Abstract
Cytoskeletal proteins play a pivotal role in cytokinesis in prokaryotes and eukaryotes. Most bacteria and a major branch of the archaea called the Euryarchaeota harbor a tubulin homolog, FtsZ, which assembles into a dynamic polymeric ring structure required for cytokinesis. However, Crenarchaeota, another branch of the archaea, lack FtsZ and instead use Cdv proteins, which are homologs of the ESCRT-III-like system involved in vesicular sorting and cytokinesis in eukaryotes, for cell division. Recently, a group of Crenarchaeota that grow in non-extreme environments was found to be sufficiently divergent to warrant its own branch of the archaea called the Thaumarchaeota. Notably, Thaumarchaeota have both Cdv and FtsZ homologs, which begs the question of which system is used for cell division. In this issue of Molecular Microbiology, Pelve and colleagues tackle this question. They found that cells of the thaumarchaeon Nitrosopumilus maritimus likely divide using the Cdv system and not FtsZ, based on localization of Cdv proteins but not FtsZ to division sites. The authors also provide evidence that the cell cycle during growth of N. maritimus differs significantly from those of other archaea.
Despite the diversity represented by the three domains of life, surprisingly few distinct modes of cytokinesis exist. In the Eukarya domain, for instance, nearly all animal and fungal cells use an actin-myosin ring to separate daughter cells (Pollard, 2010). Plant cells use actin as well as microtubules to position a cell plate at the division site but not for driving growth of the cell plate (Smith, 1999). Many bacteria and some archaea also use tubulin, in the form of the FtsZ protein, for cytokinesis (Margolin, 2005). However, Crenarchaeota possess neither tubulin nor FtsZ homologues, and at least one genus, Sulfolobus, uses a completely different system for cytokinesis called the Cdv system (Cell division) (Bernander & Ettema, 2010). Two of the three Cdv proteins share significant homology with the eukaryotic endosomal sorting complex required for transport (ESCRT) system, a complex of proteins that act to pinch membranes, which are involved in formation of multivesicular bodies, virus budding, and the final abscission stage of cytokinesis in eukaryotic cells (Saksena & Emr, 2009, Guizetti et al., 2011). Specifically, archaeal proteins CdvB and CdvC resemble the eukaryotic ESCRT-III and Vps4 proteins, respectively, whereas the CdvA protein does not have a eukaryotic counterpart (Lindas et al., 2008, Samson et al., 2008).
Recently, an additional branch of archaea was proposed, called Thaumarchaeota (Brochier-Armanet et al., 2008). Among the many intriguing surprises from characterizing organisms in this branch was the discovery of both cdv and ftsZ in their genomes. In this issue, Pelve et al. test experimentally whether the thaumarchaeon they study, Nitrosopumilus maritimus, uses the Cdv system, FtsZ, or both to divide its cells. They find that Cdv seems to be active in cytokinesis but FtsZ is not, suggesting that this may be true for other members of the Thaumarchaeota. They also characterize the cell cycle of N. maritimus and identify key differences from the cell cycle parameters of Crenarchaeota.
Organisms belonging to the Archaea domain were historically classified, along with bacteria, as members of the now outdated prokaryotic kingdom Monera. Although archaea share morphological similarities with bacteria, the evolutionary lineage of archaea is distinct from bacteria, and many of the molecular mechanisms used by archaea more closely resemble those found in eukaryotes. These differences prompted the separation of archaea and bacteria into two separate domains (Woese & Fox, 1977). The Archaea domain encompasses two major phyla, the Crenarchaeota and the Euryarchaeota, however additional phyla, including the Thaumarchaeota phylum, have been proposed (Woese et al., 1990, Brochier-Armanet et al., 2008). The Thaumarchaeota phylum consists of archaeal species that were previously classified as mesophilic and psychrophilic crenarchaea. Creation of this new phylum was recommended based on gene content analyses and phylogenetic evidence that suggest hyperthermophilic crenarchaea and euryarchaea emerged later in evolutionary history than thaumarchaea (Brochier-Armanet et al., 2008).
