Summary
The utility of induced pluripotent stem cells (iPSCs) as models to study diseases and as sources for cell therapy depends on the integrity of their genomes. Despite recent publications of DNA sequence variations in the iPSCs, the true scope of such changes for the entire genome is not clear. Here we report the whole-genome sequencing of three human iPSC lines derived from two cell types of an adult donor by episomal vectors. The vector sequence was undetectable in the deeply sequenced iPSC lines. We identified 1058–1808 heterozygous single nucleotide variants (SNVs), but no copy number variants, in each iPSC line. Six to twelve of these SNVs were within coding regions in each iPSC line, but ~50% of them are synonymous changes and the remaining are not selectively enriched for known genes associated with cancers. Our data thus suggest that episome-mediated reprogramming is not inherently mutagenic during integration-free iPSC induction.
Keywords: Human iPS cells, Reprogramming, Episomal vectors, Integration-free, Genetic mutations, Whole Genome Sequencing
Introduction
The iPSC technology holds great promise for human stem cell biology and regenerative medicine but the reprogramming processes and the resulting iPSCs remain incompletely characterized. Specifically, it is not clear how many changes occur at the DNA level during reprogramming. With recently available technologies such as single nucleotide polymorphism (SNP) array and exome sequencing, several recent studies reported first glimpses of genetic abnormalities in human iPSCs derived from fibroblasts (Gore et al., 2011; Hussein et al., 2011; Laurent et al., 2011; Martins-Taylor K et al., 2011; Mayshar et al, 2010). Nucleotide substitutions, copy number variation (CNV) changes, and other chromosomal aberrations, which are either pre-existing or generated during reprogramming, may be selected for iPSC induction and/or expansion (Pera, 2011). In addition, the origin of starting cell types may influence the quality of derived iPSCs (Kim et al., 2010; Polo et al., 2010). A whole-genome sequencing (WGS) analysis is necessary to assess potential alterations in the entire nuclear and mitochondrial genomes, because recent WGS studies confirmed the notion that DNA mutational rates in somatic cells are lower in exons than un-transcribed regions likely due to transcription-coupled DNA repair (Lee et al., 2010; Pleasance et al., 2010). In this study, we conducted WGS analysis to determine the DNA sequences of 3 human iPSC lines.
Results
Generation of multiple iPSC lines from different tissues of the same adult donor
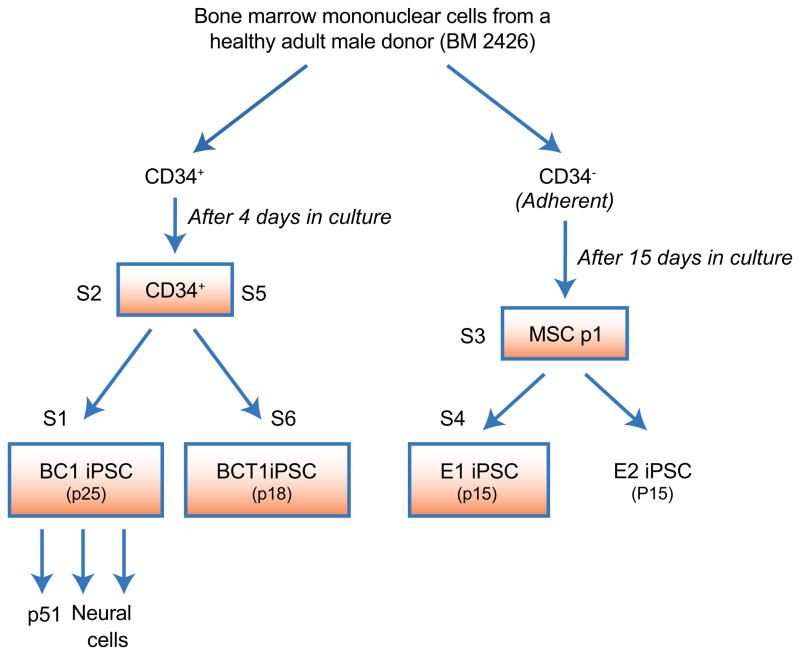
The iPSC lines were derived with plasmid vectors based on the EBNA1/OriP episomal replicon, from two cell types of a healthy donor (a 31-year-old anonymous male of mixed ethnic backgrounds). The relationship of the 4 iPSC lines and their somatic cell ancestors are shown in Figure 1. Bone marrow CD34+ cells were used to generate the BC1 iPSC line after 4-day culture with a single episomal vector (pEB-C5) expressing 5 reprogramming genes (Chou et al., 2011). The second iPSC line BCT1 was derived from the same cultured CD34+ cells by using an additional episomal vector expressing SV40 Large T antigen (SV40-LT) gene together with pEB-C5. The CD34− marrow mononuclear cells were used to establish adherent marrow stromal cells (MSCs) (Cheng et al., 2003). The established MSCs after primary and first passage (p1) in culture (15 days total) were then used to generate two iPSC lines, E1 and E2 (see Experimental Procedures). Using standard methods for characterization of expanded iPSC lines such as BC1 (Chou et al., 2011), we confirmed normal pluripotency and karyotyping (by G-banding) of iPSC lines E1 (Supplementary Figure 1), E2 (data not shown) and BCT1 (Supplemental Figure 3). For the whole-genome DNA sequencing and SNP array analyses, expanded iPSC lines were cultured under a feeder-free condition (on Matrigel) for at least one passage to reduce mouse feeder cells before total DNA was extracted. In addition, DNA was extracted by the same method from the corresponding parental cells, i.e., the cultured CD34+ cells and the p1 MSCs. DNA from these somatic samples and from iPSCs at various passages, were also used for other analyses.
