Summary
In Caulobacter crescentus, phosphorylation of key regulators is coordinated with the second messenger cyclic di-GMP to drive cell cycle progression and differentiation. The diguanylate cyclase PleD directs pole morphogenesis while the c-di-GMP effector PopA initiates degradation of the replication inhibitor CtrA by the AAA+ protease ClpXP to license S-phase entry. Here we establish a direct link between PleD and PopA reliant on the phosphodiesterase PdeA and the diguanylate cyclase DgcB. PdeA antagonizes DgcB activity until the G1-S transition when PdeA is degraded by the ClpXP protease. The unopposed DgcB activity, together with PleD activation, upshifts c-di-GMP to drive PopA-dependent CtrA degradation and S-phase entry. PdeA degradation requires CpdR, a response regulator that delivers PdeA to the ClpXP protease in a phosphorylation-dependent manner. Thus, CpdR serves as a crucial link between phosphorylation pathways and c-di-GMP metabolism to mediate protein degradation events that irreversibly and coordinately drive bacterial cell cycle progression and development.
Keywords: Caulobacter crescentus, cyclic di-GMP, phosphodiesterase, EAL, diguanylate cyclase, GGDEF, cell cycle, cell differentiation, ClpXP, proteolysis, protease adaptor
Introduction
The development of all living organisms depends on the generation of specialized cells in appropriate numbers. This requires tight regulation of proliferation-differentiation decisions by integrating cell fate determination processes with replication and cell division. For example, embryonic development and tissue homeostasis requires a strict balance between cells in a multipotent, self-renewing state and differentiated non-replicative cells (Yadirgi and Marino, 2009). Just how differentiation and proliferation processes are coordinated remains poorly understood (Bateman and McNeill, 2004; Caro et al., 2007). Many bacteria use complex developmental strategies to optimize their survival. Like their eukaryotic counterparts, bacteria tightly coordinate morphogenetic programs with growth and division, be this to facilitate the transition between a replicative and a terminally differentiated cell form (Errington, 2003; Flardh and Buttner, 2009; Kaiser, 2008), or to couple cell differentiation to cell proliferation (Curtis and Brun, 2010). In the gram-negative bacterium Caulobacter crescentus an asymmetric division produces two daughters with distinct morphologies, behavior, and replicative potential, a motile swarmer cell and a sessile stalked cell. Whereas the newborn stalked cell immediately enters S-phase to initiate chromosome replication, the swarmer cell inherits a chromosome that remains quiescent for an extended period termed the G1-phase. Concurrent with the morphological transition of the swarmer cell into a stalked cell, the replication block is suspended and cells proceed first into S-phase to double their chromosomes and then into G2-phase to undergo cell division.
C. crescentus uses two-component phosphorylation systems to integrate cellular asymmetry with the processes that determine temporal progression through the division cycle. One of the key components of C. crescentus development and cell cycle control is the essential response regulator CtrA. CtrA functions as a transcription factor for more than 100 cell cycle genes and acts as repressor of replication initiation by directly binding to the chromosomal origin of replication (Cori) where it occludes replication initiation factors (Laub et al., 2002; Laub et al., 2000; Quon et al., 1998). Upon S-phase entry, activated CtrA∼P is eliminated by two redundant mechanisms, dephosphorylation and proteolysis (Domian et al., 1997; Domian et al., 1999), thereby freeing the Cori and enabling replication to start. Cell cycle dependent degradation of CtrA (Chien et al., 2007; Jenal and Fuchs, 1998) entails a specific spatial arrangement of the proteolytic machinery. During the G1-S transition CtrA and the ClpXP protease both transiently localize to the incipient stalked cell pole where CtrA is degraded (McGrath et al., 2006; Ryan et al., 2004). Polar localization of ClpXP and CtrA are highly regulated processes, each of which is governed by dedicated localization factors. ClpXP localization requires CpdR, a single domain response regulator that itself sequesters to the incipient stalked pole subject to its phosphorylation state (Iniesta et al., 2006). CpdR is kept in an inactive, phosphorylated form by the CckA-ChpT phosphorylation cascade that also activates CtrA, thereby coupling CtrA activity and stability (Biondi et al., 2006). Delivery of CtrA to the same pole requires PopA, a protein that sequesters to the incipient stalked pole upon binding of the second messenger cyclic di-GMP (Duerig et al., 2009). PopA activation and localization coincides with an upshift of c-di-GMP during G1-S transition (Christen et al., 2010; Paul et al., 2008) arguing that periodic fluctuations of the second messenger are an integral part of the C. crescentus cell cycle clock. The only diguanylate cyclase (DGC) known to modulate c-di-GMP levels during the cell cycle is the response regulator PleD, which is inactive in swarmer cells but is phosphorylated during differentiation to set off pole morphogenesis (Paul et al., 2008; Paul et al., 2004). Although PleD and PopA are activated simultaneously during the cell cycle, genetic analysis failed to demonstrate PleD interference with PopA-mediated CtrA degradation (Duerig et al., 2009). Hence, although c-di-GMP controls both S-phase entry and pole development it is unclear how the second messenger serves to connect these processes and how it integrates with the CckA-ChpT-CpdR phosphorelay, implicated in modulating ClpXP localization.
Here we isolate critical components of the c-di-GMP regulatory network in C. crescentus and uncover the molecular principles that serve to coordinate development and cell cycle progression. In particular, we identify the phosphodiesterase PdeA as a key regulator of the swarmer cell program. PdeA is limited to the swarmer cell where it antagonizes the activity of the diguanylate cyclase DgcB and blocks differentiation. During the G1-S transition PdeA degradation by ClpXP unleashes DgcB activity coincident with PleD activation. DgcB and PleD together establish the sessile stalked cell program, driving PopA-mediated CtrA degradation to orchestrate pole morphogenesis with the underlying cell cycle. Finally, we demonstrate that CpdR directly controls PdeA degradation by acting as a phosphorylation-dependent adaptor protein for the ClpXP protease. These experiments reveal CpdR as a central component of an interwoven network of phosphorylation, protein degradation and c-di-GMP mediated reactions that are designed to coordinately drive cell differentiation and cell cycle progression.
Results
The phosphodiesterase PdeA and the diguanylate cyclases PleD and DgcB coordinately control C. crescentus development
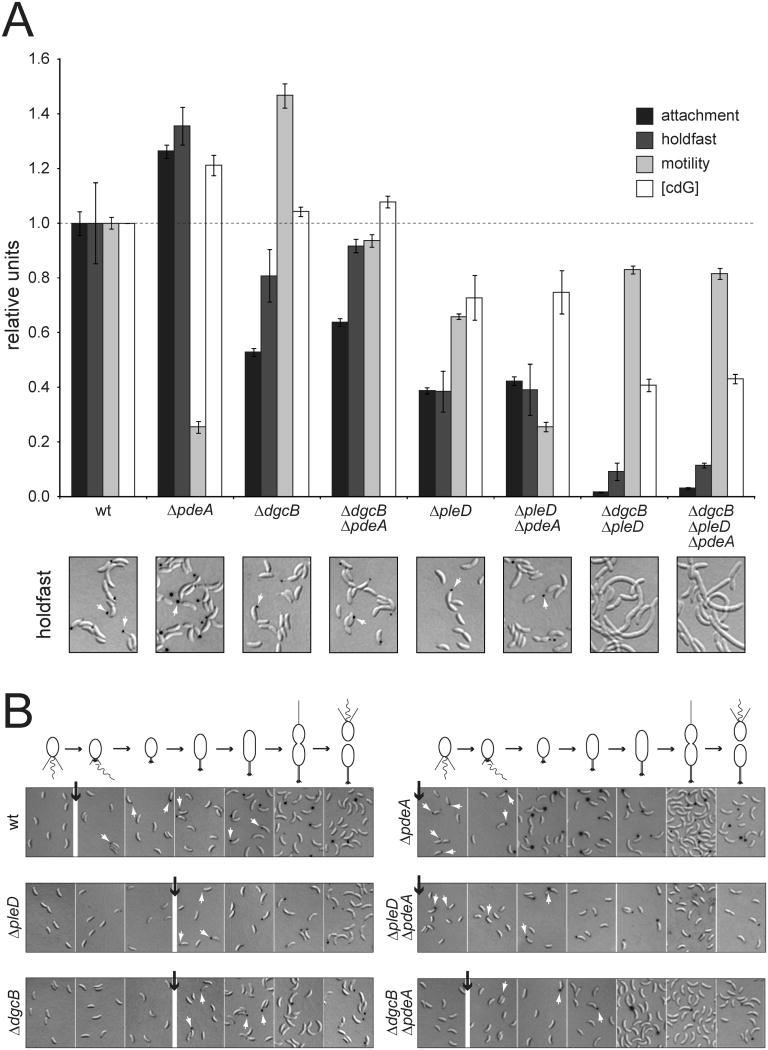
The DGC PleD and the c-di-GMP effector protein PopA have been identified as regulatory components involved in C. crescentus pole morphogenesis and cell cycle progression, respectively (Duerig et al., 2009; Paul et al., 2004). To identify additional proteins of the c-di-GMP network involved in C. crescentus cell cycle control, genes encoding potential diguanylate cyclases and phosphodiesterases were deleted individually and the resulting mutants analyzed with respect to their morphological features and cell type-specific behavior. Deletion of one of the selected genes, pdeA (CC3396), resulted in a severe loss of motility and an increased propensity to attach to surfaces, a phenotype generally associated with increased levels of c-di-GMP (Hengge, 2009) (Fig. 1A). Consistent with these observations, the pdeA gene codes for an active EAL phosphodiesterase (Christen et al., 2005) and cells lacking PdeA had increased concentrations of c-di-GMP (Fig. 1A).
