Abstract
Myelin insulates axons in the peripheral nervous system to allow rapid propagation of action potentials, and proper myelination requires the precise regulation of genes encoding myelin proteins, including PMP22. The correct gene dosage of PMP22 is critical; a duplication of PMP22 is the most common cause of the peripheral neuropathy Charcot-Marie-Tooth Disease (CMT) (classified as type 1A), while a deletion of PMP22 leads to another peripheral neuropathy, hereditary neuropathy with liability to pressure palsies. Recently, duplications upstream of PMP22, but not containing the gene itself, were reported in patients with CMT1A like symptoms, suggesting that this region contains regulators of PMP22. Using chromatin immunoprecipitation analysis of two transcription factors known to upregulate PMP22—EGR2 and SOX10—we found several enhancers in this upstream region that contain open chromatin and direct reporter gene expression in tissue culture and in vivo in zebrafish. These studies provide a novel means to identify critical regulatory elements in genes that are required for myelination, and elucidate the functional significance of non-coding genomic rearrangements.
INTRODUCTION
Myelin is a lipid-rich electrical insulator of axons and is required for rapid propagation of action potentials along axons. In the peripheral nervous system, each myelin sheath is composed of a Schwann cell wrapping around a single axon, creating layers of membrane containing myelin structural proteins, including the membrane protein peripheral myelin protein 22 (PMP22) (1). The genes encoding myelin structural proteins are upregulated early in development and are required for the correct formation of the myelin sheath. A 1.4 Mb duplication of chromosome 17, containing the PMP22 gene, causes the most common type of the demyelinating peripheral neuropathy Charcot-Marie-Tooth disease, type 1A (CMT1A) (2–4), while a deletion of one copy of the same region causes hereditary neuropathy with liability to pressure palsies (HNPP) (5,6). Although the human 1.4 Mb duplication contains many genes besides PMP22, point mutations in PMP22 also cause CMT1A and HNPP (7). Additional evidence for PMP22 as the disease-causing gene within the 1.4 Mb duplication was also provided by rodent models for CMT1A, showing that transgenic overexpression of Pmp22 (8–13) causes a CMT1A-like peripheral neuropathy. These rodent models have been used to demonstrate that lowering expression of Pmp22 improves myelination (12,14,15). Therefore, a better understanding of PMP22 transcriptional regulation will provide a mechanistic basis to modulate the levels of PMP22, and provide the basis for identifying novel treatments for CMT1A and HNPP patients.
PMP22 is expressed from two alternate promoters, P1 transcribing exon 1A and P2 transcribing exon 1B, with the same protein produced with the translation start site in exon 2. The region immediately upstream of the Pmp22 P1 promoter contributes to the regulation of this locus in Schwann cells (16–19). However, the P1 promoter cannot direct transgenic expression in peripheral nerve similar to endogenous PMP22, suggesting that other regulatory elements are involved (20). Two transgenic studies found overlapping upstream regions that lead to Schwann cell-specific expression (20–22). Transgenic reporter gene expression in mice showed that this upstream region recapitulates the later expression of endogenous Pmp22 and was named the ‘late myelination Schwann cell-specific element’ (LMSE) (20). A region that directs earlier developmental expression was recently found in the largest intron of PMP22 at 11 kb downstream of the translation start site (23). Given the extremely high expression level of PMP22 in peripheral nerve, these studies indicate that PMP22 is coordinately regulated by multiple enhancers.
While proximal regulatory elements often are the first to be identified, there are many examples of genes that are regulated by more distal enhancers. Interestingly, copy number variation (CNV) analysis in patients with a mild form of CMT identified genomic duplications upstream of but not containing the PMP22 gene (24,25). The two duplicated regions overlap by 168 kb (NCBI build 36 Chr17:15,143,663–15,311,619) and contain the TekT3 gene, which is primarily expressed in the testes (26) and is involved in sperm motility (27). Because duplication of these upstream genomic segments results in a phenotype similar to the overexpression of PMP22, without duplication of the gene itself, it was proposed that this large region contains PMP22 regulatory element(s) (24,25).
The 11 kb enhancer is upregulated by two transcription factors: early growth response 2 (EGR2/KROX20, hereafter referred to as EGR2) and SOX10. EGR2 is required for Pmp22 expression (28,29) and also regulates many other myelin genes during early development (30–32). SOX10 regulates Pmp22 as well (23) and is also more widely required at several stages of Schwann cell development (33–36). There is evidence that EGR2 and SOX10 work together as activators of myelin genes: SOX10 binds near EGR2 at myelin gene loci more often than would be expected by chance (37), and SOX10 and EGR2 function synergistically at other myelin-related loci: the Connexin 32, Myelin Basic Protein and Myelin Protein Zero loci (38–41). Here, we use chromatin immunoprecipitation (ChIP) of EGR2 and SOX10 combined with massively parallel sequencing (ChIP-seq) to identify three enhancers in the 168 kb distal region. Furthermore, we provide evidence that the enhancers display hallmarks of enhancer elements, including open chromatin, activity in luciferase assays and activity in the motor nerves of zebrafish.
RESULTS
Identification of EGR2 and SOX10 in regions distal to TekT3
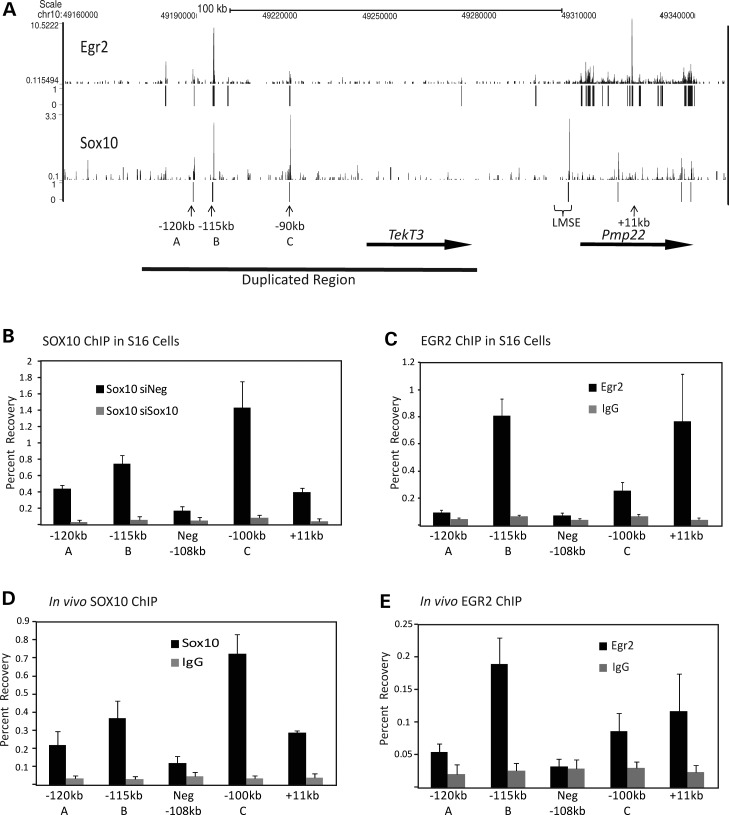
Our previous ChIP-chip analysis of Pmp22 had examined an area of 200 kb surrounding the gene (23). To gain a more comprehensive analysis of EGR2- and SOX10-binding sites in this locus, we used ChIP assays in P15 rat sciatic nerve followed by massively parallel sequencing (in vivo ChIP-seq). The sciatic nerve is highly enriched in Schwann cells, and EGR2 and SOX10 are selectively expressed in this cell type (30,33,42). ChIP-seq identified several peaks of both EGR2 and SOX10 binding far upstream of Pmp22 and TekT3 and within the 168 kb region duplicated in patients (Fig. 1). Peak analysis was performed using the MOSAiCS program (43) controlling the false discovery rate at 0.05. MOSAiCS uses a model-based approach, where the background distribution for unbound regions takes into account systematic biases, such as mappability and guanine/cytosine (GC) content, and the peak regions are described with a two component negative binomial mixture model. A strong peak of EGR2 was identified at the previously identified 11 kb enhancer within PMP22, but due to the stringency of the MOSAiCS program the SOX10 peak at 11 kb was not identified (23). Additionally, a strong peak of SOX10 was identified at the previously identified LMSE element (20). Three peaks of overlapping EGR2 and SOX10 were identified at −120, −115 and −91 kb in relation to the Pmp22 translation start site (Fig. 1A).
Figure 1.
