Abstract
Clinical observations and epidemiologic studies suggest that the incidence of head and neck squamous cell carcinoma (HNSCC) correlates with dental hygiene, implying a role for bacteria-induced inflammation in its pathogenesis. Here we begin to explore the pilot hypothesis that specific microbial populations may contribute to HNSCC pathogenesis via epigenetic modifications in inflammatory- and HNSCC-associated genes. Microbiomic profiling by 16S rRNA sequencing of matched tumor and adjacent normal tissue specimens in 42 individuals with HNSCC demonstrate a significant association of specific bacterial subpopulations with HNSCC over normal tissue (P < 0.01). Furthermore, microbial populations can separate tumors by tobacco status (P < 0.008), but not by alcohol status (P = 0.41). If our subhypothesis regarding a mechanistic link from microorganism to carcinogenesis via inflammation and consequent aberrant DNA methylation is correct, then we should see hypermethylation of relevant genes associate with specific microbiomic profiles. Methylation analysis in four genes (MDR1, IL8, RARB, TGFBR2) previously linked to HNSCC or inflammation shows significantly increased methylation in tumor samples compared with normal oral mucosa. Of these, MDR1 promoter methylation associates with specific microbiomic profiles in tumor over normal mucosa. Additionally, we report that MDR1 methylation correlates with regional nodal metastases in the context of two specific bacterial subpopulations, Enterobacteriaceae and Tenericutes (P < 0.001 for each). These associations may lead to a different, and potentially more comprehensive, perspective on the pathogenesis of HNSCC, and support further exploration of mechanistic linkage and, if so, novel therapeutic strategies such as demethylating agents and probiotic adjuncts, particularly for patients with advanced or refractory disease.
INTRODUCTION
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common cancer worldwide, accounting for more than 500 000 new cases each year. This malignancy affects the squamous cells lining the oral cavity, hypopharynx, pharynx and larynx. Although the main cause for this cancer stems from exposure to tobacco and alcohol, infection with high-risk human papillomavirus (HPV) subtypes and Epstein-Barr virus, as well as oral hygiene, are important etiologic factors in subsets of the cases (1). Patients often present with metastatic nodal disease, and 25% of cases will develop second malignancies 5 years post-diagnosis. Unfortunately, because patients do not manifest the disease until advanced stages, clinical outcome is poor, with <50% surviving 5 years. Despite growing research efforts in this field and increasing knowledge of the impact of HPV, there has been non-exponential improvement in the treatment and no improvement in early diagnosis for this disease.
The microbiome in the gastrointestinal tract is a complex and dynamic system interacting with the host organism (2). The host organism benefits from this symbiotic relationship by harnessing the microbiome's trophic services in its energy metabolism (3). The native microbiome of mammals can also play a role in the metabolism of drugs and foods, altering their availability and use for the host (4). Regardless of the function and benefit of the microbiome to its host, some bacterial populations can be the source of pathogenic infections or carcinogenesis (5). Helicobacter pylori (H. pylori) in the stomach is a well-known example, which can cause gastritis, gastric ulcers and, in some cases, gastric cancer (6).
When H. pylori was linked with gastric cancer and its eradication was associated with regression, a paradigm-shifting model of carcinogenesis was created. The overreaction of host cells to pathogenic intestinal bacteria can cause chronic inflammation and the release of stimulatory and mutagenic cytokines, which results in altered gene expression. Furthermore, chronic inflammation has the potential to induce several mediators, such as nuclear factor-kappaB, that can lend to carcinogenesis through enhanced cell growth advantage and apoptotic resistance (7). There is substantial evidence for a significant association between chronic inflammation and malignant transformation (8). For example, inflammatory responses can cause both somatic genetic and epigenetic alterations, which ultimately disrupts the cellular homeostasis necessary in preventing malignant transformation (7,9).
The inflammation associated with bacteria has also been demonstrated to lead to the upregulation of an enzyme that causes DNA mutations in the genome (10). These types of inflammation-induced DNA damage can cause aberrant de novo DNA methylation during carcinogenesis (11). Because the hypermethylation events discussed in this context are observed in response to chronic inflammation, it is feasible that they could serve as early cancer indicators and would therefore act as effective tumor markers for diagnostic screening (11). Decreased expression of multidrug resistance gene 1 (MDR1) and bacteria-triggered inflammation has both been linked to inflammatory bowel disease, ulcerative colitis and gastric cancer (12,13). Interestingly, MDR1 has been implicated as potentially relevant in the context of tobacco-related diseases (such as HNSCC). This gene is highly expressed in bronchial epithelial cells in order to handle toxic substances crossing the cellular membrane (14). Therefore, silencing of MDR1 results in chronic obstructive pulmonary disease from decreased detoxification of cigarette smoke and/or other toxic inhalants (15).
To explore the relation between inflammation, somatic promoter methylation and oral microbiota, we chose in this paper to focus on MDR1 and three other genes related to HNSCC or oral inflammation: IL8, RARB and TGFBRII. IL8 codes for an inflammatory cytokine with differential methylation in epithelial oral cells of individuals with and without chronic periodontitis (16). In multiple studies, RARB shows aberrant promoter methylation in HNSCC (17). Downregulation of TGFBR2 and the loss of the chromosome on which it resides occur frequently in HNSCC tumors (1,16,18,19).
