Abstract
Two novel gull-specific quantitative PCR (qPCR) assays were developed using 16S rRNA gene sequences from gull fecal clone libraries: a SYBR green assay targeting Streptococcus spp. (gull3) and a hydrolysis TaqMan assay targeting Catellicoccus marimammalium (gull4). The objectives of this study were to compare the host specificity of a previous C. marimammalium qPCR assay (gull2) with that of the new markers and to examine the presence of the three gull markers in environmental water samples from different geographic locations. Most of the gull fecal samples tested (n = 255) generated positive signals with the gull2 and gull4 assays (i.e., >86%), whereas only 28% were positive with gull3. Low prevalence and abundance of tested gull markers (0.6 to 15%) were observed in fecal samples from six nonavian species (n = 180 fecal samples), whereas the assays cross-reacted to some extent (13 to 31%) with other (nongull) avian fecal samples. The gull3 assay was positive against fecal samples from 11 of 15 avian species, including gull. Of the presumed gull-impacted water samples (n = 349), 86%, 59%, and 91% were positive with the gull2, the gull3, and the gull4 assays, respectively. Approximately 5% of 239 non-gull-impacted water samples were positive with the gull2 and the gull4 assays, whereas 21% were positive witg the gull3 assay. While the relatively high occurrence of gull2 and gull4 markers in waters impacted by gull feces suggests that these assays could be used in environmental monitoring studies, the data also suggest that multiple avian-specific assays will be needed to accurately assess the contribution of different avian sources in recreational waters.
INTRODUCTION
Enacted in 2000, the Beaches Environmental Assessment and Coastal Health (BEACH) Act has a goal to improve the quality of coastal waters designated for recreational uses such as swimming, bathing, surfing, and boating (http://water.epa.gov/lawsregs/lawsguidance/beachrules/act.cfm). From a microbiological standpoint, water quality is measured by estimating the levels of fecal pollution using fecal indicator bacteria (FIB). Different fecal sources can contribute to pollution of environmental waters, each of them carrying different health risks. Of all wildlife fecal sources, several waterfowl species have been implicated as a source of fecal bacterial indicators in recreational waters (9, 21). Specifically, gulls are often seen in high numbers in inland and coastal waters, and depending on the geographic location and availability of food sources, gull colonies can be seen all year round near recreational waters, thus chronically contributing to fecal bacterial loadings (9, 16, 28). In other cases, gull roosting and nesting are seasonal (3), and so is their importance to public health.
Recently, Lu et al. (24) developed a SYBR green-based quantitative PCR (qPCR) gull-specific assay (i.e., gull2) targeting the 16S rRNA gene of Catellicoccus marimammalium. This marker has been shown to be host specific (i.e., no amplification with non targeted hosts) and to exhibit a widespread host distribution (i.e., positive to 71% of individual gulls tested) and has been detected in waters presumed to be impacted by gull fecal contamination. For example, when 1,348 water samples from southern Ontario and around Lake Ontario were tested with the SYBR green gull assay, 58% of the samples generated positive signals (25). A TaqMan-based qPCR has also been developed using the original C. marimammalium-specific primers (36), but its use has been limited to marine coastal waters. While the aforementioned studies have shown the potential value of the current C. marimammalium gull qPCR assays as part of the microbial source tracking (MST) toolbox, some limitations need to be addressed. First, a limited number of fecal samples has been used to test potential cross-amplification with other animals, and therefore further validation is needed. Second, target quantification using SYBR green assays can be hampered by the presence of double peaks, suggesting secondary amplification products, and therefore significantly limiting its application as presence/absence assays. While TaqMan-based assays are an alternative, only scarce information is available on the sequence diversity of the region targeted by TaqMan reporter probe and how it affects the sensitivity of the assays. Third, the gull2 marker was designed to target C. marimammalium, as this species was highly abundant in clone libraries generated using gull fecal samples collected in North America. The relative abundance of C. marimammalium in gull species inhabiting other regions may vary considerably; as a result, other bacterial species may potentially be better targets in environmental applications in such cases. Indeed, geographical variability has been documented for other MST markers (12, 19, 39), suggesting that multiple methods and approaches may be needed to increase the reliability of accurate source detection (20, 34).
In this study, we developed a new gull marker (i.e., gull3 assay) targeting Streptococcus spp. and a new TaqMan-based assay (gull4) targeting a smaller region of the C. marimammalium 16S rRNA gene based on additional sequencing information. Evaluation of gull-specific assays included comparison studies against the original gull2 assay by testing for host distribution and specificity of each assay against a high number of gull and nongull feces. Finally, the applicability of these assays to environmental water samples was tested by investigating the prevalence of these gull markers in gull- and non-gull-impacted water samples collected from different geographic locations across North America and Puerto Rico.
MATERIALS AND METHODS
Bacterial strains and plasmid preparation.
C. marimammalium DSMZ M35/04/3T (University of Göteborg Culture Collection, Göteborg, Sweden) and Streptococcus bovis ATCC 33317 were used for preparing plasmids used as qPCR standards. Briefly, biomass from C. marimammalium and S. bovis was harvested from Columbia SB agar (Becton Dickinson, Sparks, MD) and heart infusion agar (Becton Dickinson, Sparks, MD) plates, respectively. PCR products generated with relevant primer sets (i.e., gull2 and gull3 assays; Table 1) were cloned into pCR4 TOPO vector and transformed to TOPO10 chemically competent Escherichia coli cells as described by the manufacturer (Invitrogen, Carlsbad, CA). Individual clones were subcultured at 37°C for 18 h, and plasmids were extracted and purified using the Zyppy plasmid miniprep kit (Zymo Research Corp.).
Table 1.
