SUMMARY
The Core Binding Factor (CBF) acute myeloid leukemias (AMLs) are a prognostically distinct subgroup that includes patients with the inv(16) and t(8:21) chromosomal rearrangements. Both of these rearrangements result in the formation of fusion proteins, CBFB-MYH11 and AML1-ETO respectively, that involve members of the CBF family of transcription factors. It has been proposed that both of these fusion proteins function primarily by dominantly repressing normal CBF transcription. However, recent reports have indicted that additional, CBF-repression independent activities may be equally important during leukemogenesis. This article will focus on these recent advances.
Keywords: CBFB, RUNX1, AML1-ETO, CBFB-MYH11, AML
INTRODUCTION
The CBF family is composed of four proteins, the 3 α subunits, RUNX1 (AML1, Cbfa2), RUNX2 (Cbfα1), and RUNX3 (Cbfα3) [Ogawa et al., 1993b], and the single β subunit, CBFβ [Ogawa et al., 1993a; Wang et al., 1993]. Disruptions of both CBFβ and RUNX1 are associated with acute myeloid leukemia (AML). CBFβ is involved in the recurrent chromosomal abnormality inv(16)(p13q22) as well as the less common t(16;16)(p13q22) translocation, both of which create a fusion between the CBFB gene on 16q22, and MYH11 on 16p13, the gene that encodes smooth muscle myosin heavy chain (SMMHC) [Liu et al., 1993]. The resulting CBFB-MYH11 fusion gene, which encodes the oncoprotein CBFβ-SMMHC, is found in nearly all patients with French-American-British (FAB) classification subtype M4 with eosinophilia (M4Eo) AML [Le Beau et al., 1983; Liu et al., 1995]. RUNX1 is involved in the t(8;21) translocation that results in a fusion between RUNX1 and the gene for an E-box family protein, ETO (RUNX1T1, MTG8), to generate AML1-ETO (RUNX1-RUNX1T1)[Erickson et al., 1992], which is associated with AML subtype M2 [Rowley, 1973]. Together, the inv(16)(p13q22) and t(8:21) translocations account for approximately 20–25% of adult AML [Speck and Gilliland, 2002], making RUNX1 and CBFB the most commonly targeted genes in human AML. In addition, point mutations in RUNX1 have been found in families with a familial platelet disorder with predisposition to AML [Minelli et al., 2004; Osato, 2004] and in patients with de novo AML, particularly among those with subtype M0 [Osato, 2004; Roumier et al., 2003]. Gene expression profiling also indicates that RUNX1 inactivation is associated with a distinct M0 subgroup [Silva et al., 2009; Tang et al., 2009].
CBFβ and RUNX1 form a heterodimer and together they bind to the consensus TGTGGT DNA sequence and regulate gene expression. The RUNX1 protein contains a conserved RUNT homology domain (RHD), which is responsible for binding DNA and CBFβ [Speck and Gilliland, 2002]. CBFβ does not bind DNA directly, but stabilizes the RUNX1-DNA interaction allosterically [Tang et al., 2000] and protects RUNX1 from ubiquitination and degradation [Huang et al., 2001]. Both RUNX1 and CBFβ are master regulators of definitive hematopoiesis.
It is thought that both CBFβ-SMMHC and AML1-ETO function by dominantly repressing normal CBFβ/RUNX1 heterodimer activity. Based on this model of dominant repression, the development of new therapies for CBF leukemias has focused on disrupting this activity. However, recent work indicates that these fusion proteins may have gain of function activities as well, which could represent additional targets for future drug discovery. In this article we will review the relevant literature establishing the dominant negative model, as well as highlight recent reports that challenge this model.
MECHANISMS OF CBFβ-SMMHC INDUCED LEUKEMOGENESIS
Initial studies of Cbfb-MYH11 in mice suggest a dominant repression model. Mice heterozygous for a knocked-in Cbfb-MYH11 fusion allele (Cbfb+/MYH11) have a nearly identical phenotype [Castilla et al., 1996] as mice null for either Cbfb (Cbfb−/−) or Runx1 (Runx1−/−) [Niki et al., 1997; Okada et al., 1998; Okuda et al., 1996; Sasaki et al., 1996; Wang et al., 1996a; Wang et al., 1996b], which includes embryonic lethality from massive hemorrhaging and a complete block in definitive hematopoiesis. Subsequent in vitro studies indicate that the fusion protein CBFβ-SMMHC has a higher affinity for RUNX1 than endogenous CBFβ [Lukasik et al., 2002]. The N-terminus of the fusion protein retains the RUNX1 dimerazation residues from CBFβ, but CBFβ-SMMHC also contains a second RUNX1 high-affinity binding domain (HABD) located at the proximal end of SMMHC [Lukasik et al., 2002] (Figure 1A). As a result, CBFβ-SMMHC binds RUNX1 at two sites, and can outcompete CBFβ for RUNX1 binding. After preferentially binding RUNX1, it has been proposed that CBFβ-SMMHC represses RUNX1 transactivation by a number of different mechanisms, including sequestration to the cytoplasm [Adya et al., 1998], and recruitment of transcriptional repressors by the SMMHC tail [Lutterbach et al., 1999].