Figure 1. Relationship of iPSC lines and their parental somatic cells used in this study.
Mononuclear cells from bone marrow (BM) of a healthy adult donor (2426) were separated into CD34+ cells (~1%) and CD34-depleted (CD34−) cells. The CD34+ cells were cultured for 4 days with hematopoietic cytokines before being reprogrammed by episomal vectors (left). The BC1 iPSC line was derived by using a single plasmid pEB-C5 while the BCT1 iPSC line was derived by addition of a second plasmid pEB-Tg. The BM CD34− cells were used to establish cultures of marrow stromal cells (also called mesenchymal stem cells or MSCs) by first selecting adherent cells followed by selective expansion of MSCs for additional 13 days (after primary and first passage). The harvested MSCs after first passage (p1) were used for reprogramming similarly by episomal vectors. Two independent iPSC lines, E1 and E2, were established and expanded for 15 passages (p15). Functional Characterizations of E1 and BCT1 iPSC lines are shown in Supplemental Figures 1 and 2, while characterization of BC1 iPSC line was published previously (Chou et al., 2011). Three expanded and characterized iPSC lines, BC1, BCT1 and E1 (boxed), were also analyzed by whole genome sequencing at a deep length. This was done in pair with the parental somatic cells (also boxed). While S1/S2 samples were sequenced at BGI as one pair, S3/S4 and S5/S6 samples were sequenced at NIH as two pairs.
Deep and pair-wise whole genome sequencing of 3 iPSC lines and their parental somatic cells
To reduce false positive errors of differences in shotgun DNA sequencing due to variations such as library preparations and sequencing procedures (Ajay et al., 2011; Kinde et al., 2011), we sequenced the iPSC lines (BC1, BCT1 and E1) with their parental somatic cells at the same time in a pair-wise fashion by Illumina HiSeq2000 technology. For all 3 pairs, >1.5×109 reads per sample were generated and analyzed. Table 1 shows a summary of sequence analyses of the 3 iPSC lines. We obtained alignable DNA sequences of >130 Gb for each iPSC line and their parental cells. The amount of sequence data is equivalent to 48× or higher coverage of the haploid (nuclear) genome. Sequence analysis indicated that we achieved high quality coverage (i.e., sufficient coverage in all 6 samples to call genotypes with >99.8% accuracy) for >94% of the autosomal genome of each sample. This level of deep and pair-wise sequencing of 6 related genomes (from the same person) provides us a high level of confidence to detect true and small sequence differences.
Table 1.
Summary of sequencing 3 pairs of iPSC lines and their parental somatic cells
| Features/iPSC lines* | BC1* | BCT1* | E1* |
|---|---|---|---|
| Total nucleotides sequenced | 172 Gb | 153 Gb | 217 Gb |
| Alignable nucleotides | 165 Gb | 142 Gb | 134 Gb |
| Genome coverage (fold) | 59× | 51× | 48× |
| Total SNVs (compared to the hg19 reference) | 4,158,672 (2,193,600)** | 4,207,199 (2,237,532) | 4,231,439 (2,259,512) |
| Total SNVs (compared to parental cells) | 3,234 (2,217) | 4,850 (3,458) | 4,470 (3,053) |
| High quality, filtered SNVs | 1,058 (626) | 1,109 (662) | 1,808 (1029) |
| SNVs in intergenic regions | 676 (420) | 684 (438) | 1125 (678) |
| SNVs in conserved non-coding regions | 376 (201) | 420 (225) | 674 (352) |
| SNVs in introns | 367 (206) | 409 (229) | 664 (360) |
| SNVs in 5′ or 3′ UTRs | 11 (4) | 13 (1) | 20 (3) |
| SNVs in coding regions | 6 | 6 | 12 |
| Synonymous (S) | 3 | 4 | 5 |
| Non-synonymous (NS) | 3 | 2 | 6 |
| Nonsense | 0 | 0 | 1 |
| NS:S ratio | 1 | 0.5 | 1.4 |
| Coding region indels | 0 | 0 | 2 |
| SNVs in CpG islands | 2 | 0 | 7 |
| SNVs in sno/microRNA regions | 0 | 1 | 0 |
| SNV in mitochondrial genome | 1 (nt 89) | 0 | 0 |
| Vector sequences anywhere | none | none | none |
BC1 and BCT1 iPSC lines were derived from the same batch of CD34+ cells cultured for 4 days.
E1 iPSC line were derived from MSCs established from CD34− (adherent) cells after culture of 15 days;
Parental cultured CD34+ cells (twice) and MSCs were sequenced at similar depths in each pair together with an iPSC line; while BC1 iPSC/CD34+ pair (S1/S2) was sequenced at BGI, the E1 iPSC/MSC (S3/S4) and BCT1 iPSC/CD34+ (S6/S5) pairs were sequenced at NIH.
SNVs: single nucleotide variations. The SNV counts in parentheses are those lie within regions annotated in the UCSC hg19 “RepeatMasker” track.
Absence of episomal vector sequence in the genomes of the iPSC lines
The deep sequencing of the total DNA (5 μg or from 800,000 iPSCs) provides more definitive evidence for the lack of vector DNA (either integrated or as episome) in these iPSC lines. We did not detect the pCEP4 vector backbone sequence above background in any of the 3 sequenced iPSC lines derived by episomal vectors. This conclusion was further supported by the PCR method that would detect 0.2 copies of vector DNA per cell (or a total of ~300 copies per the cell population tested) as shown in Supplemental Figures 1 and 2, and in previous studies (Chou et al., 2011).
