Abstract
Hepatic lipase (gene: LIPC; enzyme: HL; E.C.3.1.1.3) is one of three members of the triglyceride lipase family that contributes to vascular lipoprotein degradation and serves a dual role in triglyceride hydrolysis and in facilitating receptor-mediated lipoprotein uptake into the liver. Amino acid sequences, protein structures, and gene locations for vertebrate LIPC (or Lipc for mouse and rat) genes and proteins were sourced from previous reports and vertebrate genome databases. Lipc was distinct from other neutral lipase genes (Lipg encoding endothelial lipase and Lpl encoding lipoprotein lipase [LPL]) and was located on mouse chromosome 9 with nine coding exons on the negative strand. Exon 9 of human LIPC and mouse and rat Lipc genes contained “stop codons” in different positions, causing changes in C-termini length. Vertebrate HL protein subunits shared 58%–97% sequence identities, including active, signal peptide, disulfide bond, and N-glycosylation sites, as well as proprotein convertase (“hinge”) and heparin binding regions. Predicted secondary and tertiary structures revealed similarities with the three-dimensional structure reported for horse and human pancreatic lipases. Potential sites for regulating LIPC gene expression included CpG islands near the 5″-untranslated regions of the mouse and rat LIPC genes. Phylogenetic analyses examined the relationships and potential evolutionary origins of the vertebrate LIPC gene family with other neutral triglyceride lipase gene families (LIPG and LPL). We conclude that the triglyceride lipase ancestral gene for vertebrate neutral lipase genes (LIPC, LIPG, and LPL) predated the appearance of fish during vertebrate evolution.
Keywords: vertebrates, amino acid sequence, hepatic lipase, evolution, gene duplication
Introduction
Hepatic lipase (HL; gene LIPC; E.C.3.1.1.3) is one of three members of the triglyceride lipase family that contributes to lipoprotein degradation within the circulation system.1–3 HL also regulates the metabolism of low-density lipoprotein, intermediate-density lipoprotein, and high-density lipoprotein particles and is capable of catalyzing the hydrolysis of phospholipids, triglycerides, and acyl-CoA thioesters.4,5 Endothelial lipase (EL; gene LIPG; E.C.3.1.1.3) is a related family member that plays a major role in high-density lipoprotein cholesterol metabolism in the body, catalyzing phospholipase and triglyceride lipase activities6–8 and lipoprotein lipase (LPL; gene LPL; E.C.3.1.1.34) functions in the hydrolysis of triglycerides of circulating chylomicrons and very low-density lipoproteins.9–11 These enzymes share sequence similarities (38%–44% identities) and are usually referred to as the vascular lipase gene family7,12,13 because of their contributions to plasma lipoprotein, cholesterol, and triglyceride phenotypes and to the development of coronary heart diseases in human and animal populations.14–21
The human LIPC gene is located on chromosome 15 and comprises 158.3 kb nucleotides on the direct strand with nine exons and eight introns and encodes a 449 amino acid protein subunit.3,22 Genetic variants have been described that cause HL deficiency and associated hyperlipidemia.23 Several promoter polymorphisms in linkage disequilibrium have also been identified, and the more frequent −250G > A substitution in the LIPC promoter region is associated with changes in plasma lipid concentrations and the risk of coronary artery disease in some ethnic groups.24 LIPC is expressed predominantly in the liver, where the enzyme contributes significantly to the determination of lipoprotein levels, structure, and metabolism.1–3 Studies of Lipc−/Lipc− knockout mice have supported multiple roles for HL in vascular lipoprotein metabolism, including a lipolytic role and a ligand binding function facilitating lipoprotein uptake, which influence lipoprotein particle size in the circulation.17 Following synthesis in the liver endoplasmic reticulum, rat HL is processed by the hydrolysis of the N-terminal leader peptide and acquisition of oligosaccharides within the Golgi and is then rapidly secreted and subsequently bound to heparin sulfate proteoglycans on the surface of hepatocytes.25 HL forms a dimeric subunit structure26 exhibiting similarities with EL, which behaves as a homodimer with a proposed head-to-tail conformation,27 and is subject to proprotein convertase cleavage at a site in the “hinge” region separating the N- and C-terminal enzyme domains.28 Three-dimensional studies of a related mammalian lipase (LIPP, pancreatic lipase)29,30 have enabled identification of three major structural domains for the mammalian neutral lipase family, including an N-terminal domain with a catalytic triad of serine, aspartate, and histidine residues; a “lid” domain that covers the active site and contributes to the specificity for triglyceride and phosphoglyceride substrates; and a C-terminal or “plat” domain, which contributes to lipid binding and specificity.31,32
This paper examines and reviews the gene structures and amino acid sequences for several vertebrate LIPC genes and proteins; the predicted secondary and tertiary structures for vertebrate HL enzymes; and the structural, phylogenetic, and evolutionary relationships for these genes and enzymes with those for human and mouse lipase neutral lipase gene families, LIPG (encoding endothelial lipase), and LPL (encoding lipoprotein lipase).
Methods
Vertebrate LIPC gene and HL identification
Protein BLAST (Basic Local Alignment Search Tool) analyses generated several vertebrate HL amino acid sequences from the National Center for Biotechnology Information (NCBI) database (http://blast.ncbi.nlm.nih.gov/Blast.cgi)33 (Table 1). Nonredundant protein sequence databases for vertebrate genomes were examined using the blastp algorithm, including human (Homo sapiens),34 chimpanzee (Pan troglodytes),35 orangutan (Pongo abelii) (http://genome.wustl.edu), rhesus monkey (Macaca mulatta),36 cow (Bos taurus) (http://hgsc.bcm.tmc.edu/projects/bovine), mouse (Mus musculus),37 rat (Rattus norvegicus),38 rabbit (Oryctolagus cuniculus) (http://www.broadinstitute.org/science/projects/mammals-models/rabbit/rabbit-genome-sequencing-project), opossum (Monodelphis domestica),39 chicken (Gallus gallus),40 frog (Xenopus tropicalis) (http://genome.jgi-psf.org/Xentr3/Xentr3.home.html), and zebrafish (Danio rerio) (http://www.sanger.ac.uk/Projects/D_rerio/). Predicted or previously reported vertebrate HL-like protein sequences were then subjected to analyses of protein and gene structures (Table 1).
Table 1.