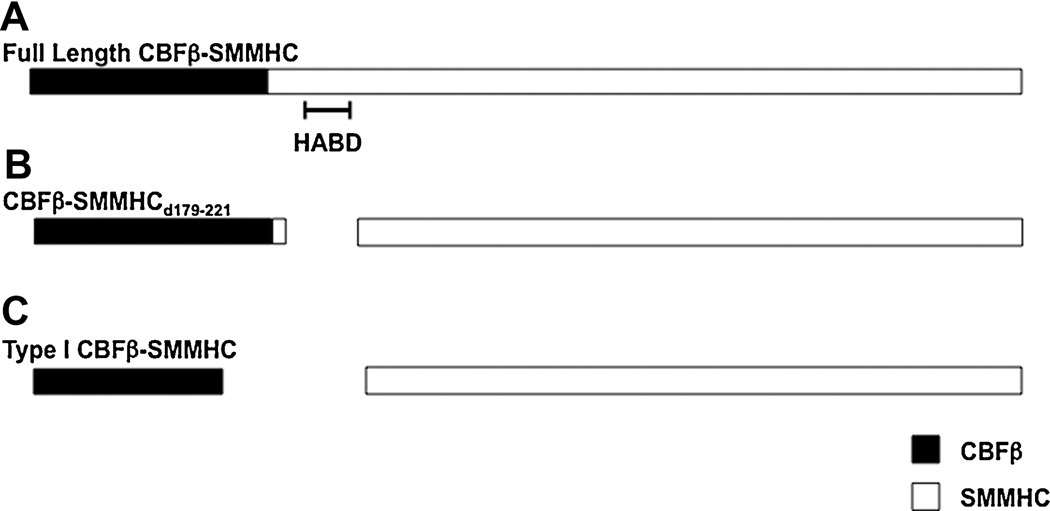
Figure 1. Diagrammatic representation of CBFβ-SMMHC variants.
Schematic of (A) full length CBFβ-SMMHC, (B) the CBFβ-SMMHCd179-221 deletion mutant, and (C) the Type I CBFβ-SMMHC fusion. The CBFβ and SMMHC are represented as black and white boxes, respectively. The high affinity binding domain (HABD) is indicated.
The HABD is predicted to be important for leukemogenesis by CBFβ-SMMHC if dominant repression of RUNX1/CBFβ is a critical step for leukemia development. To test this hypothesis, we generated knockin mice expressing a mutant Cbfb-MYH11 allele (Cbfb-MYH11d179-221, expressing CBFβ-SMMHCd179-221, Figure 1B) in which the HABD (aa 179–221) is deleted (Kamikubo, et al. manuscript under review after revision). As expected, this allele had reduced repression of CBFβ/RUNX1 functions as evidenced by in vitro studies as well as partial rescue of the embryonic lethality and definitive hematopoiesis blockage phenotypes in the Cbfb+/MYH11d179-221 embryos. Surprisingly, the decreased repression of Runx1 did not correlate with reduced or delayed leukemogenesis. Mice carrying the Cbfb-MYH11d179-221 allele developed leukemia faster than those expressing full length Cbfb-MYH11. Furthermore, we found that expression of Cbfb-MYH11d179-221 induced clonal expansion of human CD34+ cells with a similar efficiency as full length Cbfb-MYH11. Taken together, these results indicate that the HABD in CBFβ-SMMHC is not required for leukemogenesis, implying that dominant repression of RUNX1 may not be as central to CBFβ-SMMHC’s oncogenic activity as previously believed.
Consistent with these findings is the observation that the so-called type I CBFβ-MYH11 fusion, detected in a small percentage of inv(16) AML patients, produces a CBFβ-SMMHC fusion protein that lacks the HABD and a significant portion of the C-terminal segment of CBFβ (Figure 1C) [Dissing et al., 1998; Van der Reijden et al., 2001]. Consequently, the type I fusion protein has very low binding affinity for RUNX1 (Kamikubo, et al. manuscript under review after revision). The clinical course and the characteristics of leukemia with the type I fusion are indistinguishable from those with longer forms of the fusion protein, further indicating that dominant repression of RUNX1 is not strictly required for CBFβ-SMMHC to induce leukemia.
A corollary implication of this conclusion is that CBFβ-SMMHC has activities not directly related to RUNX1 repression. In fact, we have recently shown that, in primitive blood cells, which are mostly nucleated erythrocytes that arise from the initial wave of embryonic hematopoiesis, Cbfb-MYH11 blocks differentiation through a Cbfb/Runx1-repression independent mechanism [Hyde et al., 2009]. Primitive blood cells from Cbfb+/MYH11 embryos have the histological appearance of more immature precursor cells [Castilla et al., 1996], as well as continued expression of genes associated with early progenitor or stem cells, as detected by microarray analysis [Hyde et al., 2009]. Primitive blood cells from neither Cbfb−/− nor Runx1−/− embryos showed significant differentiation defects, indicating that loss of Cbfb/Runx1 activity is not responsible for the Cbfb-MYH11 induced block in differentiation. Therefore, the fusion gene must have additional, gain of function activities.
Interestingly, many of the genes whose expression was deregulated in the Cbfb+/MYH11 embryos via this novel activity were also found expressed in leukemic cells from mice and humans. In the case of the mouse leukemias, this gene set was expressed equally in cells from mice with the full length Cbfb-MYH11 allele or the Cbfb-MYH11d179-221 deletion mutant (RKH, YK, PPL, unpublished results). This finding implies that the Cbfβ/Runx1 repression independent activity described during primitive hematopoiesis is likely involved in Cbfb-MYH11 induced leukemogenesis as well.