Single Nucleotide variations in the genomes of the iPSC lines
When compared to the human reference genome sequence (hg19), we identified approximately 4.2 million single nucleotide variants (SNVs) in each of the iPSC lines as well as in their parental CD34+ cells and MSCs (Table 1). Some of these SNVs were aligned to repetitive regions of the reference genome (in parentheses). This level of sequence variations is comparable to those of other sequenced human genomes (Bentley et al., 2008; Ding et al., 2010; Lee et al., 2010; Pleasance et al., 2010; Ajay et al., 2011). When the sequences between each iPSC line and its parental cells were directly compared, 1058 to 1808 likely SNVs were identified between each iPSC line and its parental somatic cells sequenced in pairs (Table 1). All SNVs found in iPSCs were heterozygous (i.e., single-allele) changes as compared to their parental somatic cells. Importantly, none of these iPSC-associated SNVs are shared between the 3 iPSC lines. Neither did we observe any clustering of these variations in specific chromosomal regions.
SNVs in known functional elements of the genome
To investigate functional relevance of SNVs found in iPSCs after induction and expansion, we next focused on SNVs in exons, especially those not present in dbSNP (build 132) database as reported previously. High-quality sequencing revealed 6 SNVs in BC1 iPSCs residing within the coding regions of 6 different genes (Tables 1 and 2). Three of them are non-synonymous (Table 2). The paired sequencing data revealed 6 SNVs in coding regions of 6 different genes in BCT1 iPSCs (derived from the same CD34+ cells): 2 of them are non-synonymous (Tables 1 and 2). Twelve SNVs in E1 iPSCs were found in the coding regions of 12 different genes: 6 of them are non-synonymous and one is a truncation (Tables 1 and 2). In searching for small insertions or deletions (indels) in the coding sequences that are unique to the 3 iPSC lines, we only identified and confirmed a 5-nucleotide deletion in the SETD8 gene in E1 iPSCs (Table 2).
Table 2.
Sequence variants in the coding regions in BC1 and E1 iPSCs
| iPSC lines | Gene name | Chromosome | Position | Ref allele | Variant allele | Variant type |
|---|---|---|---|---|---|---|
| BC1 | CNBD1 | 8 | 88365930 | G | A | Non-synonymous |
| ITGA11 | 15 | 68650878 | T | C | Non-synonymous | |
| SLIT2 | 4 | 20490512 | C | T | Non-synonymous | |
| DHX34 | 19 | 47865827 | C | T | Synonymous | |
| DOCK1 | 10 | 128836023 | C | T | Synonymous | |
| PRPS1L1 | 7 | 18066668 | A | G | Synonymous | |
| BCT1 | ADAMTS17 | 15 | 100657067 | C | T | Non-synonymous |
| TG | 8 | 133961089 | C | T | Non-synonymous | |
| CDH4 | 20 | 60318830 | G | A | Synonymous | |
| EPPK1 | 8 | 144941146 | G | C | Synonymous | |
| MCM10 | 10 | 13213236 | C | A | Synonymous | |
| OR8K1 | 11 | 56113928 | C | T | Synonymous | |
| E1 | SDHA | 5 | 231082 | C | T | Stop |
| DHX33 | 17 | 5359463 | C | A | Non-synonymous | |
| FAM3B | 21 | 42717750 | T | A | Non-synonymous | |
| KCNH8 | 3 | 19389396 | C | A | Non-synonymous | |
| HRH1 | 3 | 11301698 | C | G | Non-synonymous | |
| IRF4 | 6 | 397216 | C | T | Non-synonymous | |
| ZNF226 | 19 | 44677036 | G | C | Non-synonymous | |
| CYP11B2 | 8 | 143999035 | C | T | Synonymous | |
| HIST3H2A | 1 | 228645201 | A | G | Synonymous | |
| HJURP | 2 | 234746286 | C | T | Synonymous | |
| PRKD2 | 19 | 47197220 | G | A | Synonymous | |
| RPL13AP6 | 10 | 112696395 | G | A | Synonymous | |
| SETD8 | 12 | 123892105 | CCAAA | -- | Del CCAAA |
In addition, there were single nucleotide variations in the 5′ or 3′ untranslated regions (UTRs), introns, and non-coding regions deemed “conserved” by the program PhyloP when run on UCSC’s 46-way multi-alignments (Table 1). All of these changes could potentially affect gene expression. Of the 1058 SNVs between BC1 iPSCs and CD34+ cells, only 2 lie in a CpG island near a promoter, and none lie within the sno/microRNA regions in the UCSC’s sno/microRNA track. For BCT1 iPSCs, there were no SNVs in the CpG islands but one in the micro RNA mir-124-2. For other changes in the E1 iPSC line, 7 of them are within CpG islands and none in the sno/microRNA regions (Table 1).
Validation of SNVs by genomic PCR and Sanger DNA sequencing
We set to confirm the presence of SNVs and indels located in the exons and selected ones in the introns or UTRs of the iPSC genomes, using genomic PCR (with specific primers shown in Supplemental Table 2) followed by Sanger sequencing. We confirmed 48 out of total 51 SNVs tested (Supplemental Table 1), for an overall confirmation rate of 94%. Two of the putative nucleotide substitutions (one in BC1 and the other in E1 iPSCs) and a single-nucleotide deletion in the ZNF479 gene in E1 iPSCs could not be confirmed by this approach, and therefore are likely to have been false positive calls. Alternatively, the unchanged alleles may have been preferentially amplified during PCR reactions, leading to confirmation failure. Thus, the vast majority of sequence changes at non-repetitive regions that are identified by deep WGS using the HiSeq2000 technology, along with appropriate filtering, are real.