In this study, we aim to uncover a potential association between the oral microbiome and HNSCC pathogenesis. While the microbiomes of normal human oral mucosa, skin and gut are being explored (20), the relationships between microbiota and cancer, host-gene effects, clinical factors and environmental exposures have not been previously investigated. Elucidation of HNSCC-specific microbial subpopulations could greatly improve our understanding of the genetic alterations that are manifestations of the bacterial populations. Identified populations could also serve as effective targets for early detection or prediction and would revolutionize HNSCC diagnosis and treatment. Thus, we sought to determine whether specific microbiomic profiles are associated with HNSCC in the context of promoter hypermethylation of genes that were previously implicated in HNSCC and/or inflammation.
RESULTS
Microbiomic profiles and DNA methylation between HNSCC and matched normal oral mucosa
We analyzed bacteria-specific 16S rRNA genes to examine microbiomic profiles of 42 paired normal-tumor specimens from HNSCC patients in the context of methylation of MDR1 and three other genes that are known to be associated with inflammation (IL8) or with HNSCC (RARB, TGFBII), assessed by combined bisulfite restriction analysis (COBRA).
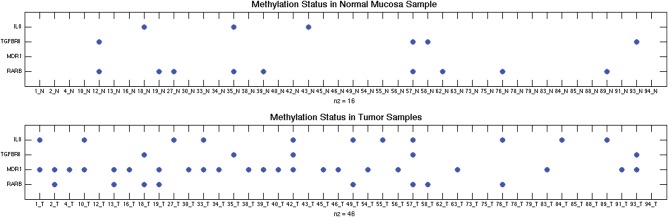
Overall, there was more promoter methylation in all four genes in the tumors compared with the matched normal tissue (P < 0.01) (Fig. 1). MDR1 shows the greatest difference between normal and tumor samples, with 0/42 methylation events in normal mucosa in comparison to 22/42 methylation events in tumor mucosa (P < 10−9).
Figure 1.
Scatter plot of methylation status of IL8, TGFBR2, MDR1 and RARB gene promoters. Each dot represents the presence of promoter hypermethylation of that gene in that particular tumor sample compared with its corresponding normal oral mucosal tissue. There are 42 paired tumor and normal mucosa samples analyzed for these four genes.
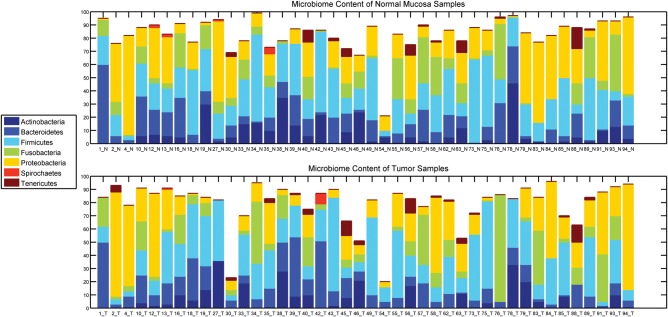
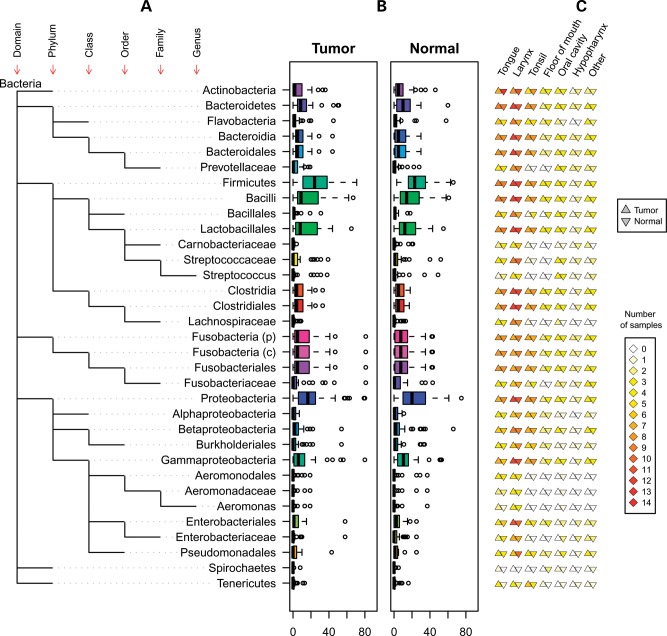
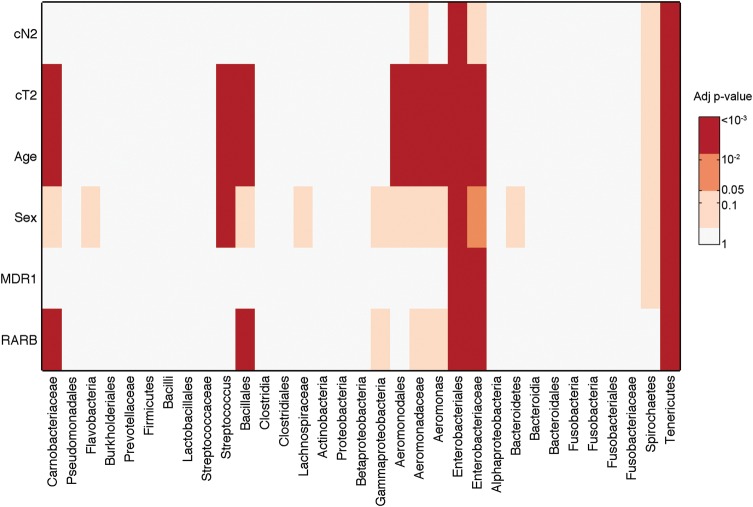
Microbiomic profiling revealed seven different phyla represented in the oropharyngeal tissues (Fig. 2). The relative quantities of the bacterial subpopulations, most striking at the phyla level, clearly differ between the HNSCC tumors and paired normal mucosae. Comparison of bacterial subpopulations in paired normal-tumor samples in the same individual reveals differences between members of each pair, even by visual inspection. Taxonomic hierarchy of the bacterial subpopulations, from phylum to genus, observed in the specimens is depicted in Figure 3A. When viewed at the group level (all tumors versus all matched normal), variations in distribution that distinguish between tumor and normal can also be observed (Fig. 3B). These distinctions remain significant despite differences in distribution of microbial subpopulations among oropharyngeal sites (Fig. 3C). Specifically, Enterobacteriaceae (family) and Tenericutes (phylum) were formally identified by multivariate analysis of variance (MANOVA) as significantly different (P < 0.001 for each) between tumor and adjacent tissue (Fig. 4).
Figure 2.
Microbial subpopulational content at the phylum level of HNSCC and matched normal mucosa. Each bar represents a sample, and the bars are split showing the microbial content quantitatively of that sample at the phylum level. There are 42 matched HNSCC (top) and normal oral mucosal (bottom) samples where a total of 7 microbial phyla identified.
Figure 3.