Vertebrate hepatic lipase (LIPC), human and mouse endothelial lipase (LIPG) and lipoprotein lipases (LPL), horse pancreatic lipase (LIPP), and sea squirt lipase genes and proteins
| Hepatic lipase Gene LIPC | Species | RefSeq ID 1 Ensembl (predicted) |
GenBank ID | UNIPROT ID | Amino acids | Chromosome location | Exons (strand) | Gene size bps | pI | Subunit MW | Signal peptide (cleavage site) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | Homo sapiens | NM_000236.2 | BC146659 | P11150 | 499 | 15:56,511,524–56,648,315 | 9 (+ve) | 136,792 | 9.22 | 55,914 | 1–24 [LG-QS] |
| Chimpanzee | Pan troglodytes | 1 XP_001172241.1 | 2 | 2 | 499 | 15:55,899,209–56,038,386 | 9 (+ve) | 139,178 | 9.27 | 56,024 | 1–24 [LG-QS] |
| Orangutan | Pongo abelii | 4 | 2 | 2 | 499 | 15:55,440,202–55,470,553 | 9 (+ve) | 430,352 | 49.16 | 456,012 | 4 |
| Rhesus | Macaca mulatta | 1 XP_001095252.1 | 2 | 2 | 499 | 7:36,761,876–36,903,612 | 9 (+ve) | 141,737 | 9.36 | 56,023 | 1–24 [HG-QS] |
| Mouse | Mus musculus | NM_008280.2 | BC021841 | P27656 | 510 | 9:70,645,935–70,782,615 | 9 (−ve) | 136,681 | 8.34 | 57,389 | 1–22 [AC-GQ] |
| Rat | Rattus norvegicus | NM_012597 | BC088160 | P07867 | 494 | 8:75,323,443–75,450,353 | 9 (−ve) | 126,911 | 8.49 | 55,752 | 1–22 [AC-GQ] |
| Rabbit | Oryctolagus cuniculus | NM_001082032.1 | AF041202 | 2 | 499 | 17:13,811,866–13,970,782 | 9 (−ve) | 158,863 | 9.08 | 55,857 | 1–23 [HG-QS] |
| Cow | Bos taurus | NM_001035410.1 | BC103072 | Q3SZ79 | 500 | 10:52,220,965–52,415,726 | 2,3 | 194,762 | 9.09 | 56,826 | 1–23 [HG-QS] |
| Dog | Canis familaris | 1 XP_535495.2 | 2 | 2 | 502 | 30:26,546,842–26,574,268 | 9 (+ve) | 427,427 | 8.53 | 56,594 | 1–24 [VG-SP] |
| Opossum | Monodelphis domestica | 1 XP_001377665.1 | 2 | 2 | 4460 | 41:162,290,356–162,337,323 | 48 (−ve) | 446,968 | 4 | 4 | 4 |
| Chicken | Gallus gallus | 1 XP_425067.2 | 2 | 2 | 4474 | 410:7,955,298–7,967,646 | 48 (−ve) | 412,349 | 4 | 4 | 4 |
| Frog | Xenopus tropicalis | NM_001114259.1 | BC158363 | B0BMB8 | 496 | 5 sc301:941,950–1,004,887 | 9 (−ve) | 62,938 | 8.47 | 56,687 | 1–21 [LT-QK] |
| Zebrafish | Danio rerio | NM_201022.1 | BC053243 | Q7T359 | 514 | 7:33,180,131–33,193,766 | 9 (−ve) | 13,636 | 8.50 | 57,933 | 1–20 [DG-AT] |
| Other lipase gene | |||||||||||
| Horse LIPP | Equus caballus | NM_001163949 | X66218 | P29183 | 465 | 1:15,534,773–15,551,621 | 12 (−ve) | 16,849 | 5.46 | 54,435 | 1–16 [VG-NE] |
| Human LIPG | Homo sapiens | NM_006033.2 | BC060825 | Q9Y5X9 | 500 | 18:45,342,677–45,367,216 | 10 (+ve) | 24,540 | 8.1 | 56,795 | 1–20 [AG-SP] |
| Mouse LIPG | Mus musculus | NM_010720.3 | BC020991 | Q9WVG5 | 500 | 18:75,102,996–75,120,628 | 10 (−ve) | 17,633 | 8.79 | 56,629 | 1–20 [AG-SI] |
| Human LIPL | Homo sapiens | NM_000237.2 | BC011353 | P06858 | 475 | 8:19,841,232–19,864,008 | 9 (+ve) | 22,777 | 8.4 | 53,163 | 1–27 [AA-AD] |
| Mouse LIPL | Mus musculus | NM_008509.2 | BC003305 | P11152 | 474 | 8:71,404,652–71,426,282 | 9 (+ve) | 21,631 | 8.0 | 53,109 | 1–27 [AA-AD] |
| Sea squirt LIP | Ciona intestinalis | 1 ENSCINT00000009034 | 2 | 2 | 460 | 07q:1,148,429–1,153,888 | 11 (−ve) | 5460 | 4.57 | 51,184 | 1–18 [NC-DT] |
Notes:
Predicted Ensembl amino acid sequence;
not available;
exon 1 missing;
incomplete sequence available;
scaffold of DNA used in sequencing frog genome. GenBank IDs are derived from NCBI sources http://www.ncbi.nlm.nih.gov/genbank/; Ensembl ID was derived from Ensembl genome database http://www.ensembl.org; UNIPROT refers to UniprotKB/Swiss-Prot IDs for individual acid lipases (http://kr.expasy.org); bps refers to base pairs of nucleotide sequences; pI refers to theoretical isoelectric points; the number of coding exons is listed.
Abbreviation: RefSeq, the reference amino acid sequence.
BLAT (BLAST-like Alignment Tool) analyses were subsequently undertaken for each of the predicted HL amino acid sequences using the University of California, Santa Cruz genome browser (http://genome.ucsc.edu/cgi-bin/hgBlat)41 with the default settings to obtain the predicted locations for each of the mammalian LIPC genes, including predicted exon boundary locations and gene sizes. BLAT analyses were also undertaken for human LPL (encoding lipoprotein lipase)9 and LIPG (encoding endothelial lipase)6–8 (see Table 1). Structures for human, mouse, and rat isoforms (splicing variants) were obtained using the AceView website to examine predicted gene and protein structures42 (http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/index.html?human).
Predicted structures and properties of vertebrate hepatic lipases
Secondary and tertiary structures for human and other vertebrate HL-like proteins were predicted using Web tools from PSIPRED v2.5 (http://bioinf.cs.ucl.ac.uk/psipred/) and SWISS MODEL (http://swissmodel.expasy.org), respectively.43,44 The structure for the human pancreatic lipase–colipase complex45 served as a reference for the predicted horse LIPP (pancreatic lipase) structure (previously reported by Bourne et al30) and the human, opossum and zebrafish LIPC tertiary structures, with modeling ranges of residues 18–465, 25–471, 4–448 and 25–485, respectively. Theoretical isoelectric points and molecular weights (http://au.expasy.org/tools/pi_tool.html), location of signal peptide cleavage sites (http://www.cbs.dtu.dk/services/SignalP/),46 and potential N-glycosylation sites (http://www.cbs.dtu.dk/services/NetNGlyc/) for vertebrate LIPC proteins were obtained using Web tools.
