Abstract
Pemetrexed, approved for the treatment of non-small cell lung cancer and malignant mesothelioma, has adverse effects including neutropenia, leucopenia, thrombocytopenia, anemia, fatigue and nausea. The results we report here represent the first genome-wide study aimed at identifying genetic predictors of pemetrexed response. We utilized expression quantitative trait loci (eQTLs) mapping combined with drug-induced cytotoxicity data to gain mechanistic insights into the observed genetic associations with pemetrexed susceptibility. We found that CTTN and ZMAT3 expression signature explained >30% of the pemetrexed susceptibility phenotype variation for pemetrexed in the discovery population. Replication using PCR and a semi-high-throughput, scalable assay system confirmed the initial discovery results in an independent set of samples derived from the same ancestry. Furthermore, functional validation in both germline and tumor cells demonstrates a decrease in cell survival following knockdown of CTTN or ZMAT3. In addition to our particular findings on genetic and gene expression predictors of susceptibility phenotype for pemetrexed, the work presented here will be valuable to the robust discovery and validation of genetic determinants and gene expression signatures of various chemotherapeutic susceptibilities.
INTRODUCTION
As a multitargeted antifolate agent, pemetrexed is used in the treatment of malignant mesothelioma and advanced non-small cell lung cancer alone or in combination with cisplatin (1,2). Pemetrexed hinders RNA and DNA synthesis by inhibiting three enzymes in the nucleotide biosynthetic pathway, including thymidylate synthase (TYMS), dihydrofolate reductase (DHFR) and glycinamide ribonucleotide formyltransferase (GART) (3). Commonly observed adverse reactions to pemetrexed treatment include neutropenia, leucopenia, thrombocytopenia, anemia, fatigue, nausea and vomiting (4). Vitamin B12 and folic acid administration have been included in pemetrexed-based therapies to decrease toxicity (5); but even with the addition of these nutrients, potentially life-threatening complications induced by pemetrexed have been observed, including grade 3/4 neutropenia (4), acute myocardial infarction (6) and acute renal failure (7,8).
Candidate gene studies have demonstrated that lower levels of TYMS, DHFR, GART and multidrug resistance-associated protein 4 (MRP4) gene expression are correlated with pemetrexed sensitivity in 18 different tumor cell lines explanted from patients (9). In addition, homocysteine and methylmalonic acid, indicators of folate and vitamin B12 deficiency, have been identified as predictive markers for pemetrexed-induced toxicity (10). In a separate study, elevated plasma homocysteine concentrations are reported to be associated with increased risk of hematological toxicity in pemetrexed-treated patients (11). However, genome-wide studies aimed at identifying genetic predictors of differential pemetrexed response spanning the range from severe toxicity to no toxicity as well as differentiating responders and non-responders have not been reported.
Previous studies have utilized human cell-based models for pharmacogenomic discovery and validation (12). Although these models come with their own limitations (e.g. tissue specificity), the experimental ease of using such models for the evaluation of genetic factors contributing to variation in response to highly toxic drugs makes them a suitable platform for pharmacogenomic discovery (12). The International HapMap human lymphoblastoid cell lines (LCLs) offer an unlimited resource of extensively genotyped cell lines for genotype–phenotype studies. Utilizing the LCL system, our group has developed a genome-wide approach that integrates genotypes (SNPs), gene expression and drug response phenotypes to identify genetic factors, including SNPs and CNVs, and gene expression signatures associated with chemotherapeutic susceptibility (13–16). Some of the genetic variants we identified using this in vitro approach have shown utility in predicting clinical patients' outcome (17,18). Furthermore, the cell model system has been used by multiple groups to functionally validate genetic variants from genome-wide association studies (GWASs) found to be associated with clinical outcomes (19,20). In this study, we present an integrative approach that assimilates recent progress in genome-wide association, the genomic regulation of gene expression and siRNA technology to validate important genes associated with susceptibility to pemetrexed toxicity (Fig. 1). We identified the expression signatures of actin-binding protein, cortactin (CTTN), and p53-associated protein, ZMAT3, as potential predictors for pemetrexed response. The results we report here are composed of a genome-wide study aimed at identifying genetic determinants of susceptibility to pemetrexed-induced toxicity. Aside from the potential clinical impact of our specific study results, the integrative approach we describe here will be valuable for future efforts aimed at the robust discovery and validation of pharmacogenomic findings.
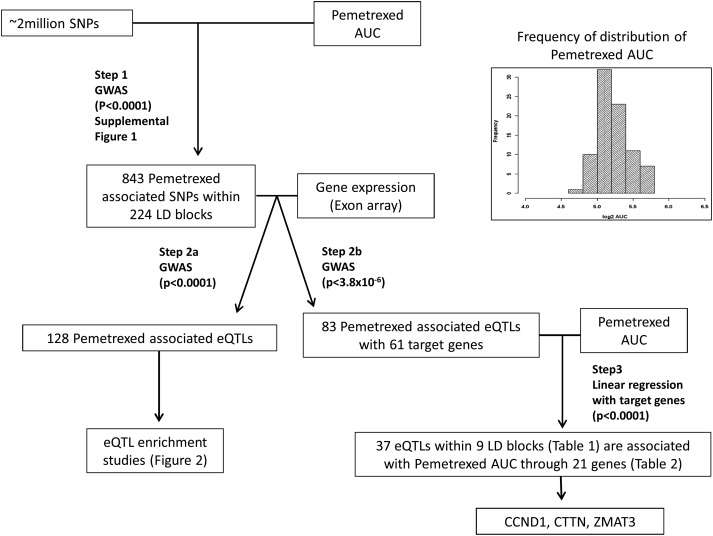
Figure 1.
Schematic diagram of our approach. Step 1: 843 SNPs (224 LD blocks) were found to be associated, at a loose threshold (P< 0.0001), with pemetrexed AUC from a GWAS analysis. Step 2: Of these 843 SNPs, 128 SNPs were identified as associated with the expression of a gene at P< 0.0001 (Step 2a), but at a more stringent threshold (P< 3.8 × 10−6), 83 eQTLs within 30 LD blocks were significant for association with 61 gene expression (Step 2b). Step 3: Of these 83 eQTLs, 37 SNPs within 9 LD blocks (see Table 1) were associated with pemetrexed AUC through association with 21 gene expression traits (see Table 2) (P< 0.0001). Based on the known function of these 21 genes and the P-value of the associations, we chose CCND1, CTTN and ZMAT3 for further replication and validation. Inset: Phenotype distribution of pemetrexed phenotype. Histogram of the distribution of AUC values for pemetrexed in the set of HapMap CEU samples.
RESULTS
Pemetrexed-induced cytotoxicity
LCLs from 84 HapMap CEU population were phenotyped for cellular sensitivity to pemetrexed using a short-term, colorimetric growth inhibition assay. The percent survival was determined at six different concentrations of pemetrexed (0.02, 0.10, 0.50, 1, 5 and 10 μm) for each cell line. Area under the curve (AUC) was then calculated from the survival curve, obtained by fitting percent survival against drug concentration, using the trapezoidal rule and was used as phenotype. The largest inter-individual variation in area under the survival curve was attributable to the first three concentrations (0.02, 0.10 and 0.50 μm); we thus used this modified AUC as phenotype. The mean (standard deviation) of the log2 AUC in the CEU samples was found to be 5.29 (±0.10 [%μm]). Figure 1 illustrates a schematic of how the data were analyzed and the inset shows the distribution of log2 AUC values in the CEU samples.
