Abstract
Soil metagenomes represent an unlimited resource for the discovery of novel biocatalysts from soil microorganisms. Three large-inserts metagenomic DNA libraries were constructed from different grassland soil samples and screened for genes conferring cellulase or xylanase activity. Function-driven screening identified a novel cellulase-encoding gene (cel01) and two xylanase-encoding genes (xyn01 and xyn02). From sequence and protein domain analyses, Cel01 (831 amino acids) belongs to glycoside hydrolase family 9 whereas Xyn01 (170 amino acids) and Xyn02 (255 amino acids) are members of glycoside hydrolase family 11. Cel01 harbors a family 9 carbohydrate-binding module, previously found only in xylanases. Both Xyn01 and Xyn02 were most active at 60°C with high activities from 4 to 10 and optimal at pH 7 (Xyn01) and pH 6 (Xyn02). The cellulase gene, cel01, was expressed in E. coli BL21 and the recombinant enzyme (91.9 kDa) was purified. Cel01 exhibited high activity with soluble cellulose substrates containing β-1,4-linkages. Activity with microcrystalline cellulose was not detected. These data, together with the analysis of the degradation profiles of carboxymethyl cellulose and barley glucan indicated that Cel01 is an endo 1,4-β-glucanase. Cel01 showed optimal activity at 50°C and pH 7 being highly active from pH range 5 to 9 and possesses remarkable halotolerance.
Electronic supplementary material
The online version of this article (doi:10.1007/s10529-011-0830-2) contains supplementary material, which is available to authorized users.
Keywords: Soil metagenome, Cellulase, Xylanase, Metagenomic libraries, Activity-based screening
Introduction
The plant cell wall constituents cellulose and the hemicellulose xylan are the most abundant polysaccharides in nature (Beg et al. 2001; Lynd et al. 2005; Polizeli et al. 2005). The structure of cellulose is based on linear polymers of β-1,4-linked d-glucose residues. The hemicellulose xylan consists of a linear β-1,4-linked xylose backbone that can be substituted by arabinofuranosyl and/or glucuronopyranosyl side chains with the degree and nature of substitution varying between tissues and species. Xylans can be modified by additional sugar side chains, methylation, acetylation, or feruloylation (Beg et al. 2001; Kulkarni et al. 1999). Both polysaccharides have an enormous potential as renewable sources for production of biofuels (Sanchez and Cardona 2008).
Enzymes degrading (hemi)cellulose are found in microbes, plants, and the digestive tracts of animals. Three classes of cellulases, including endoglucanases (EC 3.2.1.4), exoglucanases (EC 3.2.1.91), and β-glucosidases (EC 3.2.1.21) are involved in the degradation of cellulose (Lynd et al. 2002). Endo-1,4-β-xylanase (EC 3.2.1.8) and β-xylosidase (EC 3.2.1.37) can hydrolyze the backbone of xylan. Additionally, acetyl esterases (EC 3.1.1.6), α-d-glucuronidases (EC 3.2.1.1), and α-L-arabinofuranosidases (EC 3.2.1.55) cleave the side chains of xylan (Polizeli et al. 2005). Cellulases and hemicellulases have been recognized as very useful biocatalysts because of their wide-ranging versatility in industrial applications, including food technology, textile production, biofuel formation, and paper production (Collins et al. 2005; Lynd et al. 2002; Steele et al. 2009).
Soils are considered to be the most diverse microbial habitat on Earth with respect to species diversity and community size. One gram of soil may contain up to 10 billion microorganisms of possibly thousands of different species (Rosselló-Mora and Amann 2001). Soil microorganisms have been the major source for the isolation of novel biocatalysts and other biomolecules of industrial importance (Strohl 2000). So far, most soil-derived cellulases and xylanases and the corresponding genes have been recovered from cultured soil microorganisms such as Cellvibrio mixtus and Clostridium thermocellum (Fontes et al. 2000; Gilad et al. 2003). Since less than 1% of soil microorganisms are readily culturable, only a small fraction of soil microbial diversity is assessed by cultivation-dependent approaches. To expand the range of natural product discovery, culture-based methods have been complemented or replaced by culture-independent metagenomic approaches, which theoretically provide access to the collective nucleic acids of all indigenous microorganisms present in an environmental sample (Handelsman 2004; Simon and Daniel 2011). Functional metagenomics based on the direct isolation of DNA from environmental samples, generation of metagenomic libraries from the isolated DNA, and function-driven screening of the constructed libraries has led to the identification and characterization of a variety of novel biocatalysts (Simon and Daniel 2009), including proteases (Waschkowitz et al. 2009; Zhang et al. 2011), lipolytic enzymes (Nacke et al. 2011b; Yu et al. 2011) and (hemi)cellulolytic enzymes (Brennan et al. 2004; Duan et al. 2009; Shedova et al. 2009; Voget et al. 2006).