Phylogenetic studies and sequence divergence
Alignments of vertebrate HL with human and mouse EL and LPL sequences were assembled using BioEdit v.5.0.1 and the default settings.47 Alignment ambiguous regions, including the amino and carboxyl termini, were excluded prior to phylogenetic analysis, yielding alignments of 395 residues for comparisons of vertebrate HL, human and mouse EL, and LPL sequences with the sea squirt (Ciona intestinalis) lipase sequence (Table 1). Evolutionary distances were calculated using the Kimura option48 in TREECON.49 Phylogenetic trees were constructed from evolutionary distances using the neighbor-joining method50 and rooted with the sea squirt lipase sequence. Tree topology was re-examined by the bootstrap method (100 bootstraps were applied) of resampling and only values that were highly significant (≥90) are shown.51
Results and discussion
Alignments of vertebrate HL amino acid sequences
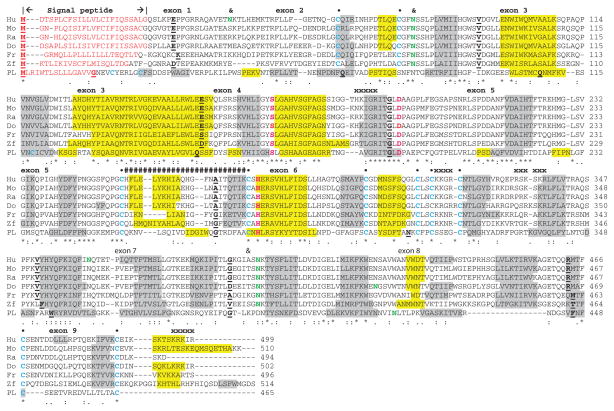
The deduced amino acid sequences for dog, frog, and zebrafish HL are shown in Figure 1 together with previously reported sequences for human HL,3,22 mouse HL,52 rat HL,53,54 and horse pancreatic lipase (LIPP)30 (Table 1). Alignments of human and other vertebrate HL sequences examined showed between 49% and 98% identities, suggesting that they are products of the same family of genes, whereas comparisons of sequence identities of vertebrate HL proteins with human and mouse EL and LPL and horse LIPP exhibited lower levels of sequence identities, EL (38% and 42%, respectively), LPL (44% and 45%, respectively), and LIPP (25%), indicating that they are members of distinct but related neutral lipase families (Table 2).
Figure 1.
Amino acid sequence alignments for vertebrate hepatic lipase (HL) and horse pancreatic lipase (LIPP) sequences. See Table 1 for sources of HL and horse LIPP sequences.
Notes: *shows identical residues for lipase subunits; similar alternative residues; dissimilar alternative residues; residues involved in N-signal peptide formation are shown in red; N-glycosylated (marked as and for human HL) and potential N-glycosylated Asn sites are in green bold; active site triad residues Ser (S), Asp (D), and His (H) are in pink bold; predicted disulfide bond Cys residues are shown in blue bold (●); α-helix for horse LIPP or predicted for vertebrate HL is in shaded yellow; β-sheet for horse LIPP or predicted for vertebrate HL is in shaded grey; bold underlined font shows residues corresponding to known or predicted exon start sites; exon numbers refer to human LIPC gene exons; #### refers to residues that correspond to the horse LIPP “lid” region; xxxxx refers to the four predicted “heparin binding” regions for human HL.
Abbreviations: Do, dog HL; Fr, frog HL; Hu, human HL; Mo, mouse HL; PL, horse pancreatic lipase; Ra, rat HL; Zf, zebrafish HL.
Table 2.
Percentage identities for vertebrate hepatic lipases, human and mouse endothelial and lipoprotein lipases, horse pancreatic lipase, and sea squirt lipase amino acid sequencesa
| Lipase gene | Human HL | Chimp HL | Rhesus HL | Mouse HL | Rat HL | Rabbit HL | Cow HL | Opossum HL | Chicken HL | Frog HL | Zebrafish HL | Human EL | Mouse EL | Human LPL | Mouse LPL | Horse LIPP | Sea squirt LIP |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Human HL | 100 | 98 | 94 | 74 | 74 | 79 | 77 | 66 | 64 | 57 | 49 | 38 | 37 | 41 | 42 | 25 | 21 |
| Chimp HL | 98 | 100 | 95 | 75 | 75 | 80 | 78 | 66 | 65 | 57 | 50 | 39 | 37 | 41 | 42 | 25 | 21 |
| Rhesus HL | 94 | 95 | 100 | 74 | 75 | 80 | 77 | 66 | 64 | 56 | 49 | 40 | 38 | 42 | 43 | 24 | 21 |
| Mouse HL | 74 | 75 | 74 | 100 | 89 | 75 | 70 | 64 | 61 | 56 | 50 | 42 | 40 | 44 | 43 | 29 | 25 |
| Rat HL | 74 | 75 | 75 | 89 | 100 | 74 | 68 | 64 | 60 | 54 | 51 | 41 | 39 | 43 | 42 | 28 | 25 |
| Rabbit HL | 79 | 80 | 80 | 75 | 74 | 100 | 76 | 68 | 64 | 56 | 50 | 39 | 38 | 41 | 40 | 25 | 21 |
| Cow HL | 77 | 78 | 77 | 70 | 68 | 76 | 100 | 63 | 62 | 54 | 49 | 40 | 37 | 40 | 40 | 23 | 23 |
| Opossum HL | 66 | 66 | 66 | 64 | 64 | 68 | 63 | 100 | 68 | 63 | 56 | 43 | 42 | 43 | 42 | 27 | 23 |
| Chicken HL | 64 | 65 | 64 | 61 | 60 | 64 | 62 | 68 | 100 | 70 | 57 | 44 | 43 | 44 | 44 | 24 | 23 |
| Frog HL | 57 | 57 | 56 | 56 | 54 | 56 | 54 | 63 | 70 | 100 | 55 | 41 | 41 | 45 | 42 | 24 | 24 |
| Zebrafish HL | 49 | 50 | 49 | 50 | 51 | 50 | 49 | 56 | 57 | 55 | 100 | 40 | 42 | 42 | 41 | 23 | 23 |
| Human EL | 38 | 39 | 40 | 42 | 41 | 39 | 40 | 43 | 44 | 41 | 40 | 100 | 80 | 44 | 45 | 25 | 25 |
| Mouse EL | 37 | 37 | 38 | 40 | 39 | 38 | 37 | 42 | 43 | 41 | 42 | 80 | 100 | 45 | 46 | 25 | 25 |
| Human LPL | 41 | 41 | 42 | 44 | 43 | 41 | 40 | 43 | 44 | 45 | 42 | 44 | 45 | 100 | 93 | 24 | 25 |
| Mouse LPL | 42 | 42 | 43 | 43 | 42 | 40 | 40 | 42 | 44 | 42 | 41 | 45 | 46 | 93 | 100 | 26 | 25 |
| Horse LIPP | 25 | 25 | 24 | 29 | 28 | 25 | 23 | 27 | 24 | 24 | 23 | 25 | 25 | 24 | 26 | 100 | 35 |
| Sea squirt LIP | 21 | 21 | 21 | 25 | 25 | 21 | 23 | 23 | 23 | 24 | 23 | 25 | 25 | 25 | 25 | 35 | 100 |
Note:
Numbers show the percentage of amino acid sequence identities.