Microbiomic content of the paired normal and tumor samples. (A) Biological classifications of the bacteria are shown. The baseline is the kingdom bacteria, and each level represents the next taxonomic rank (phylum, class, order, family, genus). (B) rRNA-based count of each microbial population in normal and tumor samples are represented by boxplots. Separate circles represent the outliers. (C) Site-specific microbial classifications are shown. Distribution of bacteria in HNSCC sites is indicated by columns of triangles to the right of the taxonomy tree, where the left triangle represents tumor samples and the right represents the corresponding normal mucosa samples.
Figure 4.
MANCOVA of bacterial subpopulations, DNA methylation status and clinical and exposure information in HNSCC and matched normal tissue. The heat map depicts –log(P-values), i.e. orange and darker are significant at least at the P < 0.05 level. The bacterial populations detected are listed on the x-axis, and the models that are considered in the analysis are listed on the y-axis. Models with variables MDR1 methylation, RARB methylation, sex, age (dichotomized at cut-off 62.5 years), clinical tumor stage (cT, dichotomized at stages 2 or more) and clinical nodal involvement (cN, dichotomized at 0 versus 1 or more). The models that were not significant (e.g. IL8) are not shown.
By factor analysis, a statistical method used to describe variability among observed variables (Supplementary Material, Methods), we found that HPV status was not associated with specific bacterial subpopulations in differentiating HNSCC from normal tissue nor was it associated with MDR1 methylation status (Supplementary Material, Fig. S1).
Significant association among specific microbial subpopulations, MDR1 methylation and regional nodal status in HNSCC tumors
Multivariate analysis of covariance (MANCOVA) was applied to clinically relevant variables and to methylation status of the four genes of interest (Supplementary Material, Table S1) to determine whether any of these measures differentiate normal oral mucosal epithelium from HNSCC in the context of specific microbiomic profiles (Supplementary Material, Tables S2 and S3). This test performs general linear modeling for multiple responses that are also likely covariates. MANCOVA verified significant differences with regard to all variables (P < 0.01). We proceeded to see which of the variables have significantly different means across the groups. First, we found that hypermethylation of the promoters of MDR1 (P < 10−9) and IL8 (P < 0.019) was significant discriminators of tumor compared with normal specimens, irrespective of microbiomic profiles. Secondly, this multivariate analysis clearly shows that hypermethylation of MDR1 is associated with specific microbial subflora, and these two factors together are also able to differentiate HNSCC from normal oral mucosa (Fig. 4). Specifically, MDR1 methylation in the context of HNSCC (versus normal oral mucosa) was associated with Enterobacteriaceae (family) and Tenericutes (phylum). In contrast, IL8 methylation in HNSCC is not correlated with specific microbial populations (Fig. 4).
General linear modeling with multiple responses also showed that MDR1 methylation has a similar association with clinical nodal stage (cN) in the context of microbial subpopulations in differentiating HNSCC versus matched normal oral epithelium (Supplementary Material, Fig. S1). Indeed, MDR1 methylation associated with advanced regional nodal metastases (any cN >1) in the context of two specific bacterial subpopulations, Enterobacteriaceae and Tenericutes (Fig. 4). Moreover, increased age-at-diagnosis, larger tumor size and RARB promoter hypermethylation were all associated with similar patterns in bacterial subpopulations (Fig. 4), although RARB methylation alone does not differentiate normal from tumor.
