Abstract
PABP1 [poly(A)-binding protein 1] is a central regulator of mRNA translation and stability and is required for miRNA (microRNA)-mediated regulation and nonsense-mediated decay. Numerous protein, as well as RNA, interactions underlie its multi-functional nature; however, it is unclear how its different activities are co-ordinated, since many partners interact via overlapping binding sites. In the present study, we show that human PABP1 is subject to elaborate post-translational modification, identifying 14 modifications located throughout the functional domains, all but one of which are conserved in mouse. Intriguingly, PABP1 contains glutamate and aspartate methylations, modifications of unknown function in eukaryotes, as well as lysine and arginine methylations, and lysine acetylations. The latter dramatically alter the pI of PABP1, an effect also observed during the cell cycle, suggesting that different biological processes/stimuli can regulate its modification status, although PABP1 also probably exists in differentially modified subpopulations within cells. Two lysine residues were differentially acetylated or methylated, revealing that PABP1 may be the first example of a cytoplasmic protein utilizing a ‘methylation/acetylation switch’. Modelling using available structures implicates these modifications in regulating interactions with individual PAM2 (PABP-interacting motif 2)-containing proteins, suggesting a direct link between PABP1 modification status and the formation of distinct mRNP (messenger ribonucleoprotein) complexes that regulate mRNA fate in the cytoplasm.
Keywords: mRNA translation, poly(A)-binding protein (PABP), poly(A)-binding-protein-interacting motif 2 (PAM2)–poly(A)binding protein C-terminal domain (PABC) interaction, posttranscriptional control, post-translational modification, RNA-binding protein
Abbreviations: AdOX, adenosine dialdehyde; DTT, dithiothrietol; eEF, eukaryotic elongation factor; eIF, eukaryotic initiation factor; eRF, eukaryotic release factor; eRF3-N, N-terminal eRF3; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; G3BP, Ras GAP (GTPase-activating protein) SH3 (Src homology 3) domain-binding protein; HRP, horseradish peroxidase; MEF, mouse embryonic fibroblast; miRNA, microRNA; MLLE, methionine-leucine-leucine-glutamate motif-containing domain; MS/MS, tandem MS; PABC, poly(A)-binding protein 1 C-terminal domain; PABP, poly(A)-binding protein; PAIP, poly(A)-interacting protein; PAM, PABP-interacting motif; PAN, poly(A) nuclease; PCNA, proliferating cell nuclear antigen; PRMT, protein arginine N-methyltransferase; PTM, post-translational modification; RRM, RNA recognition motif; SG, stress granule; TD-NEM, transcription-dependent nuclear export motif; TOB, transducer of ERBB2; TSA, trichostatin A
INTRODUCTION
Ensuring co-ordinated temporospatial and amplitudinal regulation of gene expression is crucial for the control of cell proliferation, differentiation and function. In recent years, it has become evident that rigorous post-transcriptional control in the cytoplasm is a major component of gene regulation: the majority of vertebrate genes are subject to regulated mRNA translation and/or stability [1] and their dysregulation contributes to the aetiology of a broad spectrum of diseases [2], including neurological, metabolic, reproductive and neoplastic disorders. In keeping with this, diverse mRNA-specific and global regulatory mechanisms have been described in an extensive variety of biological contexts.
PABP [poly(A)-binding protein] 1 serves as a central regulator of mRNA fate in the cytoplasm, co-ordinating the regulation of mRNA utilization and destruction [3–5]. It simultaneously binds the mRNA 3′ poly(A) tail and interacts with the translation factor eIF (eukaryotic initiation factor) 4G, part of the cap-binding complex eIF4F (eIF4E, eIF4G and eIF4A) [6], bringing the ends of the mRNA into functional proximity. This ‘closed-loop’ conformation enhances translation initiation by increasing ribosomal recruitment while also protecting the mRNA from deadenylation, decapping and degradation [7,8]. Contacts with additional factors e.g. PAIP [poly(A)-interacting protein] 1 further stabilize this closed-loop [9]. PABP1 also has other roles in post-transcriptional regulation, both positively and negatively regulating mRNA-specific translation and mRNA stability [3,4] and is part of the machinery underlying miRNA (microRNA)-mediated regulation and NMD (nonsense-mediated mRNA decay) [8,10]. In some cases, these less well characterized roles involve interactions with the basal translational machinery [e.g. eIF4G, PAIP1 and eRF (eukaryotic release factor) 3], but it is clear that additional protein and RNA interactions are also critical [3,4].
PABP1 comprises four non-identical RRMs (RNA recognition motifs), a proline-rich region and a PABC (PABP C-terminal domain) (also known as MLLE) and, although many binding sites await further definition, it is clear that each domain mediates important functional interactions. For instance, RRMs 1 and 2 bind eIF4G and PAIP1 [3,4] and provide high-affinity poly(A)-binding, while PABP1–PABP1 interactions mediated by the proline-rich region also contribute to ordered binding to poly(A) [3]. RRMs 3 and 4 bind poly(A) with reduced affinity, but also bind AU-rich RNA and mediate protein–protein interactions [3,4], e.g. eEF (eukaryotic elongation factor) 1α. Finally, the PABC domain interacts with PAM (PABP-interacting motif) 2 motif-containing proteins, e.g. eRF3, PAIP1, the negative regulator of translation PAIP2 and the PAN [poly(A) nuclease] 3 deadenylase [9], as well as some non-PAM2-containing proteins, e.g. the miRNA-pathway protein GW182 [11]. Thus it is unclear how all the roles of PABP1, and the array of protein interactions that they require, are co-ordinated. Although it seems unlikely that PABP1 can maintain all the interactions described simultaneously, since binding sites frequently overlap [4].
PTMs (post-translational modifications) offer a way to dynamically regulate multifunctional proteins, and many RNA-binding proteins, including those involved in mRNA translation, are subject to complex regulation by phosphorylation. However, other PTMs have also been identified on RNA-binding proteins (e.g. lysine acetylation or arginine methylation), although their functional consequences are less well understood. Indeed human PABP1 is asymmetrically dimethylated on Arg455 and Arg460 in the proline-rich region by the type I PRMT (protein arginine N-methyltransferase) 4 [12], but the function of these PABP1 PTMs is unknown.
In the present study we establish that PRMT4-dependent methylation is not required for PABP1 association with translation complexes or for its normal subcellular distribution. However, we find that PABP1 is highly post-translationally modified and identify 14 novel human PABP1 PTMs, including lysine, glutamate, aspartate and PRMT4-independent arginine methylations, as well as lysine acetylations. All, but one, are conserved in mouse. Interestingly, the PTM status of PABP1 is dynamically regulated during the cell cycle, apparently via modulation of lysine acetylation. Two lysine residues were both methylated and acetylated, with structural modelling implicating their modification status in differentially modulating key PABP1 protein interactions. This provides the first insight into how the multiple functions of this central regulator of post-transcriptional control may be co-ordinated.
EXPERIMENTAL
Materials
Alexa Fluor®-488- and Alexa Fluor®-546-conjugated anti-IgG antibodies were from Invitrogen, and anti-(α-tubulin) antibody, HRP (horseradish peroxidise)-conjugated goat anti-rabbit IgG antibody and biochemicals were from Sigma. The anti-PABP1 antibody has been described previously [13], and antibodies specific for PABP1 asymmetrically dimethylated at Arg455/Arg460 were from Mark Bedford (Department of Carcinogenesis, University of Texas, MD Anderson Cancer Center, Smithville, TX, U.S.A.) and Cell Signaling Technology, and ASYM24 and SYM10 antibodies were from Millipore. HRP-conjugated goat anti-mouse IgG, anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and anti-G3BP [Ras GAP (GTPase-activating protein) SH3 (Src homology 3) domain-binding protein] antibodies were from Thermo Scientific, Abcam and Becton Dickinson respectively. Calyculin A, TSA (trichostatin A), nocodazole and a ProteoExtract kit were from Calbiochem. Immobilized linear pH gradient strips, ampholytes and Protein G–Sepharose beads were from GE Healthcare.
