Abstract
Oscypek is a traditional Polish scalded-smoked cheese, with a protected-designation-of-origin (PDO) status, manufactured from raw sheep's milk without starter cultures in the Tatra Mountains region of Poland. This study was undertaken in order to gain insight into the microbiota that develops and evolves during the manufacture and ripening stages of Oscypek. To this end, we made use of both culturing and the culture-independent methods of PCR followed by denaturing gradient gel electrophoresis (PCR-DGGE) and pyrosequencing of 16S rRNA gene amplicons. The culture-dependent technique and PCR-DGGE fingerprinting detected the predominant microorganisms in traditional Oscypek, whereas the next-generation sequencing technique (454 pyrosequencing) revealed greater bacterial diversity. Besides members of the most abundant bacterial genera in dairy products, e.g., Lactococcus, Lactobacillus, Leuconostoc, Streptococcus, and Enterococcus, identified by all three methods, other, subdominant bacteria belonging to the families Bifidobacteriaceae and Moraxellaceae (mostly Enhydrobacter), as well as various minor bacteria, were identified by pyrosequencing. The presence of bifidobacterial sequences in a cheese system is reported for the first time. In addition to bacteria, a great diversity of yeast species was demonstrated in Oscypek by the PCR-DGGE method. Culturing methods enabled the determination of a number of viable microorganisms from different microbial groups and their isolation for potential future applications in specific cheese starter cultures.
INTRODUCTION
Protected-designation-of-origin (PDO) labels are granted to traditional agricultural products and foodstuffs originating in a specific area whose quality or characteristics are essentially or exclusively due to particular geographic environments (with their inherent natural and human factors) and whose production, processing, and preparation take place in a defined region (19). The quality of PDO cheeses is based on particular species and herds, grazing pastures, and ancient technologies developed and maintained in a shared manner by the human communities within the PDO area. All these factors select for specific microorganisms that have evolved through the ages and whose activity plays a pivotal role in the sensorial, safety, and preservative properties of the products (22, 46). Therefore, identification, typing, and characterization of these microorganisms are essential for selecting specific starters to control the fermentation while preserving the original sensory profiles.
Oscypek is a traditional Polish scalded-smoked cheese that has had PDO status since 2008. The cheese has the shape of a spindle, with a beautiful characteristic pattern imprinted by the carved wooden molds that give the cheese its final form. It is manufactured from raw sheep's milk without starter cultures in the Tatra Mountains region of Poland, by the process diagramed in Fig. 1. The Oscypek cheese is ready for consumption after smoking without additional ripening, although it can be stored for several weeks. The traditional manufacturing process maintained throughout centuries may have selected appropriate species and/or strains of lactic acid bacteria (LAB) that could be used as specific starters. The new cultures can also be used to complement or replace the starters currently used by the large-scale dairy industry (5). In addition, traditional cheese ecosystems may harbor LAB strains showing unique flavor-forming capabilities (4), enhanced bacteriophage resistance (31), or the production of new, broad-range antimicrobial agents (1).
Fig 1.
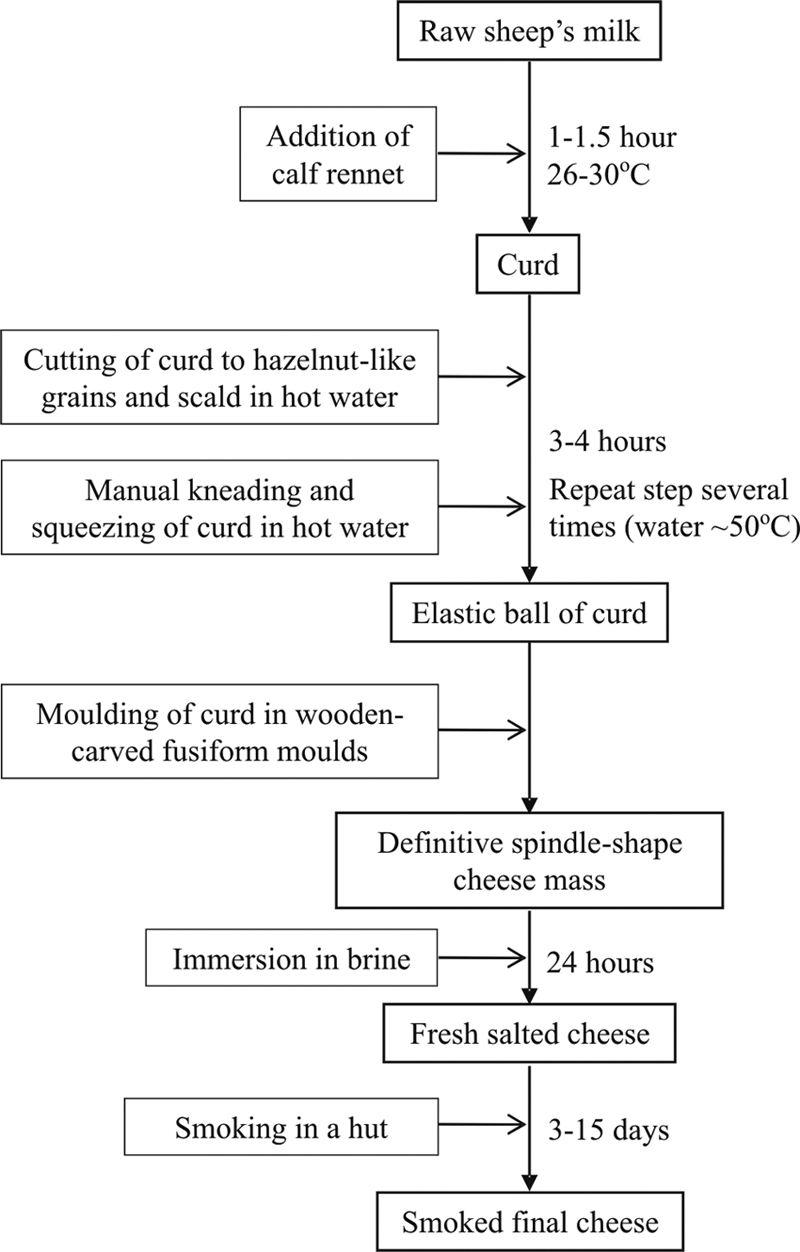
Flowchart of manufacturing and ripening stages of the traditional Polish cheese Oscypek.