Summary of oligonucleotide primers and probes for gull-specific qPCR assays
| Assay | Primer/probe sequence (5′ to 3′) | Ta (°C)a | Size (bp) | Reference or source |
|---|---|---|---|---|
| gull2 SYBR green | Forward: TGCATCGACCTAAAGTTTTGAG | 64 | 412 | 24 |
| Reverse: GTCAAAGAGCGAGCAGTTACTA | 64 | 412 | 24 | |
| gull3 SYBR green | SAG1F: ATTTAACCCATGTTAGATGC | 56 | 319 | This study |
| SAG1R: CGTCCCTTTCTGGTAAGT | 56 | 319 | This study | |
| gull4 TaqMan | qGull7F: CTTGCATCGACCTAAAGTTTTGAG | 60 | 116 | This study |
| qGull8R: GGTTCTCTGTATTATGCGGTATTAGCA | 60 | 116 | This study | |
| qGull7Pb: FAM-ACACGTGGGTAACCTGCCCATCAGA-TAMRA | 60 | 116 | This study |
Optimum PCR annealing temperatures were determined using temperature gradients.
FAM, 6-carboxyfluorescein, fluorescence reporter dye; TAMRA, 6-carboxytetramethylrhodamine, fluorescence quencher dye.
Sample collection and DNA extraction.
Two gull fecal samples collected in South Africa were used to develop 16S rRNA gene clone libraries and to design the gull3 assay. By using gull samples from a geographically distant location, we limited the potential effect of migration on the overall composition of gull fecal microbial communities, as they would be exposed to different regional environmental conditions. For geographic and host distribution of gull markers, 255 fecal samples were collected from a variety of gull species inhabiting different geographical locations, and fecal DNA extracts were tested against all three gull assays. Gull fecal specimens used in this study were collected from California (Larus californicus), Ohio (Larus delawarensis), Alaska (Larus glaucescens), Georgia (Larus atricilla), Delaware (L. atricilla and Larus smithsonianus), and South Africa (Larus cirrocephalus). Additionally, 429 fecal samples from a variety of nongull host animals (i.e., 180 fecal samples from domesticated animals and 249 samples from poultry and waterfowl) were used for testing host specificity. The potential value of each assay in environmental applications was tested against environmental water samples presumed to be impacted by gull feces (n = 349 samples) and by fecal sources other than gulls (n = 239 samples) collected from different sites in North America and Puerto Rico. Briefly, gull-impacted samples were collected from marine recreational waters in California (Doheny Beach) and Delaware (Tower Beach) and surface waters at five different locations in Alaska, whereas non-gull-impacted water samples were collected from an intensive agricultural area in Canada (Sumas watershed, British Columbia), Alaska (three lakes), and Puerto Rico (Rio Grande de Arecibo). Water samples (100 ml) were filtered onto polycarbonate membranes (0.4-μm pore size, 47-mm diameter; GE Water and Process Technologies, Trevose, PA). Fecal samples were collected aseptically with sterile spatulas, transferred to sterile tubes, and transported to the laboratory in ice coolers. Filters and fecal samples were shipped overnight on dry ice to the U.S. Environmental Protection Agency (USEPA), Cincinnati, OH, and stored at −80°C until further processing. DNA extraction from filters and fecal samples was performed using a Mo Bio PowerSoil kit (MO BIO Laboratories, Carlsbad, CA) according to the manufacturer's protocol. DNA concentration was measured using a NanoDrop ND-1000 UV spectrophotometer (NanoDrop Technologies, Wilmington, DE). DNA extracts were stored at −20°C until further processing.
Cloning and sequencing analyses.
The microbial community composition of the gull fecal samples collected in South Africa was determined as described elsewhere (23) with minor modifications. Briefly, PCR was performed using the universal bacterial primer set 8F-787R, and selected PCR products were cloned into pCR4 TOPO vector by following the manufacturer's instructions (Invitrogen, Carlsbad, CA). Individual clones were subcultured into 300 μl of Luria broth containing 50 μg/ml ampicillin and screened for inserts using M13 primers. PCR products were sequenced in both directions in the Children's Hospital DNA Core Facility (Cincinnati, OH) using M13 primers and an Applied Biosystems Prism 3730XL DNA analyzer. Raw sequences were processed using Sequencher software (Gene Codes, Ann Arbor, MI). Sequences identified as chimeric structures using Bellerophon (15) were not included in further analyses. Sequences were submitted to Greengenes for alignment using the nearest alignment space termination algorithm (6, 7). The clone libraries were compared using naive Bayesian rRNA classifier version 2.0 of the Ribosomal Database Project (RDP) with a 95% confidence threshold (4). Sequence homology searches were performed using BLASTn (http://www.ncbi.nlm.nih.gov/BLAST/) (1).
New assay development.
To develop the gull3 assay, we first generated a phylogenetic tree that included sequences generated from South African gull feces, and the SILVA database was developed using a neighbor-joining algorithm in ARB (27). Unique phylogenetic clades (i.e., >99% identity) were identified, and candidate primers were then identified for targeting clades using the primer design algorithm in ARB. In silico testing of primer specificity was performed against published data and additional avian 16S rRNA gene clone sequences generated in our laboratory (unpublished data). The assay was optimized through temperature gradients and with various concentrations of fecal DNA templates and tested for host specificity and host and geographic distribution using the fecal samples described above. Additionally, C. marimammalium 16S rRNA gene sequences were used to generate a TaqMan assay (i.e., gull4 assay) based on a DNA fragment targeted by the previously designed assay. Primers and a hydrolysis probe for this TaqMan assay were designed using Primer Express Software (Applied Biosystems, Foster City, CA) (Table 1). This newly developed qPCR assay was also tested for host distribution, host specificity, and presence in environmental waters against the aforementioned set of samples.
Quantitative PCR assays.