Cell culture and synchronization, and confocal immunofluorescence microscopy
Prmt4+/+ and Prmt4−/− MEFs (mouse embryonic fibroblasts) and HeLa cells were cultured as described previously [13,14] and, where required, supplemented with 400 nM TSA (Calbiochem)/5 mM nicotinamide or 20 μM AdOX (adenosine dialdehyde) for 24 h prior to harvest and/or 500 μM sodium arsenite for 30 min. HeLa cells were synchronized by a double-thymidine/nocodazole block [15]. Microscopic analyses were carried out as described previously [13].
OFFGEL analysis
Proteins were precipitated using a ProteoExtract kit and resolubilized into 1× OFFGEL buffer [6.8 M urea, 2 M thiourea, 62.5 mM DTT (dithiothrietol) and 5% glycerol] containing 0.5% ampholytes. Solution-phase isoelectric focusing was performed using 13 cm pH 6–11 or pH 3–10 immobilized linear pH gradient strips in an Agilent 3100 OFFGEL fractionator according to the manufacturer's instructions. Fractions were recovered and re-precipitated before being solubilized in an equal volume of 1× Laemmli sample buffer [250 mM Tris/HCl (pH 6.8), 2% (w/v) SDS, 20% (v/v) glycerol and 100 mM DTT]. Equal volumes of each fraction were analysed by PAGE and Western blotting.
Sucrose gradient polysome analysis
If appropriate, cells were incubated in 100 μg/ml cycloheximide for 15 min prior to harvesting in ice-cold PBS. Cells were lysed in polysome lysis buffer [10 mM Tris/HCl (pH 7.2), 150 mM KCl, 10 mM MgCl2, 20 mM DTT, 0.5% Nonidet P40, 100 units/ml RNasin® (Promega) and 10 nM calyculin A] containing Complete™ protease inhibitor cocktail (Roche) and either 150 μg/ml cycloheximide or 20 mM EDTA, and were incubated on ice for 10 min. Lysates were cleared for 1 min at 14000 g at 4°C and the KCl concentration of the supernatant was increased to 250 mM before applying to 10–50% (w/v) sucrose gradients prepared in gradient buffer [10 mM Tris/HCl (pH 7.2), 250 mM KCl, 10 mM MgCl2, 2.5 mM DTT, 0.5 μg/ml heparin and 0.5% Nonidet P40]. Gradients were centrifuged for 2 h at 38000 rev./min at 4°C in a Sorval TH-641 rotor and fractionated using a Superfrac fraction collector (Pharmacia) at 4°C. The absorbance of RNA at 254 nm was recorded by an inline UV monitor (Pharmacia) and proteins were precipitated from fractions using 10% trichloroacetic acid, resolubilized in 1× Laemmli sample buffer and analysed by Western blotting.
Western blotting and immunoprecipitation
Cell extracts for Western blotting were prepared by lysing cells in 1× Laemmli sample buffer and shearing genomic DNA by passing lysates through Qiashredder spin columns (Qiagen). Western blotting was performed [13] using primary antibodies at a 1:10000 dilution or at the manufacturer's recommended dilution, and HRP-conjugated secondary antibodies were used at 1:150000 (anti-rabbit IgG) or 1:20000 (anti-mouse IgG). For immunoprecipitation, extracts were prepared by lysing cells in immunoprecipitation lysis buffer [50 mM Tris/HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% Nonidet P40, 0.2% SDS, 10 mM sodium pyrophosphate, 25 mM 2-glycerophosphate, 0.5% sodium deoxycholate, 10 mM sodium orthovanadate, 5 mM sodium fluoride, 2 mM DTT and 10 nM calyculin A] containing Complete™ protease inhibitor cocktail for 5 min on ice. Extracts were clarified (14000 g for 10 min) and protein supernatant concentrations were determined by the Bradford assay. Equal amounts of protein supplemented with either anti-PABP1 or rabbit IgG (200 ng/mg of protein) and 60 μl of a 50% slurry Protein G–Sepharose beads were tumbled overnight at 4°C, washed extensively with immunoprecipitation lysis buffer at 4°C and eluted in 1× Laemmli sample buffer.
MS analysis
Excised colloidal Coomassie-Blue-stained protein bands were digested with trypsin and IMAC (immobilized metal ion chromatography) and MS samples were prepared as described previously [16]. Full scan MS survey spectra (m/z 350–1800) in profile mode were acquired in the orbitrap with a resolution of 60000 after accumulation of 500000 ions. The five most intense peptide ions from the preview scan in the Orbitrap were fragmented by collision-induced dissociation and these ions were rejected for 30 s after two occurrences. Precursor ion charge state screening was enabled and all unassigned charge states, as well as singly charged species, were rejected. The lock mass option was enabled for survey scans to improve mass accuracy and the data were acquired using Orbitrap 2.4 software and analysed using Xcalibur 2.0.7 software.
Mascot generic files were created from the raw files using raw2msm (a gift from Professor M. Mann, Max Planck Institute of Biochemistry, Munich, Germany) and searched against the SwissProt database using Mascot version 2.2 software (http://www.matrixscience.com) on a local server. The search parameters were set using trypsin as the enzyme (up to two missed cleavages allowed), and a mass tolerance of 10 p.p.m. for the precursor masses and 0.6 Da for the MS/MS (tandem MS) scans. Carbamidomethylation of cysteine was set as a fixed modification, and variable modifications included oxidation of methionine, phosphorylation of serine, threonine or tyrosine, acetylation of lysine, methylation of lysine, arginine, aspartate and glutamate, and dimethylation of lysine and arginine. All but one identified peptide had a minimum Mascot ion score of 20, and all sites of predicted PTM were manually validated by inspection of the MS/MS spectra.
Predictive modelling
Discovery Studio was used for molecular modelling (version 3.1, Accelrys). The PABP1 RRM4 structure was generated by performing sequence alignment-based molecular superimposition on to the NMR solution structure of human PABP3 RRM4 (PDB code 2D9P). The structural geometry was optimized by application of a fast Dreiding-like forcefield using the ‘Clean Geometry’ toolkit. This structure was superimposed on to the crystal structure of human PABP1 RRM2 in complex with poly(A) RNA (PDB code 1CVJ) using multiple protein tethers. RRM4 and poly(A) relational geometries were optimized as above. PABC–PAM2 models were based on crystal structures PDB code 3KUS and PDB code 3KUI [17,18]. Dimethyl or acetyl groups were added as required using Discovery Studio. The modified molecules were energy-minimized using conjugate gradients employing a CHARMM forcefield [19], until the root-mean-square gradient was less than 0.0001 kcal/mol per Å (1 Å=0.1 nm).
RESULTS
Effect of PRMT4-dependent arginine methylation on the polysomal association and subcellular distribution of PABP1
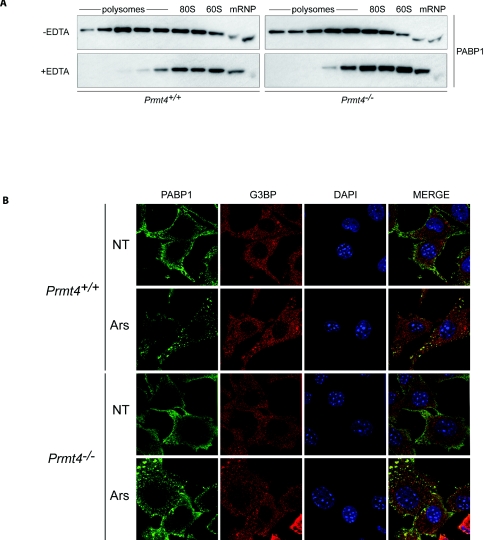
Loss of PRMT4 methyltransferase activity in mice leads to a small-size phenotype and perinatal mortality [14]. As Arg455 and Arg460 of PABP1 are major targets of PRMT4 [12,14], loss of their methylation may contribute to these phenotypes, leading us to probe their functional role. Initially, their contribution to the fundamental ability of PABP1 to promote translation was addressed using MEFs from Prmt4+/+ and Prmt4−/−mice (Supplementary Figure S1 at http://www.BiochemJ.org/bj/441/bj4410803add.htm). As expected in Prmt4+/+ MEFs under normal growth conditions, a significant proportion of PABP1 was associated with actively translating mRNAs on polysomes, although also present in mRNP fractions containing non-translating mRNAs (Figure 1A). PABP1 was redistributed upon disruption of polysomes with EDTA treatment. Importantly, this pattern was maintained in Prmt4−/− MEFs (Figure 1A). Thus although Arg455/Arg460 lie within the proline-rich region required for co-operative high-affinity binding to poly(A) tails [3,13], the unaltered polysomal association of PABP1 in Prmt4−/− MEFs suggests that neither its in vivo poly(A)-binding function nor its participation in translation complexes is significantly affected. Reduced PABP1 half-life could also disrupt protein synthesis and may be masked by PABP1 autoregulation [13]. However, PABP1 protein stability was not altered (Supplementary Figure S2 at http://www.BiochemJ.org/bj/441/bj4410803add.htm), showing that PRMT4-mediated methylation does not significantly affect the rate of PABP1 turnover.