Association of specific microbiome with tobacco but not alcohol exposure in HNSCC
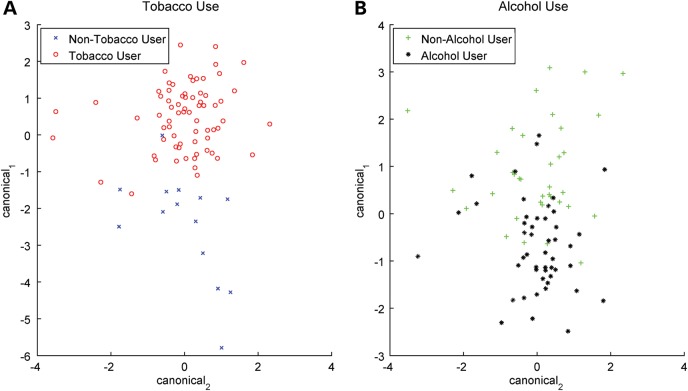
MANOVA testing, which can determine whether the mean of a variable differs significantly among canonical variables—in this case, microbial populations—was used to investigate tobacco and alcohol exposure in relationship to microbiomic profiles. Figure 5 depicts the first two canonical variables from this analysis, which are linear combinations of the original variables, maximizing the separation between bacterial subpopulations. Bacterial subpopulations significantly differentiated tobacco users from non-users (P < 0.008) (Fig. 5A). No correlation to microbial profiles was found in alcohol users (P = 0.41) (Fig. 5B). An analysis of patients who both smoke and drink alcohol compared with the rest of the cohort did not return any significant results (P = 0.17).
Figure 5.
MANOVA correlates tobacco use with microbiome but not alcohol use. The plots depict MANOVA results. The two axes are the first two canonical variables from the analysis where these variables are calculated based on linear combinations of the mean-centered original values of the microbiome in the samples analyzed. (A) The scatter plot of tobacco use canonical variables shows more separation between groups (P < 0.0036), whereas (B) alcohol use scatter plot shows two clouds that have a larger overlap (P = 0.41).
Among tobacco users compared with non-users, decreases in Flavobacteria (class) and in Tenericutes (phylum) were statistically significant. Tenericutes populations were significantly less frequent in smokers compared with non-smokers, whereas Flavobacteria and its phylum Bacteroidetes were slightly less frequent in smokers over non-smokers.
DISCUSSION
Despite research efforts in HNSCC, there has been little improvement in treatment efficacy or effective early diagnosis of this disease, and a paradigmatic shift may be required to make progress. When H. pylori in the stomach was linked with gastric cancer, a new model of carcinogenesis and a novel way to eradicate this cancer were introduced. With the knowledge of the clinical epidemiologic association between oral hygiene and HNSCC and that chronic inflammation associated with persistent presence of abnormal bacteria can induce DNA damage through aberrant de novo DNA methylation (11), we set out to determine specific microbiomic profiles which associate with promoter hypermethylation of genes relevant to HNSCC and/or to inflammation and clinico-pathologic features. For our methylation analysis, we had the option of analyzing a few well-rationalized candidate genes or performing whole-genome methylation profiling. We purposefully chose the former, based on with well-established links to inflammation and/or HNSCC pathogenesis. Methylome analysis would have almost certainly resulted in multiple testing-based errors given the number of genes interrogated, the number of microbial subpopulations assessed, and our sample sizes. Thus, we carefully selected four genes: MDR1, whose promoter methylation has been correlated with Helicobacter pylori infection (a precedent microbe) and gastric carcinogenesis (21); IL8, a well-characterized inflammatory cytokine (16); RARB, which has frequent promoter hypermethylation in HNSCC (17); and TGFBR2, which is well established in HNSCC, GI carcinogenesis and inflammation (16,18,19).
Evidence of a link between microbiota and HNSCC
In this study, we have shown that certain microbial subpopulations in HNSCC are strongly associated with MDR1 promoter methylation, thus setting the stage for future investigation for a potential mechanistic link to inflammation and tumorigenesis. First, we have shown that there are subtle regional differences in microbiomic composition (Fig. 3). Despite this, we have also shown microbiomic profiles, particularly regarding Enterobacteriaceae (family) and Tenericutes (phylum), differentiate HNSCC tumors from their paired normal counterparts (Figs 2 and 3). Secondly, we saw strong associations only between specific microbiomic profiles and MDR1 methylation, but no correlations were present for the other three genes tested. The presence of MDR1 methylation can differentiate between HNSCC and matched normal oral mucosa. Furthermore, MDR1 promoter methylation was not observed in any of our normal samples. Our observations hint that specific microbe-associated inflammation could lead to MDR1 methylation, a causation chain that has been observed in other diseases such as inflammatory bowel disease and has been associated with more aggressive disease states (21,22). In support of these findings, we see the same microbial subpopulations that associate with MDR1 promoter methylation linked with worse nodal metastases.