EGR2 and SOX10 occupancy far upstream of Pmp22. (A) ChIP-seq analysis of EGR2 and SOX10 binding was performed using antibodies directed at either EGR2 or SOX10 in rat sciatic nerve at P15. ChIP using each antibody was performed with two biological replicates. A single read, 36 bp run was performed and reads were mapped to the Rattus norvegicus genome Rn4. Sequencing of input DNA was used to assess the distribution of reads from sonicated peripheral nerve chromatin (28 million reads). Peak finding was performed using the R package MOSAiCS (43) controlling the false discovery rate at 0.05. The genomic coordinates (Rattus norvegicus genome rn4) are shown above. The transcribed regions of TekT3 and Pmp22 are shown as arrows below. The overlapping region found in the patients with CNVs (24,25) is shown as a thick line below. Upward arrows and bracket below designate areas of overlapping EGR2 and SOX10 peaks, and previously identified enhancers proximal to the Pmp22 gene. (B) SOX10 ChIP assays were performed in S16 rat Schwann cells treated with either control siRNA (siNeg) or Sox10 siRNA (siSox10). Samples were analyzed by quantitative polymerase chain reaction (PCR) using the indicated primer sets and percent recovery is calculated relative to input. Error bars represent the standard deviation of at least two biological replicates. (C) EGR2 ChIP was performed in S16 rat Schwann cells alongside an IgG control, using the same primer sets as shown in (B)–(E). Error bars represent the standard deviation of at least two biological replicates. (D) In vivo SOX10 ChIP was performed using pooled sciatic nerve from P15 mice alongside an IgG control, using the same primer sets as shown in (B)–(E). Error bars represent the standard deviation of three biological replicates. (E) In vivo EGR2 ChIP was performed as in (D).
To validate the occupancy of EGR2 and SOX10 at these sites, we used ChIP assays in both the S16 rat Schwann cell line and in vivo using rat sciatic nerve. The S16 cell line expresses near physiological levels of Pmp22 gene expression (44), and comparative analysis indicates that many myelin gene regulatory sites are occupied both in vivo and in the S16 cell line by EGR2 and SOX10 (23,41,45,46). To test the specificity of the SOX10 antibody, ChIP assays were performed in S16 cells treated with either control siRNA (siNeg) or an siRNA directed towards Sox10 (siSox10), showing that the percent recovery using the SOX10 antibody in the control-treated cells is specific to the presence of SOX10. In S16 rat Schwann cells, significant binding of SOX10 at the positive control site, 11 kb, −115 and −91 kb and a smaller amount at −120 kb was detected relative to the negative control immunoprecipitation (siSox10). In contrast, little or no SOX10 binding was detected at a negative control site lacking a SOX10 peak in the ChIP-seq (Neg −108 kb) (Fig. 1B). Although there is a small amount of recovery at the negative site, the recovery is higher at the sites of interest. In fact, SOX10 binding is detected at lower levels at several sites throughout the Pmp22 locus (data not shown). Background binding likely reflects general affinity of SOX10 for DNA, particularly since its binding site is relatively non-complex and similar analyses of other factors have identified weak binding throughout the genome (47). In vivo SOX10 ChIP performed in P15 rat sciatic nerve also detected occupancy of SOX10 at the −120, −115, −91 and +11 kb regions compared with a non-specific IgG control antibody. In contrast, there is very little occupancy of SOX10 at a negative control site (Neg −108 kb) (Fig. 1D). The relative percent recovery of SOX10 is the highest at the −91 kb site in S16 cells and in vivo, which is consistent with the relative peak heights in the ChIP-seq data.
ChIP using an antibody directed towards EGR2 in both S16 cells and in vivo shows that EGR2 binding overlaps with SOX10 at −120, −115, −91 kb, and the positive control, 11 kb, but much less at a negative control site (Neg −108 kb) (Fig. 1C and E). The relative percent recovery of EGR2, with the highest recovery of EGR2 at −115 kb, both in S16 cells and in vivo is in agreement with the ChIP-seq data, suggesting that −115 kb is the strongest EGR2-binding site. The overlapping binding of EGR2 and SOX10 at these three sites lead us to hypothesize that −120, −115 and −91 kb regions are enhancers of Pmp22, and hereafter these sites are referred to as region A (−120 kb), region B (−115 kb) and region C (−91 kb), respectively.
Open chromatin is present at the distal regions
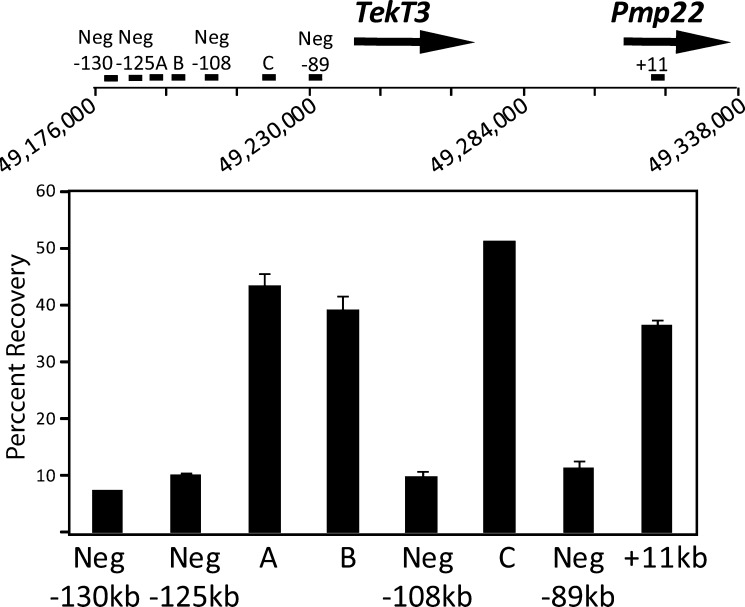
If regions A, B and C are enhancers, we expect to find common characteristics of enhancers at these sites, such as open chromatin (48). To identify regions of open chromatin, we used formaldehyde assisted isolation of regulatory elements (FAIRE) (49). This technique uses formaldehyde to separate open chromatin from nucleosomal DNA, in which formaldehyde efficiently crosslinks histone protein to DNA. Using FAIRE analysis of S16 rat Schwann cells, which have very high levels of Pmp22 expression (44), open chromatin was found at regions A, B and C in comparison to several negative sites surrounding the open regions and a positive control, 11 kb (Fig. 2). These data suggest that regions A, B and C are specifically surrounded by open chromatin, a hallmark of regulatory regions.
Figure 2.
Regions of open chromatin in the upstream region. FAIRE analysis of the indicated sites in the region upstream of Pmp22 and TekT3. The location of the primers is indicated above. The percent recovery was calculated relative to total input. Error bars represent the standard deviation of two technical replicates, the standard deviation of Neg −130 is too small to be displayed at ±0.04. The data are representative of two biological replicates.
Chromosomal context of newly identified enhancers
The three genomic regions exhibiting SOX10- and EGR2-binding are directly upstream of the TekT3 gene. Although TEKT3 expression is restricted to the testis compared with other tissues (26,27), it is possible that the 168 kb duplicated region contains enhancers of TekT3. Quantitative polymerase chain reaction (PCR) was used to determine the level of TekT3 expression in rat sciatic nerve or S16 rat Schwann cells compared with testis, a tissue where TekT3 is highly expressed (26,27). While TekT3 is highly expressed in mouse testis, TekT3 expression is >35-fold lower in both P15 rat sciatic nerve and S16 rat Schwann cells (Supplementary Material, Fig. S1). As TekT3 is expressed at very low levels in Schwann cells, it suggests that enhancers active in Schwann cells are not activating TekT3 expression.
The CNV also contains part of a predicted gene, CMT1A duplicated region transcript 4 (CDRT4) upstream of the enhancers. No function has been attributed to this gene, and while its expression is detected in fetal tissues, it is very low in adult tissues (26). Primers designed to detect Cdrt4 by qPCR were unable to detect a significant level of Cdrt4 in S16 rat Schwann cells or P15 sciatic nerve (Supplementary Material, Fig. S1). Enhancers regulated by EGR2 and SOX10 would be expected to direct expression of targets in both S16 rat Schwann cells and sciatic nerve, suggesting that these enhancers do not activate Cdrt4.
Directly upstream of Cdrt4 is Fam18b2, a gene of unknown function. While Fam18b2 expression is detected in both S16 rat Schwann cells and P15 rat sciatic nerve, its expression is unaffected by reducing SOX10 levels using siSox10 (Supplementary Material, Fig. S1). This suggests that SOX10-regulated enhancers are not activating FAM18B2 expression.