Figure 1. PRMT4-dependent arginine methylation is not required for the association of PABP1 with translation complexes or regulation of its subcellular localization.
(A) Polysomal association of PABP1 in Prmt4+/+ and Prmt4−/− MEFs. Cell lysates were fractionated following 10–50% sucrose-density-gradient centrifugation in the presence (+) or absence (−) of EDTA and immunoblotted for PABP1. Position of polysomes, 80S monosomes, 60S subunits and mRNPs are indicated. (B) Cytoplasmic localization of PABP1 and its recruitment to SGs in Prmt4+/+ and Prmt4−/− MEFs. MEFs were either left untreated (NT) or treated with 500 μM sodium arsenite (Ars), and PABP1 (green) distribution was determined by confocal microscopy. G3BP is an SG marker (red) and DNA was visualized by DAPI staining (blue).
Although predominantly diffusely cytoplasmic, the subcellular localization of PABP1 is subject to dynamic regulation, being relocalized to the nucleus and/or cytoplasmic SGs (stress granules) in response to specific stresses [13,20]. Although normally growing Prmt4−/− MEFs exhibit wider morphological variation than Prmt4+/+ MEFs in terms of cell size and shape, the predominantly cytoplasmic distribution of PABP1 was not altered by the absence of PRMT4-mediated methylation (Figure 1B). Similarly the majority of Prmt4−/− cells exhibited normal SG formation, with PABP1 being recruited to these foci (Figure 1B). We therefore conclude that PRMT4-dependent methylation does not regulate PABP1 polysome association, PABP1 stability or its subcellular distribution in normally growing or arsenite-stressed cells.
PABP1 is a substrate for PRMT4-independent arginine methylation
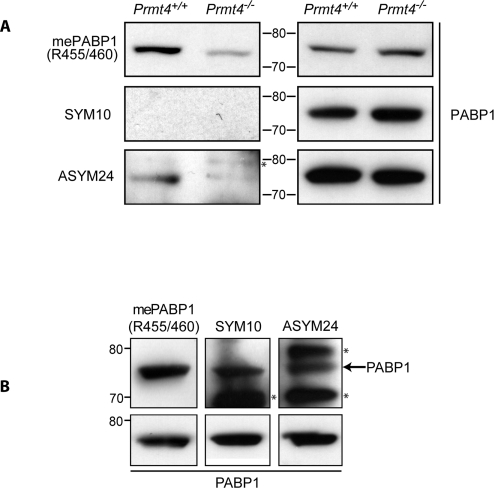
PRMT4, and other type 1 PRMTs (PRMT1, 3, 6 and 8), asymmetrically dimethylate arginine. In Prmt4−/− MEFs, the amount of asymmetrically dimethylated arginine in PABP1 is dramatically decreased, but not abrogated (Figure 2A; ASYM24, [14]), consistent with a role for other type 1 PRMTs. Since the anti-(methyl-Arg455/Arg460 PABP1) antibody (Figure 2A) detects reduced but persistent dimethylation in Prmt4−/− MEFs, the sites for these other type 1 PRMTs must include one or both of these residues. In contrast, type 2 PRMTs (PRMT5, 7 and 9) catalyse symmetrical arginine dimethylation which was not detected in PABP1 (Figure 2A; SYM10), although readily detected in other proteins present within input lysates (Supplementary Figure S3 at http://www.BiochemJ.org/bj/441/bj4410803add.htm). This suggests that PABP1 is not a Type 2 PRMT substrate in MEFs.
Figure 2. PABP1 is arginine dimethylated independently of PRMT4.
PABP1 was immunoprecipitated from Prmt4+/+ and Prmt4−/− MEFs (A) or HeLa cells (B) and Western blotted using anti-(dimethyl-Arg455/Arg460-PABP1), anti-(symmetrically dimethylated arginine) (SYM10), anti-(asymmetrically dimethylated arginine) (ASYM24) or anti-PABP1 antibodies (A, right-hand panel; B, bottom panel). The anti-(methyl-arginine) antibodies also detected additional methylated antigens (marked with *) in PABP1 immunoprecipitates. Molecular mass in kDa is indicated.
To determine whether PABP1 methylation status varies between cell type/species, PABP1 methylations were probed in HeLa cells, which are described to contain methylated PABP1 [12]. As in MEFs, PABP1 is asymmetrically dimethylated at sites including Arg455 and/or Arg460 but, in contrast, also appears to be subject to type 2 symmetrical arginine dimethylation at unknown sites (Figure 2B; SYM10). Since PABP1 arginine methylation status differs between cell types, we examined PABP1 function in HeLa cells treated with AdOX. AdOX blocked general arginine methylation, but did not affect PABP1 subcellular localization or stability (Supplementary Figure S4 at http://www.BiochemJ.org/bj/441/bj4410803add.htm), suggesting that these basal PABP1 properties are not significantly affected by arginine methylation. We were unable to examine the polysomal association of PABP1 in AdOX-treated HeLa cells due to the perturbation of polysome levels by AdOX (results not shown).
Mammalian PABP1 is highly post-translationally modified
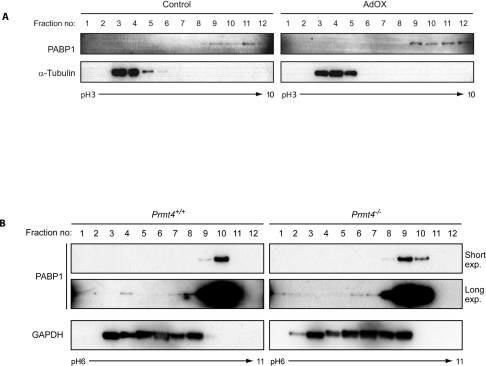
Since the pI of unmodified human PABP1 is pH 9.52, its distribution in HeLa cells (between ~pH 7.6 and 10) is consistent with multiple PTMs. This distribution was only slightly altered following AdOX treatment (Figure 3A), indicating the presence of PTMs other than arginine methylation. Similarly, in both Prmt4+/+ and Prmt4−/− MEFs a small proportion of PABP1 is detected in fractions containing proteins with significantly lower pI values (Figure 3B; e.g. fractions 1–4 represent a pI range ~pH 6.0–7.7). Since its pI distribution is not significantly affected by arginine methylation, PABP1 appears to be modified by multiple PTM species.
Figure 3. Post-translational modification of PABP1 is not restricted to arginine methylation.
(A) HeLa cells were either left untreated (Control) or treated with 20 μM AdOX. Cell extracts were subjected to OFFGEL isoelectric fractionation using a pH 3–10 linear immobilized pH gradient (IPG) and fractions immunoblotted for PABP1 and α-tubulin. (B) Cell extracts from Prmt4+/+ and Prmt4−/− MEFs were fractionated and immunoblotted as described in (A) using a pH 6–11 gradient and GAPDH as a control. A longer exposure of the PABP1 blots is shown to visualize low abundance highly modified forms of PABP1. (A) α-Tubulin [40] and (B) GAPDH [41] exhibit expected pI distributions comprising unmodified and modified forms.