Abbreviations: EL, endothelial lipase; HL, hepatic lipase; LIP, sea squirt lipase; LIPP, pancreatic lipase; LPL, lipoprotein lipase.
The amino acid sequences for human, chimp, orangutan, rhesus monkey, and rabbit HL contained 499 residues whereas mouse, rat, cow, dog, and frog HL contained 510, 494, 500, 502, and 496 amino acids, respectively (Table 1; Figure 1). Previous three-dimensional studies of horse pancreatic lipase (LIPP)30 and modeling studies of human EL29 have enabled predictions of key residues for vertebrate HL amino acid sequences (numbers refer to human HL). These included the catalytic triad for the active site (Ser168, Asp194, and His279); the hydrophobic N-terminus signal peptides (see also Table 1), which facilitate enzyme secretion into the circulation system;25 five disulfide bond-forming residues (Cys62/Cys75, Cys254/Cys277, Cys302/Cys313, Cys316/Cys321, and Cys467/Cys487); the predicted “lid” region (255–276), which covers the active site and participates in lipid substrate binding in analogous lipases;31,32 and a predicted “hinge” region for vertebrate HL (332Arg-333Ser-334Lys-335Ser) (based on sequence similarity with human EL [327Arg-328Asn-329Lys-330Arg], which contains a proprotein convertase proteolytic cleavage site).29–32 With the exception of the N-terminus signal peptides, the vertebrate HL sequences were strictly conserved or underwent conservative substitutions, which may reflect the essential nature of these residues in contributing to HL structure and function. The N-terminal region (residues 1–63) underwent major changes in the number and sequence of amino acid residues but retained a predicted signal peptide property in each case (Figure 1; Table 1). The horse LIPP sequence shared the catalytic triad residues, four of the five disulfide bonds predicted for the vertebrate HL sequences, and an N-signal peptide sequence property; however, other sequences were distinct with only 25% identical residues observed for horse LIPP and human HL.
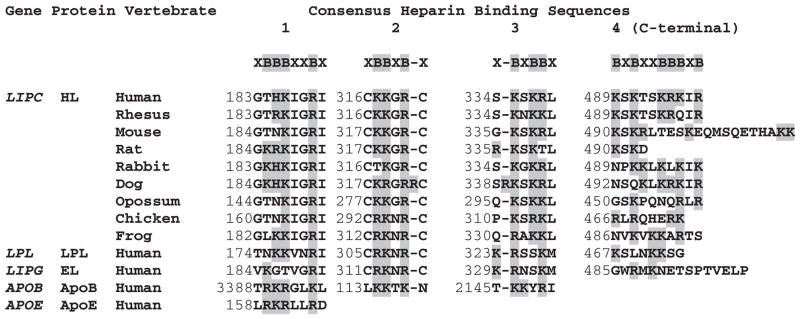
Figure 2 compares vertebrate HL sequences for four putative heparin binding sites described for human HL, which contain clusters of basic amino acid residues with different consensus sequences.56–59 These sites are apparently responsible for HL binding to heparin sulfate proteoglycans on the surface of parenchymal cell microvilli where the enzyme functions in liver lipoprotein catabolism.58,59 Several vertebrate HL sequences have also been compared with human LPL and EL putative heparin binding sites, as well as those for human apolipoproteins APOB and APOE, the major proteins of chylomicrons, low-density lipoprotein, and very low-density lipoprotein, which function as recognition signals for the cellular binding and internalization of low-density lipoprotein particles.60 Several differences from heparin binding consensus sequences were observed. For consensus sequence 1 (XBBBXXBX where B refers to a basic amino acid and X to any other amino acid), human, rhesus, and mouse HL and human LPL sequences lacked the first basic amino acid, and human EL contained only two of four basic amino acids. Consensus sequence 2 (XBBXBX) showed consistency for all vertebrate HL, human LPL and EL, and APOB sequences examined with the exception of rabbit HL, which lacked one of the B residues, and dog HL, which contained an extra B residue. Several differences were observed for consensus sequence 3 (XBXBBX), including rat and dog HL and human EL and LPL. The C-terminal heparin binding site (consensus sequence 4) showed major differences among the HL sequences examined, especially for mouse and rat HL, which lacked this motif. This may explain why mouse HL is predominantly found in the circulation system as compared with human HL, which is released into the circulation following heparin administration.61
Figure 2.
Comparative amino acid sequences for predicted heparin binding sequences for vertebrate hepatic lipase (HL) and human lipoprotein lipase (LPL), endothelial lipase (EL), apolipoprotein B (APOB), and apolipoprotein E (APOE) sequences. Four predicted heparin binding sites are shown based on previous studies56–60 and the predicted vertebrate HL sequences reported in this paper.
Abbreviations: B, basic amino acid; K, lysine; R, arginine; X, any other amino acid residue.
Four N-glycosylation sites have previously been reported for human HL at 42Asn-43Lys-44Thr, 78Asn-70Ser-71Ser, 362Asn-363Gln-364Thr, and 397Asn-398Lys-399Thr.62,63 A comparative analysis of potential N-glycosylation sites for vertebrate HL has shown that there are seven sites overall, although only two of these have been predominantly retained for the 13 vertebrate HL sequences examined (designated as sites 3 and 7) (Table 3). Site-directed mutagenesis studies of site 3 (human HL 78Asn) have demonstrated that this N-glycosylation site is required for the efficient secretion of this liver enzyme.64,65
Table 3.