Upstream PMP22 elements are active in reporter gene assays
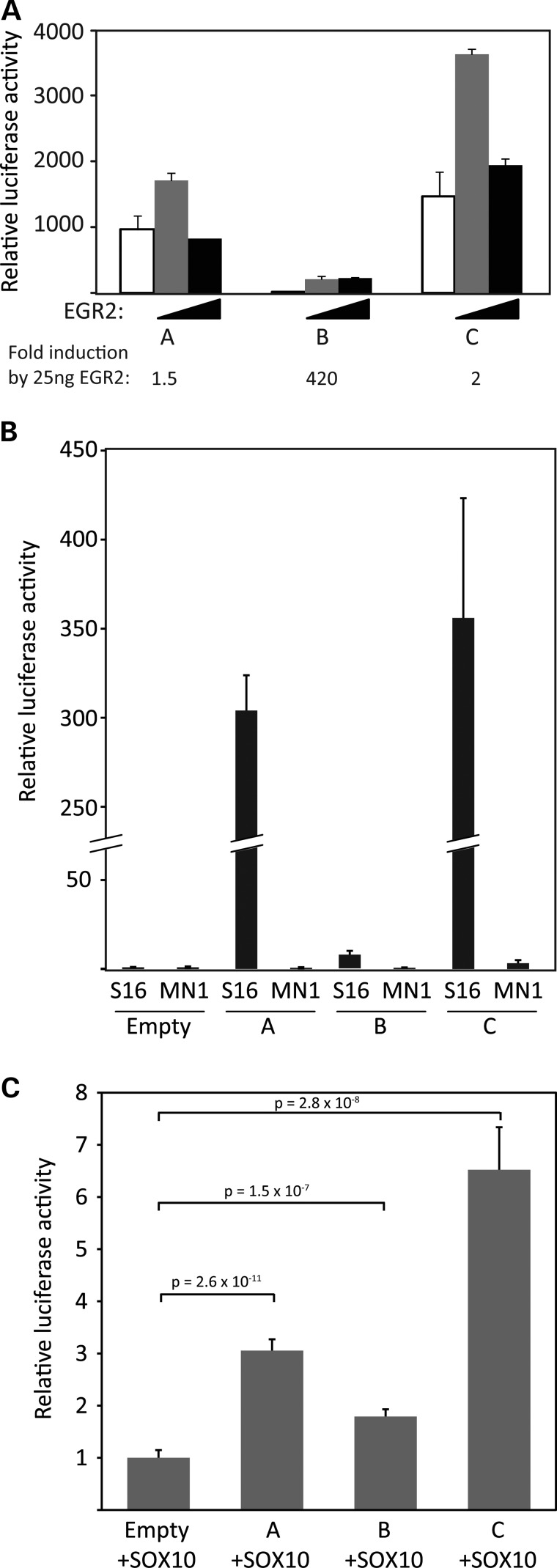
To determine whether regions A, B and/or C have enhancer activity, the DNA sequences identified with EGR2/SOX10 peaks were cloned upstream of a minimal promoter directing expression of a luciferase reporter gene. Because these regions are highly conserved, the homologous human sequences were used. Region A, region B or region C was co-transfected with EGR2 in the B16/F10 melanoma cell line, which expresses Sox10 (50) but not Egr2 (51) to test for EGR2 activation (Fig. 3A). Relative luciferase activity is determined by comparing each reporter to an empty luciferase reporter containing the minimal promoter. In this assay, region B is highly induced over 400-fold, while regions A and C have very low EGR2 induction of ∼2-fold. Control assays using an empty expression plasmid had no effect on these reporters (Supplementary Material, Fig. S2). Interestingly, regions A and C are highly active without co-transfection of EGR2, suggesting that regions A and C may be induced by an endogenous factor, making regions A and C less sensitive to further activation by EGR2 co-transfection.
Figure 3.
Reporter gene analysis of the human segments far upstream of PMP22. (A) Analysis of enhancer activity of regions A, B and C in B16/F10 cell lines (SOX10+ and EGR2–). The indicated segments of the human PMP22 gene were placed upstream of a luciferase reporter gene containing a minimal TATA element. The PMP22 reporters were co-transfected in the B16/F10 cell line with expression plasmids for EGR2: 0 ng (white), 25 ng (grey) and 50 ng (black). Results are expressed as the mean ± SD of normalized luciferase activity relative to the activity of the empty luciferase reporter. (B) Analysis of enhancer activity of regions A, B and C in S16 (SOX10+ and EGR2+) and MN1 (SOX10– and EGR2–) cell lines. PMP22 regions A, B and C were cloned upstream of a luciferase reporter gene and transfected into S16 and MN1 cells. Results are expressed as the mean ± SD of normalized luciferase activity relative to the pe1b empty vector. (C) Constructs harboring PMP22 region A, B, or C were co-transfected into cultured MN-1 cells with an additional construct to express SOX10. Luciferase assays were then employed to test the activity of each region relative to the activity of the control (‘Empty’) vector, all in the presence of SOX10. Error bars indicate standard deviations, and P-values are provided for each region.
To further investigate the activation of the putative enhancers, a second transfection assay was performed using S16 rat Schwann cells. Unlike the B16/F10 melanoma cell line, S16 rat Schwann cells express high levels of myelin genes, including Sox10 and Egr2, comparable to myelinating Schwann cells (44). S16 cells transfected with the reporters were analyzed using the dual-luciferase reporter assay. All three regions exhibit enhancer activity when compared with an empty vector (Fig. 3B). Regions A and C showed a >250-fold increase in activity, whereas region B approached a 10-fold increase in activity.
To test the enhancer activity of each genomic segment in the absence of SOX10 and EGR2, we performed similar luciferase assays in mouse immortalized motor neuron (MN-1) cells, which do not express these transcription factors (data not shown). None of the tested genomic segments displayed significant enhancer activity compared with that observed in S16 cells, or with an empty vector (Fig. 3B).
We further harnessed the absence of SOX10 in MN-1 cells to test the responsiveness of regions A, B and C to this transcription factor. Specifically, we co-transfected the luciferase expression constructs for region A, B or C with an expression construct for SOX10 and compared the luciferase activity with an empty vector in the presence of SOX10. This revealed that SOX10 significantly increased the enhancer potential of all three genomic segments (Fig. 3C). An empty expression vector not containing SOX10 had no effect (data not shown) consistent with other genomic elements that have been studied in this system (52,53). While they may have differing sensitivities in the B16/F10 and S16 assays, PMP22 regions A, B and C display strong enhancer potential in SOX10- and EGR2-postive cells and are responsive to both factors, suggesting that these regions are transcriptionally regulated by EGR2 and SOX10.
A subset of EGR2 and SOX10 consensus sequences are essential for enhancer activity
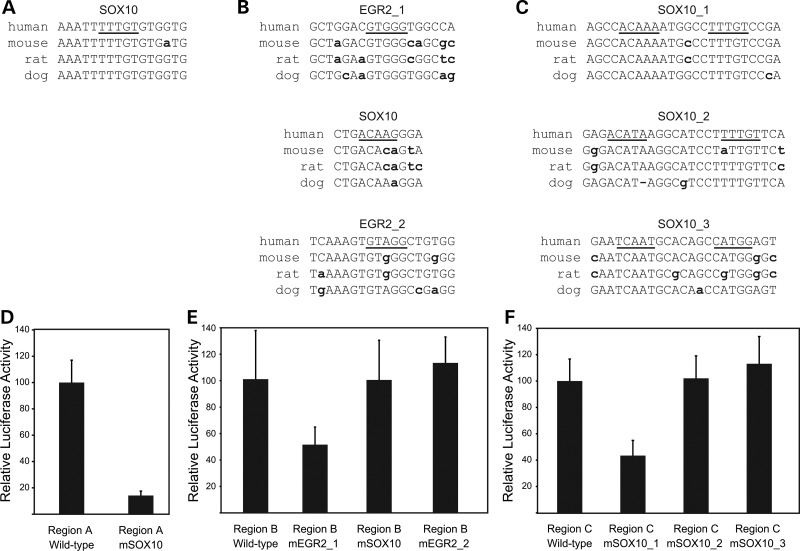
Computational analysis revealed at least one EGR2 and/or SOX10 consensus sequence motif within PMP22 regions A, B and C (see Materials and Methods). Region A harbors a single SOX10 consensus sequence that is conserved between human, mouse, rat and dog (Fig. 4A). Region B contains two EGR2 consensus sequences (EGR2 1 and EGR2 2) and one SOX10 monomeric consensus sequence (Fig. 4B). The core consensus sequence (underlined text in Fig. 4B) of EGR2_1 is conserved among all four species, while those of EGR2_2 and SOX10 have at least 1bp difference among mammals. Finally, region C has three dimeric, head-to-head SOX10 consensus sequences (SOX10_1, SOX10_2 and SOX10_3; Fig. 4C). Note that both of the core sequence consensus sites for SOX10_1 are conserved among human, mouse, rat and dog—this was not observed for SOX10_2 or SOX10_3 (Fig. 4C).