In this study, we report on the identification of one cellulase and two xylanases, which were derived from the soil metagenome. We constructed large-insert metagenomic libraries from three different grassland soil samples, which were collected in the German Biodiversity Exploratories Schorfheide-Chorin and Schwäbische Alb (Fischer et al. 2010). The library-containing Escherichia coli clones were screened for genes encoding cellulase or xylanase activity. Two novel xylanases and one cellulase were identified. Characterization of the xylanases and the purified cellulase was performed.
Materials and methods
Bacterial strains and vectors
Fosmids and plasmids used in the present study are shown in Table 1. Escherichia coli strain EPI300-T1R (Epicentre Biotechnologies, Madison, WI, USA) was used as a host for the cloning of metagenomic DNA. In addition, E. coli strains TOP10 and BL21(DE3) (Invitrogen GmbH, Karlsruhe, Germany) were employed for subcloning and expression of the targeted genes, respectively.
Table 1.
Vectors used in this study
| Type of vector | Designation | Relevant characteristicsa | Source |
|---|---|---|---|
| Fosmid | pCC1FOS | Chlr, T7 promoter, FI and pMB1 replicon | Epicentre |
| fLC01 | pCC1FOS: 16,250 bp fragment of cloned metagenomic DNA | This study | |
| fLX01 | pCC1FOS: 36,000 bp fragment of cloned metagenomic DNA | This study | |
| fLX02 | pCC1FOS: 30,200 bp fragment of cloned metagenomic DNA | This study | |
| Plasmid | pCR-XL-TOPO | Kanr, Zeor, lac promoter, pMB1 replicon | Invitrogen |
| pET101/D | Apr, T7 promoter, pMB1 replicon | Invitrogen | |
| pLC01 | pCR-XL-TOPO: 3,220 bp fragment of metagenomic DNA subcloned from fLC01 | This study | |
| pLX01 | pCR-XL-TOPO: 5,523 bp fragment of metagenomic DNA subcloned from fLX01 | This study | |
| pLX02 | pCR-XL-TOPO: 4,985 bp fragment of metagenomic DNA subcloned from fLX02 | This study | |
| pCel01 | pET101/D: 2,433 bp fragment containing bases 646–3,075 of cel01 gene and a synthetic ATG start codon under the control of the T7 promoter | This study |
a Ap r ampicillin resistance, Chl r chloramphenicol resistance, Kan r kanamycin resistance, Zeo r zeocin resistance
Soil sampling, isolation of DNA, and construction of metagenomic DNA libraries
Soil metagenome-derived genes encoding (hemi)cellulolytic enzymes were recovered from A horizons of soil samples, which had been taken from three grassland sites of the German Biodiversity Exploratories Schorfheide-Chorin (sample SEG9) and Schwäbische Alb (samples AEG3 and AEG6). Samples were collected in April and May 2008 and sampling was performed as described by Nacke et al. (2011a). Descriptions of the soil characteristics are provided in Supplementary Table S1. Names of the metagenomic libraries refer to the designation of the samples from which the libraries were derived. To generate metagenomic libraries total microbial community DNA was isolated from 10 g soil per sample. For this purpose, the MoBio Power Max Soil DNA extraction kit (MoBio Laboratories, Carlsbad, CA, USA) was used according to the instructions of the manufacturer.