The mechanism for this novel activity can only be speculated at present. One hypothesis is that CBFβ-SMMHC binds RUNX1, but does not repress its activity. Rather, perhaps through the recruitment of co-factors by the SMMHC tail, the fusion protein changes RUNX1 target gene specificity or transactivation ability. A second possibility is that CBFβ-SMMHC has activities that are completely independent of RUNX1 association, probably mediated by the SMMHC tail. Little is known about the interactions of the SMMHC tail in vivo, and it is conceivable that as yet unknown factors interact with CBFβ-SMMHC and contribute to leukemogenesis.
While the above described observation indicate that CBFβ-SMMHC has important oncogenic activities independent of RUNX1 repression, it should not be concluded that inactivation of the CBFβ/RUNX1 heterodimer does not also contribute to leukemogenesis. Mice with one Cbfb-MYH11 knockin allele, and one Cbfb null allele (Cbfb−/MYH11) show accelerated development of leukemia as compared to Cbfb+/MYH11 mice [Heilman et al., 2006]. On the other hand, Cbfb-MYH11 knockin mice with Runx1 mutations developed leukemia at rates inversely correlating with the severity of Runx1 loss (Ling Zhao and PPL, unpublished results). A possible interpretation of these findings is that CBFβ-SMMHC competes with CBFβ for leukemogenesis while partial inhibition of RUNX1 is more leukemogenic than complete RUNX1 inhibition. Of note PU.1 contribution to leukemogenesis is similarly dose dependent; mice carrying hypomorphic alleles of Pu.1 with reduced expression (20% of normal) developed AML rapidly and efficiently, while mice with homo- or heterozygous deletion of Pu.1 did not develop leukemia [Rosenbauer et al., 2004]. At present, it is not possible to weigh the relative importance of the CBF- repression dependent and independent activities. It seems likely that both pathways contribute substantially to the oncogenic effects of CBFβ-SMMHC, and consequently, could be important targets for the development of new treatments for inv(16)+ leukemia.
MECHANISMS OF AML1-ETO INDUCED LEUKEMOGENESIS
The fusion protein resulting from the t(8;21) translocation, AML1-ETO, contains the N-terminal region of RUNX1 which includes the DNA and CBFβ binding runt homology domain (RHD), joined to nearly the entire ETO protein (Figure 2A). ETO is a member of the E-box family of transcriptional factors, and contains four conserved Nervy Homology Regions (NHR). The ETO NHR domains have been shown to interact with a number of transcriptional repressors, including N-CoR, SMRT, Sin3A, and HDAC1-3 [Peterson and Zhang, 2004]. Based on the structure of the AML1-ETO protein, it has been proposed that it functions through repression of RUNX1 target genes. Because the fusion protein retains the intact RHD, it was originally presumed to share many of the same target genes as the endogenous RUNX1. However, due to the NHR domains of the ETO portion, AML1-ETO has been considered a transcriptional repressor rather than an activator. Consistent with this model, it has been shown that AML1-ETO represses expression of the tumor suppressor p14ARF, which is normally activated by RUNX1 [Linggi et al., 2002]. Through recruitment of chromatin remodeling proteins, AML1-ETO has also been shown to repress expression of the microRNA miR-223, a potential effecter of the AML1-ETO induced block in differentiation [Fazi et al., 2007]. In addition, as in the case of CBFβ-SMMHC, mice expressing a knockin allele of AML1-ETO [Okuda et al., 1998; Yergeau et al., 1997] have the same phenotype of embryonic lethality and block in definitive hematopoiesis as the Runx1−/− and Cbfb−/− mice [Niki et al., 1997; Okada et al., 1998; Okuda et al., 1996; Sasaki et al., 1996; Wang et al., 1996a; Wang et al., 1996b], which is consistent with RUNX1-ETO having dominant repressor activities.
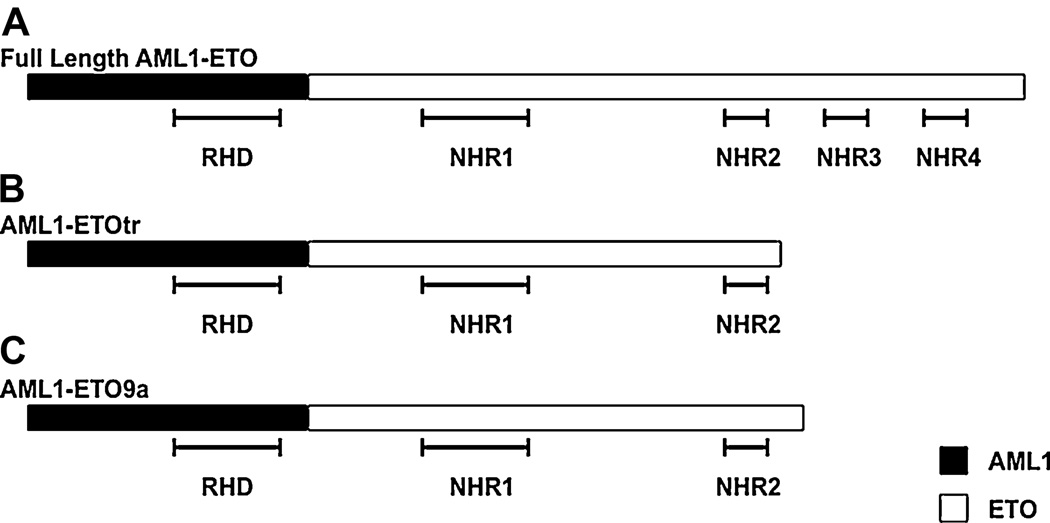
Figure 2. Diagrammatic representation of AML1-ETO constructs.