Figure 4.
Regions A, B and C have at least one EGR2 or SOX10 consensus sequence required for activity. (A–C) Computational and functional analyses of transcription factor-binding sites within PMP22 regions A, B and C. The genomic sequence of region A (A), region B (B) and region C (C) were submitted to TRANSFAC to identify potential SOX10- and EGR2-binding sites. The genomic sequence surrounding each predicted SOX10- and EGR2-binding site from human, mouse, rat and dog are depicted with underlined text indicating the TRANSFAC core sequence, and bold, lowercase text indicates nucleotides that are not conserved against human. Dashes indicate gaps in the sequence alignment. (D–F) Mutagenized (‘m’; see Supplementary Material, Fig. S3) and wild-type versions of PMP22 region A (D), region B (E) and region C (F) were transfected into S16 cell lines and tested for the ability to direct luciferase expression compared with an empty vector (data not shown). Results are expressed as the mean ± SD percentage difference of luciferase activity in mutagenized regions compared with the wild-type.
To test the function of each putative site, the consensus sequences were mutagenized (Supplementary Material, Fig. S3) in the luciferase reporter constructs, and the enhancer activity compared with the respective wild-type construct in S16 cells. Mutagenesis of the single SOX10-binding site within region A resulted in an ∼85% decrease in enhancer activity (Fig. 4D). Of the three putative binding sites within region B, only one was associated with a significant reduction (∼45%) in enhancer activity upon mutagenesis—EGR2 consensus sequence 1 (Fig. 4E). Finally, mutagenesis of SOX10 consensus sequence 1 within region C resulted in an ∼65% reduction in enhancer activity (Fig. 4F). Thus, each PMP22 region harbors at least one SOX10 or EGR2 consensus sequence that is required for the full enhancer activity of the wild-type genomic segment.
Regions A, B and C display enhancer activity along motor nerves in developing zebrafish
Zebrafish are a robust model for testing the enhancer activity of mammalian transcriptional regulatory elements (54–57). Importantly, myelinating Schwann cells develop in zebrafish embryos beginning at 48 h post fertilization (hpf). Furthermore, the zebrafish genome contains an ortholog of pmp22 that is expressed in Schwann cells (58) and a duplication of the pmp22 gene in the closely related medaka fish leads to reduced nerve conduction velocity. This suggests that pmp22 plays an important role, similar to the role in mammals, in the peripheral myelin of teleost fish (59).
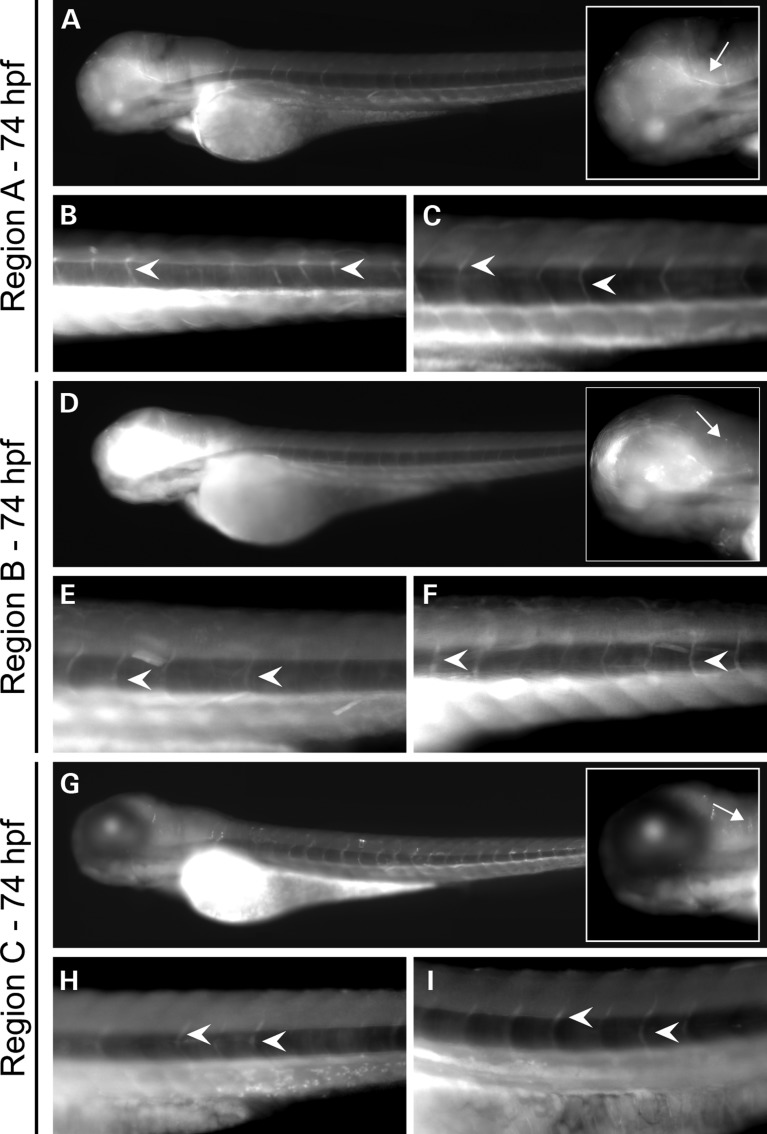
To assess the enhancer activity of PMP22 regions A, B and C in vivo, constructs containing each region directing expression of an EGFP reporter gene were injected into single-celled AB zebrafish embryos and analyzed at 74hpf. For each region, EGFP expression was observed along ventral projections consistent with Schwann cells along motor nerves along the length of the zebrafish (Fig. 5A, D, G, and arrowheads in Fig. 5B, C, E, F, H, I). This expression pattern is similar to that of motor nerve staining observed in previous studies on Schwann cells (53,56,57). Expression of EGFP along the motor nerves was consistent in the majority of transient transgenic embryos assessed for each PMP22 region—73/78 (93.50%), 91/94 (96.81%) and 79/86 (91.86%) EGFP-positive embryos for regions A, B and C, respectively. EGFP expression was also observed in the zebrafish central nervous system (arrows within insets in Fig. 5A, D, G) consistent with the known expression pattern of PMP22 and central nervous system phenotypes reported in some patients with PMP22 deletions and duplications (60–64).
Figure 5.
Regions A, B and C direct reporter gene expression in zebrafish peripheral nerves. Expression of EGFP directed by PMP22 region A (A–C), region B (D–F) and region C (G–I) in transiently transfected zebrafish embryos. Zebrafish embryos were injected with constructs harboring human PMP22 region A, B or C directing enhanced green fluorescent protein (EGFP) expression and analyzed at 74 hpf. Arrows in the insets within (A), (D) and (G) indicate expression in the central nervous system. Two higher-magnification representative images for each construct (B, C, E, F, H and I) are shown, with motor nerves indicated by arrowheads.
DISCUSSION
Using EGR2 and SOX10 transcription factor binding as indicators of myelin gene enhancers, we have identified three new enhancers upstream of PMP22. These three elements are within the boundaries of two recently identified duplications upstream of the PMP22 gene using the CNV analysis. Although the patients with the CNV have significantly milder peripheral neuropathy in comparison to CMT1A patients with the 1.4 Mb duplication (24,25), it seems likely that the duplication results in increased enhancer activity that would increase expression of the PMP22 gene. The three regions have several characteristics that suggest they are enhancers. First, all three have overlapping EGR2- and SOX10-binding and conserved consensus-binding sites, which we have shown is predictive of myelin gene enhancers (37,46). Secondly, all three genomic segments are active in EGR2- and SOX10-dependent reporter assays. Investigation of the DNA state reveals that regions A, B and C contain open chromatin, which is predictive of regulatory regions (48). Finally, regions A, B and C direct expression in structures consistent with Schwann cells in developing zebrafish, as would be expected for an enhancer of PMP22. Overall, our data indicate that the upstream regulatory regions of PMP22 should be considered when evaluating patients with CMT or HNPP for disease-associated mutations, and when developing therapies for these two diseases.