Genome-wide study of pemetrexed-induced cytotoxicity
A GWAS was performed, using quantitative trait disequilibrium test (QTDT) (21), to identify SNPs associated with cellular sensitivity to pemetrexed. SNPs with minor allele frequency >5%, no Mendelian errors and in Hardy–Weinberg equilibrium (P > 0.001) were included in the association analysis. Genomic control lambda (λGC) (22) was calculated as the median of all genome-wide observed test statistics divided by the expected median under the null hypothesis; in our study, λGC was found to be 1.02. Supplementary Material, Figure S1 is a Manhattan plot showing the results of the GWAS. We identified 843 SNPs associated with pemetrexed sensitivity at the P < 0.0001 threshold. This loose threshold was chosen to filter SNPs to carry forward into a second stage. Although this threshold clearly falls short of genome-wide significance, we used it only as an SNP selection process to identify SNPs to evaluate using increasingly more stringent statistical and increasingly more relevant biological criteria.
Using Haploview (23), we found that the top 843 pemetrexed SNP associations fell into 224 linkage disequilibrium (LD) blocks (r2< 0.8). Supplementary Material, Figure S2 illustrates the LD pattern in the region centered at the SNP found to be the most significantly associated with pemetrexed sensitivity.
Evaluation of eQTLs
In order to assess the biological relevance of the top SNP findings from our GWAS, we evaluated the effect of each of the 843 SNPs on global gene expression. Baseline gene expression using Affymetrix exon array of all CEU samples was previously published (24). We annotated the top SNPs with functional information for their relationship with baseline gene expression (see Supplementary Material, Table S1) (25). All of our top SNPs were evaluated for the likelihood of being eQTLs compared with a random set of SNPs in the genome that match the minor allele frequency distribution of the pemetrexed-associated SNPs (26). Figure 2A shows the result of an eQTL enrichment analysis we conducted on the top pemetrexed SNPs. We identified 128 SNPs associated with gene expression at P < 0.0001, which is a highly significant enrichment relative to frequency-matched SNPs for expression-associated SNPs among the top pemetrexed SNPs. See Materials and Methods for details of the enrichment analysis. We conducted additional bioinformatics analyses on these 128 expression-associated SNPs. We queried SNP Function Portal (27), which analyzes the genomic sequences containing the SNP alleles to identify potentially affected transcriptional factor-binding sites, and found that 124 of the 128 SNPs are annotated as ‘TRANSFAC Binding Site’, suggesting a potential mechanism for the transcriptional effects of these (pemetrexed-associated) SNPs.
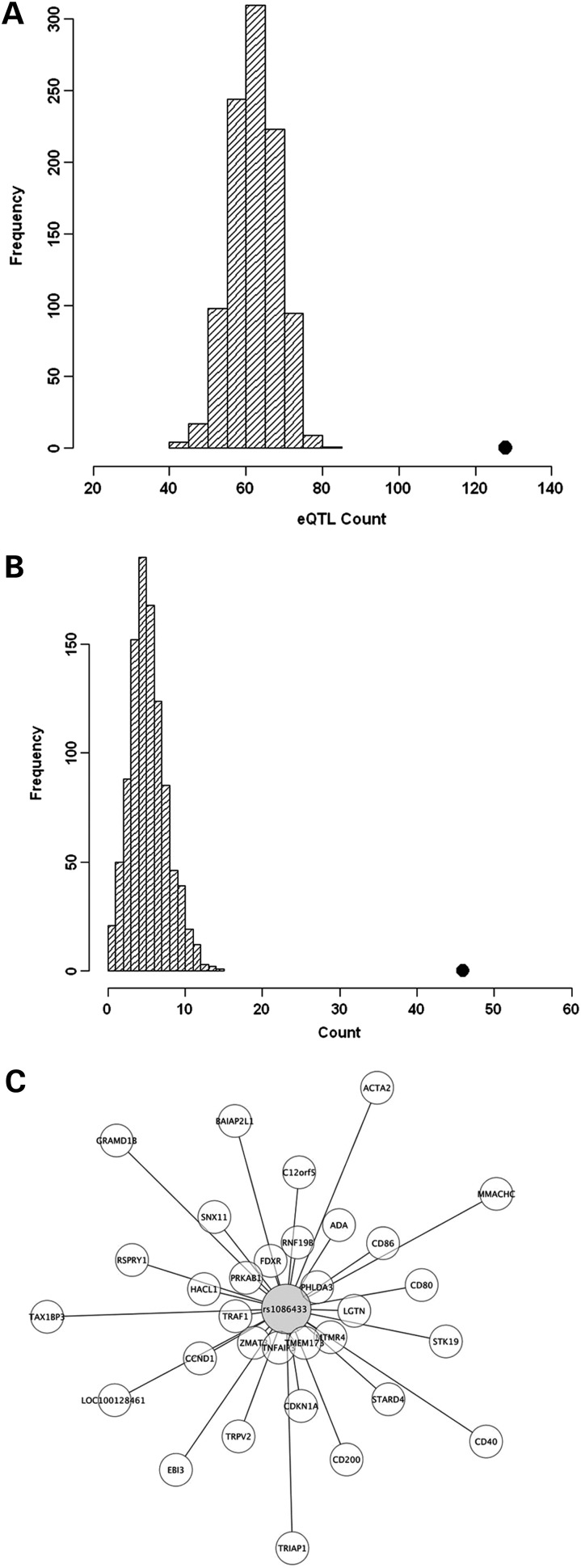
Figure 2.
eQTL enrichment analysis and master regulator enrichment analysis of all pemetrexed-associated SNPs at P< 0.0001. (A) Distribution of eQTL (expression P< 0.0001) count in 1000 simulations, each matching the MAF distribution of the chemotherapeutic susceptibility-associated SNPs (chemotherapeutic susceptibility P< 0.0001). The black dot is the observed eQTL count in the chemotherapeutic susceptibility-associated SNPs. (B) Distribution of master regulator count in 1000 simulations. A master regulator is defined to be an SNP that predicts the expression of 10 or more genes (eQTL P< 0.0001). The black dot is the observed master regulator count in the chemotherapeutic susceptibility-associated SNPs. (C) An example of a master regulator SNP (rs1086433) and 31 target genes (P< 0.0001) is shown. The distance between nodes reflects the strength of the association between the SNP genotype and gene expression levels: the closer the gene is to the SNP, the higher the correlation between the SNP genotype and gene expression.
Supplementary Material, Figure S3 shows the genomic locations of the top susceptibility loci that were observed to be eQTLs. Furthermore, we observed that the top susceptibility SNPs are more likely to be eQTLs regulating the expression of 10 or more genes than a random set of frequency-matched SNPs as illustrated in Figure 2B. These might be considered ‘master regulators’. Figure 2C illustrates an example of a master regulator, rs10864333, including 31 associated genes (P < 0.0001).
Relationship between gene expression and pemetrexed sensitivity
Target genes are defined as genes whose expression level is associated with the genotype of an SNP, the SNP referred to as an eQTL. We hypothesized that some proportion of the target genes resulting from the evaluation of the pemetrexed- and expression-associated SNPs were correlated with cellular sensitivity to pemetrexed, thereby providing us with a mechanistic hypothesis for the observed SNP associations. We conducted regression analysis between the expression of each target gene and pemetrexed AUC, as measured in the CEU cell lines. We used the Toeplitz covariance model with two diagonal bands in the regression analysis due to the relatedness of the HapMap samples (trio structure), as previously described (28). Supplementary Material, Table S2 lists the top target gene expression correlations (P < 0.0001) with pemetrexed AUC.
Identifying gene expression signatures associated with pemetrexed-induced cytotoxicity.