Figure 1. DgcB, PleD and PdeA antagonistically regulate C. crescentus polar development.
(A) Behavior of C. crescentus wild type and c-di-GMP signaling mutants (see also Figs. S1 and S2). Surface attachment (black bars), fraction of cells producing a holdfast (dark grey), colony size on motility plates (light grey), and cellular concentration of c-di-GMP (white) are indicated. Representative examples of pictures of DIC images with overlayed fluorescent holdfast staining (black spots) are shown underneath the graph for all strains. Error bars represent the standard error of the mean. Note that pleD mutants form smaller colonies on semisolid agar plates, despite of their reported hypermotility phenotype in liquid media (Aldridge et al., 2003; Burton et al., 1997). All deletions were complemented by providing a copy of the respective gene in trans (Fig. S4 and data not shown).
(B) Timing of holdfast synthesis during the C. crescentus cell cycle. Fluorescently labeled holdfast structures are shown as in (A). Cell cycle progression is shown schematically above the micrographs. Small white arrows highlight holdfasts; black arrows indicate the time point of holdfast appearance.
Although pdeA mutants were unable to spread on semisolid agar plates, they assembled a flagellum and were motile in liquid media (data not shown). This is reminiscent of c-di-GMP mediated flagellar motility control in Escherichia coli (Boehm et al., 2010) and argued that PdeA reduces c-di-GMP to modulate the cell cycle interval reserved for motility and chemotaxis. The increased surface attachment of mutants lacking PdeA correlates with an increased number of cells exhibiting a polar holdfast (Fig. 1A). In agreement with this, pdeA mutants assembled holdfast prematurely resulting in newborn swarmer cells that ectopically express an adhesive organelle (Fig. 1B). These results suggested that PdeA is a phosphodiesterase specifically required for the motile swarmer cell program and that it acts by delaying the transition to the sessile cell type.
To identify components interacting with PdeA, we isolated motile suppressors of the pdeA mutant. One suppressor allele mapped to gene CC1850, which codes for a protein with an N-terminal coiled coil and a C-terminal GGDEF domain. Biochemical studies demonstrated that CC1850 exhibits Mg2+-dependent and Ca2+-sensitive diguanylate cyclase activity and that the protein is a constitutive dimer in vitro (Fig. S1). We thus renamed this protein DgcB. A dgcB deletion strain showed increased motility and reduced attachment levels (Fig. 1A). Deletion of dgcB in a pdeA background restored motility, attachment and holdfast biogenesis indicating that the two proteins functionally interact to coordinate pole development (Fig. 1A). Importantly, dgcB pdeA double mutants restored the proper cell cycle timing of holdfast formation (Fig. 1B) indicating that DgcB is active in G1 swarmer cells and responsible for premature holdfast synthesis observed in a pdeA mutant. In contrast, PdeA does not seem to functionally interact with the DGC PleD as pleD pdeA double and pdeA single mutants showed the same motility and holdfast timing defect and attachment of cells lacking both the PdeA and PleD remained as low as observed for pleD single mutants (Fig. 1). Since PleD is only active in sessile stalked cells (Paul et al., 2008), this raised the possibility that PdeA activity is limited to swarmer cells.
PleD and DgcB are both required for optimal attachment and holdfast biogenesis. Whereas single mutants showed delayed holdfast formation, an intermediate level of surface attachment and intermediate stalk length, strains lacking both DGCs showed markedly reduced c-di-GMP levels and a complete failure to elongate stalks, synthesize holdfast, and attach to surfaces (Figs. 1, S2). Importantly, deleting pdeA in mutants lacking both cyclases did not restore attachment, holdfast formation, and c-di-GMP levels (Fig. 1A). Together this indicated that C. crescentus cell fate determination is governed by the coordinated action of the phosphodiesterase PdeA and the diguanylate cyclases PleD and DgcB.
Regulated proteolysis limits PdeA to the motile swarmer cell type
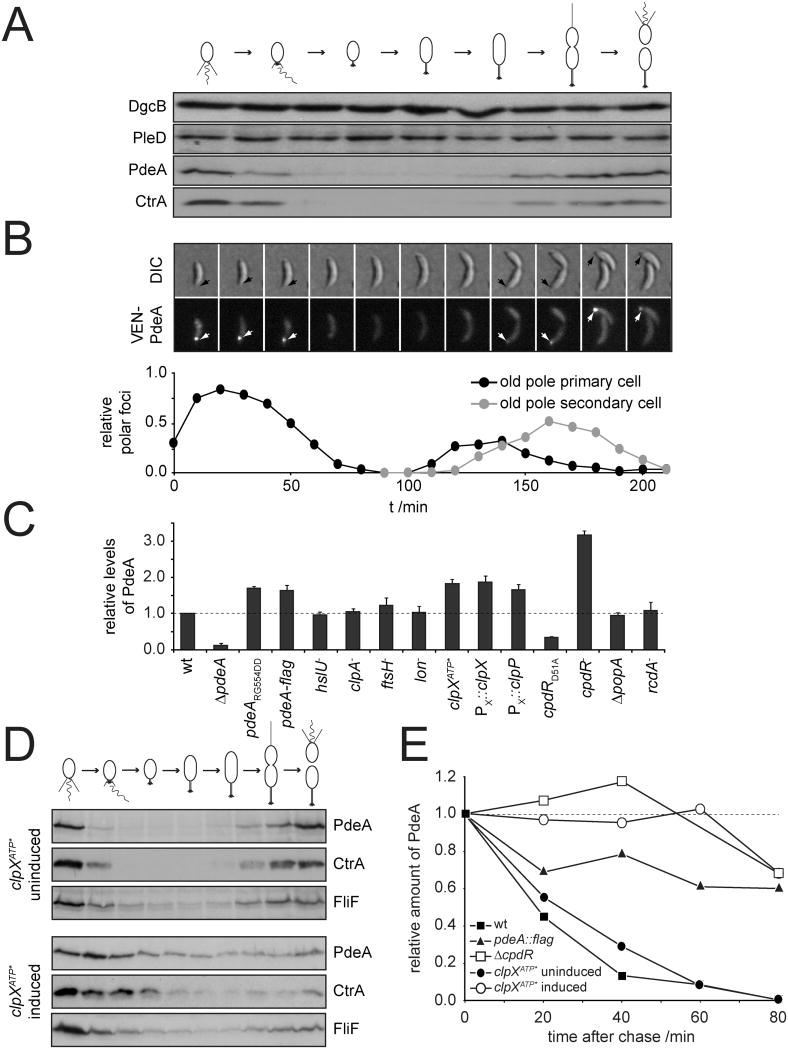
The functional analysis discussed above suggested that DgcB is active both in swarmer and in stalked cells, while PdeA seems to act exclusively in swarmer cells. With the help of polyclonal antibodies raised against purified PdeA and DgcB, we analyzed the distribution of both proteins. Whereas DgcB is present throughout the cell cycle, PdeA was only observed at the very beginning and end of the cell cycle (Fig. 2A). This indicated that PdeA is present in swarmer cells but is specifically removed prior to S-phase entry, reminiscent of the cell cycle-dependent distribution of CtrA (Domian et al., 1997) (Fig. 2A). This was confirmed by the analysis of the cellular distribution of fluorescently tagged PdeA, which transiently localized to the ClpXP occupied stalked cell pole before being cleared (Fig. 2B). To identify the protease responsible for PdeA degradation, several mutants lacking known ATP-dependent proteases were analyzed. PdeA levels were normal in hslU, clpA, ftsH, and lon mutants, but levels were increased in cells depleted for ClpP or ClpX and in cells expressing a dominant negative copy of clpX, clpXATP* (Potocka et al., 2002) (Fig. 2C). A similar increase was observed when the highly charged FLAG tag was fused to the C-terminus of PdeA or when the two amino acids at the PdeA C-terminus, Arg-Gly, were altered to Asp-Asp. This indicated that, similar to other ClpXP substrates, the C-terminus of PdeA serves as a protease recognition motif (Fig. 2C). To substantiate a specific involvement of ClpXP in cell cycle-dependent degradation of PdeA, PdeA levels were analyzed in synchronized cells containing an inducible dominant negative allele of clpX (clpXATP*). Induction of clpXATP* in synchronized swarmer cells resulted in a marked stabilization of both PdeA and CtrA during the G1-to-S transition (Fig. 2D). In contrast, stability of FliF, a protein specifically degraded by the ClpAP protease during G1-S (Grunenfelder et al., 2004), was not affected. Finally, we determined the overall stability of PdeA by pulse-chase experiments in non-synchronous populations (Fig. 2E). PdeA was degraded rapidly in wild-type cells with a half-life of about 20 minutes, but, as expected, induction of clpXATP* resulted in complete stabilization of PdeA. Likewise, PdeA-FLAG was considerably more stable than non-tagged PdeA (Fig. 2E). Together, these data indicated that PdeA is present in swarmer cells but is specifically degraded during the G1-to-S transition by the ClpXP protease complex.