While a small subset of samples had methylation in normal samples and not in the tumor samples for the three genes other than MDR1, these results might arise from preferential loss of heterozygosity (LOH) events that favor the methylated allele in these patient samples. Such a phenomenon was noted while studying somatic methylation and LOH of the 3p genes, including RASSF1A (Eng and Plass, unpublished data).
Despite their apparent use as early detection tools, hypermethylation events linked with bacteria-induced inflammation have not been extensively studied for their clinical application. Because development of early detection tools is drastically needed to improve HNSCC treatment outcome, elucidation of specific microbial consortia that could serve as effective targets for early detection or prediction would potentially revolutionize HNSCC diagnosis and treatment.
Microbiota-associated methylation is independent of HPV status
HPV infection is a strong risk factor for HNSCC (23) regardless of other factors, such as tobacco or alcohol use. Our findings suggest that specific subtypes of microbiota associate with the methylation of specific genes as well as to HNSCC, independent of HPV status. While HPV+ HNSCC appears to favor certain sites, e.g. tonsils, we have controlled for this by repeating our analysis with site-matched HPV+ and HPV− patient samples (data not shown). Using this type of site-matched analysis, HPV status remains not statistically significant in correlating with either specific microbiomes or methylation of the specific genes examined. In partial support of the latter, we previously studied tonsillar carcinomas and showed that a specific set of genes known to be somatically hypermethylated in HNSCC associated directly with alcohol and tobacco exposure, but not with HPV status (24). Intriguingly, the independence of our findings from HPV status reinforces the commonly held notion that HPV+ HNSCC has a different etiologic mechanism and biologic behavior than tobacco- and alcohol-related HNSCC and suggests that methylation of inflammatory genes is not necessary for HPV+ carcinogenesis.
Concurrent analysis of methylation status of specific genes add great value to this analysis in linking and identifying a likely candidate for a carcinogenic mechanism, namely that aberrant methylation could possibly be triggered through inflammation caused by microbial populations at the at-risk oral site. This association could lead to better understanding of the pathogenesis of HNSCC, suggesting value in future direct cause-effect mechanistic studies, and if so proven, a role for novel treatment strategies in HNSCC such as demethylating agents and probiotic adjuncts (25), in particular for HPV-negative disease which continues to carry a poor outcome.
Interaction of key risk factors of HNSCC with microbiota
We have also identified that specific microbes are observed in the context of tobacco use but not with alcohol exposure in HNSCC. Tenericutes (phylum) populations were significantly lower in smokers and Flavobacteria class and its overarching phylum Bacteroidetes were slightly lower in smokers over non-smokers. The decrement in these specific subpopulations may be due to morphological differences of these bacteria (e.g. lack of a rigid cell wall). Our correlative observations, albeit preliminary, could suggest that the carcinogenic pathways of two established HNSCC risk factors, tobacco and alcohol, may differ by modulating the oral microbiome. While tobacco exposure might favor a bacterial subpopulation-associated pathway leading perhaps to inflammation, alcohol exposure may be agnostic to bacterial subpopulations or induce carcinogenesis independent of specific microbial subpopulations. We have presented smoking and alcohol exposure variables in a straightforward manner in our analysis. Future studies including more complex features of these key risk factors would give more insight about their relation to HNSCC.
In conclusion, we have shown that epigenetic modifications in HNSCC-associated genes and clinico-pathologic features of patients associate with certain bacteria as manifested by specific microbiomic sub-profiles. We suspect that our microbiome-association with HNSCC can be generalizable to other cancers, especially those at sites where normal microbial flora resides, such as those of the colon and skin. We also suspect that oral and other microbiomes may also influence risk of diseases other than neoplasia (e.g. heart disease), where an epidemiologic link with dental hygiene is known to exist. As our knowledge of host–microbiome relationship increase, we will be able to better understand the relative role of the microbiome to health status and interventional outcomes.
MATERIALS AND METHODS
Study population
HNSCC frozen tissue and paraffin-embedded samples of paired tumor and normal oral mucosa and demographic and clinical data were collected from 42 prospectively accrued consecutive HNSCC patients (Table 1). Clinical and demographic information such as smoking and alcohol exposure status, clinical tumor stage such as tumor size (cT) and regional nodal involvement (cN), tumor site, HPV status (HPV16 and 18 status, see below for details), age, sex and metastatic status were also collected from these specimen donors (see Supplementary Material, Tables S1–S3 for full tables). The research protocol has been approved by the Cleveland Clinic Human Subjects' Protection Committee.
Table 1.