Additionally, we analyzed discordance of two somatic cell types that were both sequenced together: CD34+ cells as sample S5 and MSCs as sample S3 from the same donor (Figure 1). We found 283 possible SNVs between the two cultured somatic cell types, most of them in repetitive regions (data not shown). The subsequent PCR and Sanger sequencing failed to confirm any of the 10 randomly selected putative SNVs in non-repetitive regions; therefore, they are likely false positives. Our WGS analyses corroborate with recent findings of the noise levels even by the HiSeq2000 technology, albeit very small (Ajay et al., 2011; Kinde, et al., 2011). These data also highlight the importance of validating potential SNVs revealed by WGS analysis. In addition, our data suggest that the cell cultures of CD34+ cells and MSCs for 4–15 days did not introduce somatic mutations significantly.
Analysis of SNVs in different passages of the same iPSC line or between different iPSC lines derived from the same somatic cell population
The sequence variants were stably maintained in the BC1 iPSC line, because we detected the same variants at an earlier passage (p11) and a later passage (p51) as in the sequenced BC1 iPSCs (p25). They are also present in neural progenitor cells differentiated from BC1 iPSCs (Supplemental Table 1). Importantly, none of the confirmed 16 variants tested were detected in the parental CD34+ cells or in the sibling BCT1 iPSC line (at least at the level of <0.1%). Similar results were obtained with MSC-derived iPSCs: none of the confirmed 25 variants found in E1 iPSCs were detected in a sibling iPSC line E2 or in their common parental MSCs used for reprogramming (Supplemental Table 1). In addition, the 6 heterozygous variants found in BCT1 iPSCs were not presented in the sibling BC1 iPSC line. Therefore, none of the SNVs found by WGS and further confirmed by Sanger sequencing are shared between the 3 iPSC lines.
Analysis of the mitochondrial genome
We also obtained high quality and deep coverage of the mitochondrial genomes. No iPSC-specific substitutions were found in the BCT1 and E1 iPSCs. In the BC1 iPSC line, however, there is a single substitution at nt89 (T-> C) in the 5′ highly variable, non-coding region of the mitochondrial genome (16.5-kb). This nucleotide substitution was present in all the sequencing reads of the BC1 iPSCs (see discussion below).
Absence of CNV alterations in the iPSC lines
The WGS with deep-depth and paired-end read mapping data also provides us a new way to assess CNVs changes after reprogramming as compared to parental somatic cells. We used 3 prediction programs for CNV detection: RDXplorer (Yoon et al., 2009), CNVseq (Xie and Tammi, 2009), and BreakDancer (Chen et al., 2009), on the 3 pairs of DNA samples (iPSCs vs. respective parental somatic cells) from the same person. No tenable examples of CNV differences between an iPSC line and pair-wise sequenced parental cells were found by more than one of the three methods.
To validate the absence of new CNVs in the integration-free iPSC lines derived by episomal vectors, we also used HumanOmni2.5-Quad BeadChips that measure 2.5 million SNP markers to detect CNVs and other structural changes in iPSC lines BC1 and E1, as compared to their corresponding parental somatic cells from the same person. For BC1 iPSCs, we analyzed the iPSCs at 3 different passages: 11, 28, and 51, together with the parental CD34+ cells. For E1 iPSCs, p17 was used together with its parental MSCs. The call rates of SNP alleles for all the DNA samples were >99%, and the data met the desired quality control requirements (data not shown).
Potential CNVs were detected using cnvPartition, PennCNV (Wang et al., 2005) and Nexus and summarized in Supplemental Table 3. Using the 2.5M SNP array, we were able to detect CNVs in the range of few kb in length. The 2 smallest CNVs (2.02-kb and 2.75-kb) are shown in Supplemental Figure 3, which are shared by all the 6 samples of iPSCs and parental somatic cells. The overall numbers of CNVs in the iPSCs and their parental cells are similar to other human somatic cells, since the numbers of CNVs predicted in these cells are comparable with the median value for a larger cohort of 84 unrelated human samples analyzed using PennCNV with the same parameters (Supplemental Table 3).
The potential CNVs were further evaluated by visual examination using both GenomeStudio and Nexus, and only 45 of them were deemed to be real (Table 3). Importantly, all of these 45 CNVs are shared among all 6 tested samples (both iPSCs and parental cells), except for one that is unique to BC1p51 iPSCs. After detailed analyses, this putative CNV was found to be an incomplete deletion in a region (40.42-kb) containing many segmental duplications within chromosome 7q11.21. Visual demonstrations of the incomplete deletion are shown in Supplemental Figure 4. The putative CNV or alteration was only found in the 51st passage of the BC1 iPSCs (not sequenced), but not in other 5 samples including BC1 iPSCs of earlier passages (p11 and p28; p25 was sequenced). The exact nature of the incomplete deletion in this region remains to be determined. It is also well known that extensively cultured human ESCs and iPSCs often contain CNV alterations, although not at this locus (Laurent et al., 2011; Martins-Taylor K et al., 2011). Therefore, our SNP analysis effectively validated the WGS data: as compared to parental cells, essentially no new CNVs were introduced to the episome-mediated, integration-free iPSC lines.
Table 3.