Figure 2. Cell cycle dependent degradation of PdeA by the ClpXP protease.
(A) Immunoblots of synchronized cultures of C. crescentus were stained with anti-DgcB, anti-PleD, anti-PdeA or anti-CtrA antibodies as indicated.
(B) Subcellular localization of PdeA during the C. crescentus cell cycle. DIC and fluorescence images of synchronized C. crescentus ΔpdeA cells expressing an N-terminal Venus-PdeA fusion protein. Black arrows mark the old cell pole; white arrows indicate polar PdeA (see also Fig. S3 and Movie S1). The relative numbers of fluorescent foci at the old pole of the primary cell (black) and the old pole of the newbore secondary cell (grey) are depicted over time (n = 58).
(C) Quantification of cellular PdeA levels as determined by immunoblots from C. crescentus wild type and mutant strains. The dominant negative clpX allele (clpXATP*) was induced for 3 h, ClpX and ClpP depletion strains (PX∷clpX, PX∷clpP) were grown in the absence of xylose for 10 h prior to sample harvest. Data were normalized to wild type PdeA levels. Experiments were performed as independent triplicates. Error bars represent the standard error of the mean.
(D) Analysis of ClpX-dependent degradation of PdeA during the cell cycle. Strains containing the dominant negative clpX allele (clpXATP*) under the control of the xylose inducible promoter Pxyl were analyzed during the cell cycle in the presence (induced) and absence (uninduced) of xylose. Specific antibodies were used to determine levels of ClpXP (PdeA and CtrA) and ClpAP substrates (FliF).
(E) PdeA stability as determined by pulse/chase. The dominant negative clpX allele (clpXATP*) was induced with xylose for 3 hours prior to the radioactive labeling. All samples were normalized to radiolabeled PdeA present immediately after adding the chase solution.
CpdR, but not PopA and RcdA are required for cell cycle dependent proteolysis of PdeA
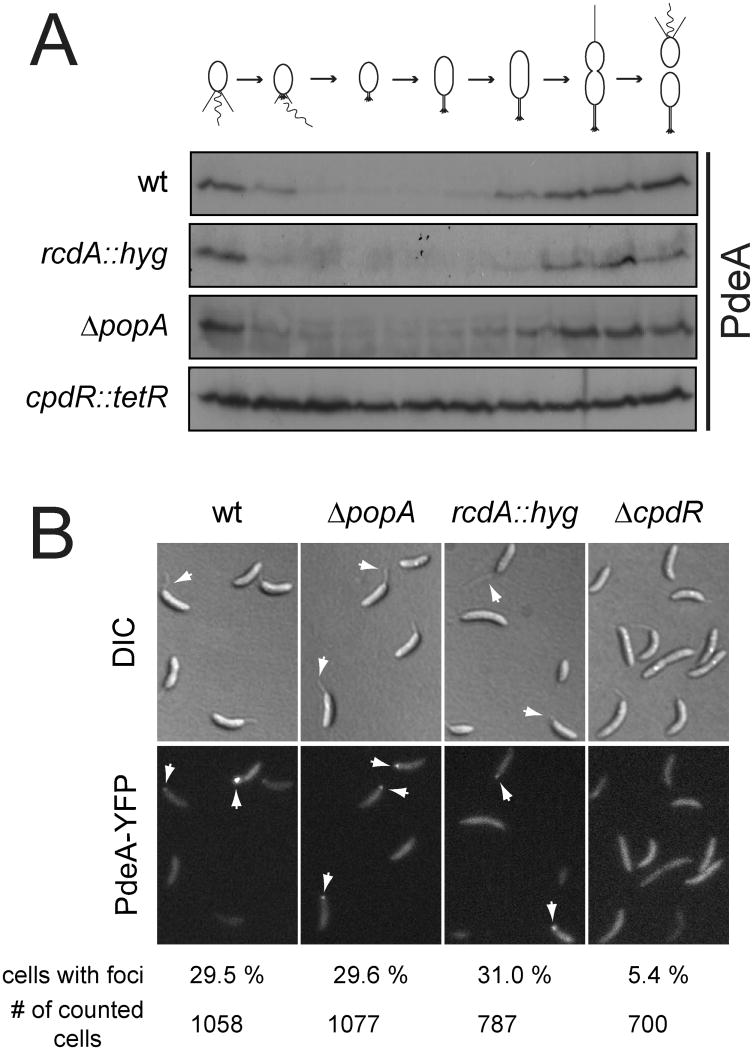
Temporal control of CtrA degradation involves several factors controlling the coordinated spatial organization of the ClpXP protease and its substrate. This includes CpdR, which is involved in recruiting ClpXP to the old cell pole upon S-phase entry (Iniesta et al., 2006) as well as PopA and RcdA, which are required for the localization of CtrA to the same subcellular site (Duerig et al., 2009; McGrath et al., 2006). To test if ClpXP degrades PdeA via the same pathway, we analyzed the dynamic distribution of PdeA throughout the cell cycle. An N-terminal fusion of PdeA to the fluorescent protein Venus was found at the flagellated pole of the newborn swarmer cell (Figs. 2B, S3A and Movie S1). During the swarmer-to-stalked cell transition VEN-PdeA disappeared from the pole coincident with its proteolytic removal (Figs. 2A, 2B, S3A and Movie S1). VEN-PdeA reappeared in the predivisional cell but upon cell constriction quickly localized to the pole in the stalked compartment, while retaining a dispersed distribution in the swarmer compartment (Figs. 2B, S3A and Movie S1). This suggested that VEN-PdeA redistributes to the ClpXP occupied old cell pole to be degraded upon entry of cells into S-phase. Consistent with this, fusion of YFP to the C-terminus of PdeA resulted in a protein that mimicked the spatial behavior of VEN-PdeA but persisted at the old cell pole due to shielding of the C-terminal ClpXP degradation motif (Fig. S3B).
