the low numbers of integrases observed in the sequenced halovirus metagenomes (these enzymes are needed for completing the integration of the virus nucleic acids into its host genome, typical of some lysogenic strategies) points to a low prevalence of lysogeny in the environment. In any case, since all these data come from the study of different hypersaline systems (Table 1), caution must be exerted in their comparison.
The study (26) in Mediterranean coastal samples indicated that, in spite of the high numbers of VLP measured, the percentage of prokaryotic abundance losses per day due to viral lysis was lower than 5%. Viruses then have a small effect in controlling the whole prokaryotic abundance in the systems that are considered to be under “steady state.” However, stability in numbers and even in species does not necessarily mean steady state, since, as discussed below, microbial populations in (at least some) salterns seem to be undergoing cycling of genotypes.
METAGENOMICS: DIVERSITY AND TENTATIVE HOST ASSIGNMENT
Since there is no universal marker present in all viral genomes that would allow for the cataloguing of viral diversity in a way analogous to the rRNA approach (1), one of the most effective ways to describe uncultured viral diversity is the characterization of viral metagenomes or metaviromes (42). Without any doubt, TEM also provides a way of classifying viruses and comparing diversity among environmental samples (15), but this approach cannot provide information on genome type and size or life strategies of a particular type of virus. The main caveat of viral metagenomics is that most of the viral sequences are unique and have no matches in databases (53), and therefore most of the halovirus sequences directly retrieved from the environment are uninformative in a first approach; in a way, it is like opening a black box and finding a new black box inside. However, there are tools that allow for the retrieval of information beyond the annotation of libraries, such as PHACCS (a tool [3] which uses metagenomic data to provide a prediction of the community structure in terms of diversity, evenness, and richness), GAAS (a software that simultaneously estimates both genome relative abundances and average genome length from metagenomic sequences [4]), and other approaches described below. In addition, viral metagenomics may make possible to ascertain the relative abundance of the different viral populations encompassing the viral assemblage (23).
To the best of our knowledge, the first report of a metagenomic approach to describe the viral assemblage in hypersaline environments is that of Sabet et al. (51) in their study of Mono Lake (a “moderately hypersaline” system). Viral DNA was prepared for PFGE, and the 35- to 55-kb band was cloned into bacterial artificial chromosomes. Although the cloning efficiency was very low (as thoroughly discussed in the paper), sequence analysis of viral clones revealed homology to bacteriophage and bacterial proteins. Later, viral metagenomic libraries have been constructed and analyzed for coastal salterns in San Diego (21, 47) and Santa Pola, Alicante, Spain (54, 56) and from the hypersaline Lake Retba in Senegal (58).
San Diego salterns were studied by direct pyrosequencing of viral DNA extracted from water samples. Sequences (with an average of 100 bp) from these salterns, together with sequences from another eight biomes, were analyzed (21) by using the SEED platform (http://www.theseed.org), showing that metabolic profiles could be used as sample “signatures.” In the case of San Diego salterns, viral sequences related to metabolism of carbohydrates and DNA as well as virulence factors dominated and characterized the hypersaline environment (21). These San Diego sequences (coming from three ponds of 8, 14.5, and 30% salinity, with samples taken at various time points separated by 1 day to more than 1 year) were further used (47) to analyze the species composition of haloviruses and their hosts. The authors found that most abundant microbial species persisted over time, although with changes in their relative abundances. Below this stability, there was a variation in the microdiversity of the samples with a cycling of host genotypes. This dynamic was mirrored in the viral assemblage, in which dominant viral taxa persisted while viral genotypes were rapidly changing (47).