Predicted N-glycosylation sites for vertebrate hepatic lipases. Numbers refer to amino acids in the acid sequences, including N-asparagine, K-lysine, I-isoleucine, M-methionine, H-histidine, S-serine, R-arginine, T-threonine, Q-glutamine, and V-valine. Note that seven potential sites were identified, including four confirmed sites for human LIPC (HL) (sites 1, 3, 5, and 7). High- (yellow highlighted) and lower probability N-glycosylation sites were identified using the NetNGlyc 1.0 Web server (http://www.cbs.dtu.dk/services/NetNGlyc/)
| Vertebrate | Species | Site 1 | Site 2 | Site 3 | Site 4 | Site 5 | Site 6 | Site 7 | No. of sites |
|---|---|---|---|---|---|---|---|---|---|
| Human | Homo sapiens | 42NKT | 78NSS | 362NQT | 397NKT | 4 | |||
| Chimpanzee | Pan troglodytes | 42NKT | 78NSS | 362NQT | 397NKT | 4 | |||
| Orangutan | Pongo abelii | 42NKT | 78NSS | 362NQT | 3 | ||||
| Rhesus | Macaca mulatta | 42NKT | 78NSS | 362NQT | 397NKT | 4 | |||
| Mouse | Mus musculus | 79NSS | 398NKT | 2 | |||||
| Rat | Rattus norvegicus | 79NSS | 398NKT | 2 | |||||
| Rabbit | Oryctolagus cuniculus | 78NSS | 397NKT | 2 | |||||
| Cow | Bos taurus | 67NHS | 78NSS | 363NQT | 398NET | 4 | |||
| Dog | Canis familaris | 79NSS | 400NKT | 2 | |||||
| Opossum | Monodelphis domestica | 39NSS | 143NGT | 358NKT | 378NFT | 4 | |||
| Chicken | Gallus gallus | 55NAS | 374NKT | 2 | |||||
| Frog | Xenopus tropicalis | 78NES | 394NKT | 2 | |||||
| Zebrafish | Danio rerio | 75NSS | 395NKT | 2 |
Predicted secondary and tertiary structures for vertebrate hepatic lipases
Predicted secondary structures for vertebrate HL sequences were compared with the previously reported secondary structure for horse LIPP (pancreatic lipase)30 (Figure 1). α-Helix and β-sheet structures for the vertebrate HL protein sequences were examined and found to be similar for several regions with the horse LIPP secondary structures. Consistent structures were predicted near key residues or functional domains, including the β-sheet and α-helix structures near the active site residues (human HL numbers used) Ser168, Asp 194, and His279; the “lid” domain (residues 255–276); and the “hinge” region, which commences with an α-helix and concludes with a β-sheet (residues 333–339). Figure 3 describes predicted tertiary structures for human, opossum, and zebrafish HL protein sequences and shows significant similarities for these polypeptides with horse pancreatic lipase (LIPP).30 The three LIPP and HL domains were readily apparent, including the N-terminal “lipase” domain with the active site triad residues buried under the “lid” domain observed for horse LIPP. The “lid” has previously been shown to contribute to the preference for triglyceride and phopholipid substrates of vascular lipases HL and LPL.31,65 A “hinge” region was also observed for these vertebrate HL proteins, separating the “lipase” and “plat” domains, with the latter having a “sandwich-like” β-pleated sheet structure. The “plat” domain for HL and LPL has been shown to be essential for binding these enzymes to lipoprotein micelles and also contributes to preferences in lipoprotein binding.29 These comparative studies for other vertebrate HL proteins suggest that these properties and key sequences are substantially retained for all of the vertebrate sequences examined.
Figure 3.
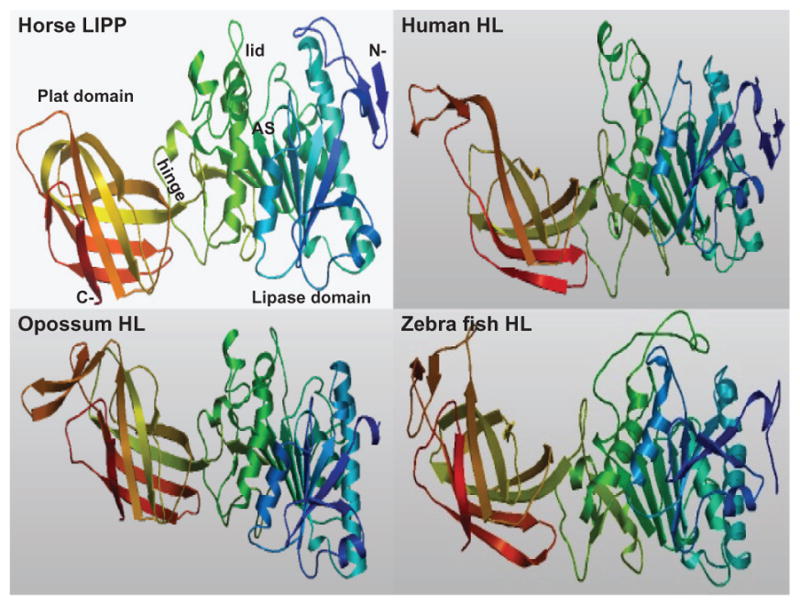
Predicted Tertiary Structures for Horse LIPP and for Human, Opossum and Zebrafish LIPC Predicted horse LIPP and human, opossum and zebrafish LIPC tertiary structures were obtained using SWISS MODEL methods; the rainbow color code describes the tertiary structures from the N- (blue) to C-termini (red color); the horse LIPP tertiary structure shows the N- and C-termini, the ‘lipase’, ‘lid’ (in yellow) and ‘plat’ domains which are separated by a ‘hinge’ region; and the active site region (AS) for horse LIPP is identified ( based on the horse LIPP structure reported by Bourne et al.30).
Predicted gene locations and exonic structures for vertebrate LIPC genes
Table 1 summarizes the predicted locations for vertebrate LIPC genes based on BLAT interrogations of several vertebrate genomes using the reported sequences for human,6,7 mouse,66 and rat HL38 and the University of California, Santa Cruz genome browser.41 The predicted primate LIPC genes were transcribed on the positive strand, whereas other vertebrate LIPC genes were transcribed on the negative strand. Figure 1 summarizes the predicted exonic start sites for vertebrate LIPC genes with each having nine coding exons, in identical or similar positions to those predicted for the human LIPC gene.67
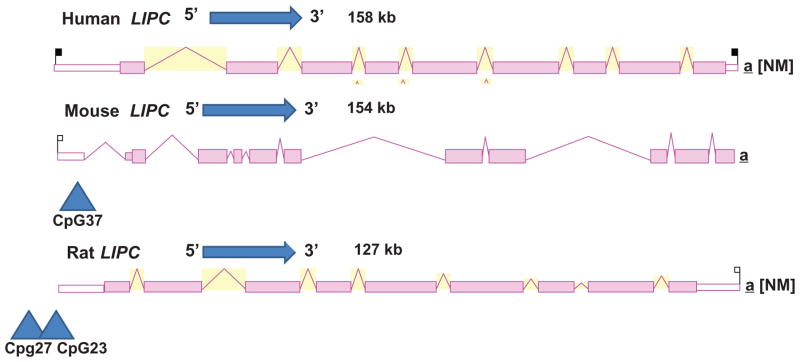
Figure 4 illustrates the predicted structures of mRNA for human, mouse, and rat LIPC transcripts for the major transcript isoform in each case.42 The transcripts were 127–158 kbs in length with nine introns present for these LIPC mRNA transcripts. Figure 5 examines the predicted amino acid and nucleotide sequence for the C-terminus end of exon 9 human, mouse, and rat LIPC sequences. It is proposed that exon 9 has undergone nucleotide substitutions or deletions/insertions that have introduced a termination codon for the rat LIPC gene encoding an incomplete C-terminus for rat HL and an extended C-terminus for mouse HL. The significance of these differences in rodent LIPC structure has been previously discussed in terms of the observed changes in HL binding to heparin sulfate proteoglycans on liver parenchymal cells where the enzyme functions in liver lipoprotein catabolism.57–59
Figure 4.