Combining genotypes, cytotoxicity (AUC) and the expression of target genes for eQTLs, we narrowed down the list of genes to carry forward for further functional validation using the following strategy: among the 128 expression-associated SNPs (P < 0.0001), 83 within 30 LD blocks were significant for association with gene expression (at P < 3.8 × 10−6, based on the total number of tested genes). Of these, 37 eQTLs within 9 LD blocks (Table 1) were associated with pemetrexed AUC along with the correlation of 21 target gene expressions with pemetrexed AUC (Table 2) (P < 0.0001). All 21 target genes resulted from trans associations. Among the top pemetrexed SNPs (P < 0.0001), we did identify 20 potential putative cis-acting eQTLs (at a loose threshold P< 0.01 with the expression of nearby gene within 4 Mb) for which the (cis) target gene was also correlated with pemetrexed AUC (P < 0.05); see Supplementary Material, Table S3. Based on the known function of these 21 genes and the P-value of the associations, we chose CCND1, CTTN and ZMAT3 for further replication.
Table 1.
List of tag SNPs from the 37 eQTLs within 9 LD blocks associated with pemetrexed-induced cytotoxicity
| Chromosome | Tag SNP | P_cytotoxicity | Number of SNPs in LD | Average r2 |
|---|---|---|---|---|
| CHR1 | rs4908741 | 3×10−5 | 19 | 0.9787 |
| CHR1 | rs4908739 | 2×10−6 | 9 | 0.9239 |
| CHR13 | rs1408612 | 1×10−4 | 1 | |
| CHR13 | rs9544725 | 4×10−5 | 1 | |
| CHR21 | rs2832447 | 4×10−5 | 1 | |
| CHR3 | rs10935252 | 1×10−5 | 3 | 0.9637 |
| CHR7 | rs6467410 | 1×10−4 | 1 | |
| CHR7 | rs7792457 | 1×10−4 | 1 | |
| CHR9 | rs664832 | 6×10−6 | 1 |
Table 2.
List of the 21 genes associated with pemetrexed-induced cytotoxicity (genes chosen for replication in 52CEPH are indicated by an asterisk)
| Gene name | P_cytotoxicity |
|---|---|
| PSTPIP2 | 1.12 × 10−7 |
| CCND1* | 3.68 × 10−7 |
| ZMAT3* | 5.25 × 10−7 |
| CTTN* | 1.14 × 10−6 |
| C20orf112 | 1.26 × 10−6 |
| LOC284804 | 1.26 × 10−6 |
| HSPG2 | 6.56 × 10−6 |
| FMNL3 | 9.58 × 10−6 |
| TRAF1 | 1.01 × 10−5 |
| MTMR4 | 1.67 × 10−5 |
| FBXO16 | 1.71 × 10−5 |
| ZNF395 | 1.71 × 10−5 |
| PHLDA3 | 2.38 × 10−5 |
| NRG2 | 2.91 × 10−5 |
| ADA | 2.92 × 10−5 |
| TUBA1A | 5.21 × 10−5 |
| TUBA1B | 5.21 × 10−5 |
| RNF19B | 5.33 × 10−5 |
| C20orf59 | 8.61 × 10−5 |
| LOC100129042 | 8.88 × 10−5 |
| RP1-21O18.1 | 8.88 × 10−5 |
Replication of gene expression–pemetrexed AUC associations in LCLs
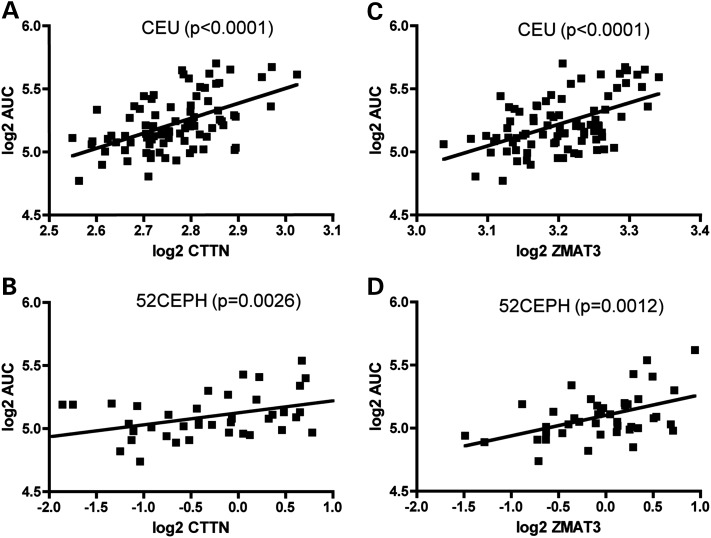
To replicate the correlation between gene expression and pemetrexed response, we quantified three gene expression levels using real-time PCR and pemetrexed cytotoxicity assay in a sample set consisting of 52 independent Caucasian LCLs from the Centre d' Etude du Polymorphisme Humain (52 CEPH). The significant correlation between ZMAT3 and pemetrexed-induced cytotoxicity (AUC) and that between CTTN and pemetrexed-induced cytotoxicity (AUC) were both successfully replicated (P = 0.0014 and P = 0.0026, respectively; Fig. 3). Consistent in both the discovery and the replication sample sets, higher CTTN or ZMAT3 expression levels were found to be associated with resistance to pemetrexed (or higher percent survival). However, CCND1 mRNA level was not associated with pemetrexed cytotoxicity (AUC) in the 52 CEPH validation cell lines (P = 0.86) (data not shown). We, therefore, focused on CTTN and ZMAT3 for additional functional validation.
Figure 3.
CTTN and ZMAT3 expression levels of LCLs were significantly associated with pemetrexed-induced cytotoxicity (AUC) in both discovery CEU population and CEPH validation population. Log2-transformed CTTN expression and log2-transformed pemetrexed AUC correlation in CEU population (A) and 52 CEPH validation set (B). Log2-transformed ZMAT3 expression and log2-transformed pemetrexed AUC correlation in CEU population (C) and 52 CEPH validation set (D).
Linear modeling to quantify the impact of the two-gene expression signature
A linear model was constructed with log2-transformed AUC as the dependent variable. Independent variables included log2-transformed CTTN and ZMAT3 expression levels. The adjusted r2 estimate was calculated for the linear model. In the CEU population, the two-gene expression signature explained 30.6% of the susceptibility phenotype variation for pemetrexed. Furthermore, our two-gene model was found to be significantly better than the null model (P = 1.4 × 10−7). The two gene expression traits were modestly correlated in the CEU samples (Pearson correlation 0.69). We constructed a similar linear model using quantitative PCR expression levels in the independent replication set of 52 CEPH samples. This analysis yielded 29.6% as the proportion of the variance in the phenotype explained by the two genes alone.
Table 3 summarizes the results of this analysis in the discovery CEU samples and in the replication CEPH samples. In particular, the two-gene expression signature from the multivariate analyses provided a more robust predictive model than the single-gene signatures from the univariate analyses; the percent of phenotypic variation explained by the two-gene model had less variability between the discovery and the replication sets.
Table 3.
Linear model analysis of CTTN and ZMAT3 in CEU and 52 CEPH
| Population | Genes | %Phenotype explained by genes |
|---|---|---|
| CEU | CTTN | 26.9 |
| CEU | ZMAT3 | 26.2 |
| CEU | CTTN + ZMAT3 | 30.6 |
| 52 CEPH | CTTN | 17.8 |
| 52 CEPH | ZMAT3 | 20.5 |
| 52 CEPH | CTTN + ZMAT3 | 29.6 |
We examined the association of CTTN and ZMAT3 (separately) with other cellular phenotypes: ZMAT3 and CTTN were associated with growth rate (P = 0.01 and P = 0.003, respectively). However, after conditioning on growth rate, ZMAT3 and CTTN were still significantly associated with pemetrexed-induced cytotoxicity (P = 9.49 × 10−6 and P = 7.54 × 10−6, respectively). We checked for the effects of EBV transformation on CTTN and ZMAT3 expression. Consistent with a recent comprehensive study (29) of the effects of cell transformation on gene regulation, the expression level for either gene was not among those found to be significantly associated with baseline EBV copy number.