The copy number variants (CNVs) helped direct our attention to these upstream enhancers but in retrospect, analyses of rodent models of CMT1A are consistent with the presence of additional regulatory elements far upstream of PMP22. While CMT1A patients only have one extra copy of PMP22, multiple copies of the PMP22 transgene are required to recapitulate the demyelinating phenotype in rat (10) and mouse (8,9,11,13) models. The rodent models contain multiple copies of either yeast artificial chromosomes or cosmids containing the Pmp22 gene and most lack all the upstream regulatory elements that we have identified. The rodent model requirement of multiple copies of Pmp22 is at least partly due to the lower level of transcription of the Pmp22 inserts compared with endogenous Pmp22 (11). Although this may be explained by species differences in constructs and/or position effects, we hypothesize that the lower expression can be explained, at least in part, by the lack of regions A, B and C in these model systems.
While all three upstream regions contain EGR2 and SOX10, the reporters had different activities in either the B16/F10 cells or the S16 cells. The difference in basal activities and fold inductions by EGR2 in the reporter assays may be due to the amount of SOX10 and EGR2 in the cell lines and the responsiveness of the individual reporters to either EGR2 or SOX10. Although both cell lines express SOX10, there are endogenous levels of EGR2 in S16 cells and over-expressed levels of EGR2 in co-transfection of B16/F10 cells. The sensitivity of each reporter to EGR2 activation in B16/F10 transfection assays corresponds to the relative EGR2 peak height in the ChIP-seq and qPCR analysis (Fig. 1) and to the number of conserved EGR2 sites within the element (Fig. 4). Region B has the highest recovery using the EGR2 antibody in ChIP assays and several EGR2 sites, and has the highest EGR2-dependent activation in the B16/F10 luciferase assay. In contrast, regions A and C have much lower recovery of EGR2 in ChIP and less EGR2-binding sites. This may explain why they have comparatively low EGR2-depenedent activation in the B16/F10 luciferase assay. However, the high basal activity of regions A and C in both the B16/F10 and S16 luciferase assays does correlate with the peak height of SOX10 binding in ChIP-Seq analysis. Therefore, it seems that the high basal expression of regions A and C is due to SOX10 because of the high number of conserved SOX10 sites within these elements, and this notion is supported by the increased activity of these two regions upon the addition of SOX10 to MN-1 cells (Fig. 3).
The differential sensitivity to EGR2 and SOX10 is a continuation of a trend seen in the previously identified enhancers, the −7 kb region within the LMSE and +11 kb. While the +11 kb enhancer was found to be very sensitive to EGR2 in transfection assays and had very high EGR2 recovery in ChIP, the −7 kb region, a small portion of the LMSE, was not sensitive to EGR2 but was sensitive to SOX10 (23). As SOX10 is expressed much earlier in development than EGR2 (36,65,66), SOX10 may be playing a role as a ‘pioneer factor’, meaning it is present at some regulatory regions early in development and may be preparing PMP22 to be highly upregulated immediately upon EGR2 expression.
The presence of three new far distal enhancers along with the previously identified enhancers, the LMSE (20) and the 11 kb enhancer (23), suggests that there is coordinate regulation between several enhancers to precisely regulate PMP22 expression. The long distance (>100 kb), the presence of TEKT3 between the distal enhancers and the PMP22 promoter, and most importantly, other examples of long-range interaction of regulatory regions, suggest that DNA looping is a likely model of activation (67–69). Overall, the identification of the three upstream enhancers helps explain the mechanism behind the CMT1A-like phenotype of patients with the upstream duplications (24,25). Furthermore, we now have a more thorough understanding of PMP22 regulation, which will be useful for development and analysis of therapeutic strategies for patients with CMT1A.
MATERIALS AND METHODS
Chromatin immunoprecipitation
ChIP assays were performed on pooled male and female sciatic nerves from Sprague–Dawley rat pups at postnatal day 15 or the S16 rat Schwann cell line as previously described (41,45), except that herring sperm DNA was omitted from the blocking procedure. The antibodies used in this study include EGR2 antibodies (Abcam cat. no. 43020 used in ChIP-seq and Covance cat. no. PRB-236P used in ChIP-qPCR), SOX10 (Santa Cruz Biotechnology, sc-17342X) and control IgG (normal rabbit IgG: Millipore 12-370 and normal goat IgG: Santa Cruz Biotechnology, sc-2028). Following recovery of ChIP DNA, quantitative PCR was performed in duplicate to calculate fold recovery of a given locus relative to non-specific IgG, using the comparative Ct method (70). All the primers used in this study are listed in Supplementary Material, Table S1. All experiments on rats/mice were performed in strict accordance with experimental protocols approved by the Institutional Animal Care and Use Committee, University of Wisconsin, School of Veterinary Medicine.
Construction and sequencing of Illumina libraries
ChIP samples and input controls were submitted to the University of Wisconsin-Madison DNA Sequencing Facility for ChIP-seq library preparation and sequencing. ChIP using each antibody was performed with two biological replicates. A single read, 36 bp run was performed and reads were mapped to the Rattus norvegicus genome rn4 using Bowtie (http://bowtie-bio.sourceforge.net/index.shtml) to produce SAM files for further analysis. Analysis of occupancy of SOX10 was performed in P15 rat peripheral nerve using a SOX10 antibody used previously (23,41,46). ChIP assays were performed on two biological replicates from independent pools of P15 littermates. The two SOX10 replicates provided a total of ∼12.1 million uniquely mapped reads. The two EGR2 replicates provided a total of ∼11.2 million uniquely mapped reads. In addition, sequencing of input DNA was used to assess the distribution of reads from sonicated peripheral nerve chromatin (28 million reads). Peak finding was performed using the R package MOSAiCS (43) controlling the false discovery rate at 0.05. MOSAiCS implements a model-based approach where the background distribution for unbound regions take into account systematic biases such as mappability and GC content and the peak regions are described with a two component negative binomial mixture model.
Formaldehyde assisted isolation of regulatory elements
The FAIRE assay on the S16 rat Schwann cell line was performed by first rinsing confluent cells with phosphate-buffered saline (PBS) and then cross-linking in PBS containing 1% formaldehyde for 5 min at room temperature (22°C). Glycine was added to 125 mm and incubated for 5 min at room temperature. After washing twice with cold PBS, the cells were harvested, pelleted and frozen at −80°C. The cell pellet was thawed and sonicated on ice using the Bioruptor (Diagenode) set on high (30 s on, 30 s off for 20 min) in lysis buffer (150 mm NaCl, 10% glycerol, 50 mm Tris, pH 8.0, 2% Triton, 1% sodium dodecyl sulfate, 1 mm ethylene diamine tetraacetic acid) containing protease inhibitor cocktail (Sigma, St Louis, MO, USA; 5 µl of cocktail per ml of buffer). The lysate was cleared by centrifugation and unbound DNA was extracted with phenol:chloroform and ethanol precipitated as previously described (71). The protocol only differed in that NaCl was used in the ethanol precipitation. Twenty percent of the cross-linked chromatin was used as an input control by reversing the cross-links at 65°C overnight before phenol:chloroform extraction. The samples were analyzed using quantitative PCR with the same primer sets used in ChIP (Supplementary Table S1) performed in duplicate to calculate the fold recovery of a given segment relative to the input control, using the Comparative Ct method (70).
Transfection assays
The B16/F10 (mouse melanoma) cell line was grown and transfected as described (46,72). The S16 (rat Schwann) cell line and MN1 cells were grown and transfected as described (57). The reporter constructs contain the following coordinates from human chromosome 17 (hg18Mar. 2006 build in UCSC Genome Browser), PMP22 −120 kb (A): chr17:15,253,855–15,254,150; PMP22 −115 kb (B): chr17:15,250,013–15,250,409; PMP22 −91 kb (C): chr17:15,221,688–15,222,096. The reporters used in the B16/F10 luciferase assays are cloned upstream of the pGL4 luciferase reporter containing the minimal E1B TATA promoter. The reporters used in the S16 and MN1 luciferase assays were created by PCR amplifying the regions of interest and then Gateway cloned (Invitrogen) into the pe1b_R vector, upstream of a luciferase reporter gene directed by a minimal promoter (73). For zebrafish studies, each genomic segment was Gateway-cloned (Invitrogen) into the pXIG-EGFP_F vector upstream of an EGFP reporter gene directed by the mouse C-fos minimal promoter (55). Each construct was sequenced to ensure the specificity of the cloned genomic segment.