Identification of novel PTMs in PABP1
To identify these novel PTMs, endogenous PABP1 was immunoprecipitated (Supplementary Figure S5 at http://www.BiochemJ.org/bj/441/bj4410803add.htm) from HeLa cells and analysed by MS/MS. Greater than 70% sequence coverage was achieved using combined data from three independent biological replicates (results not shown), and PTMs were only assigned after manual examination of spectra from ions indicated to be modified by Mascot version 2.2 analysis software.
A total of 14 novel human PABP1 PTMs were identified at 12 residues (Table 1) and comprised arginine methylation, as expected, but also lysine methylation and acetylation. Surprisingly, several methylated glutamate residues and a methylated aspartate residue were also detected, PTM types which are uncharacterized in eukaryotic cells. Supplementary Figure S6 (at http://www.BiochemJ.org/bj/441/bj4410803add.htm) shows MS/MS spectra for a representative example of each type of PTM detected. Comparative MS analyses from Prmt4+/+ and Prmt4−/− MEFs found that all but one of the identified PTMs were conserved in mouse, but also identified additional modifications resulting in a total of 22 PTMs (Supplementary Table S1 at http://www.BiochemJ.org/bj/441/bj4410803add.htm).
Table 1. Identified human PABP1 PTMs.
Peptides identified to contain modified residues (bold) are shown. In each case the highest recorded Mascot score for the modified (+) peptide is given. All spectra from putative modified peptides were subject to manual verification. Arginine can be mono- or di-methylated, lysine can be mono-, di- or tri-methylated and glutamate and aspartate are monomethylated.
| Domain | Amino acid | Modification | Peptide | Mascot score |
|---|---|---|---|---|
| RRM1–RRM2 spacer region | Lys95 | Acetylation | KSGVGNIFIK | + (31) |
| RRM2–RRM3 spacer region | Glu180 | Methylation | EAELGAR | + (25) |
| Glu182 | Methylation | EAELGAR | + (25) | |
| Lys188 | Acetylation | AKEFTNVYIK | + (33) | |
| RRM3 | Asp209 | Methylation | LKDLFGK | + (38) |
| Glu239 | Methylation | GFGFVSFER | + (44) | |
| RRM4 | Lys299 | Methylation | YQGVNLYVK | + (43) |
| Lys312 | Acetylation | KEFSPFGTITSAK | + (64) | |
| Lys312 | Dimethylation | KEFSPFGTITSAK | + (72) | |
| Lys361 | Dimethylation | IVATKPLYVALAQR | + (38) | |
| Proline-rich linker region | Arg493 | Dimethylation | VANTSTQTMGPRPAAAAAAATPAVR | + (99) |
| Arg493/Arg506 | Dimethylation, methylation | VANTSTQTMGPRPAAAAAAATPAVR | + (34) | |
| PABC/MLLE | Lys606 | Acetylation | SKVDEAVAVLQAHQAK | + (30) |
| Lys606 | Dimethylation | SKVDEAVAVLQAHQAK | + (39) |
Novel human PABP1 methylations were detected at Arg493 and Arg506. Intriguingly, the latter appeared to be a novel substrate of PRMT4 and was only detected in the monomethylated state in the context of Arg493 methylation. Conversely, in mouse Arg493 and Arg432 were methylated in the absence of PRMT4, consistent with the modification status observed in Figure 2(A). Surprisingly, multiple methylated glutamate residues were present in PABP1, with greater frequency in MEFs (ten sites) than in HeLa cells (three sites), which may indicate species- or cell-type-specific differences in the utilization of this PABP1 modification. However, Glu209 in mouse PABP1 was not methylated, despite the equivalent residue (Asp209) being methylated in human PABP1. Intriguingly modified forms of four of the MEF-specific methylated glutamate residues were not detectable in Prmt4−/− MEFs (Glu66, Glu134, Glu149 and Glu564), suggesting that they may be conferred, presumably indirectly, in a PRMT4-dependent manner. Their absence in HeLa cells, and the results in Figure 1, suggest that they are not required for PABP1 to participate in translation complexes, although they may contribute to mRNA-specific functions that only occur in some cells/processes.
Interestingly, human PABP1 contained four methylated lysine residues (Lys299, Lys312, Lys361 and Lys606) and four acetylated lysine residues (Lys95, Lys188, Lys312 and Lys606). Both of these PTMs can regulate protein–protein interactions and lysine acetylation can also regulate nucleic acid interactions, protein localization and turnover [21,22]. These PTMs are frequently found on regulators of chromatin remodelling and transcription, particularly histones, although lysine methylation is of emerging interest in cytoplasmic proteins [21]. Intriguingly, both Lys312 and Lys606 were detected in acetylated and methylated forms in the same analyses (Supplementary Figure S6 and Table 1), raising the possibility of complex regulation of PABP1 function. In histones, such lysine methylation/acetylation switches specify protein–protein interactions and are diagnostic for closed or open chromatin conformations [23]. To the best of our knowledge, PABP1 is the first non-histone protein to exhibit such a methylation/acetylation switch.
Modelling the effects of lysine modification on human PABP1 protein–protein interactions
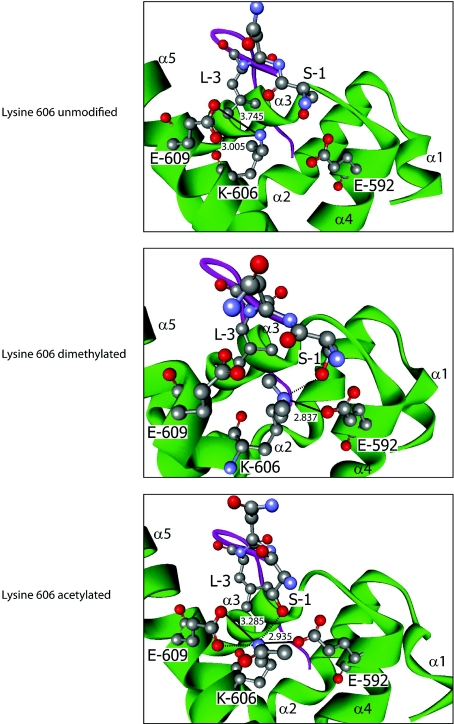
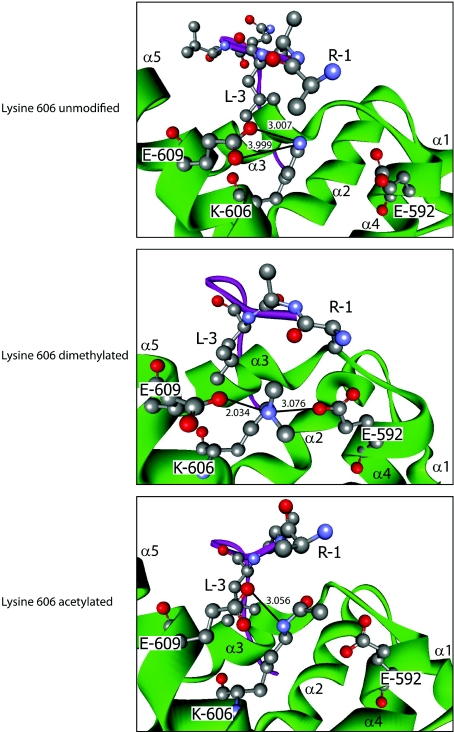
The presence of modified lysine residues within multiple functional domains of PABP1, two of which were subject to differential acetylation and methylation (Lys312 and Lys606), was of significant interest, given their ability to confer divergent functional properties. Lys606 lies within helix 5 of the 78 amino acid PABC, the only well-characterized protein interaction domain of PABP1, with high-resolution structures resolved for its interaction with PAM2 motif-containing proteins, including PAIP1, PAIP2 and eRF3 [17,18,24]. Lys606 plays a critical role in mediating the PABC–PAM2 interactions [17], suggesting that its differential modification could serve an important regulatory function. Thus Lys606 in its dimethylated and acetylated forms was modelled in silico on to the structure of the PABC domain in complex with the PAM2 motif from PAIP2 (PDB code 3KUS [17]) (Figure 4), an interaction for which the contributions of individual amino acid contacts are experimentally verified [25]. In the solved structure containing unmodified Lys606, a critical leucine residue at position 3 of the 12-amino-acid PAM2 motif projects into a hydrophobic pocket between helices 3 and 5 of the PABC domain comprising the side chains of Met584, Ile588, Ala610, Lys606 and Glu609. There the PAM2 Leu3 makes hydrophobic contacts with the aliphatic portions of Lys606 and Glu609 and the loss of these interactions reduces PABC–PAM2 affinity by ~1000-fold [17]. Lys606 also makes salt bridges with Glu609 from helix 5, which may help to stabilize the hydrophobic pocket conformation (Figure 4). Modelling revealed that dimethylation of Lys606 does not dramatically affect overall PABC conformation but, by altering local hydrophobicity, causes a rearrangement of interactions such that the hydrophobic pocket widens and Lys606 no longer salt bridges with Glu609 but, rather, salt bridges with Glu592 from helix 4 (Figure 4). This may slightly weaken the hydrophobic interaction between PAM2 Leu3 and PABC Lys606/Glu609. However, dimethylated Lys606 is also predicted to hydrogen bond with Ser1 of the PAM2 motif, an interaction that does not occur with unmodified Lys606 [17]. This suggests that Lys606 methylation may have little overall net effect or slightly weakens this interaction, as Ser1 contributes relatively little to the interaction relative to Leu3 [25].