Clinical and histopathological characteristics of the HNSCC patients analyzed
| Number of patients | 42 |
| Median age (years; range) | 62 (32–85) |
| Gender, n (%) | |
| Male | 26 (62) |
| Female | 16 (38) |
| Histological differentiation, n (%) | |
| Well | 4 (10) |
| Well-mod | 2 (5) |
| Mod | 18 (43) |
| Mod-poor | 11 (26) |
| Poor | 5 (12) |
| NOS/other | 2 (5) |
| Tumor site, n (%) | |
| Tongue | 9 (21) |
| Larynx | 12 (29) |
| Tonsil | 8 (19) |
| Floor of mouth | 3 (7) |
| Oral cavity | 4 (10) |
| Hypopharynx | 2 (5) |
| Other | 4 (10) |
| Clinical T stage, n (%) | |
| 0 | 3 (7) |
| 1 | 3 (7) |
| 2 | 10 (24) |
| 3 | 10 (24) |
| 4 | 16 (38) |
| Clinical N stage, n (%) | |
| 0 | 21 (50) |
| 1 | 8 (19) |
| 2 | 11 (26) |
| 3 | 2 (5) |
| Prior surgery, n (%) | |
| No | 7 (17) |
| Yes | 35 (83) |
| Prior XRT, n (%) | |
| No | 27 (64) |
| Yes | 15 (36) |
| Prior chemotherapy, n (%) | |
| No | 33 (79) |
| Yes | 9 (21) |
| Recurrence, n (%) | |
| No | 27 (64) |
| Yes | 15 (36) |
| Smoker, n (%) | |
| No | 7 (17) |
| Yes | 35 (83) |
| Alcohol use, n (%) | |
| No | 18 (43) |
| Yes | 24 (57) |
| HPV+, n (%) | |
| No | 35 (83) |
| Yes | 7 (17) |
DNA extraction
Fresh frozen was utilized for extraction of human genomic DNA using standard methods (see Supplementary Material, Methods).
Microbiomic analyses
DNA amplification and sequencing of rRNA genes
rRNA gene fragments were amplified with a set of primers targeting variable regions 1–4 of the prokaryotic 16S rRNA gene. Two microliters of DNA template was used from each sample, as well as HotStarTaq DNA Polymerase Kit (Qiagen) in a 7900 HT Fast Real-Time polymerase chain reaction (PCR) System (Applied Biosystems, Foster City, CA, USA). PCR was performed under the following conditions: an initial denaturation for 10 min at 95°C; followed by 35 cycles for 30 s at 94°C, 40 s at 56°C and 1 min 20 s at 72°C; lastly, the PCR was completed with 10 min at 72°C. After PCR, DNA gel extraction was performed with a 1% agarose gel using the Qiagen gel extraction kit and protocol. The DNA products were then TOPO TA cloned into pSC-A vector using the StrataClone PCR cloning protocol (Qiagen). Each sample was spread onto an ampicillin LB agar plate and grown at 37°C overnight. Approximately 96 bacterial colonies were randomly selected from a given plate and subjected to PCR amplification as described above using M13 primers. Gel electrophoresis on 1% agarose using 5 μl of the total 10 μl reaction was used to confirm the presence of the vector insert in each sample. SAP treatment was performed with the addition of 1 μl SAP and 0.25 μl of Exo1 enzyme to each PCR sample; afterwards, the samples were incubated at 37°C for 20 min followed by 82°C for 15 min. Lastly, M13 reverse primer was added and the samples were sent for Sanger sequencing. Charts displaying abundance of taxonomic groups (26) show that a final yield of ∼50 and 90 sequences show equivalent proportions of microbial subpopulations (Supplementary Material, Fig. S2).
Taxonomic assignment of rRNA sequences
To identify the bacteria represented by the sequences, we utilized a commonly used existing pipeline. Ribosomal Database Project-Release 10 was utilized for aligning and classifying the sequences to maximize the number of sequences correctly classified. Sanger sequenced samples (described above) were assigned to bacterial taxonomies by The Ribosomal Database Project (RDP) Classifier (27), a naïve Bayesian classifier and a count of bacterial populations at different levels was acquired (Fig. 3 and Supplementary Material, Table S1).
Detection of promoter methylation
Bisulfite treatment of DNA
1.0 μg DNA was obtained from each sample and placed in a volume of 50 μl. Five microliters of fresh 3 m NaOH was then added and the samples were incubated at 37°C for 25–30 min. Afterwards, 30 μl 10 mm hydroquinone and 520 μl 3 m sodium bisulfite were added to each sample and the samples were incubated at 50°C for 16 h. The DNA was then isolated using the Qiaquick gel extraction kit according to their protocol (Qiagen) and 5 μl fresh 3 m NaOH was added to each sample before incubation at room temperature for 20 min. Finally, 20 μl 3 m NaOAc (pH 5) was added and the DNA was purified again using the Qiaquick gel extraction kit, eluting with 30 μl elution buffer (Qiagen).
Combined bisulfite restriction analysis (COBRA)
Upstream regulatory sequences of MDR1 and three other selected genes that are associated with inflammation (IL8) or HNSCC (RARB, TGFBII) were amplified from bisulfite converted DNA using PCR primers specific for bisulfite-treated DNA (sequences available upon request). PCR products were digested with BstUI, a restriction enzyme that cuts at an intact CpG (NEB; Ipswich, MA, USA). Next, the products were run out on 8% polyacrylamide gel electrophoresis gels. As bisulfite treatment converts only unmethylated cytosine residues to thymidine, digested products on the COBRA gel indicate methylation, while undigested bands represent unmethylated DNA. Tumor samples were scored as methylated if they evidenced methylation in the form of digested bands that were not seen in the COBRAs of their ‘adjacent normal’ counterparts (28).