Two SYBR green-based qPCR assays (i.e., gull2 and gull3 assays) were tested against fecal and water samples. Like the gull4 assay, the gull2 assay was developed targeting the 16S rRNA gene of C. marimammalium (24). All water and fecal samples were processed as previously described (24) with the following modifications. All of the assays were performed with 0.5 to 1 ng μl−1 of DNA extracts as templates. Ten-fold dilutions of each DNA extract were used to test for PCR inhibition (32). Reaction mixtures (25 μl) contained 1× Power SYBR green master mix (Applied Biosystems, Foster City, CA), 0.2 μg/μl bovine serum albumin, and 0.2 μM each primer (final concentration) and 2 μl of the template. The amplification protocol involved an initial incubation at 50°C for 2 min, followed by 95°C for 10 min and 40 cycles of 95°C for 15 s and 64°C (gull2) or 56°C (gull3) for 1 min, followed by melting curve analysis (i.e., from 60 to 90°C in 0.1°C increments). The qPCR assays were performed using a 7900 HT fast real-time sequence detector (Applied Biosystems, Foster City, CA). All reaction mixtures were prepared in triplicate in MicroAmp Optical 96-well reaction plates with MicroAmp Optical Caps (Applied Biosystems, Foster City, CA). PCR data were analyzed using ABI's sequence detector software (version 2.2.2). PCR signals were recorded as presence/absence data and signal quantity (intensity) values. Four independent standard curves for each qPCR assay were generated by plotting threshold cycle (CT) values against the number of target copies corresponding to serially diluted plasmid standards. The target copy numbers (T) were estimated by the equation T = [D/(PL × 660)] × 6.022 × 1023, where D (g/μl) is plasmid DNA concentration and PL (bp) is plasmid length in base pairs. Each standard curve was generated from at least five 10-fold plasmid dilutions in triplicates. Percent amplification efficiencies were calculated by the instrument manufacturer's instructions (Applied Biosystems). Two no-template controls per PCR plate were used to check for cross-contamination. Disassociation curves were examined to determine the presence of potential primer dimers and other nonspecific reaction products. Signal intensity values were recorded for those reactions showing one corresponding amplification peak within the disassociation curves. PCR products were also visualized in 1% agarose gels using GelStar as the nucleic acid stain (Lonza, Rockland, ME) to confirm amplification product size.
The TaqMan qPCR assay targeting the 16S rRNA gene of C. marimammalium (gull4) was performed in 25-μl reaction mixtures containing 1× TaqMan universal PCR master mix with AmpErase uracil-N-glycosylase (Applied Biosystems, Foster City, CA), 0.2 μg/μl bovine serum albumin, 0.2 μM (final concentration) of each primer, and 6-carboxyfluorescein (FAM)-labeled hydrolysis probe. The amplification protocol involved an initial incubation at 50°C for 2 min to activate uracil-N-glycosylase, followed by 10 min of incubation at 95°C to activate AmpliTaq gold enzyme and then 40 cycles of 95°C for 15 s and 60°C for 1 min. Equipment and data analysis were performed as presented above.
Bayesian statistics.
Bayes' theorem was used to estimate the confidence of each assay at detecting gull fecal sources in environmental waters as described previously (18). Briefly, the posterior probability was calculated using the formula P(A|B) = [P(A) × P(B|A)]/[P(A) × P(B|A) + P(A′) × P(B|A′)]. This involved calculating the posterior probability [P(A|B)] by determining the ratio of true positives [P(B|A)] and false positives [P(B|A′)] in fecal samples and the ratio of water samples that tested positive [i.e., the prior probability or P(A)]. The posterior probability was also calculated by varying the prior probability from worst case scenario (i.e., negative signals in all samples or 0) to best case scenario (i.e., positive signals in all samples or 1) as described by Lamendella et al. (20).
Nucleotide sequence accession numbers.
Representative sequences were deposited in GenBank under accession numbers JN394017 to JN394075.
RESULTS AND DISCUSSION
Phylogenetic analysis of 16 rRNA gene sequences.
DNA extracts from two gull feces collected in South Africa were used to generate clone libraries in an effort to further understand the diversity of gull fecal microbial communities. South African gulls were used in this study, as they have different migration patterns than gulls from North America. By studying geographically separated gull species, we can better understand the diversity of gull microbiota in this waterfowl group and can determine if the current gull assays are useful for different gull species. While the molecular survey is only a random representation of a limited number of gulls (n = 2), this is the first study of its kind with gulls from the South African continent. A total of 354 clone sequences were analyzed in this study. Excluding sequences unclassified or classified as unknowns, 17 bacterial genera were represented in the clone library (Table 2). The gull bacterial community was composed mostly of populations closely related to Bacilli (69.2%), Gammaproteobacteria (16.7%), and Clostridia (3.1%). No sequences homologous to Bacteroidetes were found in this study, which is consistent with the overall low prevalence of members of this phylum in the avian cloacae (10, 16, 24). Within the Bacilli, 116 and 68 sequences were classified as Streptococcus spp. (32.8%) and C. marimammalium (19.2%), respectively, while the other sequences formed a clade of unclassified Lactobacillales (14.1%). In a previous study, C. marimammalium 16S rRNA gene sequences constituted 26% of the fecal clones from gull samples collected in North America (24). The detection of C. marimammalium sequences in gulls from geographically distant regions and the high percentage of positive signals detected with the gull2 and gull4 assays (Table 3) further confirm the widespread occurrence of this bacterial species in gulls. In contrast with Lu et al. (24), fewer Clostridia sequences were obtained in this study, whereas Bacilli were the most dominant bacterial class. Since sequences closely related to Streptococcus (i.e., ≥95% identity) were identified as the most dominant species, these sequences were used as potential targets for the development of new gull markers.
Table 2.
Distribution of 16S rRNA genes in the clone library of gull feces
| Class or group | % clones of total |
Genus | No. of clonesa |
||
|---|---|---|---|---|---|
| This study (n = 354) | Lu et al. (n = 282) | This study | Lu et al. | ||
| Fusobacteria | 3.1 | 0.7 | Cetobacterium | 11 | 2 |
| Bacilli | 69.2 | 37.2 | Bacillus | 0 | 3 |
| Weissella | 5 | 0 | |||
| Lactobacillus | 6 | 9 | |||
| Streptococcus | 116 | 0 | |||
| Catellicoccus | 68 | 74 | |||
| Unclassified Lactobacillales | 50 | 7 | |||
| Clostridia | 3.1 | 17.3 | Dialister | 1 | 0 |
| Subdoligranulum | 3 | 0 | |||
| Clostridium | 3 | 44 | |||
| Unknown | 2 | 2 | |||
| Erysipelotrichi | 0.9 | 0 | Turicibacter | 1 | – |
| Unknown | 2 | ||||
| Alphaproteobacteria | 0.3 | 6.7 | Rubellimicrobium | 1 | 0 |
| Paracoccus | 0 | 8 | |||
| Betaproteobacteria | 0 | 4.3 | Acidovorax | – | 6 |
| Epsilonproteobacteria | 0.3 | 0.4 | Campylobacter | 1 | 1 |
| Gammaproteobacteria | 16.7 | 11.3 | Acinetobacter | 0 | 13 |
| Enterobacter | 1 | 6 | |||
| Escherichia | 0 | 6 | |||
| Citrobacter | 3 | 0 | |||
| Shigella | 6 | 0 | |||
| Klebsiella | 17 | 5 | |||
| Unknown | 32 | 0 | |||
| Sphingobacteria | 0.6 | 0 | Unknown | 2 | – |
| Actinobacteria | 0.6 | 6.4 | Corynebacterium | 0 | 8 |
| Unknown | 2 | 3 | |||
| Mollicutes | 0.3 | 0 | Ureaplasma | 1 | – |
| Bacteroidetes | 0 | 1.1 | Bacteroides | – | 1 |
| Unknown | 5.1 | 3.2 | Unknown | 18 | 9 |
–, not found.