Mutagenesis of putative SOX10- and EGR2-binding sites
The reference human sequences for regions A, B and C were screened for putative SOX10 and EGR2 consensus-binding sites using TRANSFAC and the MATCH algorithm (74), and the ‘minimize false negatives’ setting. The V$EGR2_01 and V$SOX10_Q6 transcription factor-binding site matrices were employed for this analysis. SOX10 is known to bind to DNA as a monomer or a dimer, therefore all three regions were analyzed for both monomer and head-to-head dimer SOX10 sites. Preference in selecting binding sites to undergo further analysis was given to consensus sequences with a higher TRANSFAC probability score, and that were conserved between human, mouse and rat. Conservation was assessed via visual examination of genomic sequences using the University of California at Santa Cruz (UCSC) Human Genome Browser (version hg18).
Consensus sequences were mutagenized by site-directed mutagenesis using the QuikChange II mutatgenesis kit (Stratagene) according to the manufacturer's specifications. Each site was mutagenized such that each consensus sequence was ‘reversed’ but not ‘complemented’ (Supplementary Material, Fig. S3). This retains the GC content but should effectively abolish the binding of SOX10 and/or EGR2. For head-to-head SOX10-binding sites, both sites and the sequence between the two sites were mutagenized.
siRNA treatment
Either siRNA directed towards SOX10 (Ambion, 4390771) or a negative siRNA control (negative control #2, Ambion, AM4613) were transiently transfected into S16 cells with the Amaxa system (Lonza) using the Rat Neuron Nucleofection Solution. The transfected cells were incubated for 48 h before harvesting RNA using Tri Reagent (Ambion), and quantitative PCR was performed in duplicate to calculate the fold recovery, using the Comparative Ct method (70).
Transient transfection of zebrafish embryos
Each EGFP reporter construct (please see above) was injected into single-celled, wild-type (AB) zebrafish embryos using the Tol-2-mediated transient transfection assay previously reported (55). Embryos exhibiting EGFP expression by 24 hpf were selected and fixed in 4% paraformaldehyde at 74 hpf. Subsequently, each embryo was depigmented in 3% H2O2/0.5% 3 m sodium acetate and stored in 70% glycerol. Embryos were visualized using an Olympus model IX71 fluorescence microscope (Olympus America Inc., Center Valley, PA, USA) equipped with an F-View II digital camera (Olympus) using MicroSuite FIVE software (Olympus). All zebrafish studies were conducted in accordance with the University of Michigan Medical School's Committee on Use and Care of Animals (UCUCA).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Charcot-Marie Tooth Association and National Institutes of Health (HD41590 to J.S., NS073748 to A.A. and HD03352 core grant to Waisman Center).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Richard Quarles for providing the S16 cell line, and Marie Adams and Eric Cabot at the UW Biotechnology Center for performing Illumina sequencing.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Snipes G.J., Suter U., Welcher A.A., Shooter E.M. Characterization of a novel peripheral nervous system myelin protein (PMP-22/SR13) J. Cell Biol. 1992;117:225–238. doi: 10.1083/jcb.117.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lupski J.R., de Oca-Luna R.M., Slaugenhaupt S., Pentao L., Guzzetta V., Trask B.J., Saucedo-Cardenas O., Barker D.F., Killian J.M., Garcia C.A., et al. DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell. 1991;66:219–232. doi: 10.1016/0092-8674(91)90613-4. [DOI] [PubMed] [Google Scholar]
- 3.Timmerman V., Nelis E., Van Hul W., Nieuwenhuijsen B.W., Chen K.L., Wang S., Ben Othman K., Cullen B., Leach R.J., Hanemann C.O. The peripheral myelin protein gene PMP-22 is contained within the Charcot-Marie-Tooth disease type 1A duplication. Nat. Genet. 1992;1:171–175. doi: 10.1038/ng0692-171. [DOI] [PubMed] [Google Scholar]
- 4.Patel P.I., Roa B.B., Welcher A.A., Schoener-Scott R., Trask B.J., Pentao L., Snipes G.J., Garcia C.A., Francke U., Shooter E.M., et al. The gene for the peripheral myelin protein PMP-22 is a candidate for Charcot-Marie-Tooth disease type 1A. Nat. Genet. 1992;1:159–165. doi: 10.1038/ng0692-159. [DOI] [PubMed] [Google Scholar]
- 5.Chance P.F., Alderson M.K., Leppig K.A., Lensch M.W., Matsunami N., Smith B., Swanson P.D., Odelberg S.J., Disteche C.M., Bird T.D. DNA deletion associated with hereditary neuropathy with liability to pressure palsies. Cell. 1993;72:143–151. doi: 10.1016/0092-8674(93)90058-x. [DOI] [PubMed] [Google Scholar]
- 6.Suter U., Scherer S.S. Disease mechanisms in inherited neuropathies. Nat. Rev. Neurosci. 2003;4:714–726. doi: 10.1038/nrn1196. [DOI] [PubMed] [Google Scholar]
- 7.Reilly M.M., Murphy S.M., Laurá M. Charcot-Marie-Tooth disease. J. Peripher. Nerv. Syst. 2011;16:1–14. doi: 10.1111/j.1529-8027.2011.00324.x. [DOI] [PubMed] [Google Scholar]
- 8.Huxley C., Passage E., Manson A., Putzu G., Figarella-Branger D., Pellissier J., Fontés M. Construction of a mouse model of Charcot-Marie-Tooth disease type 1A by pronuclear injection of human YAC DNA. Hum. Mol. Genet. 1996;5:563–569. doi: 10.1093/hmg/5.5.563. [DOI] [PubMed] [Google Scholar]
- 9.Magyar J., Martini R., Ruelicke T., Aguzzi A., Adlkofer K., Dembic Z., Zielasek J., Toyka K., Suter U. Impaired differentiation of Schwann cells in transgenic mice with increased PMP22 gene dosage. J. Neurosci. 1996;16:5351–5360. doi: 10.1523/JNEUROSCI.16-17-05351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sereda M., Griffiths I., Pühlhofer A., Stewart H., Rossner M., Zimmerman F., Magyar J., Schneider A., Hund E., Meinck H., et al. A transgenic rat model of Charcot-Marie-Tooth disease. Neuron. 1996;16:1049–1060. doi: 10.1016/s0896-6273(00)80128-2. [DOI] [PubMed] [Google Scholar]
- 11.Huxley C., Passage E., Robertson A., Youl B., Huston S., Manson A., Sabéran-Djoniedi D., Figarella-Branger D., Pellissier J., Thomas P., et al. Correlation between varying levels of PMP22 expression and the degree of demyelination and reduction in nerve conduction velocity in transgenic mice. Hum. Mol. Genet. 1998;7:449–458. doi: 10.1093/hmg/7.3.449. [DOI] [PubMed] [Google Scholar]
- 12.Perea J., Robertson A., Tolmachova T., Muddle J., King R., Ponsford S., Thomas P., Huxley C. Induced myelination and demyelination in a conditional mouse model of Charcot-Marie-Tooth disease type 1A. Hum. Mol. Genet. 2001;10:1007–1018. doi: 10.1093/hmg/10.10.1007. [DOI] [PubMed] [Google Scholar]
- 13.Robertson A., Perea J., McGuigan A., King R., Muddle J., Gabreëls-Festen A., Thomas P., Huxley C. Comparison of a new pmp22 transgenic mouse line with other mouse models and human patients with CMT1A. J. Anat. 2002;200:377–390. doi: 10.1046/j.1469-7580.2002.00039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sereda M.W., Meyer zu Horste G., Suter U., Uzma N., Nave K.A. Therapeutic administration of progesterone antagonist in a model of Charcot-Marie-Tooth disease (CMT-1A) Nat. Med. 2003;9:1533–1537. doi: 10.1038/nm957. [DOI] [PubMed] [Google Scholar]
- 15.Passage E., Norreel J.C., Noack-Fraissignes P., Sanguedolce V., Pizant J., Thirion X., Robaglia-Schlupp A., Pellissier J.F., Fontes M. Ascorbic acid treatment corrects the phenotype of a mouse model of Charcot-Marie-Tooth disease. Nat. Med. 2004;10:396–401. doi: 10.1038/nm1023. [DOI] [PubMed] [Google Scholar]
- 16.Suter U., Snipes G.J., Schoener-Scott R., Welcher A.A., Pareek S., Lupski J.R., Murphy R.A., Shooter E.M., Patel P.I. Regulation of tissue-specific expression of alternative peripheral myelin protein-22 (PMP22) gene transcripts by two promoters. J. Biol. Chem. 1994;269:25795–25808. [PubMed] [Google Scholar]