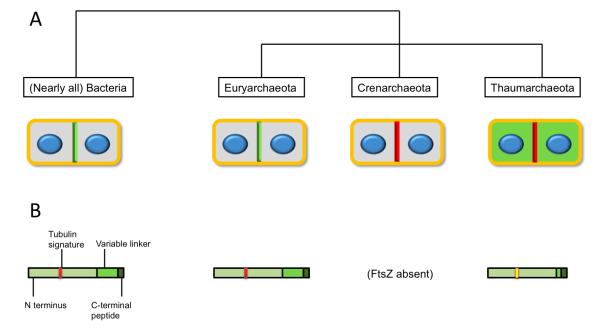
The presence or absence of ftsZ and cdv genes correlates well with archaeal phyla. Most Euryarchaea, for example, encode FtsZ but not the Cdv proteins, whereas all Crenarchaeota characterized to date lack FtsZ and, except for the Thermoproteales branch, contain Cdv proteins (Fig. 1A). Thaumarchaeota differ further, encoding both FtsZ and Cdv proteins, thus raising the question of which is used for cytokinesis in these organisms. To address this, Pelve et al. asked where the Cdv proteins localized in N. maritimus cells by using immunofluorescence microscopy with antibodies raised against CdvA, CdvC, and three CdvB homologues. They found that CdvA and CdvC localized to the cell midpoint between segregated chromosomes, mimicking the pattern observed in Sulfolobus acidocaldarius and Sulfolobus solfataricus (Lindas et al., 2008, Samson et al., 2008), as well as the typical medial FtsZ ring in bacteria (Bi & Lutkenhaus, 1991) and euryarchaea (Wang & Lutkenhaus, 1996). In addition, two of the three CdvB homologues were detected almost exclusively in cells with segregated chromosomes, although the CdvB proteins did not localize specifically between the chromosomes in these cells. In support for the Cdv system as a protein machine for cytokinesis, cells staining for CdvB also stained for CdvA and CdvC, whereas cells that lacked detectable staining for CdvB also lacked staining for CdvA and CdvC.
Fig. 1.
A. Use of FtsZ or ESCRT (Cdv) system in various prokaryotic lineages. Cells are shown at the initiation of cytokinesis, with separated daughter chromosomes in blue. A green line at the medial site of division denotes FtsZ, while a red line denotes Cdv. Green throughout the cell indicates that FtsZ is present but does not seem to localize to the site of division.
B. FtsZ protein domains present in the various prokaryotic lineages. Domains are defined in the bacterial FtsZ. The red tubulin signature motif involved in GTP binding is colored yellow for the Thaumarchaeota because of its significant divergence from consensus.
When Pelve et al. then checked for FtsZ staining in N. maritimus cells using immunofluorescence with anti-FtsZ, they saw strong fluorescence intensities, but the staining was generally uniform, with no FtsZ banding patterns between segregated chromosomes as observed with CdvA and CdvC. Moreover, FtsZ staining was observed in a majority of cells regardless of cell cycle state. Together, these results suggest that N. maritimus cells divide using the Cdv system and not FtsZ (Fig. 1A).
Pelve et al. also provide the first cell cycle characterization of a thaumarchaeon. Using flow cytometry and theoretical simulations, the authors discovered that N. maritimus cells display a long G1 phase and a relatively short post-replicative phase. This is in sharp contrast to the cell cycle organization of crenarchaeal species, which typically have a short G1 phase and long post-replicative phase. The rate of chromosome replication calculated for N. maritimus was also much slower than the rates observed in crenarchaea. The authors hypothesize that the differences in cell cycle organization and replication rate may be attributed to the distinct lifestyles of the Thaumarchaeota relative to the Crenarchaeota. Thaumarchaeota live in extremely oligotrophic environments in the deep sea, and N. maritimus has an impressively slow 33-hour generation time, which could explain the skew in its cell cycle parameters. In any case, the differences in cell cycle and cell division mechanisms between N. maritimus and Sulfolobus species further support the separation of these organisms into two separate phyla.