Table 3.
Prevalence of gull markers in various animal feces
| Animal type | Animal | Location of samples | No. of samples | No. of positive samples |
||
|---|---|---|---|---|---|---|
| SYBR green |
TaqMan, gull4 | |||||
| gull2 | gull3 | |||||
| Gull | California gull (Larus californicus) | California | 159 | 138 | 44 | 143 |
| Ring-billed gull (L. delawarensis) | Ohio | 2 | 2 | 2 | 2 | |
| Glaucous-winged gull (L. glaucescens) | Alaska | 64 | 53 | 2 | 50 | |
| Laughing gull (L. atricilla) | Georgia | 5 | 5 | 2 | 2 | |
| Laughing gull (L. atricilla) | Delaware | 3 | 2 | 3 | 3 | |
| Herring gull (L. smithsonianus) | Delaware | 20 | 16 | 16 | 18 | |
| Gray headed gull (L. cirrocephalus) | South Africa | 2 | 2 | 2 | 2 | |
| Total | 255 | 218 | 71 | 221 | ||
| Poultry and waterfowl | Chicken (houses) | Puerto Rico | 98 | 12 | 23 | 8 |
| Turkey | Puerto Rico | 5 | 0 | 5 | 0 | |
| Duck | Puerto Rico | 16 | 0 | 16 | 0 | |
| Pigeon | Puerto Rico | 11 | 1 | 2 | 2 | |
| Heron | Puerto Rico | 1 | 0 | 0 | 0 | |
| Swan | Puerto Rico | 22 | 0 | 9 | 0 | |
| Guineafowl | Puerto Rico | 11 | 0 | 1 | 0 | |
| Crane | Nebraska | 12 | 0 | 2 | 8 | |
| Snow geese | Nebraska | 10 | 0 | 0 | 4 | |
| Pelican | California | 10 | 10 | 10 | 10 | |
| Red Knot | Delaware | 17 | 1 | 0 | 1 | |
| Turnstone | Delaware | 5 | 2 | 1 | 1 | |
| Canada geese | Alaska | 25 | 6 | 0 | 4 | |
| Mallard | Alaska | 6 | 0 | 1 | 0 | |
| Total | 249 | 32 | 76 | 38 | ||
| Nonavian species | Cattle | Puerto Rico | 66 | 0 | 5 | 0 |
| Goat | Puerto Rico | 32 | 0 | 5 | 0 | |
| Monkey | Puerto Rico | 9 | 0 | 0 | 0 | |
| Fish | Puerto Rico | 13 | 1 | 0 | 0 | |
| Horse | Puerto Rico | 30 | 1 | 3 | 0 | |
| Pig | Puerto Rico | 30 | 8 | 13 | 1 | |
| Total | 180 | 10 | 27 | 1 | ||
A total of 24 sequences from genera known to contain species considered human pathogens (i.e., Campylobacter, Shigella, and Klebsiella) were identified in this study. None of the sequences retrieved were related to pathogenic species, although it should be noted that the sequencing effort associated with this study was not deep enough to efficiently retrieve sequences from pathogens, as they are considered rare members. However, bacterial pathogens have been isolated from different species of gulls (5, 13, 38). For example, several studies have also documented the presence of pathogenic Campylobacter spp. in gull excreta (29, 31, 40). On the other hand, Lu et al. (26) reported high prevalence of campylobacters in California gulls (i.e., 45% positive in 159 fecal samples) but a low occurrence of pathogenic species based on species-specific PCR assays and 16S rRNA gene sequences. Based on the latter results, the risk associated with gull fecal pollution has been estimated to be relatively low (35), suggesting that, compared to human fecal pollution sources, fecal loads from gulls and other waterfowl will have to be high in order for the risks to be significant. Nonetheless, the health risks associated with bird fecal pollution sources have been estimated by looking at a limited number of conventional pathogens and using FIB data on a limited number of waterfowl species. Waterfowl are also believed to be important reservoirs of antibiotic-resistant bacteria (2, 8) and of less studied nonbacterial pathogens, such as potentially pathogenic protozoa (14, 17) and influenza viruses (11, 37). Avian influenza viruses (H5N1) have been detected in poultry and waterfowl such as gulls, geese, ducks, swans, and shorebirds (30). While transmission to humans is believed to be relatively low, the mortality rates are relatively high (http://www.who.int/influenza/human_animal_interface/en/). Additionally, the evolutionary rate of influenza viruses is also very high, which explains the widespread occurrence of nonavian reservoirs and reported strains pathogenic to many different mammals (22, 33).
Performance of quantitative PCR.
The range of quantification (ROQ) for the C. marimammalium qPCR assays (gull2 and gull4) was 101 to 106 DNA copies per reaction. For gull3 assay, 10 copies/reaction was below the detection limit of the assay, and therefore the ROQ of the gull3 assay was determined to range from 102 to 106 DNA copies. In order to evaluate assay sensitivity, four independent standard curves were used to calculate the percent amplification efficiency average. The gull4 assay showed the greatest amplification efficiency, followed by gull2 and gull3 (i.e., average ± 1 standard deviation: 95.3 ± 0.9, 85.7 ± 2.5, and 72.8 ± 1.7, respectively). All of the no-template controls were negative, indicating the absence of cross-contamination in the qPCR experiments.