Gene structures and major splicing variant for the human, mouse, and rat LIPC transcripts. Derived from the AceView website42 http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/. Mature isoform variants (a) are shown with capped 5″- and 3″-ends for the predicted mRNA sequences; NM refers to the NCBI reference sequence; exons are in pink; the directions for transcription are shown as 5″ → 3″; blue triangles show predicted CpG island sites at or near the 5″ untranslated regions of the gene; sizes of mRNA sequences are shown in kilobases (kb).
Figure 5.
Nucleotide and amino acid sequence alignments for human, mouse, and rat LIPC genes and hepatic lipase proteins: predicted c-termini and exon 9 sequences. Identical nucleotide (nt) and amino acid (aa) sequences are shown (*). Ter (in red) refers to predicted terminating codons.
Phylogeny and divergence of hepatic lipase and other vertebrate lipase sequences
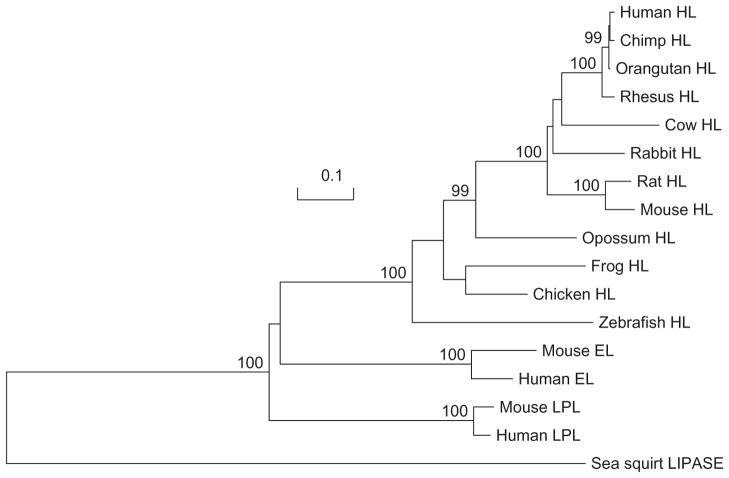
A phylogenetic tree (Figure 6) was calculated by the progressive alignment of 12 vertebrate LIPC amino acid sequences with human and mouse LIPL and LIPG sequences that was “rooted” with the Ciona intestinalis (sea squirt) lipase sequence (see Table 1). The phylogram showed clustering of the LIPC sequences into groups that were consistent with their evolutionary relatedness as well as distinct groups for human and mouse LIPL and LIPG sequences, which were distinct from the sea squirt lipase sequence. These groups were significantly different from each other (with bootstrap values of ~100/100). It is apparent from this study of vertebrate LIPC genes and proteins that this is an ancient protein for which a proposed common ancestor for the LIPC, LIPG, and LIPL neutral lipase genes may have predated the appearance of bony fish, which occurred >500 million years ago.68 This proposal is consistent with a previous report from Cohen,69 which described predicted amino acid sequences for human and pufferfish (Takifugu rubripes) LIPG, LIPL, and LIPC.
Figure 6.
Phylogenetic tree of vertebrate hepatic lipase (HL), human and mouse lipoprotein lipase (LPL) and endothelial lipase (EL), and sea squirt lipase amino acid sequences. The tree is labeled with the lipase name and the name of the animal and is “rooted” with the Ciona intestinalis (sea squirt) lipase sequence. Note the major cluster of vertebrate HL sequences, which is distinct from the human and mouse LPL and EL and the sea squirt lipase sequences. A genetic distance scale is shown. The number of times a clade (sequences common to a node or branch) occurred in the bootstrap replicates are shown. Only replicate values of 90 or more that are highly significant are shown, with 100 bootstrap replicates performed in each case. Note the significant separation of clades for the three human vascular lipases (LPL, EL, and HL).
Conclusion
These results indicate that vertebrate LIPC genes and encoded HL enzymes represent a distinct gene and enzyme family of neutral lipases that share key conserved sequences that have been reported for other neutral lipases previously studied.6–11 This enzyme has a distinct property among the neutral lipases studied in being the major liver lipase and playing a major role in the catabolism of lipoproteins in the circulation system.1–3 HL is encoded by a single gene for the vertebrate genomes studied and usually contains nine coding exons. The rat LIPC gene encoded a shorter form of this enzyme (494 residues compared with 499 amino acids for most mammalian HL sequences) due to the presence of a termination codon located in exon 9. Predicted secondary structures and tertiary structures for vertebrate HL proteins showed a strong similarity with human and horse pancreatic lipases (LIPP).29,30 Three major structural domains were apparent for vertebrate HL, including the “lipase” domain containing the catalytic triad residues; the “lid”, which covers the active site and may contribute to the substrate specificities of neutral lipases;31,64 and the “plat” domain, which contributes to lipoprotein binding.65 Phylogenetic studies using amino acid sequences for 13 vertebrate HL lipases, human and mouse LPL and EL, and an invertebrate lipase indicated that the LIPC gene has appeared early in vertebrate evolution, probably prior to the appearance of bony fish more that 500 million years ago.