The large-insert metagenomic fosmid libraries AEG3 and AEG6 were constructed by using the Copy Control Fosmid Library Production kit (Epicentre) as described by Nacke et al. (2011b). The fosmid library SEG9 has been previously generated by employing the same approach (Nacke et al. 2011b). Approx. 4,600 resulting library-containing clones per soil sample were arrayed and stored in 96-well microtiter plates. The remaining clones were collected and stored as clone pools at −80°C.
Growth conditions and activity-based screening
Escherichia coli strains were routinely grown in Luria-Bertani (LB) medium at 37°C. For activity-based screening, the arrayed library-containing E. coli clones were replica streaked on LB agar plates containing colored insoluble forms of hydroxyethyl cellulose (HECred) or xylan (xylangreen) as indicator substrates. The substrates were generated by employing the cross-linking reagent 1,4-butanediol diglycidyl ether and the dyes Cibacron Brilliant Red 3B-A (HECred) or Cibacron Brilliant Green T3GE (xylangreen) as described by Lee and Lee (1997) and Ten et al. (2005). In addition, to maintain the presence of recombinant fosmids and increase the copy number of the fosmids, the indicator agar contained 12.5 mg chloramphenicol l−1 and 0.001% arabinose, respectively. Clones showing activity with the indicator substrates were identified by the formation of clear zones (halos) after incubation for 1–14 days at 37°C under aerobic conditions.
Subcloning and sequence analysis
To subclone DNA fragments containing genes conferring (hemi)cellulolytic activity from large-insert fosmids, the recombinant fosmids from positive clones were sheared by sonication (UP200S Sonicator, Dr. Hielscher GmbH, 5 s at 30% amplitude, cycle 0.5). Subsequently, the resulting DNA fragments were separated by agarose gel electrophoresis, and fragments (2–6 kbp) were excised and extracted by using the peqGold gel extraction kit (Peqlab Biotechnologie GmbH, Erlangen, Germany). The resulting DNA fragments were ligated into pCR-XL-TOPO, and used to transform E. coli TOP10 as recommended by the manufacturer (Invitrogen). The resulting recombinant E. coli strains were screened on the corresponding indicator agar for the presence of genes conferring cellulase or xylanase activity.
The recombinant plasmids derived from positive clones were sequenced by the Göttingen Genomics Laboratory (Göttingen, Germany). The initial prediction of ORFs located on the inserts of plasmids pLC01, pLX1, and pLX2 (Table S2) was performed by using the ORF-finder program (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) provided by the National Center for Biotechnology Information (NCBI) and the Artemis program (Rutherford et al. 2000). The results were verified and improved manually by using criteria such as the presence of a ribosome-binding site, GC frame plot analysis, and similarity to known genes. All coding sequences were examined for similarities to protein families and domains using searches against the CDD databases (Marchler-Bauer et al. 2007). Signal peptides of proteins were predicted by using the SignalP 3.0 server (Bendtsen et al. 2004). Multiple alignments of deduced protein sequences were performed with ClustalW2, version 2.0.12 (Thompson et al. 1994) and examined with the Bioedit program (Hall 1999).
The nucleotide sequences of the recombinant plasmids harboring the xylanase genes xyn01 and xyn02 (pLX01 and pLX02) and the cellulase gene cel01 (pLC01) have been submitted to GenBank under accession numbers JF799945, JF799946, and JF799947, respectively.
Cloning of the cellulase-encoding gene cel01 into expression vector pET101/D
The cellulase gene cel01 was amplified from plasmid pLC01 (Table 1) without the potential signal peptide sequence by PCR. The following set of primers with a synthetic site (underlined) that contained an ATG start codon and allowed directional cloning into pET101/D using the pET101/D directional TOPO expression kit (Invitrogen) was used: 5′-GCGTTCGTTGAAACGC-3′ and 5′-CACCATGCAGGAAATGCTCGCGCCC-3′. The resulting plasmid was designated pCel01 (Table 1). The PCR reaction mixture (33 μl) contained 3.3 μl tenfold reaction buffer (Finnzymes, Espoo, Finland), 800 μM of each of the four deoxynucleoside triphosphates, 3% DMSO, 1.2 μM of each of the primers, 0.5 U Phusion hot start high-fidelity DNA Polymerase (Finnzymes), and 50 ng plasmid DNA as template. The PCR reactions were initiated at 98°C (5 min), followed by 30 cycles of 98°C (45 s), a temperature gradient ranging from 60 to 68°C (45 s), 72°C (170 s) and ended with incubation at 72°C for 5 min. The PCR product was purified using the peqGold gel extraction kit (Peqlab) and cloned into pET101/D (Invitrogen) as recommended by the manufacturer. The coding region was thereby placed under the control of the IPTG-inducible T7 promoter. In addition, sequences encoding a His6 tag and a V5 epitope provided by the vector were added to the 3′-end of the coding region.