In addition to conventional microbial characterization, a vast array of culture-independent molecular techniques is now available to address the diversity and evolution of microbial populations throughout cheese manufacture and ripening (25). Among others, techniques such as denaturing gradient gel electrophoresis (DGGE) (40), single-strand conformation polymorphism (SSCP) (14), fluorescent in situ hybridization (FISH) (18), length heterogeneity-PCR (LH-PCR) (29), quantitative real-time PCR (qPCR) (21), and terminal-restriction fragment length polymorphism (T-RFLP) (3) complement classic culturing techniques, providing a more complete picture of the cheese ecosystem. Investigators have recently begun to apply next-generation sequencing techniques, such as pyrosequencing (35), to study the diversity and dynamics of the microbial populations in natural food fermentations (24, 41, 26) and in fresh foods during storage (17). This method enables rapid insight into the population structure, dynamics, gene content, and metabolic potential of microbial communities.
In this study, both culturing and the culture-independent methods of DGGE and pyrosequencing of segments of the 16S rRNA gene were used to type major and indicator microbial populations in traditional Oscypek cheeses. This allowed us to identify the dominant cultivable bacterial species and to assess microbial diversity and dynamics during the manufacture and ripening of several cheese batches.
MATERIALS AND METHODS
Cheese samples and sampling conditions.
Cheeses manufactured by four independent cheese makers from the Tatra Mountains in Poland were sampled in September 2009. Microbiological and culture-independent molecular analyses were performed on four batches representing traditional Oscypek cheeses produced mostly from raw ewe's milk in shepherds' huts. Curd, fresh cheese (1 day), and smoked cheese that had not been ripened after smoking (3 days) were sampled according to FIL-IDF standard 50B and were kept under refrigeration until analysis.
Microbial analysis by culturing.
Twenty-gram samples of curd or cheese were mixed with 180 ml of physiological salt (PS; 0.9% sodium chloride solution) at 37°C and were homogenized in a laboratory homogenizer (H500 Pol-Eko-Aparatura; Wodzisław Sąski, Poland) for 5 min at 24,000 rpm. Serial 10-fold dilutions in PS were then plated in duplicate onto seven rich and selective media for counting different microbial groups, as follows.
Mesophilic bacteria were counted on plate count agar supplemented with 0.1% skim milk (PCMA; Merck, Darmstadt, Germany) after 72 h of incubation under both aerobic and anaerobic (in 2.5-liter jars with AnaeroGen; Oxoid Ltd., Basingstoke, Hampshire, United Kingdom) conditions at 30°C. In addition, brain heart infusion (BHI; Oxoid Ltd., Basingstoke, Hampshire, United Kingdom) agar plates supplemented with 0.2% cysteine (BHIAC) were used to count total anaerobic bacteria after incubation at 37°C for 72 h in anaerobiosis. Lactococci were grown on M17 (Oxoid) agar supplemented with 10 g lactose/liter (LM17A) and were enumerated after 48 h of incubation at 30°C. Lactobacilli were grown on de Man, Rogosa, and Sharpe agar (MRSA; Merck) adjusted to pH 5.4 and were counted after 72 h of incubation under aerobic and anaerobic conditions at 37°C. Dextran-producing leuconostocs were grown on sucrose-enriched agar supplemented with vancomycin (LMVA, containing 10 g tryptone/liter, 5 g yeast extract/liter, 10 g sucrose/liter, 20 g CaCO3/liter, 15 g agar/liter, and 200 μg vancomycin/ml) and were counted after incubation for 5 days at 21°C. Enterobacteria and coliforms were grown on MacConkey agar (MCA; Merck) and were counted after 48 h of incubation at 37°C. Finally, dilutions of the samples were plated on yeast extract-glucose-chloramphenicol agar (YGCA, containing 5 g yeast extract/liter, 20 g glucose/liter, 15 g agar/liter, and 0.1 g chloramphenicol/liter), and yeasts and filamentous molds were independently counted after 5 days of incubation at 28°C.
Molecular identification of lactic acid bacteria.
After incubation, plates were photographed, and single colonies of the different morphologies for each medium were randomly selected for identification. Bacteria were isolated from the PCMA, LM17A, BHIA, MRSA, and LMVA at different dilutions, from 10−3 to 10−6, depending on the medium. In every case, two plates of each dilution with well-separated colonies were analyzed. Isolates were purified by subculturing and were stored at −80°C in fresh medium with 15% (vol/vol) glycerol. Cryoprotected cultures were recovered in a medium corresponding to that in which they were isolated, and colonies were used as a source of DNA, which was subsequently used in PCR amplifications. Cell extracts were obtained by a mechanical procedure in a Mini-Beadbeater apparatus (BioSpec Products, Inc., Bartlesville, OK) after suspension of a single colony in 100 μl of sterile water and mixing with 50 mg of sterile glass beads (Sigma-Aldrich, Inc., St. Louis, MO). Cell extracts were separated from cellular debris by centrifugation at 13,000 × g for 10 min. Supernatants were used as a source of template DNA for PCR amplification of a large segment of the 16S rRNA gene with the universal bacterial primer 27F (5′-AGAGTTTGATYMTGGCTCAG-3′) and the universal prokaryotic primer 1492R (5′-GGTTACCTTGTTACGACTT-3′) (28).
Amplicons were purified using GenElute PCR Clean-Up columns (Sigma-Aldrich) and were sequenced with an ABI 373 DNA sequencer (Applied Biosystems, Foster City, CA) using primer 27F. On average, 850 bp was obtained and was compared with sequences in the GenBank database, using the online BLAST program (http://www.ncbi.nlm.nih.gov/BLAST/), and in the Ribosomal Database Project (http://rdp.cme.msu.edu/index.jsp). Sequences were assigned to a given species if they showed a similarity equal to or higher than 97% to the sequences of that species (47, 38).
Isolation of total microbial DNA.
Curd and cheese samples were homogenized in PS. Two-milliliter samples of homogenates were centrifuged at 9,000 × g for 1 min. For the isolation of bacterial DNA, the pellets were resuspended in 300 μl of TES buffer (25 mM Tris, 10 mM EDTA, 50 mM sucrose) containing 20 mg of lysozyme/ml (catalog no. 62971; Fluka, Sigma-Aldrich) and 15 μl of mutanolysin at 1 U/μl (catalog no. M9901; Sigma-Aldrich). Then the samples were incubated for 1 h at 37°C. After centrifugation at 9,000 × g for 1 min, the pellets were resuspended in 100 μl of Tris buffer (10 mM Tris HCl [pH 8.5]), and DNA was isolated by using the commercial Genomic Mini DNA purification kit (A&A Biotechnology, Gdynia, Poland) according to the manufacturer's recommendations. The isolation of DNA from fungi was based on the method of DNA isolation for Saccharomyces cerevisiae (45).
PCR amplification and DGGE analysis.