The viral assemblage present in the crystallizer CR-30 from the coastal solar salterns Bras del Port (Santa Pola, Alicante, Spain) has been studied by constructing and sequencing fosmid and plasmid clone libraries (54, 56). As explained above, the viral DNA from crystallizer CR-30 runs in PFGE as a single band of around 37 kb, a size appropriate for further cloning in fosmid vectors. In this way, complete viral genomes can be cloned and sequenced, as described in reference 54. This work constitutes the first example of how to reconstruct an (almost) complete genome from an uncultured halovirus. The reason for reconstructing only one genome was the low efficiency of the fosmid library, a problem previously encountered (51). The analysis of this genome (named environmental halophage-1 [EHP-1], although if this virus is infecting an archaeon this name may not be appropriate according to reference 45) revealed a GC content of 51%, lower than those of previously isolated halophages. This low GC content, together with the codon usage, allowed us to propose Hqt. walsbyi as the host. Forty open reading frames (ORFs) were detected, and most of them were annotated as hypothetical proteins. ORFs related to ribonucleotide reductases and thymidylate kinases, typical from lytic viruses, were also found, suggesting that EHP-1 could have a lytic cycle.
Two years later, new metagenomic libraries (in fosmids and in plasmids) were constructed with DNA extracted from the viral assemblage in CR-30, this time with a higher efficiency (56). Sequence analyses showed that the viral community in the crystallizer was highly diverse and very different from viral metagenomes obtained from other environments. A total of 80% of the predicted ORFs were annotated as hypothetical proteins, although most of them (70%) were conserved in the metavirome, since, in spite of not having matches in public databases, hypothetical proteins from some viral genomic fragments matched with hypothetical proteins from other viral fragments in the metavirome. Most of the predicted ORFs with a recognizable function were related to DNA metabolism, with a low proportion (0.9%) of proteins related to the lysogenic cycle. At the nucleotide level, similarities between the CR-30 metavirome and viral sequences from San Diego high-salinity ponds were observed, suggesting that haloviruses from samples with distinct origin could harbor some common features.
In addition, GC content and dinucleotide frequency (61) of viral sequences were analyzed. Sequences of genomes from putative hosts such as Hqt. walsbyi and Salinibacter ruber (most abundant Archaea and Bacteria, respectively, at the time of sampling) and other high-GC-content haloarchaea previously isolated from this crystallizer were also included in the analysis. The goal was to establish a tentative classification scheme in which viral sequences could be grouped with their putative hosts. A total of five groups were obtained based on dinucleotide frequencies. Cluster 4 included low-GC-content viral sequences and the genome of Hqt. walsbyi (at the time, the only known haloarchaeon with a GC content of around 50%), while high-GC-content viral sequences and the genome of S. ruber (with a GC content of around 70%) formed cluster 2. Cluster 1 grouped high-GC-content viral sequences with high-GC-content haloarchaea. Therefore, based both on GC content and dinucleotide frequency analysis, as a working hypothesis we proposed that viruses in cluster 4 were infecting Haloquadratum-like archaea while viruses in cluster 2 infected Salinibacter (and relatives).
Upon initial assembly of viral metagenomic data, it is generally accepted that the longer contigs correspond to the most abundant virus species. In the CR-30 metavirome (56), the longest contigs were included in the cluster of the Haloquadratum genome. In other words, according to our working hypothesis, the most abundant viruses would be the ones infecting this archaeon. Of course, this is not surprising if we consider that, in the analyzed system at the time of sampling, Haloquadratum sp. was the most abundant prokaryote.
It is worth mentioning here that one of the main problems encountered in viral metagenomics is the complete and unambiguous reconstruction of viral genomes (i.e., the assembling of contigs covering the whole viral genome). Besides the large amount of sequence data needed for this purpose, the frequent recombination among viruses of the same type (see, for instance, the case of halophilic viruses HF1 and HF2 [60]) and the modular nature of many viral genomes (27) can make the unambiguous assembly very difficult (or even impossible).