Sensitization of LCLs and lung tumor cell lines to pemetrexed by reduction in CTTN and ZMAT3 expression levels
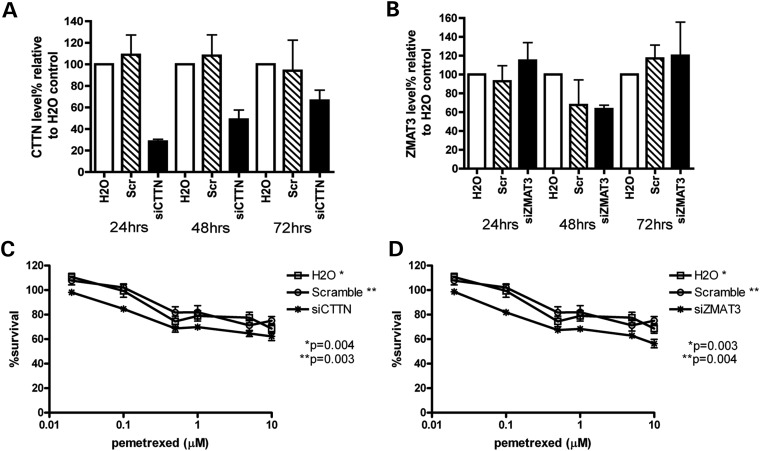
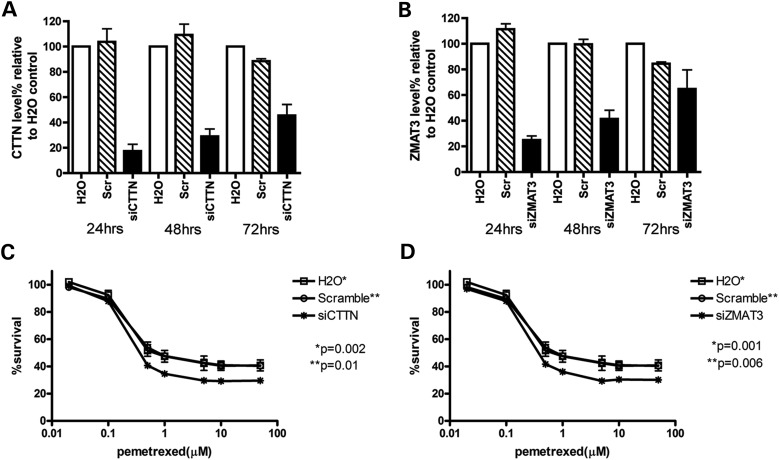
To validate the role of CTTN and ZMAT3 in cellular sensitivity to pemetrexed, we knocked down CTTN and ZMAT3 mRNA, respectively, in the LCL system using siRNA. LCL line GM12718 was randomly chosen from 52 CEPH and was transfected with H2O control, scramble siRNA control, siRNA targeting either CTTN or ZMAT3. After 18 h recovery at 37°C, transfected cells were treated in triplicates with increasing concentration of pemetrexed for 72h. Percentage of cell survival was measured using CellTiter-Glo (Promega). At 24, 48, 72h post-transfection, cells were collected for mRNA reduction measurement. Relative to H2O transfection control, mRNA levels of CTTN were reduced to 28, 48 and 66% at 24, 48 and 72h post-transfection, whereas mRNA levels of ZMAT3 were reduced to 63% at 48h. Compared with H2O or scramble control, GM12718 cells with knockdown of CTTN or ZMAT3 mRNA levels were significantly more sensitive to pemetrexed treatment (drop 5–15% in survival at different concentrations, P < 0.05) (Fig. 4). These observed knockdown effects are consistent with our correlation results in the CEU discovery and replication samples: higher CTTN or ZMAT3 level confers resistance to pemetrexed.
Figure 4.
Effect of CTTN and ZMAT3 knockdown on mRNA gene expression and LCL cellular sensitivity to pemetrexed. Following transfection with water, scramble and siRNA targeting CTTN or ZMAT3, the effect on gene expression and pemetrexed-induced cytotoxicity were determined. mRNA levels were measured 24, 48 and 72 h post-siRNA or water transfection for CTTN (A) or ZMAT3 (B). Cytotoxicity was measured 6 days after pemetrexed exposure and %cell survival is relative to no drug treatment controls following knockdown of CTTN (C) and ZMAT3 (D). Error bars indicate standard errors of six data points of two independent experiments. P-values were calculated using paired t-test comparing siCTTN or siZMAT3 %survival curve to either H2O control (*) or scramble control (**) survival curve. P-values <0.05 were considered statistically significant.
To investigate whether CTTN and ZMAT3 contribute to pemetrexed drug response in an NSCLC cell line, A549 cells were transfected with siRNA against each gene. After recovering for 24h, transfected cells were treated with increasing concentration of pemetrexed in triplicates. Relative to H2O transfection control, mRNA levels at 24, 48 and 72h post-transfection of CTTN were reduced to 17, 28 and 45%, whereas mRNA levels of ZMAT3 were reduced to 25, 41 and 65%, respectively. We detected only a modest reduction of A549 cells survival rate with reduced CTTN and ZMAT3 levels at 72h (a decrease of <5% in survival compared with control, data not shown). However, after 6 days of pemetrexed exposure, we detected a consistent decrease (∼15%, P < 0.05) across three independent experiments of cell survival in the siRNA CTTN- and siRNA ZMAT3-treated samples exposed to ≥0.5 μm pemetrexed treatment (Fig. 5). These findings in the cancer cell line are consistent with our correlation results in the CEU samples and the gene knockdown results in the LCL cells.
Figure 5.
Effect of CTTN and ZMAT3 knockdown on mRNA gene expression and A549 cellular sensitivity to pemetrexed. Following transfection with water, scramble and siRNA targeting CTTN or ZMAT3, the effect on gene expression and pemetrexed-induced cytotoxicity were determined. mRNA levels were measured 24, 48 and 72 h post-siRNA or water transfection for CTTN (A) or ZMAT3 (B). Cytotoxicity was measured 6 days after pemetrexed exposure and %cell survival is relative to no drug treatment controls following knockdown of CTTN (C) and ZMAT3 (D). Error bars for cytotoxicity and mRNA levels indicate standard errors of three independent experiments. P-values were calculated using paired t-test comparing siCTTN or siZMAT3 %survival curve to either H2O control (*) or scramble control (**) survival curve. P values <0.05 were considered statistically significant.
DISCUSSION
We performed a comprehensive investigation of about 2 million SNPs throughout the human genome for their associations with sensitivity to pemetrexed in LCLs derived from Caucasian descent. SNPs identified through this genome-wide association analysis with cellular sensitivity were evaluated for their role in the regulation of transcriptional expression. At the threshold P < 3.8 × 10−6 with gene expression, 83 pemetrexed-associated putative eQTLs were identified of which 21 target gene expression traits (37 SNPs) were correlated with pemetrexed-induced cytotoxicity (P < 0.0001). We prioritized CCND1, CTTN and ZMAT3 out of the 21 target genes for further functional validation. CTTN and ZMAT3 were successfully replicated in an independent Caucasian population for correlation between gene expression and cellular sensitivity to drug, but not CCND1. Using siRNA gene knockdown, we demonstrated that reducing CTTN and ZMAT3 gene levels sensitized LCLs and non-small cell lung cancer cells (A549) to pemetrexed treatment.