The current study of N. maritimus cell division and cell cycle properties addresses an important question in archaeal cell biology, yet many puzzles remain. Notably, what function, if any, does FtsZ perform in N. maritimus cells? One clue comes from the observation that FtsZ orthologs from N. maritimus and other Thaumarchaeota have divergent carboxy termini relative to other FtsZs, including those from their nearest euryarchaeal relatives (Fig. 1B). The short C termini are highly negatively charged, perhaps to form electrostatic bonds with other proteins. In addition, and perhaps more importantly, whereas FtsZ and tubulin are characterized by a highly conserved GGGTG(S/T)G signature sequence in the T4 loop for GTP binding that is crucial for FtsZ polymerization, N. maritimus FtsZ has AGKAGSA at this position instead (Fig. 1B). Other thaumarchaeal FtsZs have similarly divergent tubulin/FtsZ signature sequences. This suggests that thaumarchaeal FtsZ proteins may not form GTP-dependent filaments or need extra factors to form them, which might explain the lack of an FtsZ banding pattern in the immunofluorescence experiments.
What might a non-ring forming FtsZ do? One possibility is to function in chromosome segregation. Support for this idea comes from the activity of a homolog of FtsZ called TubZ, which helps drive plasmid segregation in Bacillus thuringiensis and other related Bacillus species (Ni et al., 2010, Larsen et al., 2007). Assuming that N. maritimus FtsZ can assemble into transient mitotic filaments for only a short time, particularly given the short post-replicative phase of the cell cycle, it may have made localized FtsZ hard to detect. Another possibility is that FtsZ is involved in cell wall growth of N. maritimus, as it is in bacteria (Varma & Young, 2004, Aaron et al., 2007). This might explain why it seems to be localized throughout the cell and not necessarily at the cell division site. In any case, it will be fascinating to unravel the different roles of FtsZ in the archaea and its interactions with the Cdv system.
Mitochondria of primitive eukaryotes and plant chloroplasts also use FtsZ in combination with dynamins to promote organelle fission, with dynamin becoming dominant over FtsZ in mitochondria of higher eukaryotes. Perhaps the same evolutionary forces were at work in selecting the Cdv system over FtsZ for cytokinesis in Thaumarchaeota and Crenarchaeota. Interestingly, some Euryarchaeota have homologs of Vps4 (CdvC) in addition to FtsZ (Makarova et al., 2010), and it will be illuminating to know which system these species have chosen for their cytokinesis.
The question of why the Cdv system might be preferred over FtsZ for cytokinesis in Thaumarchaeota can best be addressed by further understanding the molecular mechanism of Cdv-mediated cytokinesis. Yeast two-hybrid, pull-down, and co-crystallization studies in Sulfolobus spp. have uncovered a number of protein-protein interactions among its ESCRT homologues that closely resemble the associations found in the eukaryotic ESCRT system, further emphasizing the degree of conservation shared between the Cdv and ESCRT machines (Samson et al., 2008). Recently, Cdv proteins from Sulfolobus were also shown to deform liposomes, demonstrating that these proteins can indeed pinch membranes, which could facilitate constriction at division sites (Samson et al., 2011). Whether N. maritimus FtsZ can also pinch membranes like E. coli FtsZ (Allard & Cytrynbaum, 2009, Osawa et al., 2008) remains an open question. Interestingly, CdvA was recently shown to assemble into filaments on DNA in a DNA-dependent manner (Moriscot et al., 2011), by analogy to the bacterial DNA partition protein ParA2. As CdvA is also part of the cytokinesis machine, perhaps it acts like the bacterial FtsK protein, which coordinates cytokinesis with chromosome segregation (Bigot et al., 2007). These findings and those of Pelve et al. have provided valuable insight into the mechanism of Cdv-based cell division in archaea. Future genetic and biochemical studies of N. maritimus and other archaea, particularly those that have multiple candidates for cell division proteins, should certainly yield additional surprises.
Acknowledgements
Work in our laboratory is supported by a grant from the NIGMS (GM61074) and the Human Frontier Science Program.