Figure 4. Effects of Lys606 modification on PABP1 PABC–PAIP2 PAM2 interaction.
Modelling of the interaction between human PABP1 PABC (green) and the PAIP2 PAM2 motif (purple) when lysine606 is dimethylated (middle panel) or acetylated (bottom panel), based on the structure derived using unmodified PABC (top panel). Where key residues are visualized, they are numbered according to their position in full-length PABP1 or within the PAIP2 PAM2 motif (1–12). α-Helices of PABC are labelled α1–5. Atoms are coloured according to the following: red, oxygen; grey, carbon; and blue, nitrogen. Hydrogen atoms are not depicted. Where appropriate, bonds are depicted as follows: salt bridge, thick black line; hydrogen bond, broken black line; numerical values, bond distance (in Å) (hydrogen bonds ≤2.5 Å). A hydrophobic pocket is formed by Met584, Ile588, Ala610 (not depicted), and Lys606 and Glu609. The hydrophobic contacts between PAM2 Leu3 and the aliphatic portions of Lys606 and Glu609 are not depicted for clarity.
In contrast, modelling supports a more profound effect of Lys606 acetylation. Structural predictions for the PAIP2 PAM2–PABC complex containing acetylated Lys606 (Figure 4) indicate that the hydrophobic pocket conformation and its contacts with Leu3 are retained, with Lys606 maintaining the interaction with Glu609. However, acetylated Lys606 also mediates an additional interaction with Glu592, thereby stabilizing the PAM2-binding surface of the PABC domain, as well as a hydrogen bond with PAM2 Ser1. These additional interactions are likely to enhance the PABC–PAM2 interaction, strongly suggesting that PABP1 binding to PAIP2 is favoured by Lys606 acetylation.
These results led us to hypothesize that Lys606 modification status could differentially affect interactions with individual PAM2 motifs because, while crystal structures revealed a high degree of structural homology between complexes, they also demonstrated local rearrangement of residue side chains dependent on the specific PAM2 sequence [17,18,24]. We therefore modelled the effects of Lys606 PTM on the PABC interaction with the N-terminal eRF3 PAM2 (eRF3-N) motif as it binds PABC with similar affinity [25] but differs at four of the 12 amino acids. Uniquely, eRF3 has a second overlapping PAM2 motif that increases the affinity of the eRF3–PABC interaction ~3-fold [25]. The eRF3-N PAM2 motif has the critical Leu3 residue which hydrophobically contacts PABC Lys606 (PDB code 3KUI [18]) (Figure 5), but replaces the arginine and glutamine residues at positions 1 and 2 in PAIP2, for serine and asparagine respectively. Unlike the PAIP2–PAM2 interaction, modelling the effect of Lys606 PTMs on the eRF3-N–PABC structure suggests that, in the presence of eRF3-N, acetylated Lys606 cannot maintain both salt bridges with Glu609 and, consequently, restricts access of eRF3-N Arg1 to the PABC hydrophobic-binding surface, causing increased flexion in the PAM2 peptide and likely reducing the stability of the PABC–PAM2 interaction. Conversely, when Lys606 dimethylated PABC is bound to eRF3-N, one of its salt bridges with Glu609 is lost but a new salt bridge is formed with Glu592 in helix 3, thereby stabilizing the conformation of the hydrophobic pocket and the hydrophobic contacts made by Leu3, favouring an eRF3-N interaction.
Figure 5. Effects of Lys606 modification on PABP1 PABC–eRF3-N PAM2 interaction.
Modelling of the interaction between human PABP1 PABC (green) and the eRF3-N PAM2 motif (purple) when Lys606 is dimethylated (middle panel) or acetylated (bottom panel), based on the crystal structure of unmodified PABC [18] (top panel). Key residues are numbered according to their position in full-length PABP1 or the eRF3-N PAM2 motif (1–12). Depiction, labels and colour scheme are as in Figure 4.
Taken together these models predict that Lys606 dimethylation and acetylation favour interaction with eRF3 and PAIP2 [or other proteins containing highly similar PAM2 sequence (e.g. ataxin-2)] respectively, providing mechanistic insight into the co-ordination of multiple protein interactions with the PABC domain.
Modelling the effects of lysine modification on protein and RNA interactions of human PABP1 RRM4
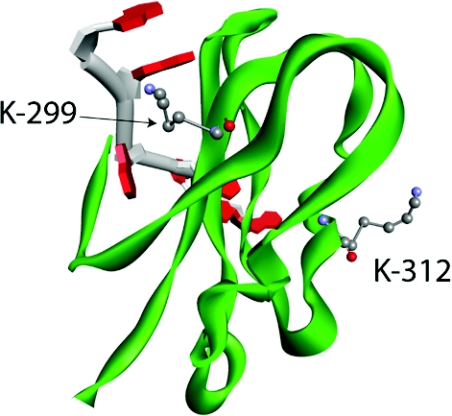
RRM4 contains a lysine residue (Lys312) that was both methylated and acetylated, in addition to several other methylated lysine residues (Lys299 and Lys361), suggesting potential for regulat-ing RRM4 function. Although there is no crystal structure for human PABP1 RRM4, a solution NMR structure for RRM4 of human tPABP (PABPc3), which differs in only one of 71 amino acids, is derived (PDB code 2D9P) allowing its use as a template for predicting the structure of human PABP1 RRM4. Not surprisingly, PABP1 RRM4 mapped very closely on to the tPABP RRM4 structure (results not shown). Since RRMs bind both RNA and proteins, the predicted human PABP1 RRM4 structure was superimposed on to the crystal structure of RRM2 of human PABP1 bound to RNA (PDB code 1CVJ) [26] as these RRMs exhibit a high degree of structural similarity (Supplementary Figure S7 at http://www.BiochemJ.org/bj/441/bj4410803add.htm). Superimposing these structures allowed the spatial positioning of Lys312 to be modelled (Figure 6 and and Supplementary Figure S7), showing its position at a solvent-exposed location in helix 1, part of the convex dorsal surface of RRM4 which is directly opposite to the RNA-binding surface, placing it in an ideal position to mediate protein–protein interactions. Indeed, the analogous dorsal surfaces of RRMs 1 and 2 are predicted to be the site of key PABP1 protein interactions, such as eIF4G and PAIP-1. Interestingly, in this regard, although few protein partners of RRM4 have been identified, both Lys312 and Lys299 directly flank the core PABP1 TD-NEM (transcription-dependent nuclear export motif) (residues 296–317) implicated in eEF1α binding and PABP1 nucleocytoplasmic transport [27]. Thus it is tempting to speculate that the modification of at least one of these residues could alter protein interactions mediated by RRM4, including the TD-NEM motif.