Our comprehensive genome-wide analysis led to the identification of 843 SNPs that were enriched for expression quantitative trait loci (eQTLs) compared with frequency-matched SNPs. Particularly, among the top susceptibility SNPs, we identified 128 eQTLs and 46 ‘master regulators’, defined as eQTLs associated with 10 or more gene expression traits. It has been shown that annotating SNPs with information on how a locus affects transcriptional expression has great utility in enhancing the ability of GWAS to identify SNPs likely to be reproducible (30). Furthermore, SNPs associated with chemotherapeutic drug susceptibility have been found to be significantly enriched for eQTLs (26). In the current study, we found that SNPs associated with transcript levels as eQTLs are important regulators of pemetrexed-induced response, as demonstrated by the observed enrichment for eQTLs among the top susceptibility loci. To gain insights into how gene expression signatures contribute to pemetrexed response, we built a screening strategy and verified two gene expression traits as significant predictors of response to pemetrexed treatment, namely CTTN and ZMAT3.
Actin-binding protein CTTN promotes cell migration and metastasis by facilitating actin filaments branching within cell motility structures, such as at lamellipodia and invadopodia (31). The CTTN gene maps to chromosome 11q13 region, the amplification of which is frequently observed in different cancers and is linked to poor prognosis in head and neck squamous cell carcinoma (HNSCC) patients and breast cancer patients (32–34). Elevated CTTN has been reported in seven different human malignancies (32). High expression levels of CTTN correlate with poor patient survival rate, high tumor metastasis and high tumor recurrence in HNSCC (35–37), estrogen receptor-negative breast cancers (38), gastric cancer (39) and colorectal adenocarcinoma (40). Furthermore, CTTN over-expression also increased drug resistance of HNSCC cells to geftinib, an epidermal growth factor receptor (EGFR) inhibitor (41), due to its participation in EGFR endocytosis (42,43). Our data also showed that high expression of CTTN was correlated with pemetrexed resistance. In contrast, CTTN levels were not correlated with the outcome of breast cancer patients being treated with anthracycline-based chemotherapy (44), suggesting that CTTN affects response to some chemotherapeutic agents but not others. In the case of lung cancers, two recent studies have shed some light on the possible association between CTTN and lung adenocarcinoma: in 90 cases of lung adenocarcinoma (a subtype of non-small cell lung cancer) patients, tumors with CTTN localized on the matrix contacting side of invadopodia were correlated with poor patient outcome (45). A second study showed that microRNA-182 may have reduced invasiveness and proliferation rate of lung adenocarcinoma cell line A549 by reducing the expression level of CTTN (46). Along with the results from these two studies, our results propose a new role of CTTN in NSCLC cell response to pemetrexed. Even though the exact mechanism by which CTTN influences cell response to pemetrexed is not clear, our findings indicate that the expression level of CTTN is associated with an eQTL, suggesting that patients with a given genotype may be more likely to be resistant to pemetrexed.
In addition to CTTN, we also validated the role of ZMAT3 (a zinc finger protein, also known as Wig-1 or PAG608) as a protein whose expression correlates with cellular resistance to pemetrexed. ZMAT3 has been shown to be a p53-regulated protein due to the presence of a functional p53-binding motif in the ZMAT3 promoter region and the induction of ZMAT3 in response to p53 activation (47). ZMAT3 stabilizes p53 mRNA by binding to an AU-rich element in the 3′-UTR (48), thereby taking part in a positive feedback loop with p53. However, accumulating evidence has demonstrated that ZMAT3 may have p53-independent roles. First, ZMAT3 is expressed in all cell types, including cells lacking p53 (47). Second, an oncogenic role of ZMAT3 is also suggested due to its location in a region amplified in nine different human malignancies (47). Third, tumor cell growth is inhibited through both over-expression and knockdown of ZMAT3 (49,50). Importantly, ZMAT3 has been shown to bind microRNA-like short RNAs, suggesting a potential role of ZMAT3 in microRNA function, and may influence cell response to chemotherapeutic drug through the regulation of other key microRNAs (51). The suggested p53-independent role of ZMAT3 may explain our observation that decreased expression of ZMAT3, a p53-positive effector/regulator, increases cell death in response to a chemotherapeutic drug.
Pemetrexed is frequently used in combination with cisplatin as a first-line treatment for patients with advanced non-squamous NSCLC (1). Thus, identifying genetic signatures associated with both pemetrexed and cisplatin response rather than signatures associated with pemetrexed alone has broader clinical applications. In fact, in our CEU discovery population, higher CTTN and ZMAT3 expression levels were also significantly associated with cellular resistance to cisplatin (P = 0.01 and P = 0.002 respectively, www.pacdb.org) (52), suggesting that CTTN and ZMAT3 may be predictive of patient response to the combination of pemetrexed and cisplatin.
In summary, we report here a genome-wide study whose purpose is to identify genetic predictors of pemetrexed therapeutic response. We demonstrated that eQTLs are important regulators of cellular sensitivity to pemetrexed exposure likely through their influence on the expression levels of target genes such as CTTN and ZMAT3. Identifying siRNA knockdown in both LCLs and cancer cell lines that impacts cellular sensitivity to pemetrexed implies that germline genetic variants may influence tumor response to pemetrexed. The expression levels of CTTN and of ZMAT3 are effective biomarkers with potential clinical relevance for predicting NSCLC patients' response to pemetrexed treatment.
MATERIALS AND METHODS
Cell lines and drugs
LCLs derived from 30 CEPH trios (n = 90) from Utah residents with ancestry of Northern and Western Europe (I) (HapMap CEU phase I) were used as the discovery set CEU in the association study (panel HAPMAPPT01). Six cell lines of CEU population were excluded from the study due to low viability (<85%). They included GM10855, GM10856, GM11829, GM12236, GM12716 and GM12750. An additional 52 non-HapMap CEU phase II CEPH (52CEPH) cell lines were used to validate gene expression (53). All LCL lines were purchased from the Coriell Institute for Medical Research (Camden, NJ, USA) and maintained in RPMI 1640 media (Mediatech, Herndon, VA, USA) supplemented with 15% fetal bovine serum (HyClone, Logan, UT, USA) and 1% l-glutamine (Mediatech). The LCLs were diluted to a concentration of 350 000 cells/ml three times a week and maintained at 37°C in 95% humidified atmosphere with 5% CO2.
A non-small cell lung cancer cell line A549 was purchased from ATCC (www.atcc.org). A549 cells were maintained in RPMI 1640 media (Mediatech) supplemented with 15% fetal bovine serum (HyClone) and 1% l-glutamine (Mediatech). The A549 line was diluted to 20–30% confluency (∼6 × 103/cm2) three times a week and maintained at 37°C in 95% humidified atmosphere with 5% CO2.
Pemetrexed disodium (CAS: 150399-23-8) was a gift from the Eli Lilly Corporation (Indianapolis, IN, USA). Phosphate-buffered saline (PBS), pH 7.4, was purchased from Life Technologies, Inc. (Carlsbad, CA, USA).
Cytotoxicity assay
In CEU and 52 CEPH cell lines, AlamarBlue® (Life Technologies) was used to measure cellular sensitivity to pemetrexed. LCLs in exponential growth phase with >85% viability, as determined utilizing the Vi-Cell XR viability analyzer (Beckman Coulter, Fullerton, CA, USA), were plated in triplicate at a density of 1 × 105 cells/ml in 96-well round-bottom plates (Corning, Inc., Corning, NY, USA). Twenty-four hours after plating, drug stock was prepared by dissolving pemetrexed in PBS and sonicated at room temperature for 5min. Cell lines were treated with either vehicle PBS or a concentration of pemetrexed (0, 0.02, 0.1, 0.5, 1, 5, 10 µm in PBS) for 72h. AlamarBlue® was added 24h prior to absorbance reading at wavelengths of 570 and 600 nm using the Synergy-HT multi-detection plate reader (BioTek, Winooski, VT, USA). Percent survival was quantified using manufacturers' protocol (http://www.biotek.com/products/). Final percent survival was ascertained by averaging at least six replicates from two independent experiments.