Purified DNA was used as a template for PCR amplification of the V3 variable region of the prokaryotic 16S rRNA gene with primer F357 (5′-TACGGGAGGCAGCAG-3′), to which a 39-bp GC sequence was linked to give rise to GC-F357, and primer R518 (5′-ATTACCGCGGCTGCTGG-3′) (36). Group-specific amplification of 16S rRNA for the lactococcus-enterococcus-streptococcus group and the lactobacillus-leuconostoc-pediococcus group was performed with the primer pairs LB2-GC (5′-GATTYCACCGCTACACATG-3′) with LAC3 (5′-AGCAGTAGGGAATCTTCGG-3′) and LB2-GC with LB1 (5′-AGCAGTAGGGAATCTTCCA-3′), respectively (16). Finally, the D1 domain of the 26S rRNA gene of fungi was amplified with primers NL1-GC (5′-GCCATATCAATAAGCGGAGGAAAAG-3′) and LS2 (5′-ATTCCCAAACAACTCGACTC-3′) (9). PCR amplification conditions were carried out as reported previously (9, 16, 36). Amplicons were analyzed electrophoretically in 8% polyacrylamide gels with 40 to 60% and 30 to 50% formamide denaturing gradients for bacteria and fungi, respectively, by using a DCode device (Bio-Rad, Richmond, CA). Electrophoresis proceeded at 75 V for 17 h for bacteria and at 130 V for 4.5 h for yeasts and molds. Bands were identified by their migration behavior compared to that on a control ladder (20), as well as by DNA isolation, reamplification with the same primers without the GC clamps, sequencing, and comparison of the sequences with the databases.
Amplicon preparation for pyrosequencing.
Primers for the target area were selected to span a region of 250 to 500 bp (a combination of the average mean read length and the maximum amplicon size for a GS FLX amplicon sequencing run as recommended by 454 Life Sciences, Roche Applied Sciences). A 294-nucleotide sequence of the V5 and V6 regions of the 16S rRNA gene (with respect to Escherichia coli 16S rRNA gene positions 786 to 1079) was amplified from the isolated DNA by PCR.
Fusion primers were designed in which a proprietary primer sequence (Adaptor) of the Roche GS FLX sequencing technology and a sample-specific 10-nucleotide key sequence (Multiplex Identifier [MID]) (italicized) were included in order to differentiate between distinct samples. The forward primer was 5′-CCTATCCCCTGTGTGCCTTGGCAGTCTCAGGATTAGATACCCTGGTAGT-3′ (where the underlined sequence corresponds to the forward primer E786F) (10), and the reverse primer was 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGATATCGCGAGTCACGACACGAGCTGACGAC-3′ (where the underlined sequence is the primer equivalent of E. coli positions 1061 to 1079 in Lactococcus lactis IL1403).
For each sample, a 50-μl PCR mixture was prepared containing 1× PCR buffer, 200 μM deoxynucleoside triphosphate mixture (Fermentas, St. Leon-Rot, Germany), 0.4 μM each primer, and 1.25 U of Ex Taq polymerase (Takara Bio Inc., Otsu, Shiga, Japan). To each reaction mixture, 1 μl of the extracted template DNA was added. The PCR conditions used were 95°C for 5 min and 30 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min, followed by one cycle at 72°C for 7 min.
Amplicon quality check and quantity measurement.
Amplicon samples were purified using Ampure XP beads (Beckman Coulter Inc., High Wycombe, United Kingdom) and were run on an Agilent Bioanalyzer, model 2100, to check sample quality. The quantity of amplicon DNA was estimated using a Quant-iT PicoGreen assay (Invitrogen, Carlsbad, CA). Prior to sequencing, the samples were diluted to the same concentration and were pooled.
Ultradeep amplicon sequencing on a 454 GS FLX pyrosequencer.
DNA amplicons were sequenced on a GS FLX pyrosequencer (454, Roche). Samples were clonally amplified by emulsion PCR. DNA-carrying beads were loaded onto the medium region of a picotiter plate (PTP) (1/16 of a plate for each sample) and were sequenced in one direction on a GS FLX Titanium instrument.
Sequence reads for each amplicon were extracted from the resulting standard flowgram files (SFF) into FASTA format and were split into separate pools based on the MIDs by using the sfffile command.
Sequences obtained from pyrosequencing were then processed with Mothur software, version 1.7.2 (44), in which the algorithms mentioned below are implemented. In the first step, reads were trimmed so as to analyze only regions with average scores over 50 bases and a window of at least 35 bases. Then reads either shorter than 150 bases, or with an ambiguous base call (an “N”), or containing a homopolymeric track longer than 8 bases were removed. Afterwards, unique reads were aligned to a SILVA-compatible alignment database. Based on this alignment, potential chimeric sequences were removed using the ChimeraSlayer algorithm (23). Finally, the remaining sequences were clustered into operational taxonomic units (OTUs) and were assigned to taxonomic branches.
The structures of the communities of all samples were compared using a weighted and an unweighted Unifrac algorithm (30) and the Yue and Clayton measure (50).
RESULTS
Basic microbial analysis.
The results of the counting of majority and indicator microbial populations of the different traditional Oscypek cheese samples analyzed in this work are summarized in Table 1. Counts of total mesophilic bacteria under aerobic (PCMA) and anaerobic (PCMA and BHIAC) conditions showed no statistical differences, similarly to counts of lactobacilli in aerobiosis and anaerobiosis (both in MRSA). In general, mesophilic bacterial counts matched those obtained in LM17A, suggesting that Lactococcus spp. were the dominant microorganisms in all cheeses (at a level near 1.0 × 109 CFU/g). High numbers of Lactobacillus species were also observed in all cheese samples, though at levels 1 or 2 logarithmic units lower than those of lactococci. Dextran-producing Leuconostoc spp. were present in all samples at variable levels (ranging from 5.72 to 7.82 log10 CFU per g). Yeasts and molds were also present in all cheese samples in numbers ranging from 5 to 6 log10 CFU per g. Finally, enterobacterial populations were present at levels similar to those of yeasts and molds, and their number showed a tendency to decrease during cheese processing from curd to smoked-cheese samples.
Table 1.