Preparation of cell extracts
Cells from 500 ml cultures were harvested by centrifugation at 10,000×g and 4°C for 10 min. To prepare cell extracts of the cellulolytic strain (E. coli BL21/pCel01) and xylanolytic strains (E. coli TOP10/pLX01 and E. coli TOP10/pLX02) the resulting cell pellets were washed twice with LEW buffer (Macherey and Nagel, Düren, Germany) or 50 mM sodium phosphate buffer (pH 7), respectively. Subsequently, the cells were resuspended in 2 ml per g cell pellet of the same buffer. The cells were disrupted by using a French press (1.38 × 108 Pa) and the extract was cleared by centrifugation at 18,000×g and 4°C for 30 min. The resulting supernatant (crude cell-free extract) was used for further analyses.
Purification of cellulase
The purification of His6-tagged cellulase from cell extracts was performed by nickel affinity chromatography using Protino-Ni-2000 prepacked columns as recommended by the manufacturer (Macherey and Nagel). Ultrafiltration employing Vivaspin concentrators (exclusion limits 100,000 and 50,000 Da; Sartorius AG, Göttingen, Germany) was used to change buffer systems and to further purify the cellulase. The purity of the resulting protein preparations was analyzed by SDS–PAGE. Detection of V5 epitope-tagged proteins by Western blot hybridization was performed as described by Waschkowitz et al. (2009).
Protein and enzyme assays
Protein was measured by the Bradford method. To analyze cellulase and xylanase activity released reducing sugars were measured as d-glucose and d-xylose equivalents, respectively (Miller 1959) and measured at 575 nm. Barley glucan (Megazyme, Bry, Ireland), carboxymethyl cellulose (Sigma–Aldrich), HEC (Sigma–Aldrich), laminarin (Sigma–Aldrich), lichenan (Megazyme), xylan from birch wood (Sigma–Aldrich), xylan from oat spelt (Sigma–Aldrich), xylan from larch wood (Sigma–Aldrich), xylan from beech wood (Sigma–Aldrich), and microcrystalline cellulose (Serva, Heidelberg, Germany) were used as substrates. The standard assay mixture contained purified enzyme (0.4 μg) or crude cell extract and 1% barley glucan (cellulase) or 1% xylan from birch wood (xylanase) in 50 mM sodium phosphate buffer adjusted to pH 7 (final volume 0.5 ml). To determine substrate specificity barley glucan was replaced by the above-mentioned substrates at a final concentration of 1%. Enzyme activity was measured after incubation of the reaction mixture at 40°C for 20 min. All enzyme assays were performed in triplicate. One unit (U) of enzyme activity was defined as the amount of enzyme required to release 1 μmol of reducing sugar per min. Enzyme activities were determined from 20 to 70°C and pH 4–10. The pH range was determined with 50 mM sodium acetate buffer (pH 4.0–6.0), 50 mM sodium phosphate buffer (pH 6.0–8.0), 50 mM Tris/HCl buffer (pH 8.0–9.0), and 50 mM glycine/NaOH buffer (pH 9.0–10.0). The thermal stability of Cel01 was analyzed after incubation of the enzyme at 30–60°C. The halotolerance of Cel01 was determined by measuring residual activity using the standard assay after incubation of the enzyme in 3 M KCl or 4 M NaCl at 4°C for 12 h. Metal ions and chemical agents were added to the standard assay mixture to investigate their effects on enzyme activity.
The mode of action of the cellulase was determined by incubating barley glucan and CMC (each 2%) with 3 μg purified enzyme in 0.2 M Na2HPO4/0.1 M citric acid buffer (pH 7) at 40°C. Reaction products were separated on a silica 60 TLC plate developed in 1-propanol/ethylacetate/H2O (6:1:3, by vol.). After spraying the plates with ethanol/sulphuric acid (9:1, v/v), sugars were visualized by heating (120°C for 20 min).