Genuine CNVs detected in the iPSCs and parental somatic cells
| Chr | Copy number | Start | End | Length | Method of Detection |
|---|---|---|---|---|---|
| 1 | 0 | 8105122 | 8114476 | 9354 | PennCNV, CNVPartition, Nexus |
| 1 | 1 | 190093507 | 190134936 | 41429 | PennCNV, CNVPartition, Nexus |
| 1 | 1 | 195089923 | 195146893 | 56970 | PennCNV, CNVPartition, Nexus |
| 2 | 3 | 95884787 | 95890722 | 5935 | CNVPartition |
| 3 | 3 | 77913197 | 77920280 | 7083 | PennCNV, CNVPartition |
| 3 | 1 | 113584277 | 113599333 | 15056 | PennCNV, CNVPartition |
| 4 | 1 | 11983684 | 11993214 | 9530 | PennCNV, CNVPartition |
| 5 | 0 | 33143879 | 33149247 | 5368 | CNVPartition |
| 6 | 1 | 79020533 | 79092458 | 71925 | CNVPartition |
| 6 | 1 | 93631870 | 93635080 | 3210 | CNVPartition |
| 6 | 1 | 113012462 | 113023906 | 11444 | CNVPartition |
| 7 | 1 | 49144752 | 49150490 | 5738 | CNVPartition |
| 7 | 1 | 139841470 | 139847922 | 6452 | CNVPartition |
| 7 | 1 | 145424894 | 145427315 | 2421 | CNVPartition |
| 7 | 1 (deletion) * | 66762793 | 66803212 | 40419 | CNVPartition, Nexus |
| 7 | 3 | 151512420 | 151745955 | 233536 | Nexus |
| 7 | 0 | 20717299 | 20720053 | 2754 | PennCNV, CNVPartition, Nexus |
| 8 | 1 | 63378480 | 63385988 | 7508 | CNVPartition |
| 8 | 1 | 39351738 | 39499553 | 147815 | PennCNV, CNVPartition |
| 9 | 1 | 580454 | 598622 | 18168 | PennCNV, CNVPartition |
| 9 | 1 | 4491352 | 4494129 | 2777 | PennCNV, CNVPartition |
| 10 | 1 | 91988554 | 91991990 | 3436 | CNVPartition |
| 10 | 1 | 26725084 | 26730905 | 5821 | PennCNV, CNVPartition |
| 10 | 3 | 46561933 | 47173875 | 611942 | PennCNV, CNVPartition, Nexus |
| 11 | 0 | 102254658 | 102261874 | 7216 | CNVPartition |
| 12 | 1 | 12424251 | 12433136 | 8885 | CNVPartition |
| 12 | 1 | 121202830 | 121215756 | 12926 | CNVPartition |
| 12 | 0 | 128624494 | 128628185 | 3691 | CNVPartition |
| 12 | 1 | 103185002 | 103193085 | 8084 | Nexus |
| 12 | 3 | 34211024 | 34744278 | 533254 | PennCNV, CNVPartition, Nexus |
| 13 | 3 | 62488915 | 62512304 | 23389 | CNVPartition, Nexus |
| 13 | 0 | 31430761 | 31435768 | 5007 | PennCNV, CNVPartition, Nexus |
| 13 | 1 | 36968549 | 37018275 | 49726 | PennCNV, CNVPartition, Nexus |
| 15 | 1 | 22615864 | 22635687 | 19823 | CNVPartition |
| 15 | 1 | 79809091 | 79958158 | 149067 | PennCNV, CNVPartition, Nexus |
| 15 | 1 | 22010825 | 22104326 | 93,502 | PennCNV, Nexus |
| 16 | 0 | 163625 | 165653 | 2028 | CNVPartition, Nexus |
| 16 | 1 | 20421133 | 20440897 | 19764 | PennCNV, CNVPartition |
| 16 | 1 | 33849764 | 33929519 | 79755 | PennCNV, CNVPartition |
| 16 | 3 | 44973739 | 45446855 | 473116 | PennCNV, CNVPartition, Nexus |
| 17 | 1 | 76309874 | 76312824 | 2950 | CNVPartition |
| 17 | 1 | 46346285 | 46353510 | 7225 | PennCNV, CNVPartition |
| 19 | 1 | 23974942 | 23977215 | 2273 | CNVPartition |
| 19 | 3 | 21623408 | 21639452 | 16,045 | PennCNV |
| 20 | 0 | 55019459 | 55021582 | 2124 | Nexus |
Asterisk (*) denotes a deletion only found in BC1 p51 (also see Supplemental Figure 4). All others were shared in all 6 samples.
Discussion
We report here results of WGS analyses of 3 iPSC lines and their parental somatic cells that were reprogrammed by episomal vectors. We confirmed that the episomal vector DNA did not integrate in the genome or persist in the 3 characterized iPSC lines, nor did it alter the structures of the iPSC genomes at detectable levels. The 3 fully sequenced iPSC lines derived from two different cell types of the same person would provide valuable references or standards with DNA sequence information, for future studies of genomic integrity and epigenomics (such as DNA methylation) of iPSCs before and after differentiation.
The present study corroborates with a recent report that ~6 SNVs or small sequence changes were found in exonic regions of examined iPSC lines derived from fibroblasts by various reprogramming methods (Gore et al., 2011). However, our deep DNA sequencing of the whole genome also allowed us to detect SNVs and other sequence changes in non-exonic regions (>98%) of the nuclear genome and in the mitochondrial genome. We found 1058–1808 sequence changes, mostly SNVs, per genome in the 3 iPSCs after induction and expansion. Currently it is unclear exactly where and when these iPSC-associated SNVs arose. At least two possibilities can be envisioned, which are not mutually exclusive. First, these SNVs are simply normal mitotic mutations during iPSC induction (presumably from a single cell) and/or subsequent expansion before WGS. Second, a founder somatic cell from adult tissues that has been reprogrammed to a clonal iPSC line may already contain most if not all these SNVs. Both are related to the fact that somatic DNA mutations occur along mitotic cell divisions both in vivo and in vitro.