Schematic of (A) full length AML1-ETO, (B) the AML1-ETOtr truncation mutant, and (C) the naturally occurring AML1-ETO9a isoform. The RUNX1 and ETO domains are represented as black and white boxes, respectively. The RUNT homology domain (RHD) and Nervy homology regions (NHR) are indicated.
Despite the attractiveness of this model, there is increasing evidence that AML-ETO mediated leukemogenesis is more complex than simple repression of RUNX1 target genes. AML1-ETO has also been shown to effect activation of some target genes, such as p21 [Peterson et al., 2007b], BCL-2 [Klampfer et al., 1996] and the differentiation blocking microRNA, miR-24 [Zaidi et al., 2009], as well as regulate genes that are not targets of endogenous CBFβ/RUNX1 [Gardini et al., 2008; Shimada et al., 2000]. Consistent with these findings, it has been demonstrated that AML1-ETO, but not RUNX1, preferentially binds promoters with duplicated RUNX1 consensus sites [Okumura et al., 2008]. In addition, immunoflourescent staining of Kasumi-1 cells, a cell line derived from an AML1-ETO+ AML patient, shows that RUNX1 and AML1-ETO are associated with different chromosomal regions [Bakshi et al., 2008], implying that AML1-ETO regulates different target genes than RUNX1.
Given these findings, it is perhaps not surprising that multiple studies have shown that DNA binding by AML1-ETO is required for leukemogenesis [Kwok et al., 2009; Roudaia et al., 2009; Yan et al., 2009]. However, whether interaction with CBFβ is also required has yet to be resolved. Using in vitro techniques, Matheny et al [Matheny et al., 2007], identified point mutations in AML1-ETO (Y113A and T161A) that specifically disrupted CBFβ binding without affecting DNA binding. These point mutants were combined, expressed in mouse bone marrow cells, and transplanted into recipient mice. Unlike the wildtype AML1-ETO, the mutant AML1-ETO (Y113A/T161A) did not induce leukemia in cooperation with TEL-PDGFβR [Roudaia et al., 2009], indicating that CBFβ binding is required for leukemogenesis.
In contrast, Kwok et al [Kwok et al., 2009], tested two different point mutants of AML1-ETO (M106V and A107T) that by immunoprecipitation and western blot, showed severely reduced CBFβ binding. These constructs, when expressed in mouse hematopoietic cells, retained serial replating ability, similar to the wildtype AML1-ETO. In addition, they found that knockdown of Cbfb by short hairpin RNA (shRNA) did not effect AML-ETO’s serial replating ability. From these results, the authors concluded that interaction with CBFβ is dispensable for AML-ETO’s leukemogenic activity.
One possible explanation for these contradictory results is that serial replating ability may not precisely correlate with leukemogenic potential. CBFβ binding may not be required for the former, but still required for the latter. In addition, it may be that AML-ETO can function properly with a very minimal amount of CBFβ binding. The M106V and A107T mutants [Kwok et al., 2009] may weakly associate with Cbfβ, such that it was barely detectable by immunoprecipitation [Fig 1D, in Kwok et al., 2009], but would be enough to stabilize AML1-ETO’s DNA binding. Follow up studies by Park et al are consistent with this possibility [Park et al., 2009]. Similarly, shRNA knockdown of Cbfβ may not have been complete, and the remaining CBFβ contributed to the serial replating activity. Further experimentation will be needed to clarify the role of CBFβ in leukemia induction by AML1-ETO. Because this interaction has been proposed as a target for the development of new therapies, resolution of this issue could have important consequences.
AML1-ETO repression of RUNX1 target gene expression has also been questioned by recent findings indicating that recruitment of co-repressors by the ETO domain may not be required for leukemogenesis. Deletion mutants of the ETO co-repressor binding NHR domains have shown that NHR1, 3, and 4 are dispensable for leukemogenesis [Kwok et al., 2009; Yan et al., 2009]. In addition, it has been shown that loss of NHR3 and 4 either in a truncation mutation (Figure 2B) [Yan et al., 2004] or in a naturally occurring splice isoform (AML1-ETO9a) (Figure 2C) [Yan et al., 2006] results in accelerated leukemogenesis. These findings indicate that, rather than contributing to leukemogenesis, NHR3 and 4 actually inhibit the oncogenic activity of AML-ETO.
These findings raise interesting questions as to the relevance of the multiple other AML-ETO isoforms expressed in patient samples. In addition to the AML1-ETO9a isoform described above, nine other isoforms have been described in patients or cell lines [Peterson et al., 2007a]. Often, multiple isoforms are found in a single sample. It will be interesting to determine the relative leukemic potential of the various isoforms, and if their differential expression has any correlation with prognosis.