To test if any of the regulators involved in CtrA degradation are required for PdeA turnover, we analyzed PdeA levels during the cell cycle in the respective mutant strains. PdeA was degraded in rcdA and popA mutants, but was completely stabilized in a mutant lacking CpdR (Figs. 2C, 2E, 3A). Moreover, in the presence of CpdRD51A, a non-phosphorylatable, constitutively active CpdR variant shown to destabilize other ClpXP substrates (Iniesta et al., 2006), PdeA levels were severely reduced (Fig. 2C). Because specific factors involved in CtrA localization are not required for PdeA degradation, we tested whether PdeA localization is dispensable for its turnover during G1-S or if an alternative degradation pathway exists for PdeA degradation. PdeA-YFP was readily detected at the stalked pole in wild-type cells and in mutants lacking PopA or RcdA, but was delocalized upon deletion of cpdR (Fig. 3B). This change was independent of the ability of CpdR to localize ClpXP as cells depleted of ClpX or ClpP showed normal PdeA localization (Fig. S5). Together, these data indicated that polar localization of PdeA is independent of the protease and of factors necessary for CtrA localization and suggested that PdeA exploits CpdR directly to regulate its localization and degradation.
Figure 3. Polar localization and degradation of PdeA requires CpdR.
(A) Cell cycle-dependent degradation of PdeA requires CpdR but not PopA or RcdA. Synchronized cultures of C. crescentus wild type and mutant strains were analyzed by immunoblots with anti-PdeA antibodies.
(B) Localization of PdeA to the cell pole requires CpdR but not PopA or RcdA. PdeA-YFP localization was analyzed in C. crescentus wild type and mutant strains (see also Figs. S4 and S5). White arrows in the DIC images mark old cell poles, arrows in the YFP channel highlight polar PdeA foci. The relative number of cells with polar foci is shown below the corresponding micrographs.
CpdR acts as phosphorylation dependent adaptor for ClpXP-mediated degradation of PdeA
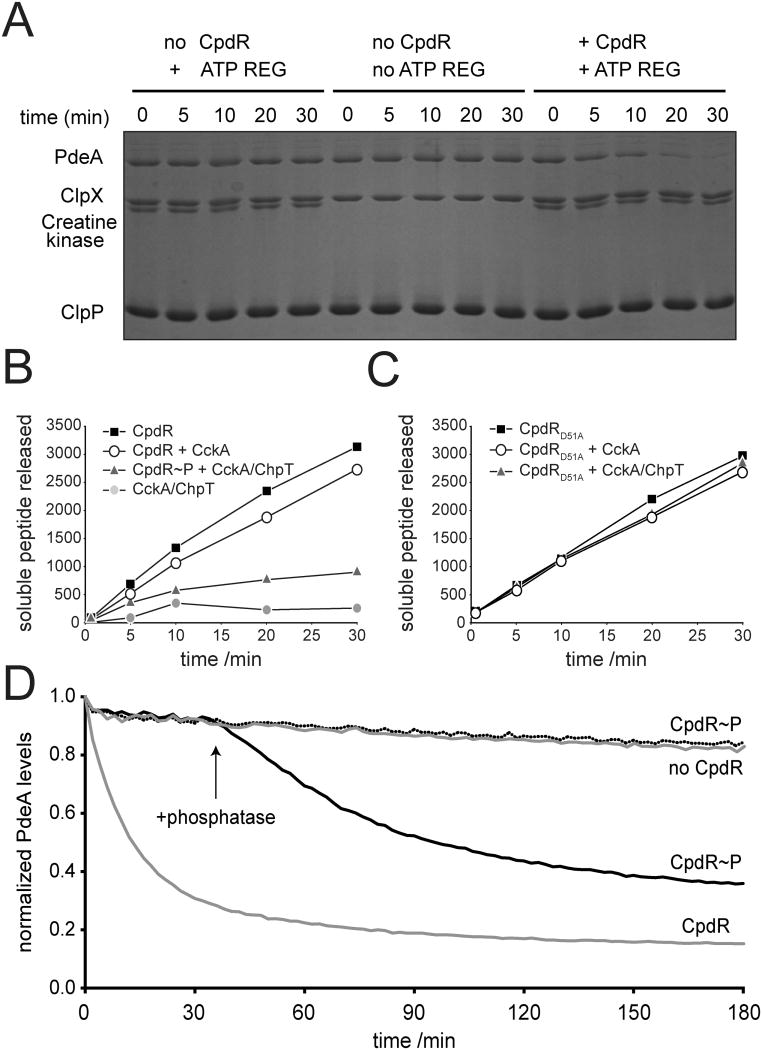
During the G1-to-S transition the unphosphorylated form of CpdR localizes to the old cell pole to recruit the ClpXP protease complex (Iniesta et al., 2006). As CpdR is important for PdeA degradation this suggested that this response regulator might initiate PdeA degradation through localization to the ClpXP containing pole. Alternatively, CpdR could act as an adaptor for PdeA delivery to the ClpXP protease. To distinguish between these possibilities, we performed in vitro degradation experiments with purified ClpXP, PdeA and CpdR. As shown in Fig. 4A, ClpXP alone was not capable of degrading PdeA; however, addition of CpdR dramatically increased degradation of PdeA in an ATP-dependent manner. This is in line with the in vivo requirement of CpdR for PdeA degradation by ClpXP and provides a mechanistic framework for CpdR playing a direct and specific role in PdeA recognition by the protease. Interestingly, CpdR mediated degradation of PdeA is enhanced by GTP (Fig. S6A), indicating that GTP binding to the allosteric GGDEF domain of the phosphodiesterase might stimulate both PdeA activity (Christen et al., 2005) and its subsequent degradation by ClpXP.
Figure 4. CpdR is a phosphorylation-dependent adaptor for PdeA degradation.
(A) CpdR is required for ClpXP mediated PdeA degradation in vitro. Purifed PdeA (1 μM) was incubated with 0.4 μM ClpX and 0.8 μM ClpP, either in the absence of CpdR, in the presence of 1 μM CpdR without addition of an ATP regeneration system, or in the presence of both 1 μM CpdR and an ATP regeneration system.
(B-C) Quantification of the soluble peptide release upon degradation of 35S labeled PdeA as a function of time. In addition to 2 μM PdeA, all reactions contain 0.2 μM ClpX, 0.4 μM ClpP, 1 mM GTP, and an ATP regeneration system (see also Figure S6).
(B) Only unphosphorylated CpdR stimulates PdeA degradation by ClpXP. CpdR: contains unphosphorylated CpdR without the phosphorelay; CpdR + CckA: contains unphosphorylated CpdR and the histidine kinase CckA without the phosphor-transfer protein ChpT; CpdR∼P + CckA/ChpT: CpdR was pre-incubated with CckA/ChpT for 10 min prior to PdeA addition; CckA/ChpT: contains the phosphorelay, but no CpdR.
(C) The non-phosphorylateable CpdRD51A stimulates PdeA degradation even in the presence of a phosphodonor. D51A: contains the non-phosphorylatable CpdRD51A variant without phosphorelay; D51A + CckA: contains CpdRD51A and the histidine kinase without the phosphor-transfer protein; D51A + CckA/ChpT: contains CpdRD51A and the CckA/ChpT phosphorelay.
(D) Dephosphorylation of CpdR drives PdeA degradation. Phosphorylation of CpdR (CpdR∼P) with the CckA/ChpT phosphorelay deactivates delivery of PdeA. Addition of the CckAH322A phosphatase (arrow) reactivates PdeA degradation. Degradation was monitored by following loss of fluorescence of a GFP-PdeA fusion protein substrate.
CpdR is regulated during the cell cycle by phosphorylation via the CckA-ChpT phosphorelay (Biondi et al., 2006) (Fig. 7A). The finding that CpdR is necessary for PdeA degradation by ClpXP in vitro led us to examine if this activity depends on the phosphorylation status of CpdR. As shown in Fig. 4B, PdeA was readily degraded by ClpXP in the presence of non-phosphorylated CpdR. When CpdR was pre-incubated with the CckA sensor histidine kinase, the ChpT phosphotransfer protein and ATP, PdeA degradation was strongly reduced. No significant reduction of PdeA degradation was observed when CpdR was pre-incubated with the phosphorelay mix lacking ChpT (Fig. 4B). Importantly, CpdRD51A, a mutant lacking the phosphoryl acceptor site facilitated PdeA degradation even in the presence of the upstream phosphorelay components (Fig. 4C). In vivo, the activation of CpdR hinges on its dephosphorylation during the swarmer to stalk transition when PdeA is rapidly turned over. As a more faithful representation of these events, we investigated if specific dephosphorylation of CpdR∼P could lead to activation of PdeA degradation in vitro. Phosphorylated CpdR was incapable of delivering GFP-PdeA for degradation; however, addition of a kinase-dead, phosphatase-active mutant of CckA (CckAH322A) (Chen et al., 2009) siphoned phosphates from CpdR∼P and rapidly restored PdeA degradation (Fig. 4D). Together, these experiments demonstrated that CpdR facilitates ClpXP-mediated PdeA degradation in vitro and that this activity is controlled by its phosphorylation state in a manner that precisely mirrors its regulation in vivo.