Evaluation of the effect of siRNA knockdown of CTTN and ZMAT3 in LCL
A CEU-derived LCL, GM12718, was diluted to 5 × 105 cells/ml 24h before transfection. siRNA transfections were performed using the Amaxa 96-well Nucleofector Shuttle System (Lonza, Gathersburg, MD, USA). Specifically, GM12718 cells (1.5 × 106) were collected and resuspended in 18 μl of SF plus supplemental (Lonza) and mixed with either 2 μl of H2O, 2 μm Alexa-488-labeled Qiagen All Star siRNA-negative control (Qiagen, Germantown, MD, USA) or 2 μm siRNA targeting either CTTN (Hs_CTTN_5) or ZMAT3 (Hs_WIG1_6) (Qiagen) per well in a 96-well nucleofection plate. Cell-siRNA mixes were then electroporated using electrical parameter DN-100 (Lonza, proprietary). After electroporation, cells were allowed to sit in 96-well plates for 5min and then were diluted with 85 μl of warm (37°C) RPMI 1640 media. After a 10 min rest, the cells were plated in 96-well round-bottom plates (Corning, Inc.) in triplicate at 10 000 cells/well and returned to the incubator. After 18h, the cells were treated with either vehicle PBS or a dose of pemetrexed (0.02, 0.1, 0.5, 1, 5, 10 µm in PBS) for 72h. Cytotoxicity effects after transfection were measured 72h post-pemetrexed drug treatment using CellTiter-Glo, a luminescent cell viability assay (Promega, Madison, WI, USA). At the time of cell titer measurement, plates were allowed to reach room temperature for 15 min. LCL cell suspension were first transferred into 96-well white round-bottom plates (Corning, Inc.). Cell suspension was mixed thoroughly by pipetting with equal volume of CellTiter Glo reagent (Corning, Inc.) and then incubated at room temperature for 30min. Luminescence at 100 nm was read using the Synergy-HT multi-detection plate reader (BioTek). Percentage of cell survival rates was made with the raw values of luminescence to gain a relative value comparing drug treatment with the control wells.
Evaluation of the effect of siRNA knockdown of CTTN and ZMAT3 in A549 cells
A549 cells were first diluted to 6 × 103/cm2 2 days before transfection. siRNA transfections were performed using Amaxa 96-well Nucleofector Shuttle (Lonza). Specifically, A549 cells (1.0 × 106) were collected and resuspended in 18 μl of SF plus supplemental (Lonza) and mixed with either 2 μl of H2O, 2 μm Alexa-488-labeled Qiagen All Star siRNA-negative control (Qiagen) or 2 μm siRNA targeting either CTTN (Hs_CTTN_5) or ZMAT3 (Hs_WIG1_6) (Qiagen) per well in a 96-well nucleofection plate. Cell-siRNA mixes were then electroporated using electrical parameter DN-100 (Lonza, proprietary). After electroporation, cells were allowed to sit in 96-well plates for 5min and then were diluted with 85 μl of warm (37°C) RPMI 1640 media. After a 10 min rest, the cells were plated in 96-well flat-bottom plates (E&K Scientific, Santa Clara, CA, USA) in triplicate at 1000 cells/well and returned to the incubator. After 24h, the cells were treated with either vehicle PBS or a dose of pemetrexed (0.02, 0.1, 0.5, 1, 5, 10, 50 µm in PBS) for 6 days. Cytotoxicity effects after transfection were measured 6 days post-pemetrexed drug treatment using CellTiter-Glo, a luminescent cell viability assay (Promega).
At the time of cell titer measurement, plates were allowed to reach room temperature for 15 min. Cells were mixed thoroughly by pipetting with equal volume of CellTiter Glo reagent (Corning, Inc.) and then incubated at room temperature for 30min. Luminescence at 100 nm was read using the Synergy-HT multi-detection plate reader (BioTek). Percentage of cell survival rates was made with the raw values of luminescence to gain a relative value comparing drug treatment with the control wells.
Quantitative PCR to measure CTTN and ZMAT3 mRNA expression levels after siRNA knockdown
Applied Biosystem TaqMan primer sets were used to quantify mRNA expression of CTTN, CCND1 and ZMAT3 (CTTN: Hs01124222_m1 for 52 CEPH validation set; Hs00193322_ml for siRNA-treated samples, ZMAT3: Hs00536976_ml, CCND1: Hs00765553_m1, Applied Biosystems, Foster City, CA, USA). A total of 5 × 106 cells were pelleted and washed in PBS at 4°C. Total RNA was extracted using the RNeasy Plus Mini Kit (Qiagen, Inc.). mRNA was reverse-transcribed to cDNA using Applied Biosystems High-Capacity Reverse Transcription Kit. Real-time PCR of each gene was compared relative to the housekeeping gene huB2M (NM_004048.2, Applied Biosystems, Cat no. 4326319E) as endogenous control on the Applied Biosystems 7500 real-time PCR system. The cycling conditions were 95°C for 20 s, then (95°C for 3 s, 60°C for 30 s) for 40 cycles using fast ramping. A relative standard curve method was used to obtain the relative gene expression. In each experiment, all the reactions were performed in triplicate. For gene expression in the 52 CEPH validation set, the average of two experiments performed on the sample cell pellet was used in the analysis. For gene expression in the siRNA-treated sample, the average of three independent experiments was used in the analysis.
Statistical analysis and visualization
GWAS was performed using QTDT (21). Regression analyses, including the relationship between pemetrexed AUC and gene expression levels, were performed in R, http://www.r-project.org/). To test for enrichment of eQTLs among the top pemetrexed-associated SNPs (P < 0.0001), we generated 1000 random sets of SNPs from the set of HapMap CEU SNPs, each with matching minor allele frequency distribution as the set of the most significantly associated SNPs. For each such random set, the number of eQTLs was calculated, yielding a distribution showing the expected count. The ratio of the difference between the observed count and the expected count to the standard deviation of the distribution provides a measure of the magnitude of the enrichment (i.e. an empirical P-value for the enrichment) (26). The package lm was used to fit linear models. For visualization of the relationships between a master regulator eQTL and its target genes, the open-source software Cytoscape was used.
SUPPLEMENTARY MATERIAL
FUNDING
This study is supported by NIH CA136765 and NIH Pharmacogenomics of Anticancer Agents grant U01GM61393.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs Heather Wheeler and Stephanie Huang for valuable discussions as well as Bonnie LaCroix and Kenneth Hecht from the Pharmacogenomics of Anticancer Agents Cell Core for cell culture assistance.
Conflict of Interest statement: None declared.