Counts of distinct microbial groups estimated under different incubation conditions during the manufacturing and ripening stages of traditional Oscypek cheeses made by four independent producers
| Microbial group (culture medium) | Count (log10 CFU/g) in the indicated samples from producer: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| I |
II |
III |
IV |
||||||
| Fresh cheese | Smoked cheese | Fresh cheese | Fresh salted cheese | Smoked cheese | Curd | Fresh cheese | Smoked cheese | Smoked cheese | |
| Total mesophilic bacteria (PCMA) | |||||||||
| Under aerobic conditions | 8.92 | 9.00 | 9.45 | 9.53 | 9.34 | 9.60 | 9.26 | 9.37 | 9.49 |
| Under anaerobic conditions | 9.03 | 8.82 | 9.43 | 9.47 | 8.42 | 9.40 | 9.29 | 9.01 | 9.49 |
| Total mesophilic bacteria (BHIAC) | 9.37 | 8.82 | 9.36 | 9.18 | 8.26 | 9.37 | 9.26 | 9.30 | 9.37 |
| Lactococci (LM17A) | 9.37 | 8.92 | 9.52 | 9.12 | 8.59 | 9.44 | 9.45 | 9.49 | 9.51 |
| Lactobacilli (MRSA) | |||||||||
| Under aerobic conditions | 7.91 | 7.03 | 7.18 | 7.30 | 7.37 | 8.42 | 8.27 | 8.10 | 7.98 |
| Under anaerobic conditions | 8.32 | 7.15 | 8.41 | 7.98 | 7.77 | 8.34 | 8.20 | 8.32 | 7.52 |
| Leuconostocs (LMVA) | >7.00 | >6.00 | 6.64 | 6.63 | 5.72 | >7.00 | >7.00 | 7.82 | 7.45 |
| Yeasts and molds (YGCA) | 5.27 | 5.34 | 5.11 | 6.56 | 5.79 | 5.65 | 6.27 | 5.76 | 4.69 |
| Enterobacteria (MCA) | 5.36 | 4.92 | 5.61 | 5.91 | 4.58 | 5.43 | 5.41 | 3.30 | <3.00 |
Identification of isolates.
From the counting plates of total mesophilic bacteria and those for the different LAB populations, 284 colonies were selected at random as representative of the different sizes and morphologies. The media used for isolation of the colonies and the identification results are summarized in Table 2. In agreement with the counting results, nearly 60% of the isolates were identified as Lactococcus lactis subsp. lactis. All these isolates but one came from counting plates of the PCMA, BHIAC, and LM17A media. As a subdominant population, lactobacilli, comprising mostly Lactobacillus casei and Lactobacillus plantarum, were identified. Most lactobacilli came from the MRSA plates, although a few were isolated from all other counting media (Table 2). Leuconostoc citreum, Leuconostoc lactis, and Leuconostoc mesenteroides constituted the majority of dextran-producing Leuconostoc species in Oscypek. The LMVA medium was rather selective for species of this genus, but surprisingly, Leuconostoc spp. were also isolated from both rich (PCMA, BHIAC) and group-selective (LM17A, MRSA) counting plates, indicating that they reach cell densities similar to those attained by lactobacilli in Oscypek. Among the minority components, three Streptococcus thermophilus isolates, one Lactococcus lactis subsp. cremoris isolate, and several isolates of distinct Enterococcus species were recovered.
Table 2.
Identification of isolates from different counting culture media
| Species | No. of isolates obtained on the following medium: |
Total no. of isolates | |||||
|---|---|---|---|---|---|---|---|
| PCMAa |
LM17A | BHIAC | MRSAb | LMVA | |||
| O2 | An | ||||||
| Lactococcus lactis subsp. lactis | 38 | 40 | 42 | 46 | 1 | 167 | |
| Lactobacillus casei | 1 | 1 | 8 | 6 | 31 | 47 | |
| Leuconostoc citreum | 7 | 4 | 6 | 3 | 9 | 29 | |
| Lactobacillus plantarum | 1 | 1 | 13 | 1 | 16 | ||
| Leuconostoc lactis | 2 | 2 | 2 | 6 | |||
| Leuconostoc mesenteroides | 4 | 4 | |||||
| Streptococcus thermophilus | 1 | 2 | 3 | ||||
| Enterococcus faecalis | 2 | 1 | 3 | ||||
| Enterococcus durans | 2 | 2 | |||||
| Enterococcus italicus | 1 | 1 | |||||
| Leuconostoc pseudomesenteroides | 1 | 1 | |||||
| Lactococcus lactis subsp. cremoris | 1 | 1 | |||||
| Bacillus simplex | 1 | 1 | |||||
| Lactobacillus parabuchneri | 1 | 1 | |||||
| Lactobacillus brevis | 1 | 1 | |||||
| Enterobacter kobei | 1 | 1 | |||||
| Total | 48 | 46 | 58 | 63 | 52 | 17 | 284 |
O2 and An indicate incubation under aerobic and anaerobic conditions, respectively.
For isolates identified on MRSA, only the numbers identified under aerobic conditions are shown.
DGGE analysis.
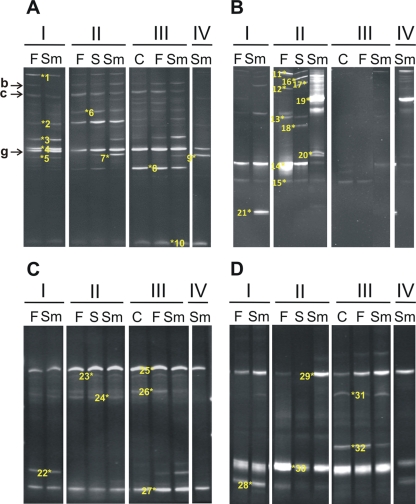
In addition to the culture-dependent approach, DGGE analysis of Oscypek cheese was performed in order to analyze the diversity of the microbial populations in the cheese by a culture-independent technique and to follow their dynamics through manufacture and ripening. The results obtained with universal and group-specific primers are all shown in Fig. 2. Universal primers were used to track the bacterial populations (Fig. 2A) and eukaryotic organisms (Fig. 2B), while specific primers were used for both the lactococcus-enterococcus-streptococcus group (Fig. 2C) and the lactobacillus-leuconostoc-pediococcus group (Fig. 2D). All the bands were identified by reamplification and sequencing (Table 3).
Fig 2.
Diversity and dynamics of bacterial, fungal, and LAB (represented by lactococci and lactobacilli) populations, as shown by analysis of their DGGE profiles. The 16S rRNA genes of bacteria and the D1 domains of the 26S rRNA genes of fungi were amplified by using universal prokaryotic (A) and eukaryotic (B) primers and specific primers for the lactococcus-enterococcus-streptococcus group (C) and the lactobacillus-leuconostoc-pediococcus group (D). The order of cheese samples is the same in all panels and corresponds to the order of samples in Table 1. Capital roman numerals represent four different producers. F, fresh; Sm, smoked; S, fresh salted; C, curd. Numbered bands were purified and identified by reamplification, sequencing, and sequence comparison. The identities of the bands are summarized in Table 3.
Table 3.