MECHANISTIC HINTS FROM POINT MUTATIONS IN RUNX1
To date, much of the research on CBF leukemias has centered on the assumption that RUNX1 directly binds the promoters of target genes in order to regulate their expression. However, there is increasing evidence that RUNX1 has DNA binding-independent activities. In some instances, RUNX1 may be recruited to the promoters of target genes through protein:protein interactions with other transcription factors [Pabst et al., 2001; Wheeler et al., 2002]. Recently, Cammenga et al [Cammenga et al., 2007] reported that point mutations in the RHD of RUNX1 found in patients with AML subtype M0 led to a gain of function activity for the RUNX1 protein. When these RHD mutants, which are not capable of binding DNA, were expressed in murine bone marrow (BM) cells, they led to an increase in serial replating efficiency and the accumulation of cells with a blast like morphology, similar to that seen with AML1-ETO. Interestingly, it was found that CBFβ interaction was not required for this activity. Although loss of RUNX1 had similar effects on serial replating as expression of the RHD mutants, it did not readily lead to immortalization of BM cells, indicating that the RHD mutants have a gain of function activity through a DNA-binding independent mechanism. From these observations, the authors argue that normal hematopoiesis requires a balance between RUNX1’s DNA binding dependent and independent activities, and that disruption of this balance leads to leukemogenesis.
This model could potentially apply to both CBFβ-SMMHC and AML1-ETO. In the case of AML1-ETO, it is clear that binding DNA is required for its leukemic activity [Kwok et al., 2009; Roudaia et al., 2009; Yan et al., 2009]. However, it is not known if the fusion protein affects RUNX1’s DNA binding independent functions, thus upsetting the balance between the two activities. Interestingly, it was recently shown by chromatin immunoprecipitation that AML1-ETO is associated with promoters lacking a known RUNX1 binding site, but enriched for sites of other hematopoiesis related transcription factors [Gardini et al., 2008]. This finding is consistent with the possibility that AML1-ETO can form complexes with other transcription factors that provide the DNA binding activity and target gene specificity.
FINAL THOUGHTS
With the development of imatinib for the treatment of chronic myeloid leukemia (CML) in patients with the BCR-ABL translocation [Druker et al., 2001a; Druker et al., 2001b; Druker et al., 1996], much attention has been focused on the development of drugs that specifically target the fusion proteins arising from other recurrent chromosomal abnormalities. However, the development of such drugs depends on a clear understanding of the molecular mechanisms of these oncogenes. In the case of the CBF leukemias, recent findings have indicated that the activity of these fusion proteins is more complex than originally thought. Both CBFβ-SMMHC and AML1-ETO appear to repress transcription of some CBFβ/RUNX1 target genes, but also activate transcription of an alternate set of target genes. The identity of the genes in this alternate set, as well as the co-factors involved in activating their transcription have yet to be determined. However, this line of inquiry promises to yield important insights into the oncogenic mechanism of both fusion proteins, and ultimately, the development of new therapies for inv(16) and t(8:21) leukemia.
ACKNOWLEDGEMENTS
This work was supported by the Intramural Research Program of National Human Genome Research Institute, National Institutes of Health.
REFERENCES
- Adya N, Stacy T, Speck NA, Liu PP. The leukemic protein core binding factor beta (CBFbeta)-smooth-muscle myosin heavy chain sequesters CBFalpha2 into cytoskeletal filaments and aggregates. Mol Cell Biol. 1998;18:7432–7443. doi: 10.1128/mcb.18.12.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi R, Zaidi SK, Pande S, Hassan MQ, Young DW, Montecino M, Lian JB, van Wijnen AJ, Stein JL, Stein GS. The leukemogenic t(8;21) fusion protein AML1-ETO controls rRNA genes and associates with nucleolar-organizing regions at mitotic chromosomes. J Cell Sci. 2008;121:3981–3990. doi: 10.1242/jcs.033431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammenga J, Niebuhr B, Horn S, Bergholz U, Putz G, Buchholz F, Lohler J, Stocking C. RUNX1 DNA-binding mutants, associated with minimally differentiated acute myelogenous leukemia, disrupt myeloid differentiation. Cancer Res. 2007;67:537–545. doi: 10.1158/0008-5472.CAN-06-1903. [DOI] [PubMed] [Google Scholar]
- Castilla LH, Wijmenga C, Wang Q, Stacy T, Speck NA, Eckhaus M, Marin-Padilla M, Collins FS, Wynshaw-Boris A, Liu PP. Failure of embryonic hematopoiesis and lethal hemorrhages in mouse embryos heterozygous for a knocked-in leukemia gene CBFB-MYH11. Cell. 1996;87:687–696. doi: 10.1016/s0092-8674(00)81388-4. [DOI] [PubMed] [Google Scholar]