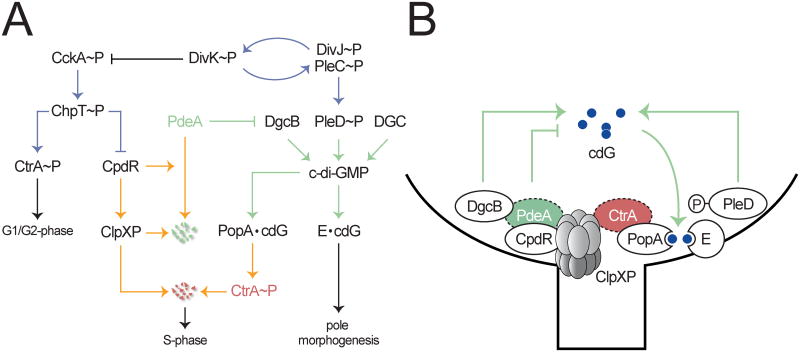
Figure 7. Model for the integration of protein phosphorylation, degradation and c-di-GMP pathways to coordinate C. crescentus pole morphogenesis with cell cycle progression.
(A) Regulatory network controlling C. crescentus pole morphogenesis and cell cycle progression. Blue lines indicate phosphorylation reactions; yellow lines indicate processes involved in the regulation of proteolysis; green lines indicate signaling via c-di-GMP. Postulated diguanylate cyclases (DGC) and c-di-GMP effector proteins (E) are indicated. Red and green protein names specify ClpXP substrates.
(B) Spatial arrangement at the incipient stalked pole of proteins involved in cell cycle control and development. PdeA and CtrA are recruited to the cell pole by CpdR and PopA. CpdR-mediated degradation of PdeA together with PleD activation increases the concentration of c-di-GMP to activate PopA as well as yet unknown effector-proteins (E) required pole morphogenesis.
Protein interaction map of PdeA and its antagonist DgcB
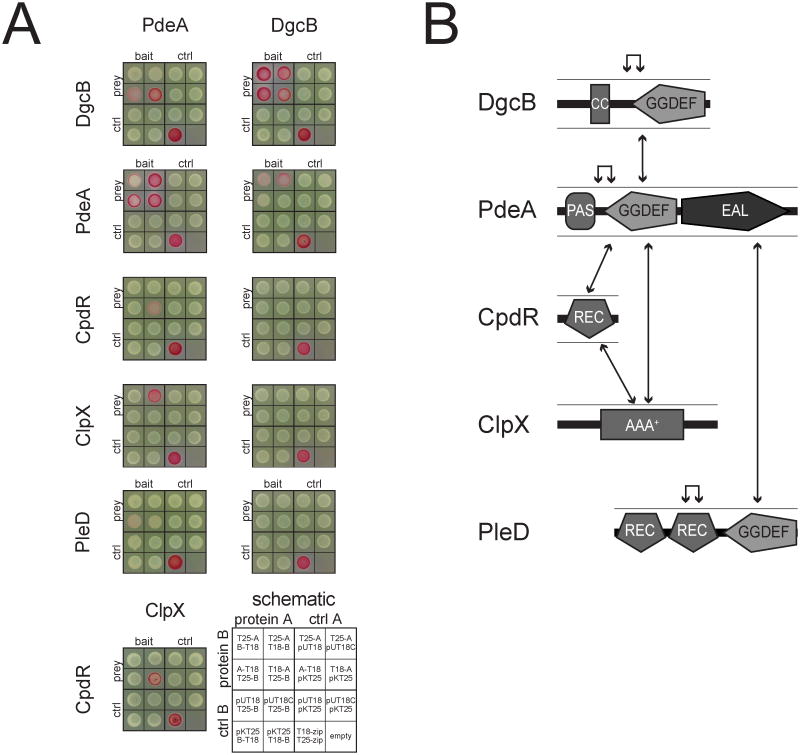
The in vitro degradation experiments with PdeA indicated that CpdR acts as facilitator or adaptor protein for ClpXP-mediated degradation of PdeA. Such a role implies that PdeA directly interacts with both CpdR and the chaperone subunit of the protease. To examine the direct interactions between CpdR, PdeA and ClpX we made use of the bacterial two-hybrid system (BacTH) that exploits complementary fragments (T18 and T25) of Bordetella pertussis adenylate cyclase (Karimova et al., 1998). As shown in Fig. 5A at least one combination of fusions scored positive for PdeA interaction with ClpX and CpdR, respectively. PdeA also strongly interacted with itself indicating an oligomerization-dependent activation mechanism. Interestingly, the BacTH assay revealed a strong interaction between PdeA and its functional counterpart DgcB (Fig. 5A). Since DgcB failed to interact with any of the other components tested, this interaction appears to be specific. PdeA-DgcB interaction was validated in vitro using purified DgcB and PdeA proteins (data not shown). Finally, a weak but reproducible interaction was observed between PdeA and PleD (Fig. 5A).
Figure 5. PdeA and DgcB directly interact and co-localize at the cell pole.
(A) Bacterial two-hybrid assays depicting PdeA and DgcB inteaction partners (Karimova et al., 1998). The loading scheme is indicated in the lower right corner.
(B) Schematic summary of the interactions shown in (A). Individual protein domains are indicated. Arrows connect interaction partners as defined in (A).
These data define a protein-protein interaction network that includes PdeA, its diguanylate cyclase antagonists, and the components mediating cell cycle dependent PdeA degradation (Fig. 5B). The observation that most of the interaction partners of PdeA localize to the cell pole during the G1-S transition led us to analyze the spatial distribution of DgcB during the cell cycle. A functional DgcB-GFP fusion protein (Fig. S4) specifically localized to the flagellated pole of newborn swarmer cells (Fig. S3C, Movie S2). During cell differentiation, DgcB is rapidly released from the incipient stalked pole only to transiently re-localize to the same pole later in the cell cycle. At the same time DgcB also localizes to the pole opposite the stalk, where it persists until the newly born swarmer progeny initiates differentiation into a sessile stalked cell (Fig. S3C, Movie S2). Since PdeA is only present in swarmer cells, the two proteins likely interact at the cell pole at this cell cycle stage. However, polar localization of PdeA and DgcB did not show interdependence (data not shown), arguing that the two proteins use distinct localization mechanisms to reach the pole. Future studies will have to address the role of PdeA and DgcB interaction and co-localization for the activities of these key regulators of C. crescentus development.