Estimation of mutational rates in somatic cells varies significantly by previous methods, which typically depend on the expression of a functional gene (Araten et al., 2005; Lynch, 2010). Recent WGS analysis provides a more definitive and selection-independent method to measure mutational frequencies in both exonic and non-exonic regions in normal primary cells as well as cultured human cell lines (Kinde et al, 2011). Based on these studies, we used the estimation of 3 to 30 mutations per haploid genome per mitotic division for somatic cells in the following discussion. Formation of a sizable iPSC colony (~1000 cells), presumably from a single somatic cell during our episome-mediated reprogramming that takes ~2 weeks and requires ~10 cell divisions. Subsequently we expanded iPSCs for 15–25 passages, with an estimated >3 cell divisions per passage, before sequencing. Therefore, it is conceivable that some of the SNVs we observed in iPSCs may have arisen during iPSC induction and subsequent expansion (the first possibility). However, the SNVs found in iPSCs could have also been inherited from a given somatic cell that was reprogrammed successfully (the second possibility). Considering that a typical human somatic cell is derived from a fertilized egg after 46–47 cell divisions during embryonic, fetal and postnatal development, each somatic cell is expected to harbor 138–1410 spontaneous mutations that differ from those in another cell in the population. Since we set the algorithms to consider a sequence change to be real if it is present in at least 10% of the reads (both nuclear and mitochondrial DNA sequences), the detected SNVs should have been either pre-existing in a founder somatic cell that was reprogrammed or acquired within ~3–4 cell divisions after iPSC induction (when a single colony was formed and picked). Otherwise, a later acquired mutation cannot be present in ≥10% of the derived iPSCs unless it offers a growth advantage in subsequent expansions and is preferentially retained. In our present study, we found 1058–1808 sequence changes, mostly SNVs, per genome in the 3 iPSC lines, which are within the expected ranges for a normal human somatic cell in adults. The present study therefore corroborates with a recent report that at least 50% of the SNVs or small sequence changes found by exome sequencing of the examined iPSC lines can be found in parental somatic cell populations (Gore et al., 2011). The somatic origin of SNVs was also supported by our data of the mitochondrial DNA sequencing. In the BC1 iPSC line where a single SNV was found in the non-coding region, the variant sequence was found in all the reads although a cell contains hundreds or thousands of mitochondrial DNA genomes. Overall, our WGS data indicate that reprogramming of iPSCs by episomal vectors is not inherently mutagenic. We predict that the level of SNVs found in the iPSC lines is likely of the same magnitude as those found in other adult somatic (stem) cells after extended proliferation.
It is possible that somatic cells harboring sequence variations that favor iPSC induction and expansion could have been selected for iPSC reprogramming. Those iPSCs with additional sequence variants generated during early passages that favor iPSC growth may be enriched during clonal expansion. Although we cannot rule out these possibilities completely, they do not seem to be likely for the 3 iPSC lines studied here, based on the following two reasons. First, bioinformatic analysis of the genes harboring non-synonymous, premature termination, and deletion variants did not reveal a recurrent pattern of mutating a single common gene or signal pathway, similar to the recent exome sequencing study (Gore et al., 2011). None of the SNVs we found (in exons and other regions) are shared among the 3 iPSCs sequenced here or with those found in the previous exome sequencing study. Second, the ratios of non-synonymous:synonymous substitutions (NS:S), which was traditionally applied to germline mutations that have evolved over a long period of evolutionary time, are 1, 0.5, and 1.4 for each iPSC line, respectively (Table 2), or 1.1 (13:12) as a group. These ratios are lower than 2.6 as reported recently for fibroblast-derived iPSC lines by exome sequencing (Gore et al., 2011), while the 3 cited cancer WGS studies reported NS:S ratios of 0.97, 1.78 and 2.64 respectively (Lee et al., 2010; Pleasance et al., 2010; Ding et al., 2010). Although the significance or relevance of using NS:S ratios in analyzing somatic mutations in cancers or iPSCs remains to be determined, the observed NS:S ratio (1:1) of SNVs in our integration-free 3 iPSC lines derived by episomal vectors did not suggest they bear such a characteristic if it is proven to be a cancer cell signature. Notably, the SNVs found in all the iPSCs sequenced by us and Gore et al. (2011) did not cluster in few genes or a group of genes encoding proteins related to a common functional pathway, such as recurrent mutations in TP53, RAS, RAF, PTEN and PIK3CA genes found in multiple cancer samples sequenced (Lee et al., 2010; Pleasance et al., 2010; Ding et al., 2010). The analysis of SNV patterns such as NS:S ratios in known functional regions further supports the notion that iPSC derivation by an improved method is not inherently tumorigenic or mutagenic. Future experimental studies, however, are still needed to determine the functional importance of the observed DNA sequence variations in iPSCs, for their growth and differentiation to various cell lineages, and whether these variations have any adverse consequences.
The robust, unbiased and increasingly affordable WGS technology for analyzing genome integrity of iPSCs and other cell types is not without limitations. Because read lengths and library fragment lengths only span a few hundred bases, and read depths are randomly distributed, accurately detecting CNVs and other structural rearrangements (such as inversions, translocations, transposon insertions, and others associated with repetitive sequences) by the current WGS technology remains difficult. Since WGS analysis alone cannot fully rule out the existence of CNV changes between iPSCs and their parental somatic cells, we also detected CNVs with the 2.5M SNP array. The data from both SNP array and WGS technologies were analyzed with multiple algorithms. We did not observe CNV changes (up to 51 passages) in these 3 iPSC lines derived by episomal vectors, while others found CNV alterations in a fraction of iPSCs derived from fibroblasts by integrating viral vectors (Hussein et al., 2011; Laurent et al., 2011; Martins-Taylor K et al., 2011). It is unclear what factors contribute to the discrepancy. The absence of detectable CNVs in our iPSC lines could be due to the fact that different types of parental somatic cells and/or reprogramming vectors were used. We used human CD34+ cells and MSCs after short cultures and episomal vectors for generating integration-free iPSCs, while the previously reported iPSCs were derived from extensively cultured fibroblasts with integrating viral vectors.