Figure 6. Modelling the spatial positioning of lysine residues subject to modification in human PABP1 RRM4.
Predicted structure of human PABP1 RRM4 (green) in complex with poly(A) RNA (backbone, grey; adenosine, red) showing the position of Lys299 and Lys312, based on the superimposition of RRM4 on to the crystal structure of RRM2 in complex with poly(A) RNA [26]. Atoms are coloured according to the following: red, oxygen; grey, carbon; and blue, nitrogen. Hydrogen atoms are not depicted.
Intriguingly, Lys299 also lies within one of the two RNP motifs of RRM4 which directly contact RNA. The predicted structure of human PABP1 RRM4 reveals that Lys299, which is analogous to Lys104 of RRM2 being at the +4 position of RNP2, adopts a similar spatial position within the RRM (Figure 6). Lys104 is required for the adenine-binding specificity of RNP2 which results from its multiple electrostatic and stacking interactions with proximal and neighbouring bases of poly(A) RNA [26]. Upon methylation, modelling shows that, although the charge state of Lys299 is unaltered, the residue becomes bulkier and more hydrophobic. This may alter the ability of Lys299 to hydrogen bond with adenine, potentially changing the RNA-binding characteristics of RRM4. Thus the modifications identified have the potential to alter both RNA and protein interactions mediated by PABP1.
Dynamic regulation of PABP1 modification status
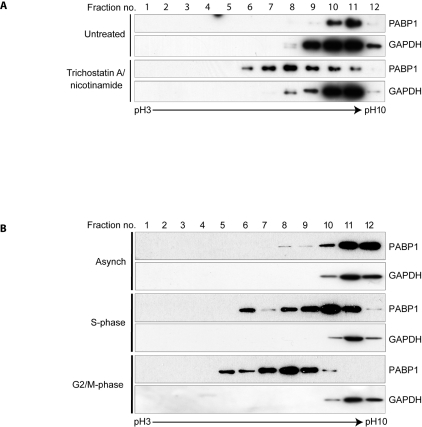
Interestingly, treatment of HeLa cells with TSA and nicotinamide, which blocks lysine deacetylation, resulted in a significant change in the proportion of PABP1 in the pI range ~pH 5.9–8.3 (Figure 7A). This suggests that, although apparently sub-stoichiometric in asynchronously growing HeLa cells, lysine acetylation could act as a major contributor to the overall charge state of human PABP1. Thus to assess whether such changes in PABP1 pI occur under physiological conditions, we examined its modification status during cell division, when extensive translational regulation is manifest [28]. In asynchronous HeLa cells the pI of the majority of PABP1 is in the pH 8.8–10 range, as expected (Figure 7B, lanes 11 and 12). However, in double-thymidine-synchronized S-phase HeLa cells PABP1 is detected with a pI range ~pH 5.9–10. The observed reduction in PABP1 pI is further enhanced in double-thymidine/nocodazole-synchronized G2/M-phase HeLa cells, where PABP1 is no longer detected in the pI range ~pH 8.8–10 with the bulk of PABP1 appearing within the pI range ~pH 5.3–8.25, consistent with a high level of de novo post-translational modification. Taken together these data reveal that PABP1 is subject to complex and dynamic post-translational modification during the mitotic cell cycle, of which lysine acetylation may be an important component.
Figure 7. The pI of PABP1 is dynamically regulated during the cell cycle and is modified by lysine acetylation.
(A) HeLa cells were either left untreated or treated with 400 nM TSA/5 mM nicotinamide. Cell extracts were fractionated using a pH 3–10 linear immobilized pH gradient (IPG) and immunoblotted for PABP1 and GAPDH. (B) HeLa cells were untreated (Asynch) or synchronized in S- or G2/M-phase and cell extracts were fractionated and immunoblotted as described in (A). (A and B) GAPDH [41] exhibits the expected pI distribution for its unmodified and modified forms.
DISCUSSION
PABP1 is a multifunctional protein which regulates different facets of post-transcriptional gene expression, suggesting that complex modulation of its RNA binding and/or protein interactions are required to co-ordinate its different functions. In the present study we have significantly advanced our understanding of PABP1 regulation by identifying numerous modifications with the potential to differentially alter its function. Molecular modelling of selected modifications, support such a role.
Two residues in PABP1 were previously identified as predominant substrates for PRMT4-dependent arginine methylation in vivo and additional putative PRMT4-dependent methylation was also identified [12–14]. However, the results of the present study argue against a PABP1-mediated deficit in global translation or change in PABP1 distribution (Figure 1) underlying the Prmt4−/− phenotype, although it remains possible that misregulation of small subsets of mRNAs may contribute. Our understanding of such mRNA-specific functions of PABP1 is in its infancy [4].
The identification of PRMT4-independent arginine methylations, including dimethylation of Arg493 within a Gly-Xaa-Arg motif (Supplementary Table S1 and Table 1) indicates that PRMT1, 3 or 6 methylate PABP1 in vivo, although they failed to do so in vitro [29]. PRMT5 can modify PABP1 in vitro [29], and the presence of symmetrical dimethylated arginine residues in HeLa cells suggests that PABP1 may be a bona fide substrate of PRMT5 (Figure 2B). Intriguingly, all of the arginine methylation sites reside within the proline-rich linker region of PABP1 that is important for ordered high-affinity poly(A)-binding. However, as RNA binding is crucial for polysome association, the nuclear export of PABP1 [13] and its distribution within the cytoplasm [13,30], these modifications do not appear to significantly affect this function (Figure 1 and Supplementary Figure S4). Interestingly, this region may also play a role in mRNA-specific regulation by individual PABP proteins [5], consistent with the idea that these modifications could modulate protein interactions important for mRNA-specific, rather than global, control. However, some PTMs may function to enable or control additional modifications, rather than influencing protein or RNA interactions directly, as there appears to be a hierarchy among the PTMs. For instance, PRMT4-dependent methylation of Arg506, which is in the same peptide as Arg493, is only detected when the latter is dimethylated.
Reduced pI forms of metazoan PABP1 in two-dimensional SDS/PAGE [31,32] have been interpreted as indicative of potential phosphorylation, and high-throughput studies have identified putative phospho-sites [15,33,34]. However, neither MS analysis following titanium-oxide-mediated enrichment for putative phosphopeptides (results not shown) nor Pro-Q Diamond staining of immunoprecipitated PABP1 detected PABP1 phosphorylation (results not shown). Thus the dramatic effect of lysine acetylation on the pI of PABP1 raises the possibility that previously observed reduced-pI forms of PABP1 are due to lysine acetylation rather than phosphorylation.
Intriguingly, PABP1 was found to be a novel methylglutamate- and methylaspartate-containing protein (Supplementary Figure S6 and Table 1), modifications that are well characterized for their role in modulating chemotactic responses in prokaryotes. Little information is available in metazoans, but human PCNA (proliferating cell nuclear antigen) and α-enolase contain several such modifications, which are altered in transformed cells, albeit with unknown functional outcomes [35,36]. PABP1 appears to be only the third such protein to be identified in humans and the first in mouse. Both PCNA and PABP1 interact with many protein partners, with glutamate methylation forming an attractive candidate for regulating interactions as it confers significant charge and hydrophobicity changes. As these PTMs are located throughout PABP1 (Table 1 and Supplementary Table S1) they could mediate complex modulation of PABP1 functions, although Prmt4−/−-dependent glutamate methylations are unlikely to be important for its basal role in global translation. Consistent with this, the solved structure [26] of RRMs 1 and 2 bound to poly(A) show that these residues do not contact poly(A) RNA.