- Dissing M, Le Beau MM, Pedersen-Bjergaard J. Inversion of chromosome 16 and uncommon rearrangements of the CBFB and MYH11 genes in therapy-related acute myeloid leukemia: rare events related to DNA-topoisomerase II inhibitors? J Clin Oncol. 1998;16:1890–1896. doi: 10.1200/JCO.1998.16.5.1890. [DOI] [PubMed] [Google Scholar]
- Druker BJ, Sawyers CL, Kantarjian H, Resta DJ, Reese SF, Ford JM, Capdeville R, Talpaz M. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N Engl J Med. 2001a;344:1038–1042. doi: 10.1056/NEJM200104053441402. [DOI] [PubMed] [Google Scholar]
- Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, Lydon NB, Kantarjian H, Capdeville R, Ohno-Jones S, Sawyers CL. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001b;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- Druker BJ, Tamura S, Buchdunger E, Ohno S, Segal GM, Fanning S, Zimmermann J, Lydon NB. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- Erickson P, Gao J, Chang KS, Look T, Whisenant E, Raimondi S, Lasher R, Trujillo J, Rowley J, Drabkin H. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood. 1992;80:1825–1831. [PubMed] [Google Scholar]
- Fazi F, Racanicchi S, Zardo G, Starnes LM, Mancini M, Travaglini L, Diverio D, Ammatuna E, Cimino G, Lo-Coco F, Grignani F, Nervi C. Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell. 2007;12:457–466. doi: 10.1016/j.ccr.2007.09.020. [DOI] [PubMed] [Google Scholar]
- Gardini A, Cesaroni M, Luzi L, Okumura AJ, Biggs JR, Minardi SP, Venturini E, Zhang DE, Pelicci PG, Alcalay M. AML1/ETO oncoprotein is directed to AML1 binding regions and co-localizes with AML1 and HEB on its targets. PLoS Genet. 2008;4:e1000275. doi: 10.1371/journal.pgen.1000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilman SA, Kuo YH, Goudswaard CS, Valk PJ, Castilla LH. Cbfbeta reduces Cbfbeta-SMMHC-associated acute myeloid leukemia in mice. Cancer Res. 2006;66:11214–11218. doi: 10.1158/0008-5472.CAN-06-0959. [DOI] [PubMed] [Google Scholar]
- Huang G, Shigesada K, Ito K, Wee HJ, Yokomizo T, Ito Y. Dimerization with PEBP2beta protects RUNX1/AML1 from ubiquitin-proteasome-mediated degradation. Embo J. 2001;20:723–733. doi: 10.1093/emboj/20.4.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde RK, Kamikubo Y, Anderson S, Kirby M, Alemu L, Zhao L, Liu PP. Cbfb/Runx1-repression independent blockage of differentiation and accumulation of Csf2rb expressing cells by Cbfb-MYH11. Blood. 2009 doi: 10.1182/blood-2009-06-227413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klampfer L, Zhang J, Zelenetz AO, Uchida H, Nimer SD. The AML1/ETO fusion protein activates transcription of BCL-2. Proc Natl Acad Sci U S A. 1996;93:14059–14064. doi: 10.1073/pnas.93.24.14059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok C, Zeisig BB, Qiu J, Dong S, So CW. Transforming activity of AML1-ETO is independent of CBFbeta and ETO interaction but requires formation of homo-oligomeric complexes. Proc Natl Acad Sci U S A. 2009;106:2853–2858. doi: 10.1073/pnas.0810558106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Beau MM, Larson RA, Bitter MA, Vardiman JW, Golomb HM, Rowley JD. Association of an inversion of chromosome 16 with abnormal marrow eosinophils in acute myelomonocytic leukemia. A unique cytogenetic-clinicopathological association. N Engl J Med. 1983;309:630–636. doi: 10.1056/NEJM198309153091103. [DOI] [PubMed] [Google Scholar]
- Linggi B, Muller-Tidow C, van de Locht L, Hu M, Nip J, Serve H, Berdel WE, van der Reijden B, Quelle DE, Rowley JD, Cleveland J, Jansen JH, Pandolfi PP, Hiebert SW. The t(8;21) fusion protein, AML1 ETO specifically represses the transcription of the p14(ARF) tumor suppressor in acute myeloid leukemia. Nat Med. 2002;8:743–750. doi: 10.1038/nm726. [DOI] [PubMed] [Google Scholar]
- Liu P, Tarl:e SA, Hajra A, Claxton DF, Marlton P, Freedman M, Siciliano MJ, Collins FS. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science. 1993;261:1041–1044. doi: 10.1126/science.8351518. [DOI] [PubMed] [Google Scholar]
- Liu PP, Hajra A, Wijmenga C, Collins FS. Molecular pathogenesis of the chromosome 16 inversion in the M4Eo subtype of acute myeloid leukemia. Blood. 1995;85:2289–2302. [PubMed] [Google Scholar]
- Lukasik SM, Zhang L, Corpora T, Tomanicek S, Li Y, Kundu M, Hartman K, Liu PP, Laue TM, Biltonen RL, Speck NA, Bushweller JH. Altered affinity of CBF beta-SMMHC for Runx1 explains its role in leukemogenesis. Nat Struct Biol. 2002;9:674–679. doi: 10.1038/nsb831. [DOI] [PubMed] [Google Scholar]