The diversity of virus-host systems in hypersaline Lake Retba (Table 1) was analyzed using a polyphasic approach that included TEM (see above) and the construction and analysis of a viral metagenomic library (58). As happened with the metaviromes from the Santa Pola crystallizer, the metavirome from Lake Retba was dominated by sequences with no matches in databases. However, the identifiable virus sequences were most similar to viral metagenomes from the above-mentioned San Diego high- and medium-salinity ponds, as well as to a cellular metagenome enriched in Haloquadratum that had been previously constructed with water from CR-30 (31). As previously explained, Haloquadratum cells frequently contain a high number of haloviruses in their cytoplasm. In spite of the vast dominance of spindle-shaped viruses in Lake Retba, no sequence similarities were found to archaeal viruses from geothermal environments, where this morphology is also very common. These authors (31), based on the predominance of unusual viral morphologies in their samples, together with the similarities between hypersaline metaviromes, suggest that “similar viruses have adapted to thrive in geographically disparate hypersaline environments.” However, the available metagenomic data do not allow identifying similar genomes in the different analyzed settings, although they do permit the identification of common traits among their viromes.
Very recently (Boujelbene et al., unpublished data), the viral assemblage present in a solar saltern in Sfax (Tunisia) has been analyzed using a metagenomic approach. Viral DNA from three ponds (two medium-salinity ponds and one crystallizer) was cloned in fosmids and end sequenced to analyze the changes in the viral assemblage along the salinity gradient. In addition, a fourth sample, taken 5 months before from the same crystallizer, was also studied to follow temporal changes. BLASTn comparisons among the 4 samples revealed that viral diversities were highly different in each pond, although medium-salinity ponds were more closely related to each other than to sequences from the crystallizer. Interestingly, matches among the two crystallizer samples were lower than expected, suggesting a temporal recycling of viral genotypes, as observed in the San Diego salterns (47). As happened with the metaviromes described above, a high percentage of sequences did not match in the databases, although a considerable number of hits were against the CR-30 metavirome.
Unfortunately, we cannot easily correlate TEM and metagenomic data from natural virus communities. For instance, although the lemon-shaped viruses are most likely infecting the halophilic archaea, more specifically the square archaeon, we cannot “fish” their sequences from the bulk metaviromic sequences by comparison with known cultured spindle-shaped virus from other (halo)archaea. In addition, there is no sequence similarity between spindle-shaped viruses infecting hyperthermophilic and hyperhalophilic archaea, while some bacterial phages have sequence similarity with archaeal head-tail viruses. In any case, sequence data are necessary to classify viruses, since, as in the case of Halorubrum pleomorphic virus 1 (HRPV-1) and HHPV-1 (49), very similar viruses can have different genome types (dsDNA and single-stranded DNA [ssDNA], respectively). Again, a complete analysis of viruses, either cultured or not, can be accomplished only with a polyphasic approach.
In any case, as happens with marine viruses present in different sites in four major oceanic regions that display a “distinct marineness quality” (2), there seems to be a “hypersalineness” common to viral assemblages present in hypersaline environments that are thousands of kilometers away from each other (such as the ones is California, Tunisia, Senegal, and Spain, discussed above). This finding is in agreement with the work by Dinsdale et al. (21) that carried out a functional metagenomic profiling of nine different biomes (subterranean, hypersaline, marine, freshwater, coral associated, microbialites, aquaculture, fish associated, terrestrial animal associated, and mosquito associated) and found differences between them that predicted the biogeochemical conditions of each environment. This difference between halophilic viruses and those from fresh and marine waters was also demonstrated by the lytic failure observed while viruses from nonextreme systems were used to infect the prokaryotic community present in Lake Retba samples (12). The authors interpreted these results based on the differences among viruses infecting Bacteria (that dominate many marine and freshwater habitats) and Archaea (that dominated the Lake Retba community). As explained above, the viral assemblage in this hypersaline sample was dominated by spindle-shaped viruses, which are most likely infecting (square) Archaea. However, many high-salt systems, although dominated by Archaea, also contain high numbers of Bacteria (5) or Archaea other than Haloquadratum-like species (17, 32), as well as viruses with different morphologies.
METATRANSCRIPTOMICS: TOWARD FUNCTION ASSIGNMENT
As discussed above, metagenomics can provide information of the diversity and potentialities of the viral communities but does not give information about what they actually do or even if they are active.