Evaluation of the gull-specific assays.
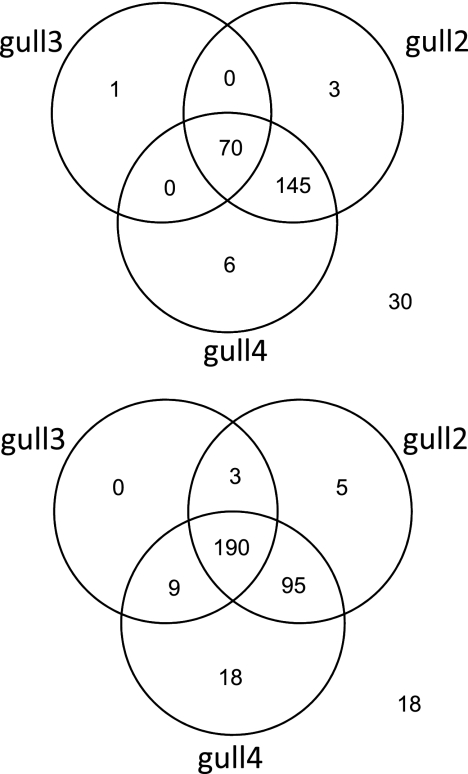
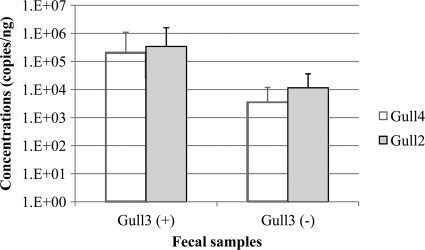
The gull-specific assays designed to target the 16S rRNA gene of C. marimammalium (gull2 and gull4) and Streptococcus spp. (gull3) were tested against individual gull fecal samples (n = 255). Most gull fecal samples were positive with the gull2 (86% or 218/255) and gull4 (87% or 221/255) assays, whereas only 28% (71/255) were positive with the gull3 assay (Table 3). By combining the results from the three assays, the detection levels increased only to 88% (225 of 255) (Fig. 1A). Only one sample was negative with gull2 and gull4 but positive with the gull3 assay, whereas the rest of the gull3-positive samples (n = 70) were positive with both gull2 and gull4 assays. The limit of detection for the gull2 and gull4 assays was one order of magnitude lower than that for the gull3 assay. Interestingly, compared to samples that were positive with all of the assays, signal intensities of gull2 and gull4 assays were relatively lower in many samples that tested negative by the gull3 assay and positive to either one of the other assays (Fig. 2). These data suggest that while Lactobacillales are the important members of the gull fecal community, their overall abundance is dynamic, possibly due to changes in age, dietary regime, or physiological status of the host.
Fig 1.
Venn diagram for gull-specific assay positives against gull feces (top) and water impacted with gull fecal contamination (bottom). Numbers outside the circles represent numbers of samples that tested negative with all assays.
Fig 2.
Mean copy numbers of target markers in gull fecal DNA in samples positive and negative with the gull3 assay. To calculate mean concentrations, below detection limits were treated as zero, and double-peak values from the gull2 assay were not considered. Error bars represent one standard deviation.
Markers targeting C. marimammalium (gull2 and gull4) were detected in the feces of all gull species tested. Lu et al. (24) reported a 71% detection of the gull2 marker in 58 individual gull fecal samples collected from different geographical locations in North America: Florida (L. atricilla), West Virginia (L. delawarensis), Ohio (L. delawarensis), Georgia (L. atricilla and L. delawarensis), and Ontario, Canada (L. delawarensis). In this study, high prevalence of the gull2 marker was also obtained in additional gull species tested, regardless of the locations at which the samples were collected. The relatively lower prevalence of the gull3 marker was more evident in the Glaucous-winged gull (L. glaucescens) excreta (2 of 64 samples). The prevalence of gull3 was also relatively low in California gulls (L. californicus) (i.e., 27%). However, on average, the prevalence of the gull3 marker was higher in the other four Larus species tested (i.e., about 78%) (Table 3). In most gull species, the gull3 marker signal had lower intensity than gull2 and gull4, suggesting that Streptococcus spp. are not as numerically dominant in gull feces as C. marimammalium. While additional studies using samples from other geographic locations and other gull species are needed to determine if our observations are sufficiently widespread, overall these data suggest that the gull3 marker might be better suited to detect fecal contamination from some specific gull species than others.
To test host specificity, a total of 429 individual fecal samples were collected from 20 different animals (i.e., 14 avian species and 6 nonavian species) (Table 3). For nonavian fecal samples (n = 180), relatively low cross-amplification was observed (i.e., 5.5%, 15%, and 0.6% for gull2, gull3, and gull4 assays, respectively). However, considerable numbers of pig fecal samples were positive with the gull2 and gull3 assays (i.e., 27% and 43%, respectively), although the numbers of DNA copies from pig feces were more than four orders of magnitude lower than those of gull feces (data not shown). The gull4 qPCR assay cross-reacted with only one of the nonavian samples, indicating that this assay is more specific to gull feces than the gull2 assay.