Bacterial and eukaryotic species identified from Oscypek cheese by the DGGE technique
| Panel | Band no. | Microorganism | GenBank accession no. of closest relative(s)a | Identity (%) |
|---|---|---|---|---|
| Bacteria | ||||
| A | 1 | Lactococcus garvieae | AP009333 | 100 |
| A | b | Lactobacillus plantarum | AB362982 | 100 |
| A | c | Leuconostoc citreum/Leuconstoc mesenteroides | AB362721/AB596940 | 99 |
| A | 2, 6 | Streptococcus uberis/Streptococcus parauberis | AM946015/CP002471 | 99/99 |
| A | 3, g | Lactococcus lactis | NC_013656 | 100 |
| A | 4, 5 | Streptococcus vestibularis | AEVI01000085 | 98 |
| A | 7 | Tetragenococcus halophilus | AP012046 | 99 |
| A | 8, 10 | Streptococcus thermophilus | FR875178 | 100 |
| A | 9 | Streptococcus salivarius | CP002888 | 100 |
| C | 22, 23, 26, 27 | Lactococcus lactis | CP003132/JF733789 | 100 |
| C | 24 | Lactococcus raffinolactis | NR_044359 | 99 |
| C | 25 | Lactococcus garvieae | NC_015930 | 100 |
| D | 28 | Leuconostoc mesenteroides/Leuconostoc pseudomesenteroides | AB671574/AF515228 | 98 |
| D | 29 | Lactobacillus plantarum/Lactobacillus paraplantarum | NC_014554/HE600693 | 100 |
| D | 30 | Leuconostoc lactis | AB548870 | 100 |
| D | 31 | Leuconostoc citreum | AB682757 | 98 |
| D | 32 | Lactobacillus helveticus/Lactobacillus crispatus | NC_010080/NC_014106 | 99/99 |
| Yeasts and molds | ||||
| B | 11 | Candida pararugosa | GQ222346 | 99 |
| B | 12 | Geotrichum silvicola | DQ377646 | 100 |
| B | 13 | Torulaspora spp. | EF063125 | 95 |
| B | 14 | Saccharomyces spp. | EU441887 | 97 |
| B | 15 | Phialemonium spp. | AB278184 | 96 |
| B | 16 | Candida zeylanoides | EU879957 | 98 |
| B | 17 | Yarrowia lipolytica | EU327102 | 100 |
| B | 18 | Kluyveromyces marxianus | EU669470 | 100 |
| B | 19, 20, 21 | Debaryomyces hansenii | GQ458041 | 100 |
BLAST analysis against the nonredundant nucleotide collection of the GenBank database was performed.
As many as 13 different bands corresponding to the bacterial V3 variable region of the 16S rRNA gene were observed in the different cheese samples (Fig. 2A). It should be noted that the cheese samples analyzed came from four batches produced in different households by independent producers. The main band in all of the samples belonged to Lactococcus lactis (band g). In agreement with counting and identification results, bands of Lactobacillus plantarum (band b) and Leuconostoc citreum/Leuconostoc mesenteroides (band c) were clearly present in almost all samples. In addition, bands corresponding to Lactococcus garvieae (band 1), Streptococcus vestibularis (bands 4 and 5), Tetragenococcus halophilus (band 7), Streptococcus thermophilus (bands 8 and 10), and Streptococcus salivarius (band 9) were observed in some cheese samples.
The composition and evolution of yeast and mold populations, analyzed by DGGE with primers NL1-GC and LS2, are shown in Fig. 2B. All the bands were identified by reamplification and sequencing. A great variability in both the number and the intensity of the bands was observed among samples from different batches and producers. Thick bands corresponding to Saccharomyces spp. (band 14) and Debaryomyces hansenii (bands 19, 20, and 21) were observed in some samples. In addition, other dairy-associated yeasts (such as Candida pararugosa, Geotrichum silvicola, Candida zeylanoides, Yarrowia lipolytica, and Kluyveromyces marxianus) were occasionally present in some other samples (Fig. 2B).
The DGGE results corresponding to the LAB groups of lactococci-enterococci-streptococci and lactobacilli-leuco-nostocs-pediococci are presented in Fig. 2C and D, respectively. Bands of Lactococcus lactis and Lactococcus garvieae were present in all samples. It is worth noting that the Lactococcus garvieae band was shown to be more intense than other bands in most of the samples. The latter species, which was also shown with the universal primers, was never identified among the isolates. In addition to these two Lactococcus species, a band belonging to Lactococcus raffinolactis (band 24) was identified in cheese samples from producers I and II. With primers LB2-GC and LB1, three out of the five bands produced belonged to the Leuconostoc species. Bands of Leuconostoc lactis (band 30) and Lactobacillus plantarum/Lactobacillus paraplantarum (band 29) were present in all curd and cheese samples, while bands of Leuconostoc mesenteroides/Leuconostoc pseudomesenteroides (band 28), Leuconostoc citreum (band 31), and Lactobacillus helveticus/Lactobacillus crispatus (band 32) were observed only occasionally.
16S rRNA-based pyrosequencing analysis of Oscypek.
Pooled 16S rRNA gene amplicons of bacterial communities from three cheese samples were sequenced. The numbers of reads after sequencing were 35,179 for curd, 59,451 for fresh cheese, and 46,894 for smoked cheese, all from producer III. These reads were later cleaned, and putative chimeric sequences were removed (14.2% of all cleaned reads). Further processing involved clustering and assignment to taxonomic branches. Good's coverage estimator was 99% for all three samples, indicating that we managed to capture the majority of the bacterial biodiversity in each sample.
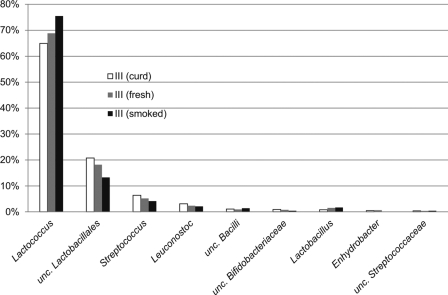
Reads were assigned to four different phyla: Firmicutes, Actinobacteria, Proteobacteria, and Bacteroidetes (see Table S1 in the supplemental material). Out of 40 genera, we have identified 9 major genera or taxonomic groups represented by at least 0.4% of the total pool across the samples (Fig. 3). Six of them belong to Lactobacillales (Firmicutes), constituting 97% of all clean reads. The remaining three were unclassified Bacilli, unclassified Bifidobacteriaceae (Actinobacteria), and Enhydrobacter (Proteobacteria). These were represented by ca. 2.5% of all clean reads. The majority of the taxonomic spectrum identified in the samples consisted of auxiliary genera represented by at most tens of reads. By use of pyrosequencing data, no deeper assignment (i.e., to the level of individual species) was possible; however, it should be noted that ca. 20% of all sequences were assigned to “unclassified” branches. This indicates the strong presence of strains that have not yet been thoroughly annotated.
Fig 3.
Major taxonomic groups in traditional Oscypek by the percentage of assigned reads. Only genus-level reads that represented at least 0.4% of the total pool of reads across the three samples are shown. unc., unclassified.
Statistical comparison of the three individual samples from producer III showed very high similarity between the memberships and structures of the samples. The Yue and Clayton measure in all-versus-all comparison is near its maximum value (that is, 1); however, in agreement with the experiments conducted, the curd and the fresh cheese, and the fresh and smoked cheeses, are slightly more similar to each other than are the curd and the smoked cheese.