In an attempt to have a picture of the active viral community in the crystallizer CR-30, we analyzed the expression of the viral genomes in the same sample used for constructing the viral metagenome described in reference 55. The viral metagenomic library was used for constructing a microarray (a Virochip) in which the different spots were pieces of the viral metagenome cloned in plasmids (55). Since this viral metagenome had been previously analyzed, the identity of every spot in the chip was known (including its sequence and its position in the classification schema discussed above). Total RNA was extracted from the cellular fraction of the natural sample, reversely transcribed, labeled, and hybridized against the Virochip. The results of this hybridization indicated that, in the natural sample, Salinibacter and high-GC-content haloarchaea-related viruses (clusters 1 and 2) showed higher activity than those related with Haloquadratum (cluster 4). This would support the hypothesis that square Archaea dominated the prokaryotic community at the time of sampling due to a lower activity in their viruses (or to effective ways of avoiding viral infection). In a second step, the natural sample was submitted to two stress conditions (dilution and UV radiation) known to induce the lytic cycle in some viruses. Total RNA was extracted from the stressed samples and, after cDNA synthesis, hybridized with the Virochips and compared with a nonstressed control. Under both stress conditions, spots in the Virochip corresponding to archaeal viruses (those related to Hqt. walsbyi and high-GC-content haloarchaea) increased their levels of expression compared to the untreated samples, which could indicate an increase of virulence of these viruses. In fact, direct counts indicated a decrease in the numbers of Archaea and an increase in the number of free viruses, with respect to the untreated sample (55).
Therefore, a large fraction of the viruses in the salterns (at least those in CR-30) seem to be active, not just chemically preserved in the environment, since around 60% the spots in the Virochip (obtained from free viruses) corresponded with viruses being expressed inside the cells. This contrasts with the low viral mortality reported for these systems, although it is compatible with the chronic infection model. However, the fast cyclic changes reported (47) in hosts and viruses would require a lytic interaction between them.
MICRODIVERSITY OF HOSTS AND VIRUSES
High-salt systems have been described as “taxonomically simplified environments” (31) in which a few taxa dominate the communities. However, beyond this low diversity of species compared to that of other aquatic habitats, a considerable level of microdiversity has been found in extremely halophilic prokaryotes (18, 31, 39, 43, 44). This microdiversity, i.e., diversity below the species level, is therefore not reflected at the 16S rRNA gene level, which is frequently used to describe prokaryotic diversity in nature. Only the analysis of genomes from different isolates or cellular metagenomes from the environment can retrieve such a degree of variation. This has been indeed the case for some of the prokaryotes most frequently retrieved from salterns, such as the haloarchaeon Hqt. walsbyi (13, 18, 22, 31) or the hyperhalophilic bacterium S. ruber (41, 43, 44). In both cases, in spite of the high degree of conservation of the ribosomal operon, variable areas have been detected in their genomes that very frequently code for proteins that could have a role in phage recognition and evasion. Accordingly, the most divergent proteins in viruses are those responsible for host receptor recognition, while the structural proteins are way more conserved (even between viruses infecting hosts from different domains) (49).
The results of the San Diego saltern study (47) fit with the picture of taxonomic simplicity and high microdiversity drawn from genome and specific metagenome analysis. In order to reconcile these observations with the well known “kill-the-winner” interaction between host and genomes, a refined model has been proposed in which the winner is not a given species but a specific lineage inside the species (a sort of Red Queen dynamic in which things have to run to remain the same [47, 48]).
To integrate the kill-the-winner model with the new wealth of genomic and metagenomic data, Rodríguez-Valera et al. (48) have proposed the constant-diversity dynamics (CDD) model, according to which the diversity of prokaryotic populations is preserved by phage predation. In crystallizer CR-30, Haloquadratum has been the most abundant prokaryote for years. If the CDD model is true, then a cycling of genotypes (well below the 16S-based definition of phylotype level) should be occurring that, in turn, implies a change in viruses that are “killing the winner” (the most successful Haloquadratum viral lineages at a given time). If this is the case, then the putative Haloquadratum virus should display a high number of changes. In the analysis of the CR-30 metavirome (56), when assembling the environmental viral sequences into contigs, single nucleotide polymorphisms (SNPs) were found, which indicated the presence of very similar sequences that were finally assembled as a consensus contig. In other words, there were very closely related viral genotypes that most likely would be infecting very closely related hosts. If we analyze the numbers of SNPs in cluster 4 viruses (those that according to our classification schema are the ones most likely infecting Haloquadratum), we see a frequency of 1.3% changes per nucleotide, while in the Salinibacter and high-GC haloarchaea-related viruses (clusters 1 and 2), this number is considerably lower (0.4%).