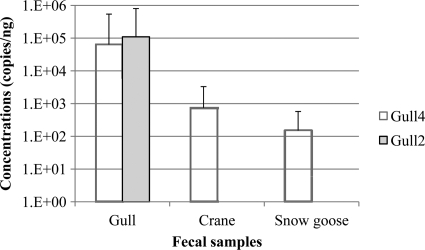
Host specificity tests against nongull avian samples revealed some interesting patterns. For example, approximately 13% and 15% of nongull avian fecal samples were positive for the gull2 and gull4 assays, respectively, whereas twice as many (31%) were positive with the gull3 assay. Specifically, the assays cross-reacted to some extent with chicken, crane, snow geese, and Canada geese, although, in general, the signal intensity was lower than that in gull samples (Fig. 3). In contrast, not only were all pelican fecal samples (n = 10) positive with all three assays, but abundance of the markers was also relatively high. It should be noted that the prevalence of the gull3 marker in nongull avian fecal samples was similar to that in gull feces. Moreover, of the 14 different nongull avian species tested in this study, 10 of them were positive against the gull3 assay, suggesting that the gull3 assay has a potential to be a general avian marker. However, the value of this marker to detect different avian pollution sources is questionable and may be restricted to waters in which swine and ruminants are not suspected primary sources of pollution in light of the cross-amplification signals detected in this study (e.g., agricultural areas nearby recreational waters in which manure is applied). We also noted a discrepancy between gull2 and gull4 assays and the signals associated with crane and snow geese samples. None of the crane and snow geese fecal samples showed cross-amplification with the gull2 assay, whereas the gull4 marker was detected in 67% and 40% of crane and snow geese fecal samples, respectively (Table 3). This is somewhat puzzling, as both gull2 and gull4 markers were designed using the C. marimammalium 16S rRNA gene. Sequencing analysis of snow geese and crane fecal clone libraries demonstrated that there was a small number of sequences nearly identical to C. marimammalium (i.e., an average of <2%; data not shown). Greater detection limits of the gull4 assay than of the gull2 assay can potentially explain the differences in detection rates.
Fig 3.
Mean copy numbers of markers in fecal DNA extracts. To calculate mean concentrations, the below detection limit values were treated as zero, and double-peak values from the gull2 assay were not considered. Error bars represent one standard deviation. The gull2 marker was not detected in any of crane and snow goose fecal samples.
Gull PCR-based signals in water samples.
Of the presumed gull-impacted water samples tested (n = 349), 86%, 59%, and 91% were positive with the gull2, the gull3, and the gull4 assays, respectively (Table 4). Most gull-impacted water samples tested in this study were collected from California recreational water samples (n = 338). Evidence of gull fecal contamination was demonstrated in the California water samples. In fact, few samples from California were negative with all assays (5%) or positive for only one assay (<7%), whereas more than half (56%) were positive by all three assays (Fig. 1B). The gull4 assay showed the highest prevalence, possibly due to its higher sensitivity (i.e., detection limits and amplification efficiency). The amplification efficiency of the gull4 assay ranged from 95% to 98%, which was higher than other assays (i.e., less than 90% for the gull2 and the gull3 assays). Of the presumed non-gull-impacted water samples (n = 239), approximately 5% of the samples were positive with the gull2 and the gull4 assays, whereas 21% were positive with the gull3 assay. Most samples that tested positive for all of the markers were collected from the Sumas watershed in western Canada, presumably contaminated by poultry and some livestock fecal sources. Interestingly, a relatively high rate of the water samples from Sumas watersheds were positive with the gull3 assay (i.e., 41%; n = 64) (Table 4), perhaps to some extent due to cross-amplification. Water samples collected from a watershed in Puerto Rico primarily contaminated by human (wastewater and septic tanks) and animal (mostly cattle, although birds such as chicken and duck are also present) fecal sources were tested for the gull assays. Of the 138 water samples tested, 19 were positive by the gull3 assay, although there were no positive signals by the gull2 and the gull4 assays in any samples. These monitoring results are comparable with specificity results showing the relatively higher cross-amplification of avian fecal samples against the gull3 assay.
Table 4.
Detection of gull markers in water samples by gull-specific assays
| Sampling location(s) | Sample type | Sampling period | No. of water samples | No. of samples positive with: |
Presumed primary fecal contamination sourcea | ||
|---|---|---|---|---|---|---|---|
| gull2 | gull3 | gull4 | |||||
| California beach | Freshwater and seawater | May to September 2008 | 338 | 293 | 202 | 312 | Gull |
| Delaware beach | Freshwater and seawater | May 2011 | 6 | 5 | 4 | 6 | Gull |
| Anchorage, AK | Freshwater | October 2010 | 5 | 1 | 1 | 1 | Gull |
| Anchorage, AK | Freshwater | October 2010 | 3 | 0 | 0 | 0 | Unknown |
| Toronto, Canada | Water treatment plant intake (Lake Ontario offshore) | December 2009 | 9 | 0 | 0 | 0 | Unknown |
| Toronto, Canada | Sewage treatment plant effluent | December 2009 | 3 | 0 | 0 | 0 | Human, gull |
| Toronto, Canada | Sewage treatment plant effluent and CSO samplesb | December 2009 | 6 | 0 | 0 | 0 | Human |
| Sumas watershed, BC, Canada (agriculture-impacted sites) | Freshwater | April 2007 to December 2007 | 64 | 12 | 26 | 10 | Chicken, some livestock |
| Sumas Watershed, BC, Canada (reference site) | Freshwater | April 2007 to December 2007 | 16 | 1 | 5 | 2 | Wildlife |
| Puerto Rico | Freshwater | September 2010 to January 2011 | 138 | 0 | 19 | 0 | Domesticated animals (including chicken) |
There is historical knowledge that host animals are present at these sites a significant part of the year.
CSO, combined sewer overflow.
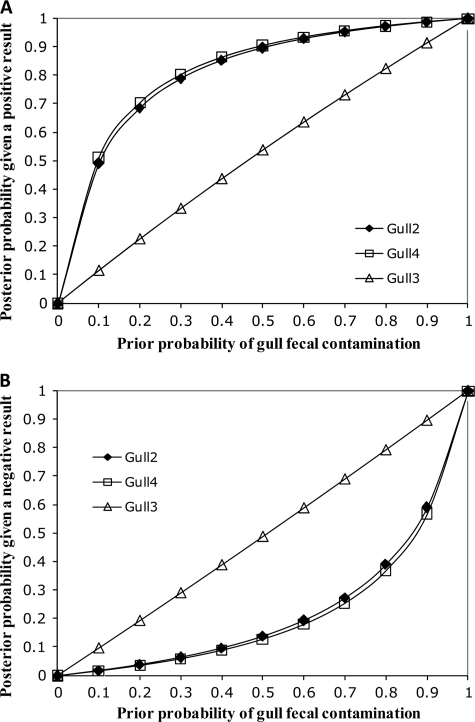
Since all gull-specific assays showed some false-positive (cross-amplification with some of the nongull fecal samples) and false-negative (no signals with some of the gull feces) signals, we conducted Bayesian statistics to determine which assays were more reliable for environmental monitoring. First, the deterministic Bayesian values with gull-impacted water samples examined in this study were estimated as described by Kildare et al. (18) (Table 5). Using this approach, the predictive positive values of the gull2 and gull4 assays were estimated at 0.98 and 0.99, suggesting a very high confidence level for water samples that tested positive. In contrast, lower confidence levels were determined for the gull3 assay. This is in agreement with the overall higher sensitivity and specificity rates exhibited by the gull2 and gull4 assays than the gull3 assay. Bayesian statistical models also showed that the gull2 and the gull4 assays are associated with a lower probability of false-negative signals in water samples under a wide range of prior probabilities of gull fecal contamination than the gull3 assay (Fig. 4). All together, these data indicate that the gull2 and the gull4 assays have more desirable properties for their use as MST markers for environmental application than the gull3 assay.