- 17.Saberan-Djoneidi D., Sanguedolce V., Assouline Z., Levy N., Passage E., Fontes M. Molecular dissection of the Schwann cell specific promoter of the PMP22 gene. Gene. 2000;248:223–231. doi: 10.1016/s0378-1119(00)00116-5. [DOI] [PubMed] [Google Scholar]
- 18.Hai M., Bidichandani S.I., Patel P.I. Identification of a positive regulatory element in the myelin-specific promoter of the PMP22 gene. J. Neurosci. Res. 2001;65:508–519. doi: 10.1002/jnr.1181. [DOI] [PubMed] [Google Scholar]
- 19.Sinkiewicz-Darol E., Kabzińska D., Moszyńska I., Kochański A. The 5′ regulatory sequence of the PMP22 in the patients with Charcot-Marie-Tooth disease. Acta Biochim. Pol. 2010;57:373–377. [PubMed] [Google Scholar]
- 20.Maier M., Castagner F., Berger P., Suter U. Distinct elements of the peripheral myelin protein 22 (PMP22) promoter regulate expression in Schwann cells and sensory neurons. Mol. Cell Neurosci. 2003;24:803–817. doi: 10.1016/s1044-7431(03)00246-x. [DOI] [PubMed] [Google Scholar]
- 21.Orfali W., Nicholson R.N., Guiot M.C., Peterson A.C., Snipes G.J. An 8.5-kb segment of the PMP22 promoter responds to loss of axon signals during Wallerian degeneration, but does not respond to specific axonal signals during nerve regeneration. J. Neurosci. Res. 2005;80:37–46. doi: 10.1002/jnr.20425. [DOI] [PubMed] [Google Scholar]
- 22.Maier M., Berger P., Nave K.A., Suter U. Identification of the regulatory region of the peripheral myelin protein 22 (PMP22) gene that directs temporal and spatial expression in development and regeneration of peripheral nerves. Mol. Cell Neurosci. 2002;20:93–109. doi: 10.1006/mcne.2002.1116. [DOI] [PubMed] [Google Scholar]
- 23.Jones E.A., Lopez-Anido C., Srinivasan R., Krueger C., Chang L.W., Nagarajan R., Svaren J. Regulation of the PMP22 gene through an intronic enhancer. J. Neurosci. 2011;31:4242–4250. doi: 10.1523/JNEUROSCI.5893-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weterman M.A., van Ruissen F., de Wissel M., Bordewijk L., Samijn J.P., van der Pol W.L., Meggouh F., Baas F. Copy number variation upstream of PMP22 in Charcot-Marie-Tooth disease. Eur. J. Hum. Genet. 2010;18:421–428. doi: 10.1038/ejhg.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang F., Seeman P., Liu P., Weterman M.A., Gonzaga-Jauregui C., Towne C.F., Batish S.D., De Vriendt E., De Jonghe P., Rautenstrauss B., et al. Mechanisms for nonrecurrent genomic rearrangements associated with CMT1A or HNPP: rare CNVs as a cause for missing heritability. Am. J. Hum. Genet. 2010;86:892–903. doi: 10.1016/j.ajhg.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inoue K., Dewar K., Katsanis N., Reiter L.T., Lander E.S., Devon K.L., Wyman D.W., Lupski J.R., Birren B. The 1.4-Mb CMT1A duplication/HNPP deletion genomic region reveals unique genome architectural features and provides insights into the recent evolution of new genes. Genome Res. 2001;11:1018–1033. doi: 10.1101/gr.180401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roy A., Yan W., Burns K.H., Matzuk M.M. Tektin3 encodes an evolutionarily conserved putative testicular microtubules-related protein expressed preferentially in male germ cells. Mol. Reprod. Dev. 2004;67:295–302. doi: 10.1002/mrd.20025. [DOI] [PubMed] [Google Scholar]
- 28.Nagarajan R., Svaren J., Le N., Araki T., Watson M., Milbrandt J. EGR2 mutations in inherited neuropathies dominant-negatively inhibit myelin gene expression. Neuron. 2001;30:355–368. doi: 10.1016/s0896-6273(01)00282-3. [DOI] [PubMed] [Google Scholar]
- 29.Le N., Nagarajan R., Wang J.Y., Araki T., Schmidt R.E., Milbrandt J. Analysis of congenital hypomyelinating Egr2Lo/Lo nerves identifies Sox2 as an inhibitor of Schwann cell differentiation and myelination. Proc. Natl Acad. Sci. USA. 2005;102:2596–2601. doi: 10.1073/pnas.0407836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Topilko P., Schneider-Maunoury S., Levi G., Baron-Van Evercooren A., Chennoufi A.B., Seitanidou T., Babinet C., Charnay P. Krox-20 controls myelination in the peripheral nervous system. Nature. 1994;371:796–799. doi: 10.1038/371796a0. [DOI] [PubMed] [Google Scholar]
- 31.Le N., Nagarajan R., Wang J.Y., Svaren J., LaPash C., Araki T., Schmidt R.E., Milbrandt J. Nab proteins are essential for peripheral nervous system myelination. Nat. Neurosci. 2005;8:932–940. doi: 10.1038/nn1490. [DOI] [PubMed] [Google Scholar]
- 32.Decker L., Desmarquet-Trin-Dinh C., Taillebourg E., Ghislain J., Vallat J.M., Charnay P. Peripheral myelin maintenance is a dynamic process requiring constant Krox20 expression. J. Neurosci. 2006;26:9771–9779. doi: 10.1523/JNEUROSCI.0716-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuhlbrodt K., Herbarth B., Sock E., Hermans-Borgmeyer I., Wegner M. Sox10, a novel transcriptional modulator in glial cells. J. Neurosci. 1998;18:237–250. doi: 10.1523/JNEUROSCI.18-01-00237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Britsch S., Goerich D.E., Riethmacher D., Peirano R.I., Rossner M., Nave K.A., Birchmeier C., Wegner M. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 2001;15:66–78. doi: 10.1101/gad.186601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schreiner S., Cossais F., Fischer K., Scholz S., Bosl M.R., Holtmann B., Sendtner M., Wegner M. Hypomorphic Sox10 alleles reveal novel protein functions and unravel developmental differences in glial lineages. Development. 2007;134:3271–3281. doi: 10.1242/dev.003350. [DOI] [PubMed] [Google Scholar]
- 36.Finzsch M., Schreiner S., Kichko T., Reeh P., Tamm E., Bösl M., Meijer D., Wegner M. Sox10 is required for Schwann cell identity and progression beyond the immature Schwann cell stage. J. Cell Biol. 2010;189:701–712. doi: 10.1083/jcb.200912142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jang S.W., Srinivasan R., Jones E.A., Sun G., Keles S., Krueger C., Chang L.W., Nagarajan R., Svaren J. Locus-wide identification of Egr2/Krox20 regulatory targets in myelin genes. J. Neurochem. 2010;115:1409–1420. doi: 10.1111/j.1471-4159.2010.07045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bondurand N., Girard M., Pingault V., Lemort N., Dubourg O., Goossens M. Human Connexin 32, a gap junction protein altered in the X-linked form of Charcot-Marie-Tooth disease, is directly regulated by the transcription factor SOX10. Hum. Mol. Genet. 2001;10:2783–2795. doi: 10.1093/hmg/10.24.2783. [DOI] [PubMed] [Google Scholar]
- 39.Denarier E., Forghani R., Farhadi H.F., Dib S., Dionne N., Friedman H.C., Lepage P., Hudson T.J., Drouin R., Peterson A. Functional organization of a Schwann cell enhancer. J. Neurosci. 2005;25:11210–11217. doi: 10.1523/JNEUROSCI.2596-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.LeBlanc S.E., Jang S.W., Ward R.M., Wrabetz L., Svaren J. Direct regulation of myelin protein zero expression by the Egr2 transactivator. J. Biol. Chem. 2006;281:5453–5460. doi: 10.1074/jbc.M512159200. [DOI] [PubMed] [Google Scholar]
- 41.Jang S.W., Svaren J. Induction of myelin protein zero by early growth response 2 through upstream and intragenic elements. J. Biol. Chem. 2009;284:20111–20120. doi: 10.1074/jbc.M109.022426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zorick T.S., Syroid D.E., Arroyo E., Scherer S.S., Lemke G. The transcription factors SCIP and Krox-20 mark distinct stages and cell fates in Schwann cell differentiation. Mol. Cell Neurosci. 1996;8:129–145. doi: 10.1006/mcne.1996.0052. [DOI] [PubMed] [Google Scholar]
- 43.Kuan P., Chung D., Thomson J., Stewart R., Keles S. J. Am. Stat. Assoc. 2011;106:891–903. doi: 10.1198/jasa.2011.ap09706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hai M., Muja N., DeVries G.H., Quarles R.H., Patel P.I. Comparative analysis of Schwann cell lines as model systems for myelin gene transcription studies. J. Neurosci. Res. 2002;69:497–508. doi: 10.1002/jnr.10327. [DOI] [PubMed] [Google Scholar]