Acknowledgments
This project was supported by National Institutes of Health Grants P01 HL028972 and P51 RR013986. In addition, this investigation was conducted in facilities constructed with support from Research Facilities Improvement Program Grant Numbers 1 C06 RR13556, 1 C06 RR15456, and 1 C06 RR017515. We gratefully acknowledge Dr Bharet Patel from Griffith University for his assistance with the phylogenetic analyses.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Martin GA, Busch SJ, Meredith GD, et al. Isolation and cDNA sequence of human postheparin plasma hepatic triglyceride lipase. J Biol Chem. 1988;263:10907–10914. [PubMed] [Google Scholar]
- 2.Datta S, Luo CC, Li WH, et al. Human hepatic lipase. Cloned cDNA sequence, restriction fragment length polymorphisms, chromosomal localization, and evolutionary relationships with lipoprotein lipase and pancreatic lipase. J Biol Chem. 1988;263:1107–1110. [PubMed] [Google Scholar]
- 3.Cai SJ, Wong DM, Chen SH, Chan L. Structure of the human hepatic triglyceride lipase gene. Biochem. 1989;28:8966–8971. doi: 10.1021/bi00449a002. [DOI] [PubMed] [Google Scholar]
- 4.Zambon A, Austin MA, Brown BG, et al. Effect of hepatic lipase on LDL in normal men and those with coronary artery disease. Arterioscler Thromb. 1993;13:147–153. doi: 10.1161/01.atv.13.2.147. [DOI] [PubMed] [Google Scholar]
- 5.Santamarina-Fojo S, Haudenschild C, Amar M. The role of hepatic lipase in lipoprotein metabolism and atherosclerosis. Curr Opin Lipidol. 1998;9:211–219. doi: 10.1097/00041433-199806000-00005. [DOI] [PubMed] [Google Scholar]
- 6.Jaye M, Lynch KJ, Krawiec J, Marchadier D, et al. A novel endothelial-derived lipase that modulates HDL metabolism. Nature Genetics. 1999;21:424–428. doi: 10.1038/7766. [DOI] [PubMed] [Google Scholar]
- 7.Hirata K, Dichek HL, Cioffi JA, et al. Cloning of a unique lipase from endothelial cells extends the lipase gene family. J Biol Chem. 1999;274:14170–14175. doi: 10.1074/jbc.274.20.14170. [DOI] [PubMed] [Google Scholar]
- 8.Holmes RS, VandeBerg JL, Cox LA. Vertebrate endothelial lipase: comparative studies of an ancient gene and protein in vertebrate evolution. Genetica. 2011 doi: 10.1007/s10709-011-9549-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wion KL, Kirchgessner TG, Lusis AJ, et al. Human lipoprotein lipase complementary DNA sequence. Science. 1987;235:1638–1641. doi: 10.1126/science.3823907. [DOI] [PubMed] [Google Scholar]
- 10.Dichek HL, Fojo SS, Beg OU, et al. Identification of two separate allelic mutations in the lipoprotein lipase gene of a patient with the familial hyperchylomicronemia syndrome. J Biol Chem. 1991;266:473–477. [PubMed] [Google Scholar]
- 11.Benlian P, De Gennes JL, Foubert L, et al. Premature atherosclerosis in patients with familial chylomicronemia caused by mutations in the lipoprotein lipase gene. New Eng J Med. 1996;335:848–854. doi: 10.1056/NEJM199609193351203. [DOI] [PubMed] [Google Scholar]
- 12.Ma K, Cilingiroglu M, Otvos JD, et al. Endothelial lipase is a major genetic determinant for high-density lipoprotein concentration, structure and metabolism. Proc Natl Acad Sci U S A. 2003;100:2748–2753. doi: 10.1073/pnas.0438039100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown RJ, Rader DJ. Lipases as modulators of atherosclerosis in murine models. Curr Drug Targets. 2007;8:1307–1319. doi: 10.2174/138945007783220614. [DOI] [PubMed] [Google Scholar]
- 14.Guerra RJ, Wang S, Grundy M, Cohen JC. A hepatic lipase (LIPC) allele associated with high plasma concentrations of high density lipoprotein cholesterol. Proc Natl Acad Sci U S A. 1997;94:4532–4537. doi: 10.1073/pnas.94.9.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murtomaki S, Tahvanainen E, Antikainen M, et al. Hepatic lipase gene polymorphisms influence plasma HDL levels. Results from Finnish EARS: participants of the European Atherosclerosis Research study. Arterioscler Thromb Vasc Biol. 1997;17:1877–1879. doi: 10.1161/01.atv.17.10.1879. [DOI] [PubMed] [Google Scholar]
- 16.de Andrade FM, Silveira FR, Arsand M, et al. Association between −250G/A polymorphism of the hepatic lipase gene promoter and coronary artery disease and HDL-C levels in a southern Brazilian population. Clin Genet. 2004;65:390–395. doi: 10.1111/j.0009-9163.2004.00243.x. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez-Navarro H, Nong Z, Amar MJA, et al. The ligand-binding function of hepatic lipase modulates the development of atherosclerosis in transgenic mice. J Biol Chem. 2004;279:45312–45321. doi: 10.1074/jbc.M406495200. [DOI] [PubMed] [Google Scholar]
- 18.Eller P, Schgoer W, Mueller T, et al. Hepatic lipase polymorphism and increased risk of peripheral arterial disease. J Intern Med. 2005;258:344–348. doi: 10.1111/j.1365-2796.2005.01549.x. [DOI] [PubMed] [Google Scholar]
- 19.Chasman DI, Paré G, Mora S, et al. Forty-three loci associated with plasma lipoprotein size, concentration, and cholesterol content in genome-wide analysis. PLoS Genet. 2009;5:e1000730. doi: 10.1371/journal.pgen.1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hodoglugil U, Williamson DW, Mahley RW. Polymorphisms in the hepatic lipase gene affect plasma HDL-cholesterol levels in a Turkish population. J Lipid Res. 2010;51:422–430. doi: 10.1194/jlr.P001578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baylin A, Ruiz-Narvaez E, Jensen MK, et al. Association between hepatic lipase-514 C/T promoter polymorphism and myocardial infarction is modified by history of hypercholesterolemia and waist circumference. Nutr Metab Cardiovasc Dis. 2010;20:498–504. doi: 10.1016/j.numecd.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ameis D, Stahnke G, Kobayashi J, et al. Isolation and characterization of the human hepatic lipase gene. J Biol Chem. 1990;265:6552–6555. [PubMed] [Google Scholar]
- 23.Hegele RA, Tu L, Connelly PW. Human hepatic lipase mutations and polymorphisms. Hum Mutat. 1992;1:320–321. doi: 10.1002/humu.1380010410. [DOI] [PubMed] [Google Scholar]
- 24.Meng L, Ruixing Y, Yiyang L, et al. Association of LIPC −250G >A polymorphism and several environmental factors with serum lipid levels in the Guangxi Bai Ku Yao and Han populations. Lipids Health Dis. 2010;9:28. doi: 10.1186/1476-511X-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cisar LA, Bensadoun A. Characterization of the intracellular processing and secretion of hepatic lipase in FU5AH rat hepatoma cells. Biochim Biophys Acta. 1987;927:305–314. doi: 10.1016/0167-4889(87)90094-2. [DOI] [PubMed] [Google Scholar]
- 26.Hill JS, Davis RC, Yang D, et al. Human hepatic lipase subunit structure determination. J Biol Chem. 1996;271:22931–22936. doi: 10.1074/jbc.271.37.22931. [DOI] [PubMed] [Google Scholar]
- 27.Griffon N, Jin W, Petty TJ, et al. Identification of the active form of endothelial lipase, a homodimer in a head-to-tail conformation. J Biol Chem. 2009;284:23322–23330. doi: 10.1074/jbc.M109.037002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin W, Fuki IV, Seidah NG, et al. Proprotein convertases are responsible for proteolysis and inactivation of endothelial lipase. J Biol Chem. 2005;280:36551–36559. doi: 10.1074/jbc.M502264200. [DOI] [PubMed] [Google Scholar]