Table 5.
Bayesian statistics for the three qPCR assays against gull-impacted water samplesa
| Assay | Conditional probability | Sensitivity | Specificity | Prevailing rate |
|---|---|---|---|---|
| gull2 | 0.98 | 0.85 | 0.90 | 0.86 |
| gull3 | 0.63 | 0.28 | 0.76 | 0.59 |
| gull4 | 0.99 | 0.87 | 0.91 | 0.91 |
The conditional probability [i.e., posterior probability or P(A|B) in the Bayesian formula] was calculated using a Bayesian statistical model. The sensitivity is the ratio of positive signals in gull fecal samples. It is numerically identical to P(B|A) in the Bayesian formula. The specificity is the ratio of negative signals in nongull fecal samples. It is numerically identical to 1 − P(B|A′) in the Bayesian formula. The prevailing rate is the ratio of positive signals in water samples. It is numerically identical to P(A) in the Bayesian formula.
Fig 4.
Probability of gull fecal contamination using a Bayesian statistical model. (A) Posterior probability of contamination given a positive qPCR result using three different gull-specific assays over a range of prior probabilities; (B) posterior probability of contamination given a negative qPCR result using three different gull-specific assays over a range of prior probabilities.
In summary, the gull2 and the gull4 assays exhibited higher specificity to gull feces than the gull3 assay, which cross-amplified with a greater number of nongull fecal samples. The gull3 assay was originally designed for the detection of gull fecal contamination. Based on the host specificity, host distribution, and environmental monitoring potential data, the gull3 assay may further support the presence of gull and other sources of avian fecal pollution only in a limited number of scenarios. Overall, gull-specific assays showed a higher level of cross-amplification with other avian species than nonavian hosts. This suggests that when several waterfowl species are present in recreational waters, multiple waterfowl assays will be needed to accurately assess the contribution of each avian source. The relatively high occurrence of gull markers in waters impacted by gull feces suggests that, combined, these assays could be used in environmental monitoring studies. However, additional studies are needed to better understand the correlation of each gull marker with fecal indicator bacteria and human pathogen levels in both fecal and water samples before such molecular signals can be used in microbial risk assessment. While initial studies should focus on public health issues, the availability of robust waterfowl assays will also be relevant to animal health risk assessments in light of the importance of migratory birds in the transmission of animal pathogens.
ACKNOWLEDGMENTS
We thank Jill Hoelle and Laura Boczek for growing C. marimammalium, Brandon Iker for technical assistance, Neil Leat for providing South African gull fecal samples, John Pearce for water and gull fecal samples, and Hans Schreier, University of British Columbia, for providing Sumas watershed water samples.
The U.S. Environmental Protection Agency, through its Office of Research and Development, funded and managed, or partially funded and collaborated in, the research described herein. This work has been subjected to the agency's administrative review and has been approved for external publication. Any opinions expressed in this paper are those of the authors and do not necessarily reflect the views of the agency; therefore, no official endorsement should be inferred. Any mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Footnotes
Published ahead of print 6 January 2012
REFERENCES
- 1. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bonnedahl J, et al. 2010. Characterization, and comparison, of human clinical and black-headed gull (Larus ridibundus) extended-spectrum beta-lactamase-producing bacterial isolates from Kalmar, on the southeast coast of Sweden. J. Antimicrob. Chemother. 65:1939–1944 [DOI] [PubMed] [Google Scholar]
- 3. Burger J. 1979. Competition and predation: herring gulls versus laughing gulls. Condor 81:269–277 [Google Scholar]
- 4. Cole JR, et al. 2009. The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Craven SE, et al. 2000. Determination of the incidence of Salmonella spp., Campylobacter jejuni, and Clostridium perfringens in wild birds near broiler chicken houses by sampling intestinal droppings. Avian Dis. 44:715–720 [PubMed] [Google Scholar]
- 6. DeSantis TZ, et al. 2006. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34:W394–W399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. DeSantis TZ, et al. 2006b. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dolejska M, Bierosova B, Kohoutova L, Literak I, Cizek A. 2009. Antibiotic-resistant Salmonella and Escherichia coli isolates with integrons and extended-spectrum beta-lactamases in surface water and sympatric black-headed gulls. J. Appl. Microbiol. 106:1941–1950 [DOI] [PubMed] [Google Scholar]
- 9. Edge TA, Hill S. 2007. Multiple lines of evidence to identify the sources of fecal pollution at a freshwater beach in Hamilton Harbour, Lake Ontario. Water Res. 41:3585–3594 [DOI] [PubMed] [Google Scholar]
- 10. Fogarty LR, Voytek MA. 2005. Comparison of Bacteroides-Prevotella 16S rRNA genetic markers for fecal samples from different animal species. Appl. Environ. Microbiol. 71:5999–6007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fouchier RAM, et al. 2005. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J. Virol. 79:2814–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fremaux B, Gritzfeld J, Boa T, Yost CK. 2009. Evaluation of host-specific Bacteroidales 16S rRNA gene markers as a complementary tool for detecting fecal pollution in a prairie watershed. Water Res. 43:4838–4849 [DOI] [PubMed] [Google Scholar]