Simultaneous activation of a diguanylate cyclase and degradation of a phosphodiesterase coordinate pole development and S-phase entry
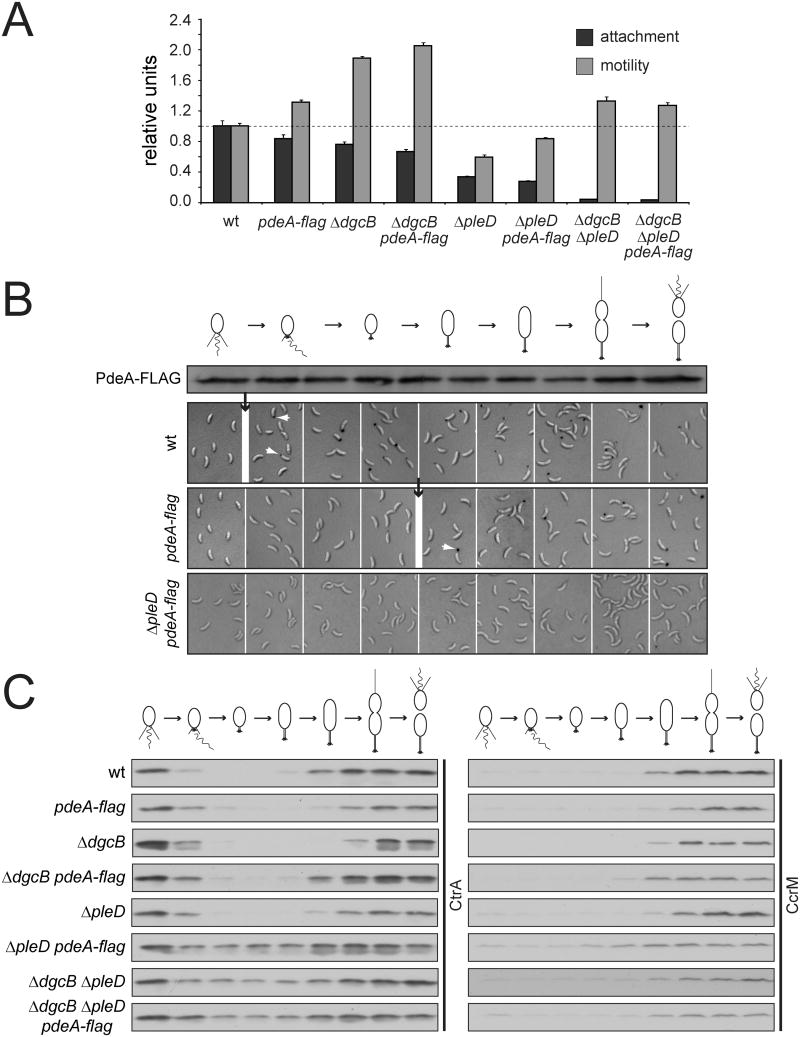
The epistasis data shown in Fig. 1 strongly indicated that DgcB and PleD together are responsible for the increase in c-di-GMP concentration necessary for the motile to sessile transition. The data also suggested that the phosphodiesterase PdeA specifically antagonizes the diguanylate cyclase DgcB, but does not interfere with PleD, because their activities are temporally separated during the cell cycle. This led us to propose that the upshift of c-di-GMP observed during G1-S results from at least two simultaneous events, PleD phosphorylation and PdeA degradation. To test the contribution of each of these events we analyzed strains expressing a stable PdeA mutant (pdeA-flag) and found a reduction in surface attachment propensity and an increase in cell motility both in C. crescentus wild type and mutants lacking either DgcB or PleD (Fig. 6A). As expected from the epistasis experiments (Fig. 1), a mutant lacking both diguanylate cyclases was not affected by the pdeA-flag allele. Because reduced surface attachment correlates with reduced levels of holdfast formation we examined the timing of holdfast appearance and found that holdfast formation was substantially delayed in wild type expressing pdeA-flag (Fig. 6B), as was loss of motility (data not shown). This is similar to the behavior of a dgcB mutant (Figs. 1), arguing that loss of DgcB and stabilization of its phosphodiesterase antagonist produces the same developmental delay. Expression of the pdeA-flag allele had an even more dramatic effect in a mutant lacking PleD, where holdfast formation was barely detectable during the first cell cycle of newly differentiated swarmer cells (Fig. 6B). This strongly argues for a model where the simultaneous activation of the diguanylate cyclase PleD and degradation of the phosphodiesterase PdeA are responsible for the correct timing of pole morphogenesis during the C. crescentus cell cycle.
Figure 6. Pole development and cell cycle progression requires PdeA degradation.
(A) Attachment and motility of C. crescentus wild type and mutant strains expressing a stabilized form of PdeA. The mean of eight (attachment) and four (motility) independent colonies is depicted. Data are presented as relative values of the wild type. Error bars represent the standard error of the mean.
(B) Cell cycle dependent holdfast formation in strains expressing a stabilized form of PdeA. Small white arrows highlight labeled holdfasts; black arrows indicate the time point of holdfast appearance. Distribution of stabilized PdeA-FLAG during the cell cycle is indicated in the immunoblot stained with anti-PdeA antibodies.
(C) Cell cycle-dependent degradation of CtrA in strains with altered c-di-GMP metabolism. Synchronized swarmer cells of wild type and mutants were followed throughout the cell cycle. CtrA protein levels were analyzed in immunoblots. Immunoblots with an anti-CcrM antibody are shown as control for cell cycle progression (see also Figure S7).
We next asked if interference with PdeA degradation has an effect on CtrA degradation during the G1-S transition. As observed earlier, cell cycle dependent degradation of CtrA was unaltered in a pleD mutant (Fig. 6C). Likewise, CtrA degradation was not affected significantly in cells expressing the pdeA-flag allele (Fig. 6C). However, CtrA degradation during G1-S was severely affected in a dgcB pleD double mutant or in pleD single mutants expressing the pdeA-flag allele (Fig. 6C). Inefficient degradation of CtrA during the G1-S transition correlated with an increased overall stability of CtrA as determined in non-synchronized cultures of the respective mutant strains (Fig. S7). Together these data suggested that an increase in c-di-GMP concentration promotes both S-phase entry and pole morphogenesis in a coordinated fashion. The necessary c-di-GMP upshift results from two simultaneous processes, PleD activation and PdeA degradation, both of which are orchestrated by interlinked phosphorylation pathways that together determine progression of C. crescentus cells through the asymmetric division cycle (Fig. 7).
The finding that CpdR is required for the degradation of both CtrA and PdeA raised the possibility that during G1-S transition, CpdR could act on CtrA stability solely through modulation of c-di-GMP levels. However, a cpdR pdeA double mutant failed to degrade CtrA excluding the possibility that persisting PdeA primarily accounts for stabilized CtrA in the ΔcpdR background (data not shown). This is consistent with the observation that PopA localization to the incipient stalked pole is not affected in mutants lacking CpdR (Duerig et al., 2009). Finally, expression of the strong heterologous diguanylate cyclase YdeH from E. coli (Boehm et al., 2009) in the cpdR mutant failed to restore CtrA degradation (data not shown). From this we conclude that the single domain response regulator CpdR is a bifunctional protein that operates as protease localization factor and at the same time acts as specific adaptor protein for certain ClpXP substrates like PdeA.
Discussion
The coupling of cell morphogenesis and proliferation allows C. crescentus to generate specialized cell types to optimize survival. Our results uncover how differentiation and cell cycle progression are coordinated in this organism by tight cohesion of phosphorylation and c-di-GMP signaling networks. Signaling through this network culminates in successive protein degradation events that robustly and irreversibly commit cells to S-phase and to a sessile lifestyle. At the top of this regulatory cascade are sensor histidine kinases, which drive and maintain the oscillatory cell cycle program. In particular, the CckA/ChpT phosphorelay is responsible for the activation and stabilization of CtrA in swarmer and predivisional cells (Biondi et al., 2006) (Fig. 7). CtrA stability control is governed through phosphorylation-mediated inactivation of CpdR, which maintains the ClpXP protease in a delocalized state in these cell types (Iniesta et al., 2006). We show here that in addition to stabilizing CtrA, the CckA pathway also stabilizes the PdeA phosphodiesterase via CpdR phosphorylation. Our data demonstrate that CpdR in its non-phosphorylated form directly facilitates ClpXP-dependent degradation of PdeA. Thus, the accumulation of non-phosphorylated CpdR during the G1-to-S transition mediates the rapid degradation of the replication initiation inhibitor CtrA by distinct mechanisms. First, CpdR affects polar localization of the ClpXP protease at the G1-S transition; second, CpdR mediated delivery of PdeA to the polar ClpXP complex contributes to the upshift in c-di-GMP, the activation of PopA, and thus the recruitment of CtrA to the ClpXP occupied cell pole (Fig. 7).
The phosphate flux through the CckA-ChpT pathway reverses prior to S-phase entry contributing to CtrA and CpdR dephosphorylation and, ultimately, replication initiation. This activity is coordinated with a second phosphorylation pathway involved in G1-S transition that triggers the synthesis of c-di-GMP through phosphorylation of the PleD diguanylate cyclase (Aldridge et al., 2003; Paul et al., 2004) (Fig. 7). The cell-type specific activity of this pathway relies on the spatial dynamic behavior of two sensor histidine kinases, PleC and DivJ, which position to opposite poles of the Caulobacter predivisional cell and differentially segregate into the daughter progeny (McAdams and Shapiro, 2003). PleC is a phosphatase in swarmer cells but during cell differentiation adopts strong kinase activity and, together with the newly synthesized DivJ kinase, promotes a rapid upshift of c-di-GMP through the activation of PleD (Aldridge et al., 2003; Paul et al., 2004). Reversal of PleC activity is implemented by the essential single domain response regulator DivK that acts as an allosteric activator of both PleC and DivJ autokinase activity in sessile stalked cells (Paul et al., 2008). At the same time, activated DivK downregulates the CckA-ChpT pathway to contribute to the removal of active CtrA during G1-S transition (Biondi et al., 2006; Tsokos et al., 2011) and through this mechanism helps to adjust the activity of the two cell type-specific phosphorylation pathways during the C. crescentus cell cycle (Fig. 7).