Results and discussion
Construction of metagenomic DNA libraries
Two different grassland soil samples derived from the Schwäbische Alb (AEG3 and AEG6) were used for the construction of two large-insert metagenomic DNA libraries using the fosmid pCC1FOS as vector. In addition, one previously constructed metagenomic library (SEG9) derived from a grassland soil sample of the Schorfheide-Chorin was employed for subsequent screening. In all cases, DNA was directly isolated from the soil samples without previous enrichment or extraction of microbial cells. The DNA yields ranged from 25.3 to 56.7 μg per g soil (Supplementary Table 1). These yields were in the same range as described for the isolation of DNA from other soil samples (Waschkowitz et al. 2009). The three metagenomic libraries contained 147,888 (SEG9), 121,520 (AEG3), and 95,160 (AEG6) clones. The total numbers of recovered metagenomic library-containing E. coli clones are higher than in most other studies in which fosmids were used as vectors for the construction of large-insert metagenomic libraries (Couto et al. 2010; Hong et al. 2007; Jeon et al. 2009; Lim et al. 2005). The quality of the three different libraries was controlled by determination of the average insert sizes and the percentage of insert-bearing E. coli clones. The average insert sizes were 23.3 (SEG9), 26.9 (AEG3), and 28.4 (AEG6) kbp. These values roughly corresponded to those published for other soil-derived large-insert metagenomic libraries (Donato et al. 2010; Heath et al. 2009; Nacke et al. 2011b). The libraries harbored 9.42 Gbp of cloned soil DNA. Assuming an average prokaryotic genome size of 5 Mbp (Hårdeman and Sjöling 2007), the metagenomic libraries represented approximately 1,900 prokaryotic genomes.
Activity-based screening for cellulase and xylanase genes
The screen for genes conferring cellulase or xylanase activity was based on the ability of library-bearing E. coli clones to form halos when grown on agar medium containing dye-labeled hydroxyethyl cellulose (HECred) or xylan (xylangreen) as indicator substrates. As function-driven screening is the only strategy that harbors the potential to discover entirely novel and functional genes from metagenomes (Simon and Daniel 2009), similar screens have been employed to identify genes encoding (hemi)cellulolytic enzymes from various environments such as soil (Kim et al. 2007, 2008), cow rumen and manure (Ferrer et al. 2005; Li et al. 2009; Zhao et al. 2010), and sludge from a biogas reactor (Jiang et al. 2010).
To perform the above-mentioned activity-based approach approximately 4,600 clones per library were arrayed and screened for the targeted activities. Positive E. coli clones were collected after incubation on the indicator agar for 1–14 days at 37°C. In order to confirm that the (hemi)cellulolytic activity of the positive clones was fosmid-encoded the recombinant fosmids were isolated and used to transform E. coli. The resulting E. coli strains were screened again on indicator agar. Two different recombinant fosmids, fLX01 and fLX02, conferred a stable xylanolytic phenotype and one (fLC01) a cellulolytic phenotype. The fosmids fLX01, fLX02, and fLC01 were derived from libraries AEG3, AEG6, and SEG9, respectively (Table 1). Studies on recovery of genes encoding xylanases and cellulases from soil-derived metagenomic large-insert libraries are rare. For example, function-driven screening of 10,000 clones derived from a metagenomic phage forest soil library resulted in the recovery of one cellulolytic clone (Wang et al. 2009). In addition, one clone producing cellulase activity on carboxymethyl cellulose (CMC) was identified by screening of 70,000 clones, which harbored a metagenomic library constructed from Korean soil (Kim et al. 2008). This clone also contained a xylanase gene, which was identified by sequence analysis of the cloned DNA.