In summary, we have conducted the first deep whole-genome sequencing of 3 independently derived and functionally characterized iPSC lines from a single donor, to comprehensively document the number and type of sequence variations in these iPSC lines. Unbiased whole genome DNA sequencing is a definitive way to identify vector DNA integration and mutations in iPSC lines. The data presented in this report suggest that the genome of an iPSC line derived by episomal vectors could be largely intact: less than one SNV per megabase of DNA when compared to the parental somatic cells, despite clonal selection and many cycles of mitotic cell divisions during iPSC reprogramming and subsequent expansion. The existence of these DNA sequence variations in thousands as compared to the starting somatic cell population, however, suggests that each established iPSC line needs to be characterized thoroughly at the genomic DNA level before it is used for comprehensive functional studies and clinical applications.
Experimental Procedures
Anonymous adult human somatic cells used for reprogramming
Human primary mononuclear cells (MNCs) obtained from bone marrow and blood of anonymous donors were collected and processed at AllCells, LLC (Emeryville, CA). The consent form signed by the healthy adult male donor coded as BM2426 is available upon request. The practice of obtaining bone marrow aspirates and blood from adult donors was approved by Institutional Review Board (IRB) at AllCells. Use of anonymous human samples for laboratory research including iPSC derivation was approved by IRB and the Institutional Stem Cell Research Oversight (ISCRO) committee at Johns Hopkins University.
Human MNCs from the marrow donor BM2426 were isolated using a standard gradient protocol using Ficoll-Paque Plus (p=1.077). The human MNCs expressing a high-level of the CD34 surface marker (CD34+) were purified using the MACS magnet system and CD34 isolation beads (Miltenyi, Auburn, CA). The CD34-depleted (CD34−) MNCs were used to establish marrow stromal cells (also called mesenchymal stem cells or MSCs) by a standard protocol (Cheng, et al., 2003; Mali, et al., 2010). In brief, total un-fractionated CD34− MNCs were first cultured for 2 days in DMEM (low glucose) plus 10% FBS in standard adherent tissue culture flasks. After discarding hematopoietic cells that remained in suspension at day 2, MSCs as adherent cells were then selectively expanded until sub-confluence before harvest by trypsin digestion. The resulting adherent cells (called passage zero or p0) were replated under the same condition, expanded and harvested at sub-confluence as p1 (15 days in culture). The cultured p1 MSCs were used for cell reprogramming as well as DNA analysis as described below.
Human iPSC lines derived from adult marrow CD34+ cells and MSCs from BM2426
The BC1 iPSC line was derived from the BM2426 CD34+ cells (after in a hematopoietic culture for 4 days) by a single episomal vector pEB-C5 as previously described (Chou et al., 2011). The BCT1 iPSC line was derived from the same cultured marrow CD34+ cells as BC1, except that the second episomal vector pEB-Tg expressing SV40-LT transiently was also used in addition to pEB-C5. For reprogramming MSCs, 0.5×106 cells were nucleofected by up to 5 μg DNA plasmid using Lonza/Amaxa’s recommended MSC solution and electroporation parameter as we previously used in the DNA transposon vector study (Mali, et al., 2010). In this study, we used episomal vectors (such as combination #6) for reprogramming MSCs as described previously (Chou et al., 2011; Yu et al., 2009). Four days after transfection by 2 or 3 plasmids, human embryonic stem cell medium was added in the presence of sodium butyrate as we previously described (Mali et al., 2010; Chou et al., 2011). The efficiency of iPSC derivation from adult MSCs was ~1 per 106 transfected MSCs even when 3 episomal vectors were used. Among various clones we picked and expanded, two, E1 and E2, were fully characterized by the functional assays such as pluripotency and karyotyping. G-banded karyotyping was conducted by a certified cytogeneticist (Cheng et al., 2003; Mali, et al., 2010). In brief, at least 20 metaphases for each sample were counted and partially analyzed. At least 5 of the 20 spreads were fully analyzed in detail. Resolution of 300–450 bands was obtained. This analysis rules out mosaicism of greater than 14% with 95% confidence.
Whole Genome Sequencing (WGS) and analysis
Whole genome DNA libraries suitable for sequencing on Illumina’s sequencing platform were generated from 5 μg of genomic DNA using the TruSeq Sample Prep Kit from Illumina. The DNA was sheared to approximately 450 bp using a Covaris E210. Size selection was achieved on a Pippin Prep using 1.5% agarose cassettes (Sage Science). The libraries were sequenced on HiSeq2000 DNA Sequencers (Illumina). While samples of S1 (BC1 iPSC) and S2 (CD34+ cells) were sequenced at BGI with 90 bp paired-end reads, samples S3 (MSC), S4 (MSC-derived E1 iPSC), S5 (CD34+ cells again) and S6 (BCT1 iPSC) were sequenced at NISC with 101 bp paired-end reads (Figure 1). Details of WGS analyses are provided in supplemental materials.
Genomic DNA PCR and Sanger Sequencing
Sequence variants found by whole genome sequencing were confirmed by PCR – Sanger sequencing using ABI 3100 Genetic Analyzer (Applied Biosystems). In brief, 20 ng genomic DNA samples from iPSC lines, their parental cells and an unrelated control blood sample were used for PCR reaction with primers located about 100 to 200 bp at either side of the selected sequence variants (Supplemental Table 2). PCR reactions were cleaned with Exo-SAP-IT (Affymetrix) and followed by sequencing reactions using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). The sequence data was analyzed with Sequencher 4.10.1.