HPV status detection by PCR-based assays
HPV16 and HPV18 amplifications were performed using HotStart Taq (Qiagen) according to the manufacturer's instructions. Briefly, 10 μl HotStart was combined with 1 μl of forward primer (10 pm), 1 μl reverse primer (10 pm), 7 μl water and 1 μl HNSCC patient DNA. The PCR conditions were as follows: 95°C for 10 min; 35 cycles of 95°C for 30 s, 56°C for 30 s and 72°C for 30 s; followed by 72°C for 10 min. PCR products were analyzed on an 8% polyacrylamide gel. Primer sequences are available upon request.
Statistical analyses
MANOVA was used to test hypotheses of whether dependent variables are affected by the difference in observed microbial populations, e.g. tumor versus non-tumor. Matlab (Mathworks, Natick, MA, USA) anova1 and manova1 functions were used. The plots to depict MANOVA statistical results were also generated in Matlab utilizing the canonical variable from MANOVA results using various plotting functions. The Matlab package ffmanova was used to perform general linear modeling with multiple responses (MANCOVA). An overall P-value for each model term is calculated by the 50–50 MANOVA method, which handles collinear responses. In these analyses, we assumed that the microbial data follow a normal distribution, with some outliers (Fig. 3 and Supplementary Material, Fig. S3). This library uses rotation testing to compute adjusted single response P-values according to family-wise error rates and false discovery rates. The results were plotted for further investigation.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported, in part, by the National Institutes of Health (R01DE21544). C.E. is the Sondra J. and Stephen R. Hardis Chair of Cancer Genomic Medicine at the Cleveland Clinic, was a Doris Duke Distinguished Clinical Scientist, and is an American Cancer Society Clinical Research Professor, generously funded, in part, by the F.M. Kirby Foundation. Funding to pay the Open Access publication charges for this article was provided by the National Institutes of Health, USA.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the research participants of this study and whose contribution made this work possible. We are grateful to William Shannon and Elena Deych (Genome Center, Washington University School of Medicine and HMP), David Serre (Genomic Medicine Institute, Cleveland Clinic and HMP) and Emily Pontzer (Genomic Medicine Institute) for critical review of drafts of this manuscript. We are grateful to the members of the UK laboratory for their thoughtful discussions.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Leemans C.R., Braakhuis B.J., Brakenhoff R.H. The molecular biology of head and neck cancer. Nat. Rev. Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. doi:10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 2.Hooper L.V., Gordon J.I. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–1118. doi: 10.1126/science.1058709. doi:10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 3.Savage D.C. Gastrointestinal microflora in mammalian nutrition. Annu. Rev. Nutr. 1986;6:155–178. doi: 10.1146/annurev.nu.06.070186.001103. doi:10.1146/annurev.nu.06.070186.001103. [DOI] [PubMed] [Google Scholar]
- 4.Nicholson J.K., Holmes E., Wilson I.D. Gut microorganisms, mammalian metabolism and personalized health care. Nat. Rev. Microbiol. 2005;3:431–438. doi: 10.1038/nrmicro1152. doi:10.1038/nrmicro1152. [DOI] [PubMed] [Google Scholar]
- 5.Berg R.D. The indigenous gastrointestinal microflora. Trends Microbiol. 1996;4:430–435. doi: 10.1016/0966-842x(96)10057-3. doi:10.1016/0966-842X(96)10057-3. [DOI] [PubMed] [Google Scholar]
- 6.Amieva M.R., El-Omar E.M. Host-bacterial interactions in Helicobacter pylori infection. Gastroenterology. 2008;134:306–323. doi: 10.1053/j.gastro.2007.11.009. doi:10.1053/j.gastro.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 7.Bartsch H., Nair J. Chronic inflammation and oxidative stress in the genesis and perpetuation of cancer: role of lipid peroxidation, DNA damage, and repair. Langenbecks Arch. Surg. 2006;391:499–510. doi: 10.1007/s00423-006-0073-1. doi:10.1007/s00423-006-0073-1. [DOI] [PubMed] [Google Scholar]
- 8.Hold G.L., El-Omar E.M. Genetic aspects of inflammation and cancer. Biochem. J. 2008;410:225–235. doi: 10.1042/BJ20071341. doi:10.1042/BJ20071341. [DOI] [PubMed] [Google Scholar]
- 9.Hussain S.P., Hofseth L.J., Harris C.C. Radical causes of cancer. Nat. Rev. Cancer. 2003;3:276–285. doi: 10.1038/nrc1046. doi:10.1038/nrc1046. [DOI] [PubMed] [Google Scholar]
- 10.Chiba T., Marusawa H. A novel mechanism for inflammation-associated carcinogenesis; an important role of activation-induced cytidine deaminase (AID) in mutation induction. J. Mol. Med. 2009;87:1023–1027. doi: 10.1007/s00109-009-0527-3. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki H., Toyota M., Kondo Y., Shinomura Y. Inflammation-related aberrant patterns of DNA methylation: detection and role in epigenetic deregulation of cancer cell transcriptome. Methods Mol. Biol. 2009;512:55–69. doi: 10.1007/978-1-60327-530-9_5. doi:10.1007/978-1-60327-530-9_5. [DOI] [PubMed] [Google Scholar]