Molecular analyses
The insert sizes of the three recombinant fosmids, fLX01, fLX02 and fLC01, recovered from the positive E. coli clones were 36,000, 30,200, and 16,250 bp, respectively. In order to identify genes conferring xylanase or cellulase activity subcloning of the recombinant fosmids was carried out by cloning of fosmid-derived DNA fragments (2–6 kbp) into the plasmid vector pCR-XL-TOPO. Subsequently, the corresponding E. coli clones were screened on HECred-containing and xylangreen-containing indicator agar. In this way, recombinant plasmids carrying the desired DNA fragments for all three fosmids were recovered.
The insert sizes of the three plasmids, pLC01, pLX01, and pLX02 (Table 1) derived from the corresponding fosmids fLC01, fLX02, and fLX02, were 3,220, 5,523, and 4,985 bp, respectively. The inserts of pLC01, pLX01, and pLX02 were sequenced and analyzed. Each of the three plasmids harbored a putative gene that exhibited similarities to known genes encoding cellulases (pLC01) or xylanases (pLX01 and pLX02) (Supplementary Table 2). Cellulase or xylanase activity was detected in crude extracts of corresponding recombinant E. coli strains E. coli TOP10/pLC01, E. coli TOP10/pLX01, and E. coli TOP10/pLX02 (data not shown).
Analysis and classification of cellulase-encoding gene
Cellulases and xylanases are subdivided in different glycoside hydrolase families based on amino acid sequence similarity (Cantarel et al. 2009; Henrissat 1991). The amino acid sequence deduced from the putative cellulase gene cel01 of pLC01 comprised 831 amino acids with a calculated molecular mass of 90.4 kDa. A potential signal peptide of 21 amino acids was predicted at the N-terminus of Cel01. The amino acid sequence of the putative signal peptide showed the typical orientation of signal peptides with three distinct parts (N, H, and C domains) (Pugsley 1993). Protein domain analyses suggested that Cel01 belongs to the glycoside hydrolase family 9, as it contains a catalytic domain (amino acids 350–814) that is typical for family 9 members (Fig. 1). The multiple sequence alignment revealed that Cel01 contained putative catalytic aspartate and glutamate residues (D418, D421, and E808), which act as nucleophile (aspartate) or proton donor (glutamate) during substrate hydrolysis (Parsiegla et al. 2002) (Fig. 2a). In addition to the catalytic domain of family 9 glycoside hydrolases, Cel01 harbored a family 9 carbohydrate-binding module (CBM9, amino acids 46–231) and a N-terminal cellulase domain (Cel-N-term, amino acids 255–338) (Fig. 1). To our knowledge, family 9 carbohydrate-binding modules have previously been detected only in xylanases. The protein sequence of Cel01 was most similar (50% identity) to a cellulase from Sorangium cellulosum ‘So ce 56’ (Schneiker et al. 2007), but not over the entire length. The cellulase from Sorangium cellulosum possesses a similar modular structure as Cel01, but lacks a family 9 carbohydrate-binding module. Most of the metagenome-derived cellulase genes that have been identified by activity-based screening belonged to glycoside hydrolase families 5 and 9 (Duan and Feng 2010). Metagenome-derived family 9 cellulases were isolated from an aquatic community (Pottkämper et al. 2009), elephant dung (Wang et al. 2009), and an enrichment culture of an alkaline lake (Grant et al. 2004) but not from other soils with the exception of compost soil (Pang et al. 2009). In summary, molecular analysis indicated that Cel01 is an extracellular cellulase belonging to glycoside hydrolase family 9. In addition, the enzyme exhibited a modular structure, which is not known from other cellulases.
Fig. 1.
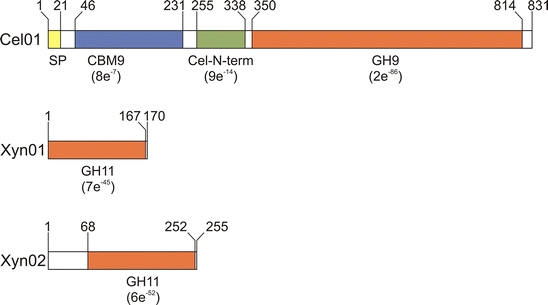
Domain structure of Cel01, Xyn01, and Xyn02. The recorded E values for the family 9 carbohydrate-binding module (cd00005), the cellulase N-terminal domain (cd02850), the family 9 glycoside hydrolase catalytic domain (pfam00759), and the family 11 glycoside hydrolase catalytic domain (pfam00457) are given in parentheses. Abbreviations: SP signal peptide, CBM9 family 9 carbohydrate-binding module, Cel-N-term cellulase N-terminal domain, GH9 family 9 glycoside hydrolase, GH11 family 11 glycoside hydrolase
Fig. 2.