- 13. Girdwood RWA, Fricker CR, Munro D, Shedden CB, Monaghan P. 1985. The incidence and significance of Salmonella carriage by gulls (Larus spp.) in Scotland. J. Hyg. (Lond.) 95:229–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Graczyk TK, et al. 1998. Giardia sp. cysts and infectious Cryptosporidium parvum oocysts in the feces of migratory Canada geese (Branta canadensis). Appl. Environ. Microbiol. 64:2736–2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huber T, Faulkner G, Hugenholtz P. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319 [DOI] [PubMed] [Google Scholar]
- 16. Jeter SN, et al. 2009. Bacteroidales diversity in ring-billed gulls (Larus delawarensis) residing at Lake Michigan beaches. Appl. Environ. Microbiol. 75:1525–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kassa H, Harrington BJ, Bisesi MS. 2004. Cryptosporidiosis: a brief literature review and update regarding Cryptosporidium in feces of Canada geese (Branta canadensis). J. Environ. Heal. 66:34–39 [PubMed] [Google Scholar]
- 18. Kildare BJ, et al. 2007. 16S rRNA-based assays for quantitative detection of universal, human-, cow-, and dog-specific fecal Bacteroidales: a Bayesian approach. Water Res. 41:3701–3715 [DOI] [PubMed] [Google Scholar]
- 19. Lamendella R, Santo Domingo JW, Oerther DB, Vogel JR, Stoeckel DM. 2007. Assessment of fecal pollution sources in a small northern-plains watershed using PCR and phylogenetic analyses of Bacteroidetes 16S rRNA gene. FEMS Microbiol. Ecol. 59:651–660 [DOI] [PubMed] [Google Scholar]
- 20. Lamendella R, et al. 2009. Evaluation of swine-specific PCR assays used for fecal source tracking and analysis of molecular diversity of swine-specific Bacteroidales populations. Appl. Environ. Microbiol. 75:5787–5796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lévesque B, Brousseau P, Bernier F, Dewailly E, Joly J. 2000. Study of the bacterial content of ring-billed gull droppings in relation to recreational water quality. Water Res. 34:1089–1096 [Google Scholar]
- 22. Li KS, et al. 2004. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature 430:209–213 [DOI] [PubMed] [Google Scholar]
- 23. Lu J, Santo Domingo JW. 2008. Turkey fecal microbial community structure and functional gene diversity revealed by 16S rRNA gene and metagenomic sequences. J. Microbiol. 46:469–477 [DOI] [PubMed] [Google Scholar]
- 24. Lu J, Santo Domingo JW, Lamendella R, Edge T, Hill S. 2008. Phylogenetic diversity and molecular detection of gull feces. Appl. Environ. Microbiol. 74:3969–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lu J, et al. 2011. Distribution and potential significance of a gull fecal marker in urban coastal and riverine areas of southern Ontario, Canada. Water Res. 45:3960–3968 [DOI] [PubMed] [Google Scholar]
- 26. Lu J, Ryu H, Santo Domingo JW, Griffith J, Ashbolt N. 2011. Molecular detection of Campylobacter spp. in California gull (Larus 1 californicus) excreta. Appl. Environ. Microbiol. 77:5034–5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ludwig W, et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McLellan SL, Salmore AK. 2003. Evidence for localized bacterial loading as the cause of chronic beach closings in a freshwater marina. Water Res. 37:2700–2708 [DOI] [PubMed] [Google Scholar]
- 29. Moore JE, et al. 2002. Occurrence of Campylobacter spp. and Cryptosporidium spp. in seagulls (Larus spp.). Vector Borne Zoonotic Dis. 2:111–114 [DOI] [PubMed] [Google Scholar]
- 30. Olsen B, et al. 2006. Global patterns of influenza A virus in wild birds. Science 312:384–388 [DOI] [PubMed] [Google Scholar]
- 31. Quessy S, Messier S. 1992. Prevalence of Salmonella spp., Campylobacter spp. and Listeria spp. in ring-billed gulls (Larus delawarensis). J. Wildl. Dis. 28:526–531 [DOI] [PubMed] [Google Scholar]
- 32. Ryu H, et al. 2011. Application of leftover sample material from waterborne protozoa monitoring for the molecular detection of Bacteroidales and fecal source tracking markers. J. Microbiol. Methods 86:337–343 [DOI] [PubMed] [Google Scholar]
- 33. Salomon R, Webster RG. 2009. The influenza virus enigma. Cell 136:402–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Santo Domingo JW, Bambic DG, Edge TA, Wuertz S. 2007. Quo vadis source tracking? Towards a strategic framework for environmental monitoring of fecal pollution. Water Res. 41:3539–3552 [DOI] [PubMed] [Google Scholar]
- 35. Schoen ME, Ashbolt NJ. 2010. Assessing pathogen risk to swimmers at non-sewage impacted recreational beaches. Environ. Sci. Technol. 44:2286–2291 [DOI] [PubMed] [Google Scholar]
- 36. Sinigalliano CD, et al. 2010. Traditional and molecular analyses for fecal indicator bacteria in non-point source subtropical recreational marine waters. Water Res. 44:3763–3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Snoeck CJ, et al. 2011. Reassortant low-pathogenic avian influenza H5N2 viruses in African wild birds. J. Gen. Virol. 92:1172–1183 [DOI] [PubMed] [Google Scholar]
- 38. Steele CM, Brown RN, Botzler RG. 2005. Prevalences of zoonotic bacteria among seabirds in rehabilitation centers along the Pacific coast of California and Washington, U.S.A. J. Wild. Dis. 41:735–744 [DOI] [PubMed] [Google Scholar]
- 39. Stoeckel DM, Harwood VJ. 2007. Performance, design, and analysis in microbial source tracking studies. Appl. Environ. Microbiol. 73:2405–2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Whelan CD, Monaghan P, Girdwood RW, Fricker CR. 1988. The significance of wild birds (Larus sp.) in the epidemiology of Campylobacter infections in humans. Epidemiol. Infect. 101:259–267 [DOI] [PMC free article] [PubMed] [Google Scholar]