As seen in Fig. 3, during processing, shifts in the microbial community structure of Oscypek were found in all four major groups: Lactococcus, unclassified Lactobacillales, Streptococcus, and Leuconostoc. The number of Lactococcus spp. increased at the expense of the latter three bacterial groups. Additional differences were more subtle.
DISCUSSION
An understanding of the microbial composition of cheeses made from raw milk is essential for the quality and safety of the final product, key factors in PDO cheeses. Besides culturing, a vast array of molecular techniques is now applied to address the question of microbial diversity and population dynamics throughout the cheese manufacturing and ripening processes (25). However, despite the increasing number of polyphasic studies, including both culture-dependent and culture-independent approaches, to our knowledge no data on 16S rRNA-based pyrosequencing of the microbiome of traditional cheeses have been published. Moreover, a single report on the microbial analysis of kefir by a high-throughput sequencing approach has been released recently (13).
In addition, little has been demonstrated concerning the microbial communities responsible for the ripening of traditional Oscypek cheese. In fact, a single report on the microbiological analysis of Oscypek and its intermediary products has been published (32). This study was performed only by culture-dependent techniques, which included determination of total microbial counts using a broth medium and selective media for lactobacilli, lactococci, streptococci, enterococci, and yeasts.
In the present work, the biodiversity of traditional Oscypek cheeses was analyzed by culturing, PCR-DGGE fingerprinting, and 454 pyrosequencing. The first two methods revealed the predominant microorganisms, belonging to the genera Lactococcus, Lactobacillus, Leuconostoc, and Streptococcus. Besides Lactococcus lactis, strains of lactobacilli and leuconostoc species could be considered possible adjunct-culture candidates for the design of specific starter cultures for Oscypek cheese. There were also microorganisms identified by one method only, either the culture-dependent method (single isolates of Enterococcus durans, Enterococcus italicus, Lactobacillus parabuchneri, Bacillus simplex, and Enterobacter kobei) or the culture-independent DGGE approach (Lactococcus garvieae, several Streptococcus species other than S. thermophilus, and Tetragenococcus halophilus, Lactococcus raffinolactis, and Lactobacillus helveticus/Lactobacillus crispatus). Although the use of different selective media has been shown to increase the chance of identification of particular organisms (12), culturing methods are limited by the fact that a great number of microorganisms are difficult or impossible to cultivate (2). This nonrecoverable microbiota can be detected by culture-independent techniques, such as DGGE. The sensitivity of the DGGE technique can be slightly improved by specific primers detecting less-abundant bacteria (43), which was the case in this study, where Lactococcus raffinolactis was identified only with primers specific for the lactococcus-enterococcus-streptococcus group, while Lactobacillus helveticus/Lactobacillus crispatus, Leuconostoc citreum, Leuconstoc mesenteroides/Leuconostoc pseudomesenteroides, and Leuconostoc lactis were identified exclusively with primers specific for the lactobacillus-leuconostoc-pediococcus group. However, one of the drawbacks of the DGGE technique is the fact that the number of rRNA genes per chromosome equivalent differs widely in bacteria (27). In addition, heterogeneous copies of rRNA gene operons exist for both bacteria (37) and fungi (49), so that different bands may belong to the same species.
Simultaneously, three samples of traditional Oscypek from producer III were analyzed by a high-throughput next-generation sequencing technique (454 pyrosequencing). In addition to the most abundant bacterial groups (e.g., Lactococcus, Lactobacillus, Leuconostoc, Streptococcus, and Enterococcus) found by culture-dependent and -independent methods, other bacteria, belonging to the families Bifidobacteriaceae and Moraxellaceae (mostly Enhydrobacter), were identified. They represented, respectively, 0.71% or 0.45% of the total pool of reads; thus, they are minor but significant elements of the bacterial community of Oscypek, since the contributions of the majority of other branches did not exceed 0.1%. Taking into account the traditional conditions of Oscypek processing (raw milk, production in shepherds' huts), it is not surprising to find bacteria known to belong to gut microbiota or the manufacturing environment (workers' hands, water, etc.). Actinobacteria and Proteobacteria, which are widely distributed in nature, have been found using pyrosequencing of tagged 16S rRNA gene amplicons in traditionally fermented food such as pearl millet slurries (24).
Interestingly, a relatively high ratio (ca. 20%) of reads was assigned to “unclassified” taxonomic branches. This result indicates a strong presence of strains that have not yet been sequenced or annotated but at the same time reveals the weakness in the current state of the field. In our studies, taxonomic assignment was possible only up to the genus level because of two factors: the shortness of the fragment sequenced (294 bp) and the limits of database classification (48). Further advances, including strain-level resolution in taxonomic assignment, will depend on whole-genome sequencing strategies (39).
Compared with the predominant bacteria, other groups identified in traditional Oscypek were detected at relatively low abundances. Among the organisms that were revealed or whose percentages of assigned reads increased in the fresh cheese (immersed in brine) from producer III, there were some salt-tolerant bacteria related to marine environments, such as Sanguibacter, Flavobacteriaceae, Tetragenococcus, or Chromohalobacter spp. Some of these groups include recognized pathogens, suggesting the necessity of improvement of the safety conditions of the cheese. The family Flavobacteriaceae includes many marine species (6). Different marine bacterial species have already been identified in other traditional cheese ecosystems by culture-independent approaches (34, 15). However, genera belonging to the family Flavobacteriaceae, except for the genus Flavobacterium, are generally adapted to nonmarine ecosystems, including the soil, water, and animal habitats (7). Flavobacteria have been shown to be associated with the spoilage of food and food products (11). It is worth noting that the percentage of reads assigned to Flavobacteriaceae decreased slightly in the smoked cheese compared to the fresh sample from producer III. Also of note is the percentage of reads for Actinobacteria (0.987%), which seemed to be members of the family Bifidobacteriaceae and possibly to belonging to Bifidobacterium species. To our knowledge, this is the first report of bifidobacteria from a traditional cheese. Since they might have great industrial potential or probiotic significance, efforts will be made in the near future at the selective recovery in culture of bifidobacteria from Oscypek.
The counts of microbial populations of different Oscypek cheese samples analyzed in this work demonstrated that the number of enterobacteria decreased from curd to smoked-cheese samples. Thus, our data suggest that the smoking process increases the quality and safety of the traditional Oscypek. These results are supported by the observation that cold smoking reduces the amount of Listeria monocytogenes in smoked salmon (42). During warm smoking of Oscypek cheese, phenolic compounds are produced from polyphenols present in the wood used by shepherds for smoking (33). Because of the bacteriostatic and/or bactericidal properties of volatile phenols, these compounds are assumed to be responsible for decreasing the microbial population during the smoking process (32). Another probable explanation, taking into account the selective reduction in the levels of particular bacterial groups, could be the effect of microbial competition, due particularly to acidification and/or the production of antimicrobial compounds by LAB species.