- 29.Winkler FK, D’Arcy A, Hunziker W. Structure of human pancreatic lipase. Nature. 1990;343:771–774. doi: 10.1038/343771a0. [DOI] [PubMed] [Google Scholar]
- 30.Bourne Y, Martinez C, Kerfelec B, et al. Horse pancreatic lipase. The crystal structure refined at 2.3-A resolution. J Mol Biol. 1994;238:709–732. doi: 10.1006/jmbi.1994.1331. [DOI] [PubMed] [Google Scholar]
- 31.Dugi KA, Dichek HL, Santamarina-Fojo S. Human hepatic and lipoprotein lipase: the loop covering the catalytic site mediates lipase substrate specificity. J Biol Chem. 1995;270:25396–25401. doi: 10.1074/jbc.270.43.25396. [DOI] [PubMed] [Google Scholar]
- 32.Broedl UC, Jin W, Fuki IV, Glick JM, Rader DJ. Structural basis for endothelial lipase tropism for HDL. FASEB J. 2004;18:1891–1893. doi: 10.1096/fj.03-1307fje. [DOI] [PubMed] [Google Scholar]
- 33.Altschul F, Vyas V, Cornfield A, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 34.International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 35.Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 36.Rhesus Macaque Genome Sequencing and Analysis Consortium. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 37.Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 38.Rat Genome Sequencing Project Consortium. Genome sequence of the brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 39.Mikkelsen TS, Wakefield MJ, Aken B, et al. Genome of the marsupial Monodelphis domestica reveals innovation in noncoding sequences. Nature. 2007;447:167–177. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- 40.International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 41.Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Res. 2003;12:994–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thierry-Mieg D, Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7:S12. doi: 10.1186/gb-2006-7-s1-s12. http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/index.html?human. [DOI] [PMC free article] [PubMed]
- 43.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 44.Kopp J, Schwede T. The SWISS-MODEL repository of annotated three-dimensional protein structure homology models. Nucleic Acids Res. 2004;32:D230–D234. doi: 10.1093/nar/gkh008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Egloff MP, Marguet F, Buono G, et al. The 2.46 Å resolution structure of the pancreatic lipase-colipase complex inhibited by a C11 alkyl phosphonate. Biochemistry. 1995;34:2751–2762. doi: 10.1021/bi00009a003. [DOI] [PubMed] [Google Scholar]
- 46.Emmanuelsson O, Brunak S, von Heijne G, Nielson H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protocols. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 47.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge: Cambridge University Press; 1983. [Google Scholar]
- 48.Van De Peer Y, de Wachter R. TreeCon for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Sci. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- 49.Saitou N, Nei M. The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 50.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 51.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 52.Chang SF, Netter HJ, Will H. Characterization of cDNA encoding the mouse hepatic triglyceride lipase and expression by in vitro translation. FEBS Lett. 1991;289:69–72. doi: 10.1016/0014-5793(91)80910-u. [DOI] [PubMed] [Google Scholar]
- 53.Komaromy MC, Schotz MC. Cloning of rat hepatic lipase cDNA: evidence for a lipase gene family. Proc Natl Acad Sci U S A. 1987;84:1526–1530. doi: 10.1073/pnas.84.6.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sensel MG, Legrand-Lorans A, Wang ME, Bensadoun A. Isolation and characterization of clones for the rat hepatic lipase gene upstream regulatory region. Biochim Biophys Acta. 1990;1048:297–302. doi: 10.1016/0167-4781(90)90071-9. [DOI] [PubMed] [Google Scholar]
- 55.Stahnke G, Sprengel R, Augustin J, Will J. Human hepatic triglyceride lipase: cDNA cloning, amino acid sequence and expression in a cultured cell line. Differentiation. 1987;35:45–52. doi: 10.1111/j.1432-0436.1987.tb00150.x. [DOI] [PubMed] [Google Scholar]
- 56.Sendak RA, Berryman DE, Gellman G, et al. Binding of hepatic lipase to heparin, Identification of specific-heparin-binding residues in two distinct positive clusters. J Lipid Res. 2000;41:260–268. [PubMed] [Google Scholar]
- 57.Perret B, Mabile L, Martinez L, et al. Hepetic lipase: structure/function relationship, synthesis, and regulation. J Lipid Res. 2002;43:1163–1169. [PubMed] [Google Scholar]
- 58.Sanan DA, Fan J, Bensadoun A, Taylor JM. Hepatic lipase is abundant on both hepatocyte and endothelial cell surfaces in the liver. J Lipid Res. 1997;38:1002–1013. [PubMed] [Google Scholar]
- 59.Breedveld B, Schoonderwoerd AJ, Verhoeven R, et al. Hepatic lipase is localized at the parenchymal cell microvilli in rat liver. Biochem. 1997;321:425–430. doi: 10.1042/bj3210425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boerwinkle E, Brown SA, Rohrbach K, et al. Role of apolipoprotein E and B gene variation in determining response of lipid, lipoprotein, and apolipoprotein levels to increased dietary cholesterol. Am J Hum Genet. 1991;49:1145–1154. [PMC free article] [PubMed] [Google Scholar]
- 61.Peterson J, Bengtsson-Olivecrona G, Olivecrona T. Mouse preheparin plasma contains high levels of hepatic lipase with low affinity for heparin. Biochim Biophys Acta. 1986;878:65–70. doi: 10.1016/0005-2760(86)90344-9. [DOI] [PubMed] [Google Scholar]
- 62.Wolle J, Jansen H, Smith LC, Chan L. Functional role of N-linked glycosylation in human hepatic lipase: asparagine-56 is important for both enzyme activity and secretion. J Lipid Res. 1993;34:2169–2176. [PubMed] [Google Scholar]
- 63.Ben-Zeev O, Stahnke G, Liu RC, et al. Lipoprotein lipase and hepatic lipase: the role of asparagines linked glycosylation in the expression of a functional enzyme. J Lipid Res. 1994;35:1511–1523. [PubMed] [Google Scholar]
- 64.Kobayashi J, Applebaum-Bowden D, Dugi KA, et al. Analysis of protein structure-function in vivo. Adenovirus-mediated transfer of lipase lid mutants in hepatic lipase-deficient mice. J Biol Chem. 1996;271:26296–26301. [PubMed] [Google Scholar]
- 65.Wong H, Davis RC, Nikazy J, et al. Domain exchange: characterization of a chimeric lipase of hepatic lipase and lipoprotein lipase. Proc Natl Acad Sci U S A. 1991;88:11290–11294. doi: 10.1073/pnas.88.24.11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.The MGC Project Team. The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC) Genome Res. 2004;14:2121–2127. doi: 10.1101/gr.2596504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clark HF, Gurney AL, Abaya E, et al. The secreted protein discovery initiative (SPDI), a large-scale effort to identify novel human secreted and transmembrane proteins: a bioinformatics assessment. Genome Res. 2003;13:2260–2264. doi: 10.1101/gr.1293003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Donoghue PCJ, Benton MJ. Rocks and clocks: calibrating the tree of life using fossils and molecules. Trends Genet. 2007;22:424–431. doi: 10.1016/j.tree.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 69.Cohen JC. Endothelial lipase: direct evidence for a role in HDL metabolism. J Clin Invest. 2003;111:318–321. doi: 10.1172/JCI17744. [DOI] [PMC free article] [PubMed] [Google Scholar]