SNP Array Analysis
Genotyping was performed using the HumanOmni2.5_Quad v1.0 DNA analysis BeadChip kit (Illumina, Inc.) and 300 ng of genomic DNA per the Illumina “infinium assay” protocol (Gunderson et al., 2005). CNVs were detected using the Illumina GenomeStudio ‘in-house’ algorithm, cnvPartition v3.1.6, PennCNV and Nexus 5.1. Details of CNV analyses are provided as supplemental materials.
Supplementary Material
Highlights.
Deep whole-genome sequencing of 3 human iPSC lines generated with episomal vectors
No vector sequence found in the nuclear and mitochondrial genomes
Single nucleotide & copy number variation occurs at a normal frequency
No evidence for selective enrichment of variation at functionally relevant loci
Acknowledgments
We thank Raman Sood and Blake Carrington for PCR and Sanger sequencing for confirming mutations, Xianmin Zeng in The Buck Institute in California for providing DNA from neural progenitor cells derived from BC1 iPSCs. This study was supported in part by Johns Hopkins University and NIH grants (RC2 HL101582, U01 HL099775, and R01 HL073781), an award from the NIH Center for Regenerative Medicine (NCRM), and The Intramural Research Program of The National Human Genome Research Institute at NIH.
Footnotes
Data deposition:
The raw SNP data from the HumanOmni2.5_Quad v1.0 array are deposited at http://research.nhgri.nih.gov/. The 3 lists (A, B, and C) of all CNV calls we analyzed by each of the three methods (PennCNV, cnvPartition and Nexus) are also available at the website.
The raw sequencing data reported in this study have been submitted to the Sequence Read Archive (SRA) at NCBI (accession number of SRA048525), which can be accessed at http://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ajay SS, Parker SC, Ozel Abaan H, Fuentes Fajardo KV, Margulies EH. Accurate and comprehensive sequencing of personal genomes. Genome Res. 2011;21:1498–505. doi: 10.1101/gr.123638.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araten DJ, Golde DW, Zhang RH, Thaler HT, Gargiulo L, Notaro R, Luzzatto L. A quantitative measurement of the human somatic mutation rate. Cancer Res. 2005;65:8111–7. doi: 10.1158/0008-5472.CAN-04-1198. [DOI] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, McGrath SD, Wendl MC, Zhang Q, Locke DP, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nature Methods. 2009;6:677–81. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Hammond H, Ye Z, Zhan X, Dravid G. Human adult marrow cells support prolonged expansion of human embryonic stem cells in culture. Stem Cells. 2003;20:121–132. doi: 10.1634/stemcells.21-2-131. [DOI] [PubMed] [Google Scholar]
- Chou BK, Mali P, Huang X, Ye Z, Dowey SN, Resar LMS, Zou C, Zhang YA, Tong J, Cheng L. Efficient human iPS cell derivation by a non-integrating plasmid from blood cells with unique epigenetic and gene expression signatures. Cell Research. 2011;21:518–529. doi: 10.1038/cr.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Ellis MJ, Li S, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore A, Li Z, Fung HL, Young JE, Agarwal S, Antosiewicz-Bourget J, Canto I, Giorgetti A, Israel MA, Kiskinis E, et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson KL, Steemers FJ, Lee G, et al. A genome-wide scalable SNP genotyping assay using microarray technology. Nat Genet. 2005;37:549–554. doi: 10.1038/ng1547. [DOI] [PubMed] [Google Scholar]
- Hussein SM, Batada NN, Vuoristo S, Ching RW, Autio R, Närvä E, Ng S, Sourour M, Hämäläinen R, Olsson C, et al. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A. 2011;108:9530–5. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent LC, Ulitsky I, Slavin I, Tran H, Schork A, Morey R, Lynch C, Harness JV, Lee S, Barrero MJ, et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell. 2011;8:106–118. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Jiang Z, Liu J, Haverty PM, et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature. 2010;465:473–477. doi: 10.1038/nature09004. [DOI] [PubMed] [Google Scholar]
- Lynch M. Rate, molecular spectrum, and consequences of human mutation. Proc Natl Acad Sci USA. 2010;107:961–968. doi: 10.1073/pnas.0912629107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Chou BK, Ye Z, Zou J, Yen J, Dowey S, Brodsky RA, Ohm JE, Yu W, Baylin SB, et al. Butyrate greatly enhances derivation of human induced pluripotent stem by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells. 2010;28:713–720. doi: 10.1002/stem.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayshar Y, Ben-David U, Lavon N, Biancotti JC, Yakir B, Clark AT, Plath K, Lowry WE, Benvenisty N. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell. 2010;7:521–531. doi: 10.1016/j.stem.2010.07.017. [DOI] [PubMed] [Google Scholar]
- Martins-Taylor K, Nisler BS, Taapken SM, Compton T, Crandall L, Montgomery KD, Lalande M, Xu RH. Recurrent copy number variations in human induced pluripotent stem cells. Nat Biotechnol. 2011;29:488–91. doi: 10.1038/nbt.1890. [DOI] [PubMed] [Google Scholar]
- Pera MF. Stem cells: The dark side of induced pluripotency. Nature. 2011;471:46–47. doi: 10.1038/471046a. [DOI] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–6. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, Shioda T, et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, Hakonarson H, Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Tammi MT. CNV-seq, a new method to detect copy number variation using high-throughput sequencing. BMC Bioinformatics. 2009;10:80. doi: 10.1186/1471-2105-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S, Xuan Z, Makarov V, Ye K, Sebat J. Sensitive and accurate detection of copy number variants using read depth of coverage. Genome Res. 2009;19:1586–1592. doi: 10.1101/gr.092981.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu K, Smuga-Otto K, et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.