Changes in microbial composition during the manufacture and ripening of Oscypek cheese were also observed among the predominant bacterial groups. The percentage of reads representing unclassified members of Lactobacillales, Streptococcus, Leuconostoc, and Bifidobacteriaceae diminished after brining and smoking, whereas that for Lactococcus increased. However, in our culture-dependent studies, we did not observe significant changes in microbial counts for lactococci, lactobacilli, or streptococci during the manufacturing process. It is worth remembering that the two approaches, pyrosequencing and culturing, differ in their levels of sensitivity. In contrast, a general decrease in the levels of most bacterial groups, including lactococci, lactobacilli, streptococci, and enterococci, during Oscypek cheese preparation has recently been reported by Majcher and coworkers (32). This discrepancy may be due to the individual characteristics of various cheeses and/or to the different media and culturing conditions used in the two studies. We also observed approximately 5-fold differences in the levels of yeasts and molds from Majcher's results. However, this could be explained by the fact that the experiments were performed on different culture media, Czapek agar in Majcher's work and YGCA in our studies. The great diversity of yeasts demonstrated by the PCR-DGGE method in our studies has been observed previously in various artisanal dairy products (9, 8, 21).
In conclusion, culturing and culture-independent methods, such as DGGE and pyrosequencing of the 16S rRNA gene fragments, were successfully applied to reveal the microbial composition and its evolution during the manufacture of traditional Oscypek cheese. Our results confirmed the usefulness of polyphasic molecular methods for the identification of cultivable, noncultivable, abundant, and scarce microorganisms in complex dairy food products. The culture-independent DGGE technique allowed identification and tracking of the majority of the population throughout manufacturing, while the pyrosequencing technique produced a complete inventory of the bacterial species encountered within the Oscypek ecosystem. It is expected that each bacterial species present in Oscypek cheese may contribute somehow to the ripening process. Finally, culturing methods enabled the estimation of the number of CFU within various microbial groups and the isolation of microorganisms for potential future applications in cheese starter cultures. On the basis of the counting results, bacteria belonging to lactococci, lactobacilli, and leuconostocs were proposed as possible candidates for starter cultures for Oscypek production. The high bacterial biodiversity in the different batches revealed by all culturing and culture-independent approaches may be partially explained by the heterogeneous production of traditional Oscypek.
Supplementary Material
ACKNOWLEDGMENTS
This work was done in the framework of scientific cooperation between the Polish Academy of Sciences (PAN) and the Consejo Superior Investigaciones Científicas (CSIC) and was partly supported by the Spanish Ministry of Science and Innovation (MICINN) (AGL2007-61869-ALI) and the Institute of Biochemistry and Biophysics, PAN (IBB PAN). Á. Alegría was supported by a grant from FICYT (BP08-053).
We thank our colleagues from the Laboratory of DNA Sequencing and Oligonucleotide Synthesis at IBB PAN for DNA sequencing.
Footnotes
Published ahead of print 13 January 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Alegría A, Delgado S, Roces C, López B, Mayo B. 2010. Bacteriocins produced by wild Lactococcus lactis strains isolated from traditional, starter-free cheeses made of raw milk. Int. J. Food Microbiol. 143:61–66 [DOI] [PubMed] [Google Scholar]
- 2. Amann RI, Ludwig W, Schleifer KH. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arteau M, Labrie S, Roy D. 2010. Terminal restriction fragment polymorphism and automated ribosomal intergenic spacer analysis profiling of fungal communities in Camembert cheese. Int. Dairy J. 20:545–554 [Google Scholar]
- 4. Ayad EHE, Verheul A, de Jong C, Wouters JTM, Smit G. 1999. Flavour forming abilities and amino acid requirements of Lactococcus lactis strains isolated from artisanal and non-dairy origin. Int. Dairy J. 9:725–735 [Google Scholar]
- 5. Ayad EHE, Verheul A, Engels WJ, Wouters JT, Smit G. 2001. Enhanced flavour formation by combination of selected lactococci from industrial and artisanal origin with focus on completion of a metabolic pathway. J. Appl. Microbiol. 90:59–67 [DOI] [PubMed] [Google Scholar]
- 6. Bowman JP. 2004. Psychrophilic prokaryote structural-functional relationships, biogeography and evolution within marine sediment. Cell. Mol. Biol. 50:503–515 [PubMed] [Google Scholar]
- 7. Bowman JP, Nichols DS. 2005. Novel members of the family Flavobacteriaceae from Antarctic maritime habitats including Subsaximicrobium wynnwilliamsii gen. nov., sp. nov., Subsaximicrobium saxinquilinus sp. nov., Subsaxibacter broadyi gen. nov., sp. nov., Lacinutrix copepodicola gen. nov., sp. nov., and novel species of the genera Bizionia, Gelidibacter and Gillisia. Int. J. Syst. Evol. Microbiol. 55:1471–1486 [DOI] [PubMed] [Google Scholar]
- 8. Callon C, Delbès C, Duthoit F, Montel MC. 2006. Application of SSCP-PCR fingerprinting to profile the yeast community in raw milk Salers cheese. Syst. Appl. Microbiol. 29:172–180 [DOI] [PubMed] [Google Scholar]
- 9. Cocolin L, Aggio D, Manzano M, Cantoni C, Comi G. 2002. An application of PCR-DGGE analysis to profile the yeast populations in raw milk. Int. Dairy J. 12:407–411 [Google Scholar]
- 10. Coloqhoun JA. 1997. Discovery of deep-sea actinomycetes. Ph.D. dissertation Research School of Biosciences, University of Kent, Canterbury, England [Google Scholar]
- 11. De Beer H. 2005. A taxonomic study of Chryseobacterium species in meat. Ph.D. thesis University of the Free State, Bloemfontein, South Africa [Google Scholar]
- 12. Delbès C, Ali-Mandjee L, Montel MC. 2007. Monitoring bacterial communities in raw milk and cheese by culture-dependent and -independent 16S rRNA gene-based analyses. Appl. Environ. Microbiol. 73:1882–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dobson A, O'Sullivan O, Cotter PD, Ross P, Hill C. 2011. High-throughput sequence-based analysis of the bacterial composition of kefir and an associated kefir grain. FEMS Microbiol. Lett. 320:56–62 [DOI] [PubMed] [Google Scholar]