As a result of the interplay between viruses and hosts, lateral transfer of cellular and viral genes can occur, as has been repeatedly shown (reference 53 and references therein). In addition, viral infection also leaves its imprint in prokaryotic genomes in the clustered regularly interspaced short palindromic repeat (CRISPR)/Cas system. CRISPR/Cas is a defense system against cell invaders, such as viruses and plasmids. They consist of one or more clusters of regularly interspaced short repeats and a variable set of associated (Cas) genes. When exposed to the invaders, cells harboring a functional CRISPR/Cas system may become specifically immunized by the acquisition of new spacers (i.e., sequences intervening between the repetitions) that derive from the foreign DNA sequences (8, 34). Many extremely halophilic archaea have CRISPRs in their genomes (25). Especially relevant to this review is the presence of CRISPRs in one of the two strains of Hqt. walsbyi. In a very recent study of the metavirome of a hypersaline environment (I. García-Heredia, A. B. Martin-Cuadrado, F. Santos, F. J. M. Mojica, A. Mira-Obrador, J. Antón, and F. Rodriguez-Valera, unpublished data), spacers homologous to that of Hqt. walsbyi have been found in the genomes of viruses that, according to several sequence-based analyses, were most likely infecting this haloarchaeon, showing that this viral immunity system is also operating in hypersaline environments.
Hypersaline systems have been traditionally regarded as simple and easy to analyze (26). However, in these last years, we have witnessed many changes regarding the well-established ideas about their microbiota: from the relatively high presence of active Bacteria (5), the distinctive characteristics of Hqt. walsbyi compared with the previously cultured haloarchaea (13), or the wide pangenome of the hyperhalophiles studied to date (31, 39, 41, 43, 44) to new lineages of haloarchaea present in salterns worldwide (37) and the just recently described widespread Nanohaloarchaea (24, 35). (The recent description of Nanohaloarchaea raises the question of whether they had ever been counted as VLP, due to their small size). It seems realistic to anticipate that improvements of the sequencing methods combined with single-cell technologies will allow the recovery of complete viral genomes directly from the environment, as well as unambiguous identification of their hosts. We thus must be open to new surprises in the near future, which certainly will affect the way in which we view haloviruses and their interactions with halophilic hosts.
ACKNOWLEDGMENTS
Our work on halophilic viral assemblages has been funded by the projects CGL2006-12714-CO2-01, CGL2006-12714-CO2-02, CGL2009-12651-C02-01, and CGL2009-12651-C02-02 from the Spanish Ministry of Science and Innovation that include funds from the European Community (FEDER funding) and by the project ACOMP/2009/155 from the Generalitat Valenciana.
We thank Martin Kessel for his kind help regarding the early observations of viruses infecting square haloarchaea and three anonymous reviewers for their helpful and encouraging comments. We are very grateful to all our colleagues involved in the studies discussed in this minireview.