- Lutterbach B, Hou Y, Durst KL, Hiebert SW. The inv(16) encodes an acute myeloid leukemia 1 transcriptional corepressor. Proc Natl Acad Sci U S A. 1999;96:12822–12827. doi: 10.1073/pnas.96.22.12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheny CJ, Speck ME, Cushing PR, Zhou Y, Corpora T, Regan M, Newman M, Roudaia L, Speck CL, Gu TL, Griffey SM, Bushweller JH, Speck NA. Disease mutations in RUNX1 and RUNX2 create nonfunctional, dominant-negative, or hypomorphic alleles. EMBO J. 2007;26:1163–1175. doi: 10.1038/sj.emboj.7601568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minelli A, Maserati E, Rossi G, Bernardo ME, De Stefano P, Cecchini MP, Valli R, Albano V, Pierani P, Leszl A, Sainati L, Lo Curto F, Danesino C, Locatelli F, Pasquali F. Familial platelet disorder with propensity to acute myelogenous leukemia: genetic heterogeneity and progression to leukemia via acquisition of clonal chromosome anomalies. Genes Chromosomes Cancer. 2004;40:165–171. doi: 10.1002/gcc.20030. [DOI] [PubMed] [Google Scholar]
- Niki M, Okada H, Takano H, Kuno J, Tani K, Hibino H, Asano S, Ito Y, Satake M, Noda T. Hematopoiesis in the fetal liver is impaired by targeted mutagenesis of a gene encoding a non-DNA binding subunit of the transcription factor, polyomavirus enhancer binding protein 2/core binding factor. Proc Natl Acad Sci U S A. 1997;94:5697–5702. doi: 10.1073/pnas.94.11.5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa E, Inuzuka M, Maruyama M, Satake M, Naito-Fujimoto M, Ito Y, Shigesada K. Molecular cloning and characterization of PEBP2 beta, the heterodimeric partner of a novel Drosophila runt-related DNA binding protein PEBP2 alpha. Virology. 1993a;194:314–331. doi: 10.1006/viro.1993.1262. [DOI] [PubMed] [Google Scholar]
- Ogawa E, Maruyama M, Kagoshima H, Inuzuka M, Lu J, Satake M, Shigesada K, Ito Y. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc Natl Acad Sci U S A. 1993b;90:6859–6863. doi: 10.1073/pnas.90.14.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H, Watanabe T, Niki M, Takano H, Chiba N, Yanai N, Tani K, Hibino H, Asano S, Mucenski ML, Ito Y, Noda T, Satake M. AML1(−/−) embryos do not express certain hematopoiesis-related gene transcripts including those of the PU.1 gene. Oncogene. 1998;17:2287–2293. doi: 10.1038/sj.onc.1202151. [DOI] [PubMed] [Google Scholar]
- Okuda T, Cai Z, Yang S, Lenny N, Lyu CJ, van Deursen JM, Harada H, Downing JR. Expression of a knocked-in AML1-ETO leukemia gene inhibits the establishment of normal definitive hematopoiesis and directly generates dysplastic hematopoietic progenitors. Blood. 1998;91:3134–3143. [PubMed] [Google Scholar]
- Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- Okumura AJ, Peterson LF, Okumura F, Boyapati A, Zhang DE. t(8;21)(q22;q22) Fusion proteins preferentially bind to duplicated AML1/RUNX1 DNA-binding sequences to differentially regulate gene expression. Blood. 2008;112:1392–1401. doi: 10.1182/blood-2007-11-124735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osato M. Point mutations in the RUNX1/AML1 gene: another actor in RUNX leukemia. Oncogene. 2004;23:4284–4296. doi: 10.1038/sj.onc.1207779. [DOI] [PubMed] [Google Scholar]
- Pabst T, Mueller BU, Harakawa N, Schoch C, Haferlach T, Behre G, Hiddemann W, Zhang DE, Tenen DG. AML1-ETO downregulates the granulocytic differentiation factor C/EBPalpha in t(8;21) myeloid leukemia. Nat Med. 2001;7:444–451. doi: 10.1038/86515. [DOI] [PubMed] [Google Scholar]
- Park S, Speck NA, Bushweller JH. The role of CBFbeta in AML1-ETO's activity. Blood. 2009;114:2849–2850. doi: 10.1182/blood-2009-07-231233. [DOI] [PubMed] [Google Scholar]
- Peterson LF, Boyapati A, Ahn EY, Biggs JR, Okumura AJ, Lo MC, Yan M, Zhang DE. Acute myeloid leukemia with the 8q22;21q22 translocation: secondary mutational events and alternative t(8;21) transcripts. Blood. 2007a;110:799–805. doi: 10.1182/blood-2006-11-019265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson LF, Yan M, Zhang DE. The p21Waf1 pathway is involved in blocking leukemogenesis by the t(8;21) fusion protein AML1-ETO. Blood. 2007b;109:4392–4398. doi: 10.1182/blood-2006-03-012575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson LF, Zhang DE. The 8;21 translocation in leukemogenesis. Oncogene. 2004;23:4255–4262. doi: 10.1038/sj.onc.1207727. [DOI] [PubMed] [Google Scholar]
- Rosenbauer F, Wagner K, Kutok JL, Iwasaki H, Le Beau MM, Okuno Y, Akashi K, Fiering S, Tenen DG. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat Genet. 2004;36:624–630. doi: 10.1038/ng1361. [DOI] [PubMed] [Google Scholar]
- Roudaia L, Cheney MD, Manuylova E, Chen W, Morrow M, Park S, Lee CT, Kaur P, Williams O, Bushweller JH, Speck NA. CBFbeta is critical for AML1-ETO and TEL-AML1 activity. Blood. 2009;113:3070–3079. doi: 10.1182/blood-2008-03-147207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roumier C, Fenaux P, Lafage M, Imbert M, Eclache V, Preudhomme C. New mechanisms of AML1 gene alteration in hematological malignancies. Leukemia. 2003;17:9–16. doi: 10.1038/sj.leu.2402766. [DOI] [PubMed] [Google Scholar]