- 45.Jang S.W., LeBlanc S.E., Roopra A., Wrabetz L., Svaren J. In vivo detection of Egr2 binding to target genes during peripheral nerve myelination. J. Neurochem. 2006;98:1678–1687. doi: 10.1111/j.1471-4159.2006.04069.x. [DOI] [PubMed] [Google Scholar]
- 46.Jones E.A., Jang S.W., Mager G.M., Chang L.-W., Srinivasan R., Gokey N.G., Ward R.M., Nagarajan R., Svaren J. Interactions of Sox10 and Egr2 in myelin gene regulation. Neuron Glia Biol. 2007;3:377–387. doi: 10.1017/S1740925X08000173. PMC2605513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Biggin M.D. Animal transcription networks as highly connected, quantitative continua. Dev. Cell. 2011;21:611–626. doi: 10.1016/j.devcel.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 48.Felsenfeld G., Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- 49.Giresi P.G., Kim J., McDaniell R.M., Iyer V.R., Lieb J.D. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kamaraju A.K., Bertolotto C., Chebath J., Revel M. Pax3 down-regulation and shut-off of melanogenesis in melanoma B16/F10.9 by interleukin-6 receptor signaling. J. Biol. Chem. 2002;277:15132–15141. doi: 10.1074/jbc.M200004200. [DOI] [PubMed] [Google Scholar]
- 51.Slutsky S.G., Kamaraju A.K., Levy A.M., Chebath J., Revel M. Activation of myelin genes during transdifferentiation from melanoma to glial cell phenotype. J. Biol. Chem. 2003;278:8960–8968. doi: 10.1074/jbc.m210569200. [DOI] [PubMed] [Google Scholar]
- 52.Hodonsky C.J., Kleinbrink E.L., Charney K.N., Prasad M., Bessling S.L., Jones E.A., Srinivasan R., Svaren J., McCallion A.S., Antonellis A. SOX10 regulates expression of the SH3-domain kinase binding protein 1 (Sh3kbp1) locus in Schwann cells via an alternative promoter. Mol. Cell. Neurosci. 2012;49:85–96. doi: 10.1016/j.mcn.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prasad M.K., Reed X., Gorkin D.U., Cronin J.C., McAdow A.R., Chain K., Hodonsky C.J., Jones E.A., Svaren J., Antonellis A., et al. SOX10 directly modulates ERBB3 transcription via an intronic neural crest enhancer. BMC Dev. Biol. 2011;11:40. doi: 10.1186/1471-213X-11-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fisher S., Grice E.A., Vinton R.M., Bessling S.L., McCallion A.S. Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science. 2006;312:276–279. doi: 10.1126/science.1124070. [DOI] [PubMed] [Google Scholar]
- 55.Fisher S., Grice E.A., Vinton R.M., Bessling S.L., Urasaki A., Kawakami K., McCallion A.S. Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat. Protoc. 2006;1:1297–1305. doi: 10.1038/nprot.2006.230. [DOI] [PubMed] [Google Scholar]
- 56.Antonellis A., Huynh J.L., Lee-Lin S.Q., Vinton R.M., Renaud G., Loftus S.K., Elliot G., Wolfsberg T.G., Green E.D., McCallion A.S., et al. Identification of neural crest and glial enhancers at the mouse Sox10 locus through transgenesis in zebrafish. PLoS Genet. 2008;4:e1000174. doi: 10.1371/journal.pgen.1000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Antonellis A., Dennis M.Y., Burzynski G., Huynh J., Maduro V., Hodonsky C.J., Khajavi M., Szigeti K., Mukkamala S., Bessling S.L., et al. A rare myelin protein zero (MPZ) variant alters enhancer activity in vitro and in vivo. PLoS ONE. 2010;5:e14346. doi: 10.1371/journal.pone.0014346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wulf P., Bernhardt R.R., Suter U. Characterization of peripheral myelin protein 22 in zebrafish (zPMP22) suggests an early role in the development of the peripheral nervous system. J. Neurosci. Res. 1999;57:467–478. [PubMed] [Google Scholar]
- 59.Itou J., Suyama M., Imamura Y., Deguchi T., Fujimori K., Yuba S., Kawarabayasi Y., Kawasaki T. Functional and comparative genomics analyses of pmp22 in medaka fish. BMC Neurosci. 2009;10:60. doi: 10.1186/1471-2202-10-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parmantier E., Braun C., Thomas J.L., Peyron F., Martinez S., Zalc B. PMP-22 expression in the central nervous system of the embryonic mouse defines potential transverse segments and longitudinal columns. J. Comp. Neurol. 1997;378:159–172. [PubMed] [Google Scholar]
- 61.Ohsawa Y., Murakami T., Miyazaki Y., Shirabe T., Sunada Y. Peripheral myelin protein 22 is expressed in human central nervous system. J. Neurol. Sci. 2006;247:11–15. doi: 10.1016/j.jns.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 62.Dacković J., Rakocević-Stojanović V., Pavlović S., Zamurović N., Dragasević N., Romac S., Apostolski S. Hereditary neuropathy with liability to pressure palsies associated with central nervous system myelin lesions. Eur. J. Neurol. 2001;8:689–692. doi: 10.1046/j.1468-1331.2001.00306.x. [DOI] [PubMed] [Google Scholar]
- 63.Sacco S., Totaro R., Bastianello S., Marini C., Carolei A. Brain white matter lesions in an italian family with charcot-marie-tooth disease. Eur. Neurol. 2004;51:168–171. doi: 10.1159/000077664. [DOI] [PubMed] [Google Scholar]
- 64.Sanahuja J., Franco E., Rojas-García R., Gallardo E., Combarros O., Begué R., Granés P., Illa I. Central nervous system involvement in hereditary neuropathy with liability to pressure palsies: description of a large family with this association. Arch. Neurol. 2005;62:1911–1914. doi: 10.1001/archneur.62.12.1911. [DOI] [PubMed] [Google Scholar]
- 65.Ghislain J., Charnay P. Control of myelination in Schwann cells: a Krox20 cis-regulatory element integrates Oct6, Brn2 and Sox10 activities. EMBO Rep. 2006;7:52–58. doi: 10.1038/sj.embor.7400573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reiprich S., Kriesch J., Schreiner S., Wegner M. Activation of Krox20 gene expression by Sox10 in myelinating Schwann cells. J. Neurochem. 2010;112:744–754. doi: 10.1111/j.1471-4159.2009.06498.x. [DOI] [PubMed] [Google Scholar]
- 67.Palstra R.J., de Laat W., Grosveld F. Beta-globin regulation and long-range interactions. Adv. Genet. 2008;61:107–142. doi: 10.1016/S0065-2660(07)00004-1. [DOI] [PubMed] [Google Scholar]
- 68.D'haene B., Attanasio C., Beysen D., Dostie J., Lemire E., Bouchard P., Field M., Jones K., Lorenz B., Menten B., et al. Disease-causing 7.4 kb cis-regulatory deletion disrupting conserved non-coding sequences and their interaction with the FOXL2 promotor: implications for mutation screening. PLoS Genet. 2009;5:e1000522. doi: 10.1371/journal.pgen.1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gheldof N., Smith E.M., Tabuchi T.M., Koch C.M., Dunham I., Stamatoyannopoulos J.A., Dekker J. Cell-type-specific long-range looping interactions identify distant regulatory elements of the CFTR gene. Nucleic Acids Res. 2010;38:4325–4336. doi: 10.1093/nar/gkq175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 71.Giresi P.G., Lieb J.D. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (Formaldehyde Assisted Isolation of Regulatory Elements) Methods. 2009;48:233–239. doi: 10.1016/j.ymeth.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.LeBlanc S.E., Ward R.M., Svaren J. Neuropathy-associated Egr2 mutants disrupt cooperative activation of myelin protein zero by Egr2 and Sox10. Mol. Cell Biol. 2007;27:3521–3529. doi: 10.1128/MCB.01689-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Antonellis A., Bennett W.R., Menheniott T.R., Prasad A.B., Lee-Lin S.Q., Green E.D., Paisley D., Kelsh R.N., Pavan W.J., Ward A., et al. Deletion of long-range sequences at Sox10 compromises developmental expression in a mouse model of Waardenburg-Shah (WS4) syndrome. Hum. Mol. Genet. 2006;15:259–271. doi: 10.1093/hmg/ddi442. [DOI] [PubMed] [Google Scholar]
- 74.Matys V., Fricke E., Geffers R., Gössling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.