While DivK links the two phosphorylation networks at the level of the kinase activities, our work reveals CpdR as an additional key component coordinating these two pathways. Although CpdR was originally identified as a polar recruitment factor for the ClpXP protease, we show here that CpdR also serves as a specific adaptor to deliver the PdeA phosphodiesterase to ClpXP prior to S-phase entry. Adaptor proteins for AAA+ proteases increase the stringency of substrate selection and alter priorities of target degradation (Kirstein et al., 2009). Before this work, the only characterized adaptor in C. crescentus was SspB that delivers ssrA-tagged substrates to the ClpXP protease (Chien et al., 2007). However, in contrast to the SspB adaptor case, delivery of PdeA by CpdR is dependent on the phosphorylation status of the adaptor. Although cell cycle regulated activation is unprecedented for ClpXP adaptors, phosphorylation dependent changes in adaptor function have been described before (Kirstein et al., 2006; Mika and Hengge, 2005). Thus, it appears that coupling of adaptor phosphorylation with adaptor activity is a conserved mechanism to control specific substrate delivery. The observation that CpdR itself is degraded by the ClpXP protease (Iniesta and Shapiro, 2008) suggests that the ClpXP pathway can be rapidly inactivated in S-phase by the simultaneous removal of adaptor and substrate protein.
Several lines of evidence argue that DgcB and PdeA function as antagonists and that PdeA, due to its dominance over DgcB, establishes the motile program in swarmer cells. How does PdeA “neutralize” DgcB in swarmer cells? A simple explanation would be that PdeA is catalytically more active than its antagonist. The direct physical coupling of PdeA and DgcB could enhance this effect. Similar to the concept of “metabolic channeling” (Conrado et al., 2008), such an arrangement could increase the efficiency of PdeA control over DgcB by preventing diffusion of c-di-GMP into the surrounding cytoplasm. This “futile cycle” mechanism, although seemingly wasteful, may provide for a rapid response to environmental signals that can override the internal cell cycle control. Alternatively, PdeA could directly control DgcB activity through allosteric or inhibitory effects resulting from the simple physical interaction between the two proteins. The observation that a PdeA active site mutation shows the same phenotype as a pdeA deletion mutation argues against such a scenario (data not shown). We have shown that PdeA activity is allosterically stimulated by GTP binding to its regulatory GGDEF domain (Christen et al., 2005). While the kinetic parameters suggest that PdeA is fully induced under physiological conditions, certain conditions could lead to a (local) drop in GTP that would be readily transduced into an increase in c-di-GMP by downregulating PdeA. Clearly, we are just beginning to understand how CpdR, PdeA, PleD, PopA and ClpXP collaborate to drive development and cell cycle progression. Understanding the dynamic nature of these complexes at the cell pole (Fig. 7) will be the aim of future work.
Experimental Procedures
More-detailed descriptions of experimental procedures and a list of all plasmids and strains (Table S1) are provided in the extended Experimental Procedures.
Microscopy
Fluorescence and DIC microscopy were performed on a DeltaVision Core (Applied Precision, USA)/Olympus IX71 microscope equipped with an UPlanSApo 100×/1.40 Oil objective (Olympus, Japan) and a coolSNAP HQ-2 (Photometrics, USA) CCD camera. Cells were placed on a PYE pad solidified with 1% agarose (Sigma, USA). Images were processed and analyzed with softWoRx version 5.0.0 (Applied Precision, USA) and Photoshop CS3 (Adobe, USA) software.
Bacterial two hybrid experiments
Bacterial two hybrid screens were performed according to Karimova et al. (Karimova et al., 1998). Full open reading frames or gene fragments were fused to the 3′ end of the T25 (pKT25), the 3′ end of the T18 (pUT18C) or the 5′ end of the T18 (pUT18) fragment of the gene coding for Bordetella pertussis adenylate cyclase. Two microliter of a MG1655 cyaA∷frt culture containing the relevant plasmids were spotted on a MacConkey Agar Base plate supplemented with kanamycin, ampicilin and maltose, incubated at 30°C.
Protein expression and purification
E. coli BL21 (DE3) pLys (Stratagene, USA) carrying the dgcB expression plasmid were grown in LB medium to an OD600 of 0.5 before expression was induced by adding isopropyl 1-thio-β-D-galactopyranoside (IPTG) to a final concentration of 0.5 mM. Cells were harvested by centrifugation, resuspended in 20 mM HEPES pH 7.5, 100 mM KCl, 20 mM imidazole running buffer and lysed by passage through a French pressure cell. Clarified crude extract was loaded on a HP HisTrap column (GE Healthcare, UK) attached to an ÄKTApurifier (GE Healthcare, UK). Protein was eluted by raising the imidazole concentration to 500 mM in running buffer. Further purification and buffer exchange were performed by size exclusion chromatography on a Superdex200 HR26/60 column (GE Healthcare, UK) using 20 mM HEPES pH 7.5, 100 mM KCl as running buffer. PdeA and CpdR were expressed as C-terminal fusions to a his-tagged SUMO domain in BL21DE3 plysS cells. Purification of the fusions, cleavage of the SUMO domain, and separation of the cleaved protein were performed as before (Wang et al., 2007). GFP-PdeA was purified using standard Ni-NTA protocols (Qiagen). Subsequent purification was performed using size exclusion chromatography (Sephacryl S-300 for PdeA and GFP-PdeA; Superdex-75 for CpdR) using 20mM HEPES, 100 mM KCl, 5 mM MgCl2, 5 mM Beta-Mercaptoethanol, 10% Glycerol, pH 7.4 as the buffer (H-buffer). Monomeric fractions were concentrated and stored at −80°C. Radiolabeled PdeA was produced by labeling with [35S] labeled L-methionine (Wang et al., 2007) and purification as above, with the exception of the gel-filtration step. ClpX and ClpP were purified as before (Chien et al., 2007).
Protease assay
PdeA degradation was assayed at 30°C in H-buffer (20mM HEPES, 100 mM KCl, 5 mM MgCl2, 5 mM Beta-Mercaptoethanol, 10% Glycerol, pH 7.4). For typical qualitative reactions, 1 μM of PdeA was incubated with 1mM GTP, 0.4 μM ClpX6, 0.8 μM ClpP14, 1 μM CpdR, and an ATP regeneration system (Chien et al, 2007). Aliquots were removed at indicated times and quenched by addition of SDS loading dye and immediately frozen. Samples were separated by SDS-PAGE and stained with Coomassie for visualization. For quantitative assays, degradation of radiolabeled PdeA was monitored by measurement of TCA soluble peptides (Wang et al., 2007). Degradation of GFP-PdeA was performed in the same conditions as above with the exception that GFP fluorescence was continuously monitored in 384-well plates using a Spectramax M5 (Molecular Devices). Phosphorylation of CpdR was performed as before (Chen et al., 2009).
Supplementary Material
Highlights.