REFERENCES
- 1.Rossi A., Ricciardi S., Maione P., de Marinis F., Gridelli C. Pemetrexed in the treatment of advanced non-squamous lung cancer. Lung Cancer. 2009;66:141–149. doi: 10.1016/j.lungcan.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 2.Ceresoli G.L., Zucali P.A., Gianoncelli L., Lorenzi E., Santoro A. Second-line treatment for malignant pleural mesothelioma. Cancer Treat. Rev. 2010;36:24–32. doi: 10.1016/j.ctrv.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Chattopadhyay S., Moran R.G., Goldman I.D. Pemetrexed: biochemical and cellular pharmacology, mechanisms, and clinical applications. Mol. Cancer Ther. 2007;6:404–417. doi: 10.1158/1535-7163.MCT-06-0343. [DOI] [PubMed] [Google Scholar]
- 4.Fossella F.V., Gatzemeier U. Phase I trials of pemetrexed. Semin. Oncol. 2002;29:8–16. doi: 10.1053/sonc.2002.30764. [DOI] [PubMed] [Google Scholar]
- 5.Longo-Sorbello G.S., Chen B., Budak-Alpdogan T., Bertino J.R. Role of pemetrexed in non-small cell lung cancer. Cancer Invest. 2007;25:59–66. doi: 10.1080/07357900601130748. [DOI] [PubMed] [Google Scholar]
- 6.Pio-Asin M., Segrelles-Bellmunt G., Arrondo-Velasco A., Sarobe-Carricas M. Acute myocardial infarction and pemetrexed. Farm Hosp. 2009;33:114–115. doi: 10.1016/s1130-6343(09)70999-7. [DOI] [PubMed] [Google Scholar]
- 7.Brandes J.C., Grossman S.A., Ahmad H. Alteration of pemetrexed excretion in the presence of acute renal failure and effusions: presentation of a case and review of the literature. Cancer Invest. 2006;24:283–287. doi: 10.1080/07357900600629567. [DOI] [PubMed] [Google Scholar]
- 8.Vootukuru V., Liew Y.P., Nally J.V., Jr Pemetrexed-induced acute renal failure, nephrogenic diabetes insipidus, and renal tubular acidosis in a patient with non-small cell lung cancer. Med. Oncol. 2006;23:419–422. doi: 10.1385/MO:23:3:419. [DOI] [PubMed] [Google Scholar]
- 9.Hanauske A.R., Eismann U., Oberschmidt O., Pospisil H., Hoffmann S., Hanauske-Abel H., Ma D., Chen V., Paoletti P., Niyikiza C. In vitro chemosensitivity of freshly explanted tumor cells to pemetrexed is correlated with target gene expression. Invest. New Drugs. 2007;25:417–423. doi: 10.1007/s10637-007-9060-9. [DOI] [PubMed] [Google Scholar]
- 10.Niyikiza C., Baker S.D., Seitz D.E., Walling J.M., Nelson K., Rusthoven J.J., Stabler S.P., Paoletti P., Calvert A.H., Allen R.H. Homocysteine and methylmalonic acid: markers to predict and avoid toxicity from pemetrexed therapy. Mol. Cancer Ther. 2002;1:545–552. [PubMed] [Google Scholar]
- 11.Li K.M., Rivory L.P., Clarke S.J. Pemetrexed pharmacokinetics and pharmacodynamics in a phase I/II study of doublet chemotherapy with vinorelbine: implications for further optimisation of pemetrexed schedules. Br. J. Cancer. 2007;97:1071–1076. doi: 10.1038/sj.bjc.6603995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Welsh M., Mangravite L., Medina M.W., Tantisira K., Zhang W., Huang R.S., McLeod H., Dolan M.E. Pharmacogenomic discovery using cell-based models. Pharmacol. Rev. 2009;61:413–429. doi: 10.1124/pr.109.001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang R.S., Duan S., Shukla S.J., Kistner E.O., Clark T.A., Chen T.X., Schweitzer A.C., Blume J.E., Dolan M.E. Identification of genetic variants contributing to cisplatin-induced cytotoxicity by use of a genomewide approach. Am. J. Hum. Genet. 2007;81:427–437. doi: 10.1086/519850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang R.S., Duan S., Bleibel W.K., Kistner E.O., Zhang W., Clark T.A., Chen T.X., Schweitzer A.C., Blume J.E., Cox N.J., et al. A genome-wide approach to identify genetic variants that contribute to etoposide-induced cytotoxicity. Proc. Natl Acad. Sci. USA. 2007;104:9758–9763. doi: 10.1073/pnas.0703736104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartford C.M., Duan S., Delaney S.M., Mi S., Kistner E.O., Lamba J.K., Huang R.S., Dolan M.E. Population-specific genetic variants important in susceptibility to cytarabine arabinoside cytotoxicity. Blood. 2009;113:2145–2153. doi: 10.1182/blood-2008-05-154302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gamazon E.R., Nicolae D.L., Cox N.J. A study of CNVs as trait-associated polymorphisms and as expression quantitative trait loci. PLoS Genet. 2011;7:e1001292. doi: 10.1371/journal.pgen.1001292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang R.S., Johnatty S.E., Gamazon E., Im H.K., Ziliak D., Duan S., Zhang W., Kistner E.O., Chen P., Beesley J., et al. Platinum sensitivity-related germline polymorphism discovered via a cell-based approach and analysis of its association with outcome in ovarian cancer patients. Clin. Cancer Res. 2011;17:5490–5500. doi: 10.1158/1078-0432.CCR-11-0724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ziliak D., O'Donnell P.H., Im H.K., Gamazon E.R., Chen P., Delaney S., Shukla S., Das S., Cox N.J., Vokes E.E., et al. Germline polymorphisms discovered via a cell-based, genome-wide approach predict platinum response in head and neck cancers. Transl. Res. 2011;157:265–272. doi: 10.1016/j.trsl.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ingle J.N., Schaid D.J., Goss P.E., Liu M., Mushiroda T., Chapman J.A., Kubo M., Jenkins G.D., Batzler A., Shepherd L., et al. Genome-wide associations and functional genomic studies of musculoskeletal adverse events in women receiving aromatase inhibitors. J. Clin. Oncol. 2010;28:4674–4682. doi: 10.1200/JCO.2010.28.5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen S.H., Yang W., Fan Y., Stocco G., Crews K.R., Yang J.J., Paugh S.W., Pui C.H., Evans W.E., Relling M.V. A genome-wide approach identifies that the aspartate metabolism pathway contributes to asparaginase sensitivity. Leukemia. 2010;25:66–74. doi: 10.1038/leu.2010.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abecasis G.R., Cardon L.R., Cookson W.O. A general test of association for quantitative traits in nuclear families. Am. J. Hum. Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Devlin B., Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 23.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 24.Zhang W., Duan S., Kistner E.O., Bleibel W.K., Huang R.S., Clark T.A., Chen T.X., Schweitzer A.C., Blume J.E., Cox N.J., et al. Evaluation of genetic variation contributing to differences in gene expression between populations. Am. J. Hum. Genet. 2008;82:631–640. doi: 10.1016/j.ajhg.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gamazon E.R., Zhang W., Konkashbaev A., Duan S., Kistner E.O., Nicolae D.L., Dolan M.E., Cox N.J. SCAN: SNP and copy number annotation. Bioinformatics. 2010;26:259–262. doi: 10.1093/bioinformatics/btp644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gamazon E.R., Huang R.S., Cox N.J., Dolan M.E. Chemotherapeutic drug susceptibility associated SNPs are enriched in expression quantitative trait loci. Proc. Natl Acad. Sci. USA. 2010;107:9287–9292. doi: 10.1073/pnas.1001827107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang P., Dai M., Xuan W., McEachin R.C., Jackson A.U., Scott L.J., Athey B., Watson S.J., Meng F. SNP Function Portal: a web database for exploring the function implication of SNP alleles. Bioinformatics. 2006;22:e523–e529. doi: 10.1093/bioinformatics/btl241. [DOI] [PubMed] [Google Scholar]