Footnotes
Published ahead of print 13 January 2012
REFERENCES
- 1. Amann RI, Ludwig W, Schleifer KH. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Angly FE, et al. 2006. The marine viromes of four oceanic regions. PLoS Biol. 4:e368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Angly F, et al. 2005. PHACCS, an online tool for estimating the structure and diversity of uncultured viral communities using metagenomic information. BMC Bioinformatics 6:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Angly FE, et al. 2009. The GAAS metagenomic tool and its estimations of viral and microbial average genome size in four major biomes. PLoS Comput. Biol. 5:e1000593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Antón J, Llobet-Brossa E, Rodríguez-Valera F, Amann R. 1999. Fluorescent in situ hybridization analysis of the prokaryotic community inhabiting crystallizer ponds. Environ. Microbiol. 1:517–523 [DOI] [PubMed] [Google Scholar]
- 6. Antón J, et al. 2008. Distribution, abundance and diversity of the extremely halophilic bacterium Salinibacter ruber. Saline Syst. 4:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Atanasova NS, Roine E, Oren A, Bamford DH, Oksanen HM. 2011. Global network of specific virus-host interactions in hypersaline environments. Environ. Microbiol. doi:10.1111/j.1462-2920.2011.02603.x [DOI] [PubMed] [Google Scholar]
- 8. Barrangou R, et al. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712 [DOI] [PubMed] [Google Scholar]
- 9. Bath C, Cukalac T, Porter K, Dyall-Smith MD. 2006. His1 and His2 are distantly related, spindle-shaped haloviruses belonging to the novel group, Salterprovirus. Virology 350:228–239 [DOI] [PubMed] [Google Scholar]
- 10. Baxter BK, Mangalea MR, Willcox S, Sabet S, Nagoulat M-N, Griffith JD. 2011. Haloviruses of Great Salt Lake: a model for understanding viral diversity, p 173–190 In Ventosa A, Oren A, Ma Y. (ed), Halophiles and hypersaline environments. Springer, Berlin, Germany [Google Scholar]
- 11. Bettarel Y, et al. 2011. Ecological traits of planktonic viruses and prokaryotes along a full-salinity gradient. FEMS Microb. Ecol. 76:360–372 [DOI] [PubMed] [Google Scholar]
- 12. Bettarel Y, Desnues A, Rochelle-Newall E. 2010. Lytic-failure in cross inoculation assays between phages and prokaryotes from three aquatic sites of contrasting salinity. FEMS Microbiol. Let. 311:113–118 [DOI] [PubMed] [Google Scholar]
- 13. Bolhuis H, et al. 2006. The genome of the square archaeon Haloquadratum walsbyi: life at the limits of water activity. BMC Genomics 4:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bolhuis H, Poele EM, Rodríguez-Valera F. 2004. Isolation and cultivation of Walsby's square archaeon. Environ. Microbiol. 6:349–360 [DOI] [PubMed] [Google Scholar]
- 15. Brum JF, Steward GF. 2010. Morphological characterization of viruses in the stratified water column of alkaline, hypersaline Mono Lake. Microb. Ecol. 3:636–643 [DOI] [PubMed] [Google Scholar]
- 16. Burns DG, Camakaris HM, Janssen PH, Dyall-Smith ML. 2004. Cultivation of Walsby's square haloarchaeaon. FEMS Microbiol. Let. 238:469–473 [DOI] [PubMed] [Google Scholar]
- 17. Burns DG, Camakaris HM, Janssen PH, Dyall-Smith ML. 2004. Combined use of cultivation-dependent and cultivation-independent methods indicates that members of most haloarchaeal groups in an Australian crystallizer pond are cultivable. Appl. Environ. Microbiol. 70:5258–5265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cuadros-Orellana S, et al. 2007. Genomic plasticity in prokaryotes: the case of the square haloarchaeon. ISME J. 1:235–245 [DOI] [PubMed] [Google Scholar]
- 19. Daniels LL, Wais AC. 1998. Virulence in phage populations infecting Halobacterium cutirubrum. FEMS Microbiol. Ecol. 25:129–134 [Google Scholar]