Multiple sequence alignment of conserved regions in glycoside hydrolases belonging to family 9 (a) and family 11 (b). Identical amino acid residues are shown as white letters on a dark background. Triangles indicate catalytic residues and asterisks represent residues that are involved in substrate binding. References: Cel01, Xyn01, and Xyn02 (this study); ZP_07606292, GH9 from Streptomyces violaceusniger Tu 4113; YP_004081973, GH9 from Micromonospora sp. L5; YP_001612873, cellulase from Sorangium cellulosum ‘So ce 56’; ZP_05497266, GH9 from Clostridium papyrosolvens DSM 2782; AAK24198, GH9 from Caulobacter crescentus CB15; BAA06837, xylanase I precursor from Aeromonas punctata; AAB72117, endo-beta-1,4-xylanase from Geobacillus stearothermophilus; AAQ14588, xylanase from Bacillus firmus; CAJ87325, endo-1,4-beta-xylanase B from Thermobacillus xylanilyticus; AAZ17386, endo-1,4-beta-xylanase from Paenibacillus macerans; YP_001310136, endo-1,4-beta-xylanase from Clostridium beijerinckii NCIMB 8052; AAD54767, endo-1,4-beta-xylanase from Xylanimicrobium pachnodae; ABL11222, xylanase from uncultured bacterium
Analysis and classification of xylanase-encoding genes
The gene products of the putative xylanase genes xyn01 (170 amino acids) and xyn02 (255 amino acids) exhibited calculated molecular masses of 18.6 and 27 kDa, respectively. Putative signal peptides were not found in the protein sequences of both gene products. Xyn01 and Xyn02 showed the highest similarity (78 and 69% identity, respectively) to a xylanase derived from a soil metagenomic library (Kim et al. 2008). The presence of a catalytic domain typical for members of the glycoside hydrolase family 11 indicated that Xyn01 and Xyn02 belong to this family (Fig. 1). In addition, the multiple sequence alignment showed that putative catalytic glutamate residues and tyrosine residues involved in substrate binding are present in the amino acid sequences of Xyn01 (E62 and E155, Y53 and Y64) and Xyn02 (E146 and E240, Y137 and Y148) (Fig. 2b). Genes encoding family 11 xylanases were also derived from metagenomic libraries of insect guts (Brennan et al. 2004; Warnecke et al. 2007) and single microorganisms such as Dictyoglomus thermophilum (Morris et al. 1998) and Cellulomonas pachnodae (Cazemier et al. 1999).
Initial characterization of the xylanases Xyn01 and Xyn02
Crude cell-free extracts of E. coli TOP10/pLX01 and E. coli TOP10/pLX02 were used to initially characterize Xyn01 and Xyn02. Both xylanases exhibited activity with different xylans as substrates, but no activity was recorded by employing barley glucan, CMC, HEC, laminarin, lichenan, or microcrystalline cellulose as substrates. Xyn01 showed the highest activity with xylan from larch wood whereas Xyn02 was most active with xylan from birch wood (Table 2). Larch wood xylan is a non-acetylated softwood xylan, which contains a higher 4-O-methylglucuronic acid content than the acetylated hardwood xylan from birch wood. In addition, softwood xylans are less branched than hardwood xylans (Sunna and Antranikian 1997). Thus, the observed differences in activity of both xylanases suggest that they require the presence of a particular substituent or branching extent of the xylan polymer to enhance their activity. The optimum activities of both enzymes were measured in the crude extracts from 10 to 70°C and range pH 4–10. Both enzymes were most active at 60°C and showed high activity over the entire tested pH range with optimal activities at pH 7 (Xyn01) and pH 6 (Xyn02). In addition, both xylanases retained approx. 50% activity at pH 4 and 10 (Supplementary Fig. 1). So far, the highest activities of xylanases derived from functional screens of metagenomes were found at 50°C and the retained activities at pH 4 were lower than 40% (Brennan et al. 2004; Lee et al. 2006; Li et al. 2009; Zhao et al. 2010). Only one metagenome-derived xylanase (XynA), which has been identified from a hot pool environmental DNA sample by a PCR-based approach, exhibited a higher temperature optimum than Xyn01 and Xyn02 (Sunna and Bergquist 2003).