- 14. Duthoit F, Godon JJ, Montel MC. 2003. Bacterial community dynamics during production of registered designation of origin Saler cheese as evaluated by 16S rRNA gene single-strand conformation polymorphism analysis. Appl. Environ. Microbiol. 69:3840–3848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. El-Baradei G, Delacroix-Buchet A, Ogier JC. 2007. Biodiversity of bacterial ecosystems in traditional Egyptian Domiati cheese. Appl. Environ. Microbiol. 73:1248–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Endo A, Okada S. 2005. Monitoring the lactic acid bacterial diversity during shochu fermentation by PCR-denaturing gradient gel electrophoresis. J. Biosci. Bioeng. 99:216–221 [DOI] [PubMed] [Google Scholar]
- 17. Ercolini D, et al. 2011. Monitoring of microbial metabolites and bacterial diversity in beef stored under different packaging conditions. Appl. Environ. Microbiol. 77:7372–7381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ercolini D, Hill PJ, Dodd CER. 2003. Bacterial community structure and location in Stilton cheese. Appl. Environ. Microbiol. 69:3540–3548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. European Commission 31 March 2006. On the protection of geographical indications and designations of origin for agricultural products and food stuffs. Commission regulation EC 510/06. Off. J. Eur. Union L93:12–25 [Google Scholar]
- 20. Flórez AB, Mayo B. 2006. Microbial diversity and succession during the manufacture and ripening of traditional, Spanish, blue-veined Cabrales cheese, as determined by PCR-DGGE. Int. J. Food Microbiol. 110:165–171 [DOI] [PubMed] [Google Scholar]
- 21. Friedrich U, Lenke J. 2006. Improved enumeration of lactic acid bacteria in mesophilic starter cultures by using multiplex quantitative real-time PCR and flow cytometry-fluorescence in situ hybridization. Appl. Environ. Microbiol. 72:4163–4171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guinane CM, Cotter PD, Hill C, Ross RP. 2005. Microbial solutions to microbial problems; lactococcal bacteriocins for the control of undesirable biota in food. J. Appl. Microbiol. 98:1316–1325 [DOI] [PubMed] [Google Scholar]
- 23. Haas BJ, et al. 2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21:494–504 doi:10.1101/gr.112730.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Humblot C, Guyot JP. 2009. Pyrosequencing of tagged 16S rRNA gene amplicons for rapid deciphering of the microbiomes of fermented foods such as pearl millet slurries. Appl. Environ. Microbiol. 75:4354–4361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jany JL, Barbier G. 2008. Culture-independent methods for identifying microbial communities in cheese. Food Microbiol. 25:839–848 [DOI] [PubMed] [Google Scholar]
- 26. Jung JY, et al. 2011. Metagenomic analysis of kimchi, a traditional Korean fermented food. Appl. Environ. Microbiol. 77:2264–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Klappenbach JA, Saxman PR, Cole JR, Schmidt TM. 2001. rrndb: the Ribosomal RNA Operon Copy Number Database. Nucleic Acids Res. 29:181–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–148 In Stackebrandt E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics, 1st ed John Wiley & Sons, Chichester, United Kingdom [Google Scholar]
- 29. Lazzi C, Rossetti L, Zago M, Neviani E, Giraffa G. 2004. Evaluation of bacterial communities belonging to natural whey starters for Grana Padano cheese by length heterogeneity-PCR. J. Appl. Microbiol. 96:481–490 [DOI] [PubMed] [Google Scholar]
- 30. Lozupone CA, Hamady M, Kelley ST, Knight R. 2007. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 73:1576–1585 doi:10.1128/AEM.01996-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Madera C, García P, Janzen T, Rodríguez A, Suárez JE. 2003. Characterization of proficient wild Lactococcus lactis strains resistant to phage infection. Int. J. Food Microbiol. 86:213–222 [DOI] [PubMed] [Google Scholar]
- 32. Majcher MA, Goderska K, Pikul J, Jeleń HH. 2011. Changes in volatile, sensory and microbial profiles during preparation of smoked ewe cheese. J. Sci. Food Agric. 91:1416–1423 [DOI] [PubMed] [Google Scholar]
- 33. Majcher MA, Jeleń HH. 2011. Key odorants of Oscypek, a traditional Polish ewe's milk cheese. J. Agric. Food Chem. 59:4932–4937 [DOI] [PubMed] [Google Scholar]
- 34. Maoz A, Mayr R, Scherer S. 2003. Temporal stability and biodiversity of two complex antilisterial cheese-ripening microbial consortia. Appl. Environ. Microbiol. 69:4012–4018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Margulies M, et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Muyzer G, de Waal EC, Uitterlinden AG. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Olivier A, Lee HY, Côté JC. 2005. Study of the heterogeneity of 16S rRNA genes in gamma-proteobacteria: implications for phylogenetic analysis. J. Gen. Appl. Microbiol. 51:395–405 [DOI] [PubMed] [Google Scholar]
- 38. Palys T, Nakamura LK, Cohan FM. 1997. Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int. J. Syst. Bacteriol. 47:1145–1156 [DOI] [PubMed] [Google Scholar]
- 39. Petrosino JF, Highlander S, Luna RA, Gibbs RA, Versalovic J. 2009. Metagenomic pyrosequencing and microbial identification. Clin. Chem. 55:856–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Randazzo CL, Torriani S, Akkermans ALD, de Vos WM, Vaughan EE. 2002. Diversity, dynamics, and activity of bacterial communities during production of an artisanal Sicilian cheese as evaluated by 16S rRNA analysis. Appl. Environ. Microbiol. 68:1882–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Roh SW, et al. 2010. Investigation of archaeal and bacterial diversity in fermented seafood using barcoded pyrosequencing. ISME J. 4:1–16 [DOI] [PubMed] [Google Scholar]
- 42. Rorvik LM. 2000. Listeria monocytogenes in the smoked salmon industry. Int. J. Food Microbiol. 62:183–190 [DOI] [PubMed] [Google Scholar]
- 43. Satokari RM, et al. 2003. Molecular approaches for the detection and identification of bifidobacteria and lactobacilli in the human gastrointestinal tract. Syst. Appl. Microbiol. 26:572–584 [DOI] [PubMed] [Google Scholar]
- 44. Schloss PD, et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 doi:10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sherman F, Fink GR, Hicks JB. 1986. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 46. Smit G, Smit BA, Engels WJ. 2005. Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiol. Rev. 29:591–610 [DOI] [PubMed] [Google Scholar]
- 47. Stackebrandt E, Goebel BM. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846–849 [Google Scholar]
- 48. Sundquist A, et al. 2007. Bacterial flora typing with deep, targeted, chip-based pyrosequencing. BMC Microbiol. 7:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Torres-Machorro AL, Hernández R, Cevallos AM, López-Villaseñor I. 2010. Ribosomal RNA genes in eukaryotic microorganisms: witnesses of phylogeny? FEMS Microbiol. Rev. 34:59–86 [DOI] [PubMed] [Google Scholar]
- 50. Yue JC, Clayton MK. 2005. A similarity measure based on species proportions. Commun. Stat. Theory Methods 34:2123–2131 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.