References
- Aaron M, Charbon G, Lam H, Schwarz H, Vollmer W, Jacobs-Wagner C. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol Microbiol. 2007;64:938–952. doi: 10.1111/j.1365-2958.2007.05720.x. [DOI] [PubMed] [Google Scholar]
- Allard JF, Cytrynbaum EN. Force generation by a dynamic Z-ring in Escherichia coli cell division. Proc Natl Acad Sci U S A. 2009;106:145–150. doi: 10.1073/pnas.0808657106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernander R, Ettema TJ. FtsZ-less cell division in archaea and bacteria. Curr Opin Microbiol. 2010;13:747–752. doi: 10.1016/j.mib.2010.10.005. [DOI] [PubMed] [Google Scholar]
- Bi E, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- Bigot S, Sivanathan V, Possoz C, Barre FX, Cornet F. FtsK, a literate chromosome segregation machine. Mol Microbiol. 2007;64:1434–1441. doi: 10.1111/j.1365-2958.2007.05755.x. [DOI] [PubMed] [Google Scholar]
- Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol. 2008;6:245–252. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- Guizetti J, Schermelleh L, Mantler J, Maar S, Poser I, Leonhardt H, Muller-Reichert T, Gerlich DW. Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science. 2011;331:1616–1620. doi: 10.1126/science.1201847. [DOI] [PubMed] [Google Scholar]
- Larsen RA, Cusumano C, Fujioka A, Lim-Fong G, Patterson P, Pogliano J. Treadmilling of a prokaryotic tubulin-like protein, TubZ, required for plasmid stability in Bacillus thuringiensis. Genes Dev. 2007;21:1340–1352. doi: 10.1101/gad.1546107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindas AC, Karlsson EA, Lindgren MT, Ettema TJ, Bernander R. A unique cell division machinery in the Archaea. Proc Natl Acad Sci U S A. 2008;105:18942–18946. doi: 10.1073/pnas.0809467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Yutin N, Bell SD, Koonin EV. Evolution of diverse cell division and vesicle formation systems in Archaea. Nat Rev Microbiol. 2010;8:731–741. doi: 10.1038/nrmicro2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin W. FtsZ and the division of prokaryotic cells and organelles. Nat. Rev. Mol. Cell. Biol. 2005;6:862–871. doi: 10.1038/nrm1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriscot C, Gribaldo S, Jault JM, Krupovic M, Arnaud J, Jamin M, Schoehn G, Forterre P, Weissenhorn W, Renesto P. Crenarchaeal CdvA forms double-helical filaments containing DNA and interacts with ESCRT-III-Like CdvB. PLoS One. 2011;6:e21921. doi: 10.1371/journal.pone.0021921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L, Xu W, Kumaraswami M, Schumacher MA. Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition. Proc Natl Acad Sci U S A. 2010;107:11763–11768. doi: 10.1073/pnas.1003817107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Anderson DE, Erickson HP. Reconstitution of contractile FtsZ rings in liposomes. Science. 2008;320:792–794. doi: 10.1126/science.1154520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD. Mechanics of cytokinesis in eukaryotes. Curr Opin Cell Biol. 2010;22:50–56. doi: 10.1016/j.ceb.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksena S, Emr SD. ESCRTs and human disease. Biochem Soc Trans. 2009;37:167–172. doi: 10.1042/BST0370167. [DOI] [PubMed] [Google Scholar]
- Samson RY, Obita T, Freund SM, Williams RL, Bell SD. A role for the ESCRT system in cell division in archaea. Science. 2008;322:1710–1713. doi: 10.1126/science.1165322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RY, Obita T, Hodgson B, Shaw MK, Chong PL, Williams RL, Bell SD. Molecular and structural basis of ESCRT-III recruitment to membranes during archaeal cell division. Mol Cell. 2011;41:186–196. doi: 10.1016/j.molcel.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LG. Divide and conquer: cytokinesis in plant cells. Curr Opin Plant Biol. 1999;2:447–453. doi: 10.1016/s1369-5266(99)00022-9. [DOI] [PubMed] [Google Scholar]
- Varma A, Young KD. FtsZ collaborates with penicillin binding proteins to generate bacterial cell shape in Escherichia coli. J. Bacteriol. 2004;186:6768–6774. doi: 10.1128/JB.186.20.6768-6774.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lutkenhaus J. FtsZ ring: the eubacterial division apparatus conserved in archaebacteria. Mol. Microbiol. 1996;21:313–320. doi: 10.1046/j.1365-2958.1996.6421360.x. [DOI] [PubMed] [Google Scholar]
- Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci U S A. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]