Coordinate action of the DGCs PleD and DgcB control Caulobacter cell cycle and development
The PDE PdeA opposes DgcB and thereby determines the swarmer cell specific program
Cell cycle dependent degradation of PdeA releases its antagonist DgcB
CpdR plays a dual role as protease localization factor and adaptor for PdeA
Acknowledgments
We thank Anna Duerig and Fabienne Hamburger for strains and plasmids; Samuel Steiner for strain E. coli MG1655 ΔcyaA∷frt; Pia Abel zur Wiesch for help with data analysis and for critical reading of the manuscript. This work was supported by SNF grants 31-108186 and 31003A_130469 to UJ., NIH grants GM-082899 to M.T.L., GM-049224 to T.A.B., and GM-084157 to P.C.; T.A.B. and M.T.L. are employees of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aldridge P, Paul R, Goymer P, Rainey P, Jenal U. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol Microbiol. 2003;47:1695–1708. doi: 10.1046/j.1365-2958.2003.03401.x. [DOI] [PubMed] [Google Scholar]
- Bateman JM, McNeill H. Temporal control of differentiation by the insulin receptor/tor pathway in Drosophila. Cell. 2004;119:87–96. doi: 10.1016/j.cell.2004.08.028. [DOI] [PubMed] [Google Scholar]
- Biondi EG, Reisinger SJ, Skerker JM, Arif M, Perchuk BS, Ryan KR, Laub MT. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature. 2006;444:899–904. doi: 10.1038/nature05321. [DOI] [PubMed] [Google Scholar]
- Boehm A, Kaiser M, Li H, Spangler C, Kasper CA, Ackermann M, Kaever V, Sourjik V, Roth V, Jenal U. Second messenger-mediated adjustment of bacterial swimming velocity. Cell. 2010;141:107–116. doi: 10.1016/j.cell.2010.01.018. [DOI] [PubMed] [Google Scholar]
- Boehm A, Steiner S, Zaehringer F, Casanova A, Hamburger F, Ritz D, Keck W, Ackermann M, Schirmer T, Jenal U. Second messenger signalling governs Escherichia coli biofilm induction upon ribosomal stress. Mol Microbiol. 2009;72:1500–1516. doi: 10.1111/j.1365-2958.2009.06739.x. [DOI] [PubMed] [Google Scholar]
- Burton GJ, Hecht GB, Newton A. Roles of the histidine protein kinase pleC in Caulobacter crescentus motility and chemotaxis. J Bacteriol. 1997;179:5849–5853. doi: 10.1128/jb.179.18.5849-5853.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caro E, Castellano MM, Gutierrez C. A chromatin link that couples cell division to root epidermis patterning in Arabidopsis. Nature. 2007;447:213–217. doi: 10.1038/nature05763. [DOI] [PubMed] [Google Scholar]
- Chen YE, Tsokos CG, Biondi EG, Perchuk BS, Laub MT. Dynamics of two Phosphorelays controlling cell cycle progression in Caulobacter crescentus. J Bacteriol. 2009;191:7417–7429. doi: 10.1128/JB.00992-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien P, Perchuk BS, Laub MT, Sauer RT, Baker TA. Direct and adaptor-mediated substrate recognition by an essential AAA+ protease. Proc Natl Acad Sci U S A. 2007;104:6590–6595. doi: 10.1073/pnas.0701776104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen M, Christen B, Folcher M, Schauerte A, Jenal U. Identification and Characterization of a Cyclic di-GMP-specific Phosphodiesterase and Its Allosteric Control by GTP. J Biol Chem. 2005;280:30829–30837. doi: 10.1074/jbc.M504429200. [DOI] [PubMed] [Google Scholar]
- Christen M, Kulasekara HD, Christen B, Kulasekara BR, Hoffman LR, Miller SI. Asymmetrical distribution of the second messenger c-di-GMP upon bacterial cell division. Science. 2010;328:1295–1297. doi: 10.1126/science.1188658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrado RJ, Varner JD, DeLisa MP. Engineering the spatial organization of metabolic enzymes: mimicking nature's synergy. Curr Opin Biotechnol. 2008;19:492–499. doi: 10.1016/j.copbio.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Curtis PD, Brun YV. Getting in the loop: regulation of development in Caulobacter crescentus. Microbiol Mol Biol Rev. 2010;74:13–41. doi: 10.1128/MMBR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domian IJ, Quon KC, Shapiro L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- Domian IJ, Reisenauer A, Shapiro L. Feedback control of a master bacterial cell-cycle regulator. Proc Natl Acad Sci USA. 1999;96:6648–6653. doi: 10.1073/pnas.96.12.6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, Giese B, Jenal U. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev. 2009;23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J. Regulation of endospore formation in Bacillus subtilis. Nat Rev Microbiol. 2003;1:117–126. doi: 10.1038/nrmicro750. [DOI] [PubMed] [Google Scholar]
- Flardh K, Buttner MJ. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat Rev Microbiol. 2009;7:36–49. doi: 10.1038/nrmicro1968. [DOI] [PubMed] [Google Scholar]
- Grunenfelder B, Tawfilis S, Gehrig S, M OS, Eglin D, Jenal U. Identification of the protease and the turnover signal responsible for cell cycle-dependent degradation of the Caulobacter FliF motor protein. J Bacteriol. 2004;186:4960–4971. doi: 10.1128/JB.186.15.4960-4971.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- Iniesta AA, McGrath PT, Reisenauer A, McAdams HH, Shapiro L. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc Natl Acad Sci U S A. 2006;103:10935–10940. doi: 10.1073/pnas.0604554103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iniesta AA, Shapiro L. A bacterial control circuit integrates polar localization and proteolysis of key regulatory proteins with a phospho-signaling cascade. Proc Natl Acad Sci U S A. 2008;105:16602–16607. doi: 10.1073/pnas.0808807105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U, Fuchs T. An essential protease involved in bacterial cell-cycle control. EMBO J. 1998;17:5658–5669. doi: 10.1093/emboj/17.19.5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser D. Myxococcus-from single-cell polarity to complex multicellular patterns. Annu Rev Genet. 2008;42:109–130. doi: 10.1146/annurev.genet.42.110807.091615. [DOI] [PubMed] [Google Scholar]
- Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirstein J, Moliere N, Dougan DA, Turgay K. Adapting the machine: adaptor proteins for Hsp100/Clp and AAA+ proteases. Nat Rev Microbiol. 2009;7:589–599. doi: 10.1038/nrmicro2185. [DOI] [PubMed] [Google Scholar]
- Kirstein J, Schlothauer T, Dougan DA, Lilie H, Tischendorf G, Mogk A, Bukau B, Turgay K. Adaptor protein controlled oligomerization activates the AAA+ protein ClpC. EMBO J. 2006;25:1481–1491. doi: 10.1038/sj.emboj.7601042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laub MT, Chen SL, Shapiro L, McAdams HH. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc Natl Acad Sci U S A. 2002;99:4632–4637. doi: 10.1073/pnas.062065699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laub MT, McAdams HH, Feldblyum T, Fraser CM, Shapiro L. Global analysis of the genetic network controlling a bacterial cell cycle. Science. 2000;290:2144–2148. doi: 10.1126/science.290.5499.2144. [DOI] [PubMed] [Google Scholar]
- McAdams HH, Shapiro L. A bacterial cell-cycle regulatory network operating in time and space. Science. 2003;301:1874–1877. doi: 10.1126/science.1087694. [DOI] [PubMed] [Google Scholar]
- McGrath PT, Iniesta AA, Ryan KR, Shapiro L, McAdams HH. A dynamically localized protease complex and a polar specificity factor control a cell cycle master regulator. Cell. 2006;124:535–547. doi: 10.1016/j.cell.2005.12.033. [DOI] [PubMed] [Google Scholar]
- Mika F, Hengge R. A two-component phosphotransfer network involving ArcB, ArcA, and RssB coordinates synthesis and proteolysis of sigmaS (RpoS) in E. coli. Genes Dev. 2005;19:2770–2781. doi: 10.1101/gad.353705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul R, Jaeger T, Abel S, Wiederkehr I, Folcher M, Biondi EG, Laub MT, Jenal U. Allosteric regulation of histidine kinases by their cognate response regulator determines cell fate. Cell. 2008;133:452–461. doi: 10.1016/j.cell.2008.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 2004;18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potocka I, Thein M, Osteras M, Jenal U, Alley MR. Degradation of a Caulobacter soluble cytoplasmic chemoreceptor is ClpX dependent. J Bacteriol. 2002;184:6635–6641. doi: 10.1128/JB.184.23.6635-6641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quon KC, Yang B, Domian IJ, Shapiro L, Marczynski GT. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc Natl Acad Sci USA. 1998;95:120–125. doi: 10.1073/pnas.95.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan KR, Huntwork S, Shapiro L. Recruitment of a cytoplasmic response regulator to the cell pole is linked to its cell cycle-regulated proteolysis. Proc Natl Acad Sci U S A. 2004;101:7415–7420. doi: 10.1073/pnas.0402153101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsokos CG, Perchuk BS, Laub MT. A dynamic complex of signaling proteins uses polar localization to regulate cell-fate asymmetry in Caulobacter crescentus. Dev Cell. 2011;20:329–341. doi: 10.1016/j.devcel.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KH, Sauer RT, Baker TA. ClpS modulates but is not essential for bacterial N-end rule degradation. Genes Dev. 2007;21:403–408. doi: 10.1101/gad.1511907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadirgi G, Marino S. Adult neural stem cells and their role in brain pathology. J Pathol. 2009;217:242–253. doi: 10.1002/path.2480. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.