- 20. Díez B, Antón J, Guixa-Boixareu N, Pedrós-Alió C, Rodríguez-Valera F. 2000. Pulsed-field gel electrophoresis analysis of virus assemblages present in a hypersaline environment. Int. Microbiol. 3:159–164 [PubMed] [Google Scholar]
- 21. Dinsdale EA, et al. 2008. Functional metagenomic profiling of nine biomes. Nature 452:629–632 [DOI] [PubMed] [Google Scholar]
- 22. Dyall-Smith ML, et al. 2011. Haloquadratum walsbyi: limited diversity in a global pond. PLoS One 6:e20968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Edwards RA, Rohwer F. 2005. Viral metagenomics. Nat. Rev. Microbiol. 3:504–510 [DOI] [PubMed] [Google Scholar]
- 24. Ghai R, et al. 2011. New abundant microbial groups in aquatic hypersaline environments. Sci. Rep. doi:10.1038/srep00135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grissa I, Vergnaud G, Pourcel C. 2007. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guixa-Boixareu N, Calderón-Paz JI, Heldal M, Bratbak G, Pedrós-Alió C. 1996. Viral lysis and bacterivory as prokaryotic loss factors along a salinity gradient. Aquat. Microb. Ecol. 11:215–227 [Google Scholar]
- 27. Hatfull GF. 2008. Bacteriophage genomics. Curr. Op. Microbiol. 11:447–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jiang S, Steward G, Jellison R, Chu W, Choi S. 2004. Abundance, distribution and diversity of viruses in alkaline, hypersaline Mono Lake, California. Microb. Ecol. 47:9–17 [DOI] [PubMed] [Google Scholar]
- 29. Kessel M. 1983. Double periodic component in the cell wall of a square-shaped halobacterium, p 746–747 In Bailey GW. (ed), Proceeding of the 41st annual meeting of the Electron Microscopy Society of America San Francisco Press, San Francisco, CA [Google Scholar]
- 30. Kukkaro P, Bamford DH. 2009. Virus-host interactions in environments with a wide range of ionic strengths. Environ. Microbiol. 1:71–77 [DOI] [PubMed] [Google Scholar]
- 31. Legault BA, et al. 2006. Environmental genomics of “Haloquadratum walsbyi” in a saltern crystallizer indicates a large pool of accessory genes in an otherwise coherent species. BMC Genomics 4:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Maturrano L, Santos F, Rosselló-Mora R, Antón J. 2006. Microbial diversity in Maras salterns, a hypersaline environment in the Peruvian Andes. Appl. Environ. Microbiol. 72:3887–3895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McDaniel L, et al. 2008. Metagenomic analysis of lysogeny in Tampa Bay: implications for prophage gene expression. PLoS One 3:e3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mojica FJM, Díez-Villaseñor C, García-Martínez J, Soria E. 2005. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 60:174–182 [DOI] [PubMed] [Google Scholar]
- 35. Narasingarao P, et al. 2012. De novo metagenomic assembly reveals abundant novel major lineage of Archaea in hypersaline microbial communities. ISME J. 6:81–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Noble RT, Fuhrman JA. 1998. Use of SYBR green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat. Microb. Ecol. 14:113–118 [Google Scholar]
- 37. Oh D, Porter K, Russ B, Burns D, Dyall-Smith M. 2010. Diversity of Haloquadratum and other haloarchaea in three, geographically distant, Australian saltern crystallizer ponds. Extremophiles 14:161–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Oren A, Bratbak G, Heldal M. 1997. Occurrence of virus-like particles in the Dead Sea. Extremophiles 1:143–149 [DOI] [PubMed] [Google Scholar]
- 39. Papke RT, Koening JE, Rodríguez-Valera F, Doolittle WF. 2004. Frequent recombination in a saltern population of Halorubrum. Science 306:1928–1929 [DOI] [PubMed] [Google Scholar]
- 40. Park JS, Kim H, Choi DH, Cho BC. 2003. Active flagellates grazing on prokaryotes in high salinity waters of a solar saltern. Aquat. Microb. Ecol. 33:173–179 [Google Scholar]