PABP1 contains several modified lysine residues (Table 1 and Supplementary Table S1). Although acetylation neutralizes the positive charge of lysine [21,22], consistent with the dramatic effects of TSA/nicotinamide on PABP1 pI (Figure 7), lysine methylation has no net effect on charge, but increases its hydrophobicity. One of the methylated lysine residues was Lys299, which lies within RNP2 of human PABP1 RRM4 (Table 1 and Figure 6). Although this modification type is not known to regulate RNA binding, at this location it presents the potential to do so by changing hydrophobicity, creating stearic hindrance or blocking the potential for acetylation.
The identified lysine modifications may also provide an opportunity to modify protein–protein interactions and, intriguingly, two residues, Lys312 within RRM4 and Lys606 within the PABC domain, were subject to both methylation and acetylation. The PABC interacts with multiple PAM2-containing proteins, but how binding partner specificity is conferred is unclear, despite the availability of solved structures and identification of key interacting residues [17,18,24]. However, modelling of Lys606 modifications on to these structures provides initial mechanistic insight regarding the co-ordination of partner specificity. Upon Lys606 acetylation, PABC is predicted to bind more favourably to the PAIP2 PAM2 motif (Figure 4), but less well than unmodified PABC to either of the two eRF3 PAM2 motifs (Figure 5 and results not shown). Conversely upon methylation of Lys606, PABC is predicted to provide a more favourable interaction site for the eRF3-N PAM2 motif than when unmodified, whereas binding of the PAIP2 PAM2 does not appear to be significantly affected. Therefore the mutually exclusive modification of a single residue appears to contribute to, and may be sufficient to allow, discrimination between PAM2-containing protein partners. Such discrimination between binding partners could facilitate the differential roles of the PABC domain in directing PABP1 function in such apparently diverse processes as translation initiation, termination, mRNA stabilization and deadenylation.
Not all PABC interactions may be directly affected by Lys606 modification. GW182 does not contact Lys606 [24], although it interacts with the same binding surface as PAM2-containing proteins. However, by altering eRF3 affinity, Lys606 PTMs could indirectly affect this interaction, or those of lower affinity PAM2-containing proteins [e.g. PAN3 or TOB (transducer of ERBB2) 1] which compete with eRF3 binding in vivo [37,38]. An absence of structural information regarding PABC–PAN3 and PABC–TOB1 complexes precludes prediction as to whether Lys606 PTMs may also directly regulate these interactions. Nonetheless, it is clear that a Lys606 methylation/acetylation switch could regulate PABP1 assembly into mRNPs with both PAM2 and non-PAM2-containing proteins that participate in different aspects of post-transcriptional regulation.
Most mammalian mRNAs, however, possess poly(A) tails of sufficient length to bind several PABP molecules. This may allow the association of an mRNA with multiple PABC-binding proteins, with the overall PABP1 interaction status determining the fate of that bound mRNA. It has been proposed that the relative stoichiometry of PABC-mediated interactions is the product of both direct binding competition and synergistic avidity effects [24]. However, our results suggest an alternative, wherein the PTM status of Lys606 (and other residues) may determine the relative binding affinity of individual PABC ligands, allowing regulation of mRNP composition and mRNA fate.
Although PABP1 probably exists in differentially modified states within the same cell, our cell-cycle analysis suggest that its modification status can be regulated in response to specific biological processes and/or extracellular stimuli. Indeed, the MS/MS analyses most likely reflect the overall modification status of PABP1 at different points within the cell cycle and within individual cells, as even synchronized cells contain multiple pI forms (Figure 7B). Given that enhanced PABP1 lysine acetylation causes changes analogous to cell cycle in PABP1 pI (Figure 7A), it appears likely that these modifications contribute to the dynamic cell-cycle-mediated changes in PABP pI. Such changes could potentially function to inhibit cap-dependent translation while maintaining translation of specific mRNAs during cell-cycle progression [28]. However, the large numbers of lysine acetyltransferases and deacetylases [39], for which very few specific substrates have been identified, precludes any speculation as to which may underlie the observed changes in PABP1 pI.
In summary, our finding that PABP1 is subject to complex and dynamic post-translational modification provides a framework for understanding how multiple interactions with this key regulator of protein synthesis may be regulated to co-ordinate its numerous roles. Our findings open novel technically challenging avenues of research requiring the production and extensive functional analysis of PABP1 of defined modification status. However, the central role of PABP1 in cellular biology underscores the importance of dissecting the functional consequences of individual PABP1 post-translational modifications.
Online data
AUTHOR CONTRIBUTION
Matthew Brook and Nicola Gray initiated the project, directed it throughout and wrote the paper. Matthew Brook designed the experiments, and Matthew Brook and Nicola Gray carried out data analysis and interpretation. Matthew Brook, Lora McCracken and James Reddington performed the experiments, and Matthew Brook and Nicholas Morrice carried out mass spectrometric analyses. Nicholas Morrice performed post hoc spectrum analyses to assign and validate PTM sites. Matthew Brook and Zhi-Liang Lu performed molecular modelling analyses.
ACKNOWLEDGEMENTS
We thank William Richardson for expert technical assistance, Mark Bedford for helpful discussions, and provision of the Prmt4−/−MEFs and the anti-(methyl-PABP1) antibody, Dirk Ostareck and Antje Ostareck-Lederer for discussion of unpublished work, Stephen Curry for critical data evaluation, and the members of the Gray laboratory for helpful discussions and/or critical reading of the paper.
FUNDING
This work was supported by a Medical Research Council Senior Non-Clinical Fellowship (to N.K.G.), Medical Research Council Unit and Centre funding (to N.K.G.), a Medical Research Council studentship (to J.R.) and a Biotechnology and Biological Sciences Research Council/Engineering and Physical Sciences Research Council IRColl Proteomic Technology grant (to N.M.).