- 28.Huang R.S., Kistner E.O., Bleibel W.K., Shukla S.J., Dolan M.E. Effect of population and gender on chemotherapeutic agent-induced cytotoxicity. Mol. Cancer Ther. 2007;6:31–36. doi: 10.1158/1535-7163.MCT-06-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caliskan M., Cusanovich D.A., Ober C., Gilad Y. The effects of EBV transformation on gene expression levels and methylation profiles. Hum. Mol. Genet. 2011;20:1643–1652. doi: 10.1093/hmg/ddr041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicolae D.L., Gamazon E., Zhang W., Duan S., Dolan M.E., Cox N.J. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buday L., Downward J. Roles of cortactin in tumor pathogenesis. Biochim. Biophys. Acta. 2007;1775:263–273. doi: 10.1016/j.bbcan.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 32.Weaver A.M. Cortactin in tumor invasiveness. Cancer Lett. 2008;265:157–166. doi: 10.1016/j.canlet.2008.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ormandy C.J., Musgrove E.A., Hui R., Daly R.J., Sutherland R.L. Cyclin D1, EMS1 and 11q13 amplification in breast cancer. Breast Cancer Res. Treat. 2003;78:323–335. doi: 10.1023/a:1023033708204. [DOI] [PubMed] [Google Scholar]
- 34.Schuuring E. The involvement of the chromosome 11q13 region in human malignancies: cyclin D1 and EMS1 are two new candidate oncogenes—a review. Gene. 1995;159:83–96. doi: 10.1016/0378-1119(94)00562-7. [DOI] [PubMed] [Google Scholar]
- 35.Hofman P., Butori C., Havet K., Hofman V., Selva E., Guevara N., Santini J., Van Obberghen-Schilling E. Prognostic significance of cortactin levels in head and neck squamous cell carcinoma: comparison with epidermal growth factor receptor status. Br. J. Cancer. 2008;98:956–964. doi: 10.1038/sj.bjc.6604245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gibcus J.H., Mastik M.F., Menkema L., de Bock G.H., Kluin P.M., Schuuring E., van der Wal J.E. Cortactin expression predicts poor survival in laryngeal carcinoma. Br. J. Cancer. 2008;98:950–955. doi: 10.1038/sj.bjc.6604246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ambrosio E.P., Rosa F.E., Domingues M.A., Villacis R.A., Coudry R.D., Tagliarini J.V., Soares F.A., Kowalski L.P., Rogatto S.R. Cortactin is associated with perineural invasion in the deep invasive front area of laryngeal carcinomas. Hum. Pathol. 2011;42:1221–1229. doi: 10.1016/j.humpath.2010.05.030. [DOI] [PubMed] [Google Scholar]
- 38.Hui A., Kulkarni G.V., Hunter W.L., McCulloch C.A., Cruz T.F. Paclitaxel selectively induces mitotic arrest and apoptosis in proliferating bovine synoviocytes. Arthritis Rheum. 1997;40:1073–1084. doi: 10.1002/art.1780400612. [DOI] [PubMed] [Google Scholar]
- 39.Wang X., Cao W., Mo M., Wang W., Wu H., Wang J. VEGF and cortactin expression are independent predictors of tumor recurrence following curative resection of gastric cancer. J. Surg. Oncol. 2010;102:325–330. doi: 10.1002/jso.21644. [DOI] [PubMed] [Google Scholar]
- 40.Cai J.H., Zhao R., Zhu J.W., Jin X.L., Wan F.J., Liu K., Ji X.P., Zhu Y.B., Zhu Z.G. Expression of cortactin correlates with a poor prognosis in patients with stages II-III colorectal adenocarcinoma. J. Gastrointest. Surg. 2010;14:1248–1257. doi: 10.1007/s11605-010-1247-2. [DOI] [PubMed] [Google Scholar]
- 41.Timpson P., Wilson A.S., Lehrbach G.M., Sutherland R.L., Musgrove E.A., Daly R.J. Aberrant expression of cortactin in head and neck squamous cell carcinoma cells is associated with enhanced cell proliferation and resistance to the epidermal growth factor receptor inhibitor gefitinib. Cancer Res. 2007;67:9304–9314. doi: 10.1158/0008-5472.CAN-07-0798. [DOI] [PubMed] [Google Scholar]
- 42.Timpson P., Lynch D.K., Schramek D., Walker F., Daly R.J. Cortactin overexpression inhibits ligand-induced down-regulation of the epidermal growth factor receptor. Cancer Res. 2005;65:3273–3280. doi: 10.1158/0008-5472.CAN-04-2118. [DOI] [PubMed] [Google Scholar]
- 43.Lynch D.K., Winata S.C., Lyons R.J., Hughes W.E., Lehrbach G.M., Wasinger V., Corthals G., Cordwell S., Daly R.J. A Cortactin-CD2-associated protein (CD2AP) complex provides a novel link between epidermal growth factor receptor endocytosis and the actin cytoskeleton. J. Biol. Chem. 2003;278:21805–21813. doi: 10.1074/jbc.M211407200. [DOI] [PubMed] [Google Scholar]
- 44.Dedes K.J., Lopez-Garcia M.A., Geyer F.C., Lambros M.B., Savage K., Vatcheva R., Wilkerson P., Wetterskog D., Lacroix-Triki M., Natrajan R., et al. Cortactin gene amplification and expression in breast cancer: a chromogenic in situ hybridisation and immunohistochemical study. Breast Cancer Res. Treat. 2010;124:653–666. doi: 10.1007/s10549-010-0816-0. [DOI] [PubMed] [Google Scholar]
- 45.Hirooka S., Akashi T., Ando N., Suzuki Y., Ishida N., Kurata M., Takizawa T., Kayamori K., Sakamoto K., Fujiwara N., et al. Localization of the invadopodia-related proteins actinin-1 and cortactin to matrix-contact-side cytoplasm of cancer cells in surgically resected lung adenocarcinomas. Pathobiology. 2011;78:10–23. doi: 10.1159/000322734. [DOI] [PubMed] [Google Scholar]
- 46.Zhang L., Liu T., Huang Y., Liu J. microRNA-182 inhibits the proliferation and invasion of human lung adenocarcinoma cells through its effect on human cortical actin-associated protein. Int. J. Mol. Med. 2011;28:381–388. doi: 10.3892/ijmm.2011.679. [DOI] [PubMed] [Google Scholar]
- 47.Vilborg A., Bersani C., Wilhelm M.T., Wiman K.G. The p53 target Wig-1: a regulator of mRNA stability and stem cell fate? Cell Death Differ. 2011;18:1434–1440. doi: 10.1038/cdd.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vilborg A., Glahder J.A., Wilhelm M.T., Bersani C., Corcoran M., Mahmoudi S., Rosenstierne M., Grander D., Farnebo M., Norrild B., et al. The p53 target Wig-1 regulates p53 mRNA stability through an AU-rich element. Proc. Natl Acad. Sci. USA. 2009;106:15756–15761. doi: 10.1073/pnas.0900862106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prahl M., Vilborg A., Palmberg C., Jornvall H., Asker C., Wiman K.G. The p53 target protein Wig-1 binds hnRNP A2/B1 and RNA Helicase A via RNA. FEBS Lett. 2008;582:2173–2177. doi: 10.1016/j.febslet.2008.04.065. [DOI] [PubMed] [Google Scholar]
- 50.Hellborg F., Qian W., Mendez-Vidal C., Asker C., Kost-Alimova M., Wilhelm M., Imreh S., Wiman K.G. Human wig-1, a p53 target gene that encodes a growth inhibitory zinc finger protein. Oncogene. 2001;20:5466–5474. doi: 10.1038/sj.onc.1204722. [DOI] [PubMed] [Google Scholar]
- 51.Mendez Vidal C., Prahl M., Wiman K.G. The p53-induced Wig-1 protein binds double-stranded RNAs with structural characteristics of siRNAs and miRNAs. FEBS Lett. 2006;580:4401–4408. doi: 10.1016/j.febslet.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 52.Gamazon E.R., Duan S., Zhang W., Huang R.S., Kistner E.O., Dolan M.E., Cox N.J. PACdb: a database for cell-based pharmacogenomics. Pharmacogenet. Genomics. 2010;20:269–273. doi: 10.1097/FPC.0b013e328337b8d6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang R.S., Duan S., Kistner E.O., Bleibel W.K., Delaney S.M., Fackenthal D.L., Das S., Dolan M.E. Genetic variants contributing to daunorubicin-induced cytotoxicity. Cancer Res. 2008;68:3161–3168. doi: 10.1158/0008-5472.CAN-07-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.