- Rowley JD. Identificaton of a translocation with quinacrine fluorescence in a patient with acute leukemia. Ann Genet. 1973;16:109–112. [PubMed] [Google Scholar]
- Sasaki K, Yagi H, Bronson RT, Tominaga K, Matsunashi T, Deguchi K, Tani Y, Kishimoto T, Komori T. Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor beta. Proc Natl Acad Sci U S A. 1996;93:12359–12363. doi: 10.1073/pnas.93.22.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada H, Ichikawa H, Nakamura S, Katsu R, Iwasa M, Kitabayashi I, Ohki M. Analysis of genes under the downstream control of the t(8;21) fusion protein AML1-MTG8: overexpression of the TIS11b (ERF-1, cMG1) gene induces myeloid cell proliferation in response to G-CSF. Blood. 2000;96:655–663. [PubMed] [Google Scholar]
- Silva FP, Swagemakers SM, Erpelinck-Verschueren C, Wouters BJ, Delwel R, Vrieling H, van der Spek P, Valk PJ, Giphart-Gassler M. Gene expression profiling of minimally differentiated acute myeloid leukemia: M0 is a distinct entity subdivided by RUNX1 mutation status. Blood. 2009 doi: 10.1182/blood-2009-03-211334. [DOI] [PubMed] [Google Scholar]
- Speck NA, Gilliland DG. Core-binding factors in haematopoiesis and leukaemia. Nat Rev Cancer. 2002;2:502–513. doi: 10.1038/nrc840. [DOI] [PubMed] [Google Scholar]
- Tang JL, Hou HA, Chen CY, Liu CY, Chou WC, Tseng MH, Huang CF, Lee FY, Liu MC, Yao M, Huang SY, Ko BS, Hsu SC, Wu SJ, Tsay W, Chen YC, Lin LI, Tien HF. AML1/RUNX1 mutations in 470 adult patients with de novo acute myeloid leukemia: prognostic implication and interaction with other gene alterations. Blood. 2009;114:5352–5361. doi: 10.1182/blood-2009-05-223784. [DOI] [PubMed] [Google Scholar]
- Tang YY, Shi J, Zhang L, Davis A, Bravo J, Warren AJ, Speck NA, Bushweller JH. Energetic and functional contribution of residues in the core binding factor beta (CBFbeta ) subunit to heterodimerization with CBFalpha. J Biol Chem. 2000;275:39579–39588. doi: 10.1074/jbc.M007350200. [DOI] [PubMed] [Google Scholar]
- Van der Reijden BA, de Wit L, van der Poel S, Luiten EB, Lafage-Pochitaloff M, Dastugue N, Gabert J, Lowenberg B, Jansen JH. Identification of a novel CBFB-MYH11 transcript: implications for RT-PCR diagnosis. Hematol J. 2001;2:206–209. doi: 10.1038/sj.thj.6200103. [DOI] [PubMed] [Google Scholar]
- Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe AH, Speck NA. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci U S A. 1996a;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Stacy T, Miller JD, Lewis AF, Gu TL, Huang X, Bushweller JH, Bories JC, Alt FW, Ryan G, Liu PP, Wynshaw-Boris A, Binder M, Marin-Padilla M, Sharpe AH, Speck NA. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell. 1996b;87:697–708. doi: 10.1016/s0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- Wang S, Wang Q, Crute BE, Melnikova IN, Keller SR, Speck NA. Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Mol Cell Biol. 1993;13:3324–3339. doi: 10.1128/mcb.13.6.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler JC, VanderZwan C, Xu X, Swantek D, Tracey WD, Gergen JP. Distinct in vivo requirements for establishment versus maintenance of transcriptional repression. Nat Genet. 2002;32:206–210. doi: 10.1038/ng942. [DOI] [PubMed] [Google Scholar]
- Yan M, Ahn EY, Hiebert SW, Zhang DE. RUNX1/AML1 DNA-binding domain and ETO/MTG8 NHR2-dimerization domain are critical to AML1-ETO9a leukemogenesis. Blood. 2009;113:883–886. doi: 10.1182/blood-2008-04-153742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M, Burel SA, Peterson LF, Kanbe E, Iwasaki H, Boyapati A, Hines R, Akashi K, Zhang DE. Deletion of an AML1-ETO C-terminal NcoR/SMRT-interacting region strongly induces leukemia development. Proc Natl Acad Sci U S A. 2004;101:17186–17191. doi: 10.1073/pnas.0406702101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M, Kanbe E, Peterson LF, Boyapati A, Miao Y, Wang Y, Chen IM, Chen Z, Rowley JD, Willman CL, Zhang DE. A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nat Med. 2006;12:945–949. doi: 10.1038/nm1443. [DOI] [PubMed] [Google Scholar]
- Yergeau DA, Hetherington CJ, Wang Q, Zhang P, Sharpe AH, Binder M, Marin-Padilla M, Tenen DG, Speck NA, Zhang DE. Embryonic lethality and impairment of haematopoiesis in mice heterozygous for an AML1-ETO fusion gene. Nat Genet. 1997;15:303–306. doi: 10.1038/ng0397-303. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Dowdy CR, van Wijnen AJ, Lian JB, Raza A, Stein JL, Croce CM, Stein GS. Altered Runx1 subnuclear targeting enhances myeloid cell proliferation and blocks differentiation by activating a miR-24/MKP-7/MAPK network. Cancer Res. 2009;69:8249–8255. doi: 10.1158/0008-5472.CAN-09-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]