References
- 1.Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 2.Chatterjee S., Pal J. K. Role of 5′- and 3′-untranslated regions of mRNAs in human diseases. Biol. Cell. 2009;101:251–262. doi: 10.1042/BC20080104. [DOI] [PubMed] [Google Scholar]
- 3.Brook M., Smith J. W.S., Gray N. K. The DAZL and PABP families: RNA-binding proteins with interrelated roles in translational control in oocytes. Reproduction. 2009;137:595–617. doi: 10.1530/REP-08-0524. [DOI] [PubMed] [Google Scholar]
- 4.Burgess H. M., Gray N. K. mRNA-specific regulation of translation by poly(A)-binding proteins. Biochem. Soc. Trans. 2010;38:1517–1522. doi: 10.1042/BST0381517. [DOI] [PubMed] [Google Scholar]
- 5.Gorgoni B., Richardson W. A., Burgess H. M., Anderson R. C., Wilkie G. S., Gautier P., Martins J. P., Brook M., Sheets M. D., Gray N. K. Poly(A)-binding proteins are functionally distinct and have essential roles during vertebrate development. Proc. Natl. Acad. Sci. U.S.A. 2011;108:7844–7849. doi: 10.1073/pnas.1017664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackson R. J., Hellen C. U., Pestova T. V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Livingstone M., Atas E., Meller A., Sonenberg N. Mechanisms governing the control of mRNA translation. Phys. Biol. 2010;7:021001. doi: 10.1088/1478-3975/7/2/021001. [DOI] [PubMed] [Google Scholar]
- 8.Fabian M. R., Sonenberg N., Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 9.Derry M. C., Yanagiya A., Martineau Y., Sonenberg N. Regulation of poly(A)-binding protein through PABP-interacting proteins. Cold Spring Harbor Symp. Quant. Biol. 2006;71:537–543. doi: 10.1101/sqb.2006.71.061. [DOI] [PubMed] [Google Scholar]
- 10.Nicholson P., Muhlemann O. Cutting the nonsense: the degradation of PTC-containing mRNAs. Biochem. Soc. Trans. 2010;38:1615–1620. doi: 10.1042/BST0381615. [DOI] [PubMed] [Google Scholar]
- 11.Tritschler F., Huntzinger E., Izaurralde E. Role of GW182 proteins and PABPC1 in the miRNA pathway: a sense of deja vu. Nat. Rev. Mol. Cell Biol. 2010;11:379–384. doi: 10.1038/nrm2885. [DOI] [PubMed] [Google Scholar]
- 12.Lee J., Bedford M. T. PABP1 identified as an arginine methyltransferase substrate using high-density protein arrays. EMBO Rep. 2002;3:268–273. doi: 10.1093/embo-reports/kvf052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burgess H. M., Richardson W. A., Anderson R. C., Salaun C., Graham S. V., Gray N. K. Nuclear relocalisation of cytoplasmic poly(A)-binding proteins PABP1 and PABP4 in response to UV irradiation reveals mRNA-dependent export of metazoan PABPs. J. Cell Sci. 2011;124:3344–3355. doi: 10.1242/jcs.087692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yadav N., Lee J., Kim J., Shen J., Hu M. C., Aldaz C. M., Bedford M. T. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 2003;100:6464–6468. doi: 10.1073/pnas.1232272100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dephoure N., Zhou C., Villen J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., Gygi S. P. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubois F., Vandermoere F., Gernez A., Murphy J., Toth R., Chen S., Geraghty K. M., Morrice N. A., MacKintosh C. Differential 14-3-3 affinity capture reveals new downstream targets of phosphatidylinositol 3-kinase signaling. Mol. Cell Proteomics. 2009;8:2487–2499. doi: 10.1074/mcp.M800544-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozlov G., Menade M., Rosenauer A., Nguyen L., Gehring K. Molecular determinants of PAM2 recognition by the MLLE domain of poly(A)-binding protein. J. Mol. Biol. 2010;397:397–407. doi: 10.1016/j.jmb.2010.01.032. [DOI] [PubMed] [Google Scholar]
- 18.Kozlov G., Gehring K. Molecular basis of eRF3 recognition by the MLLE domain of poly(A)-binding protein. PLoS ONE. 2010;5:e10169. doi: 10.1371/journal.pone.0010169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brooks B. R., Bruccoleri R. E., Olafson B. D., States D. J., Swaminathan S., Karplus M. Charmm: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 20.Salaun C., MacDonald A. I., Larralde O., Howard L., Lochtie K., Burgess H. M., Brook M., Malik P., Gray N. K., Graham S. V. Poly(A)-binding protein 1 partially relocalizes to the nucleus during herpes simplex virus type 1 infection in an ICP27-independent manner and does not inhibit virus replication. J. Virol. 2010;84:8539–8548. doi: 10.1128/JVI.00668-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Egorova K. S., Olenkina O. M., Olenina L. V. Lysine methylation of nonhistone proteins is a way to regulate their stability and function. Biochemistry (Moscow) 2010;75:535–548. doi: 10.1134/s0006297910050019. [DOI] [PubMed] [Google Scholar]
- 22.Yang X. J., Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol. Cell. 2008;31:449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Izzo A., Schneider R. Chatting histone modifications in mammals. Briefings Funct. Genomics. 9:429–443. doi: 10.1093/bfgp/elq024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozlov G., Safaee N., Rosenauer A., Gehring K. Structural basis of binding of P-body-associated proteins GW182 and ataxin-2 by the Mlle domain of poly(A)-binding protein. J. Biol. Chem. 2010;285:13599–13606. doi: 10.1074/jbc.M109.089540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozlov G., De Crescenzo G., Lim N. S., Siddiqui N., Fantus D., Kahvejian A., Trempe J. F., Elias D., Ekiel I., Sonenberg N., et al. Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase. EMBO J. 2004;23:272–281. doi: 10.1038/sj.emboj.7600048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deo R. C., Bonanno J. B., Sonenberg N., Burley S. K. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98:835–845. doi: 10.1016/s0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
- 27.Khacho M., Mekhail K., Pilon-Larose K., Pause A., Cote J., Lee S. eEF1A is a novel component of the mammalian nuclear protein export machinery. Mol. Biol. Cell. 2008;19:5296–5308. doi: 10.1091/mbc.E08-06-0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vasudevan S., Tong Y., Steitz J. A. Cell-cycle control of microRNA-mediated translation regulation. Cell Cycle. 2008;7:1545–1549. doi: 10.4161/cc.7.11.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng D., Cote J., Shaaban S., Bedford M. T. The arginine methyl-transferase CARM1 regulates the coupling of transcription and mRNA processing. Mol. Cell. 2007;25:71–83. doi: 10.1016/j.molcel.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 30.Kumar G. R., Shum L., Glaunsinger B. A. Importin α-mediated nuclear import of cytoplasmic poly(A) binding protein occurs as a direct consequence of cytoplasmic mRNA depletion. Mol. Cell. Biol. 2011;31:3113–3125. doi: 10.1128/MCB.05402-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drawbridge J., Grainger J. L., Winkler M. M. Identification and characterization of the poly(A)-binding proteins from the sea urchin: a quantitative analysis. Mol. Cell. Biol. 1990;10:3994–4006. doi: 10.1128/mcb.10.8.3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma S., Musa T., Bag J. Reduced stability of mitogen-activated protein kinase kinase-2 mRNA and phosphorylation of poly(A)-binding protein (PABP) in cells overexpressing PABP. J. Biol. Chem. 2006;281:3145–3156. doi: 10.1074/jbc.M508937200. [DOI] [PubMed] [Google Scholar]
- 33.Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 34.Olsen J. V., Vermeulen M., Santamaria A., Kumar C., Miller M. L., Jensen L. J., Gnad F., Cox J., Jensen T. S., Nigg E. A., et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signaling. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 35.Hoelz D. J., Arnold R. J., Dobrolecki L. E., Abdel-Aziz W., Loehrer A. P., Novotny M. V., Schnaper L., Hickey R. J., Malkas L. H. The discovery of labile methyl esters on proliferating cell nuclear antigen by MS/MS. Proteomics. 2006;6:4808–4816. doi: 10.1002/pmic.200600142. [DOI] [PubMed] [Google Scholar]
- 36.Zhou W., Capello M., Fredolini C., Piemonti L., Liotta L. A., Novelli F., Petricoin E. F. Mass spectrometry analysis of the post-translational modifications of α-enolase from pancreatic ductal adenocarcinoma cells. J. Proteome Res. 2011;9:2929–2936. doi: 10.1021/pr901109w. [DOI] [PubMed] [Google Scholar]
- 37.Siddiqui N., Mangus D. A., Chang T. C., Palermino J. M., Shyu A. B., Gehring K. Poly(A) nuclease interacts with the C-terminal domain of polyadenylatebinding protein domain from poly(A)-binding protein. J. Biol. Chem. 2007;282:25067–25075. doi: 10.1074/jbc.M701256200. [DOI] [PubMed] [Google Scholar]
- 38.Funakoshi Y., Doi Y., Hosoda N., Uchida N., Osawa M., Shimada I., Tsujimoto M., Suzuki T., Katada T., Hoshino S. Mechanism of mRNA deadenylation: evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev. 2007;21:3135–3148. doi: 10.1101/gad.1597707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peserico A., Simone C. Physical and functional HAT/HDAC interplay regulates protein acetylation balance. J. Biomed. Biotechnol. 2010;2011:371832. doi: 10.1155/2011/371832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hammond J. W., Cai D., Verhey K. J. Tubulin modifications and their cellular functions. Curr. Opin. Cell Biol. 2008;20:71–76. doi: 10.1016/j.ceb.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanllorenti P. M., Rosenfeld J., Ronchi V. P., Ferrara P., Conde R. D. Two dimensional non equilibrium pH gel electrophoresis mapping of cytosolic protein changes caused by dietary protein depletion in mouse liver. Mol. Cell. Biochem. 2001;220:49–56. doi: 10.1023/a:1010853719232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.