- 12.Veereman Wauters G., Ferrell L., Ostroff J.W., Heyman M.B. Hyperplastic gastric polyps associated with persistent Helicobacter pylori infection and active gastritis. Am. J. Gastroenterol. 1990;85:1395–1397. [PubMed] [Google Scholar]
- 13.Wen S., Felley C.P., Bouzourene H., Reimers M., Michetti P., Pan-Hammarstrom Q. Inflammatory gene profiles in gastric mucosa during Helicobacter pylori infection in humans. J. Immunol. 2004;172:2595–2606. doi: 10.4049/jimmunol.172.4.2595. [DOI] [PubMed] [Google Scholar]
- 14.van der Deen M., de Vries E.G., Visserman H., Zandbergen W., Postma D.S., Timens W., Timmer-Bosscha H. Cigarette smoke extract affects functional activity of MRP1 in bronchial epithelial cells. J. Biochem. Mol. Toxicol. 2007;21:243–251. doi: 10.1002/jbt.20187. doi:10.1002/jbt.20187. [DOI] [PubMed] [Google Scholar]
- 15.van der Deen M., Timens W., Timmer-Bosscha H., van der Strate B.W., Scheper R.J., Postma D.S., de Vries E.G., Kerstjens H.A. Reduced inflammatory response in cigarette smoke exposed Mrp1/Mdr1a/1b deficient mice. Respir Res. 2007;8:49. doi: 10.1186/1465-9921-8-49. doi:10.1186/1465-9921-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen X.J., Rawls J.F., Randall T., Burcal L., Mpande C.N., Jenkins N., Jovov B., Abdo Z., Sandler R.S., Keku T.O. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes. 2010;1:138–147. doi: 10.4161/gmic.1.3.12360. doi:10.4161/gmic.1.3.12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen K., Sawhney R., Khan M., Benninger M.S., Hou Z., Sethi S., Stephen J.K., Worsham M.J. Methylation of multiple genes as diagnostic and therapeutic markers in primary head and neck squamous cell carcinoma. Arch. Otolaryngol. Head Neck Surg. 2007;133:1131–1138. doi: 10.1001/archotol.133.11.1131. doi:10.1001/archotol.133.11.1131. [DOI] [PubMed] [Google Scholar]
- 18.Vannucci L., Stepankova R., Kozakova H., Fiserova A., Rossmann P., Tlaskalova-Hogenova H. Colorectal carcinogenesis in germ-free and conventionally reared rats: different intestinal environments affect the systemic immunity. Int. J. Oncol. 2008;32:609–617. [PubMed] [Google Scholar]
- 19.Zhu Y., Michelle Luo T., Jobin C., Young H.A. Gut microbiota and probiotics in colon tumorigenesis. Cancer Lett. 2011;309:119–127. doi: 10.1016/j.canlet.2011.06.004. doi:10.1016/j.canlet.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costello E.K., Lauber C.L., Hamady M., Fierer N., Gordon J.I., Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. doi:10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tahara T., Shibata T., Nakamura M., Yamashita H., Yoshioka D., Okubo M., Maruyama N., Kamano T., Kamiya Y., Nakagawa Y., et al. Effect of MDR1 gene promoter methylation in patients with ulcerative colitis. Int. J. Mol. Med. 2009;23:521–527. doi: 10.3892/ijmm_00000160. [DOI] [PubMed] [Google Scholar]
- 22.Van Brussel J.P., Jan Van Steenbrugge G., Van Krimpen C., Bogdanowicz J.F., Van Der Kwast T.H., Schroder F.H., Mickisch G.H. Expression of multidrug resistance related proteins and proliferative activity is increased in advanced clinical prostate cancer. J. Urol. 2001;165:130–135. doi: 10.1097/00005392-200101000-00032. doi:10.1097/00005392-200101000-00032. [DOI] [PubMed] [Google Scholar]
- 23.D'Souza G., Kreimer A.R., Viscidi R., Pawlita M., Fakhry C., Koch W.M., Westra W.H., Gillison M.L. Case-control study of human papillomavirus and oropharyngeal cancer. N. Engl. J. Med. 2007;356:1944–1956. doi: 10.1056/NEJMoa065497. doi:10.1056/NEJMoa065497. [DOI] [PubMed] [Google Scholar]
- 24.Bennett K.L., Lee W., Lamarre E., Zhang X., Seth R., Scharpf J., Hunt J., Eng C. HPV status-independent association of alcohol and tobacco exposure or prior radiation therapy with promoter methylation of FUSSEL18, EBF3, IRX1, and SEPT9, but not SLC5A8, in head and neck squamous cell carcinomas. Genes Chromosomes Cancer. 2010;49:319–326. doi: 10.1002/gcc.20742. [DOI] [PubMed] [Google Scholar]
- 25.Morris J.D., Diamond K.A., Balart L.A. Do probiotics have a role in the management of inflammatory bowel disease? J. La State Med. Soc. 2009;161:155–159. [PubMed] [Google Scholar]
- 26.Ondov B.D., Bergman N.H., Phillippy A.M. Interactive metagenomic visualization in a Web browser. BMC Bioinformatics. 2011;12:385. doi: 10.1186/1471-2105-12-385. doi:10.1186/1471-2105-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. doi:10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiong Z., Laird P.W. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. doi:10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.