- 41. Pasić L, et al. 2009. Metagenomic islands of hyperhalophiles: the case of Salinibacter ruber. BMC Genomics 10:570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Paul JH, Sullivan MB. 2005. Marine phage genomics: what have we learned? Curr. Op. Biotechnol. 16:299–307 [DOI] [PubMed] [Google Scholar]
- 43. Peña A, et al. 2011. From genomics to microevolution and ecology: the case of Salinibacter ruber, p 109–122 In Ventosa A, Oren A, Ma Y. (ed), Halophiles and hypersaline environments. Springer, Berlin, Germany [Google Scholar]
- 44. Peña A, et al. 2010. Fine-scale evolution: genomic, phenotypic and ecological differentiation in two coexisting Salinibacter ruber strains. ISME J. 4:882–895 [DOI] [PubMed] [Google Scholar]
- 45. Pina M, Bize A, Forterre P, Prangishvili D. 2011. The archeoviruses. FEMS Microbiol. Rev. 35:1035–1054 [DOI] [PubMed] [Google Scholar]
- 46. Porter K, Russ BE, Dyall-Smith MD. 2007. Virus-hosts interactions in salt lakes. Curr. Op. Microbiol. 10:418–424 [DOI] [PubMed] [Google Scholar]
- 47. Rodríguez-Brito B, et al. 2010. Viral and microbial community dynamics in four aquatic environments. ISME J. 4:739–751 [DOI] [PubMed] [Google Scholar]
- 48. Rodríguez-Valera F, et al. 2009. Explaining microbial population genomics through phage predation. Nat. Rev. Microbiol. 7:828–836 [DOI] [PubMed] [Google Scholar]
- 49. Roine E, et al. 2010. New, closely related haloarchaeal viral elements with different nucleic acid types. J. Virol. 84:3682–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Roine E, Oksanen HM. 2011. Viruses from the hypersaline environment, p 153–172 In Ventosa A, Oren A, Ma Y. (ed), Halophiles and hypersaline environments. Springer, Berlin, Germany [Google Scholar]
- 51. Sabet S, Chu W, Jiang SC. 2006. Isolation and genetic analysis of haloalkaliphilic bacteriophages in a North American soda lake. Microb. Ecol. 51:543–554 [DOI] [PubMed] [Google Scholar]
- 52. Sandaa RA, Skjoldal EF, Bratbak G. 2003. Virioplankton community structure along a salinity gradient in a solar saltern. Extremophiles 7:347–351 [DOI] [PubMed] [Google Scholar]
- 53. Santos F, Antón J. 2011. Viral metagenomics and the regulation of prokaryotic communities, p 33–44 In Marco D. (ed), Metagenomics: current innovations and future trends. Caister Academic Press, Norfolk, VA [Google Scholar]
- 54. Santos F, et al. 2007. Metagenomic approach to the study of halophages: the environmental halophage 1. Environ. Microbiol. 9:1711–1723 [DOI] [PubMed] [Google Scholar]
- 55. Santos F, et al. 2011. Metatranscriptomic analysis of extremely halophilic viral communities. ISME J. 5:1621–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Santos F, Yarza P, Briones C, Parro V, Antón J. 2010. The metavirome of a hypersaline environment. Environ. Microbiol. 12:2965–2976 [DOI] [PubMed] [Google Scholar]
- 57. Schapira M, Buscot Leterme M-JSC, Pollet T, Chapperon C, Seuront L. 2009. Distribution of heterotrophic bacteria and virus-like particles along a salinity gradient in a hypersaline coastal lagoon. Aquat. Microb. Ecol. 54:171–183 [Google Scholar]
- 58. Sime-Ngando T, et al. 2011. Diversity of virus-host systems in hypersaline Lake Retba, Senegal. Environ. Microbiol. 13:1956–1972 [DOI] [PubMed] [Google Scholar]
- 59. Suttle CA. 2007. Marine viruses—major players in the global ecosystem. Nat. Rev. Microbiol. 5:801–812 [DOI] [PubMed] [Google Scholar]
- 60. Tang SL, Nuttall S, Dyall-Smith M. 2004. Haloviruses HF1 and HF2: evidence for a recent and large recombination event. J. Bacteriol. 168:2810–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Willner D, Vega Thurber R, Rohwer F. 2009. Metagenomic signatures of 86 microbial and viral metagenomes. Environ. Microbiol. 11:1752–1766 [DOI] [PubMed] [Google Scholar]
- 62. Wommack KE, Colwell RR. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 641:69:114. [DOI] [PMC free article] [PubMed] [Google Scholar]