Table 2.
Determination of xylanase activity in crude cell-free extracts derived from E. coli TOP10 cells harboring the plasmids pLX01 and pLX02
| Plasmid | Specific activity (U mg−1) | |||
|---|---|---|---|---|
| Xylan (oat spelt) | Xylan (birch wood) | Xylan (larch wood) | Xylan (beech wood) | |
| Cloning vector | 0.008 ± 0.004 | 0.014 ± 0.006 | ND | ND |
| pLX01 | 4.859 ± 0.051 | 4.569 ± 0.040 | 6.005 ± 0.158 | 3.362 ± 0.082 |
| pLX02 | 0.168 ± 0.002 | 0.355 ± 0.003 | 0.337 ± 0.017 | 0.321 ± 0.013 |
ND no activity detected
Expression and purification of Cel01
To facilitate purification of the cellulase Cel01 the corresponding gene was amplified by PCR without the signal peptide sequence. The latter necessitated the addition of a start codon to the 5′ end of the coding region. The resulting PCR product was cloned into the expression vector pET101/D, thereby placing the genes under control of the IPTG-inducible T7 promoter and adding sequences encoding a His6 tag and a V5 epitope. The resulting construct pCel01 (Table 1) was transformed into E. coli BL21 and production of Cel01 was induced by addition of 0.5 mM IPTG. After incubation for 12 h at 37°C, cells were harvested and crude cell-free extracts were prepared. The production of His6-tagged Cel01 in the cell extracts was confirmed by Western-Blot analysis using antibodies against the V5 epitope (Fig. 3) and detection of cellulase activity. Subsequently, His6-tagged Cel01 was purified from cell-free extracts by metal ion affinity chromatography and ultrafiltration. The specific cellulase activity of the final enzyme preparation was 780 ± 11.5 U mg−1 with barley glucan as substrate. SDS–PAGE of the purified enzyme revealed that Cel01 has a MW of approx. 90 kDa (Fig. 3). The observed molecular mass is in good agreement with the one deduced from the sequence of His6-tagged version of Cel01 (91.9 kDa).
Fig. 3.
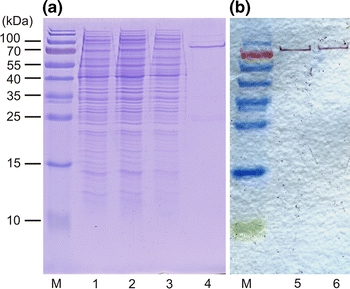
Analysis of Cel01 production and purification by SDS PAGE (a) and Western blot analysis (b). His6-tagged Cel01 was purified from cell extract of E. coli BL21/pCel01 by nickel affinity chromatography. a Lanes: M, marker proteins; 1, crude extract of E. coli BL21/pCel01; 2, flow through fraction; 3, wash fraction; 4, eluate (further purified by ultrafiltration); b Lanes: M, marker proteins 5, crude extract; 6, purified Cel01
Properties of purified Cel01
The purified Cel01 exhibited the highest cellulolytic activity by employing soluble substrates containing β-1,4-linkages such as barley glucan (780 ± 11.5 U mg−1), lichenan (516 ± 12.9 U mg−1), CMC (90 ± 2.0 U mg−1), and HEC (47.7 ± 2.5 U mg−1). No activity with xylan from birch wood, oat spelt, larch wood or beech wood, microcrystalline cellulose, and laminarin (β-1,3/β-1,6-linkages) was detected. Thus, Cel01 is most likely an endo 1,4-β-glucan hydrolase. This was confirmed by the time courses of CMC and barley glucan hydrolysis, which were analyzed by thin layer chromatography (Fig. 4). At first, CMC and barley glucan degradation resulted only in formation of high-molecular-mass products (Fig. 4). During prolonged incubation (CMC, 30 min to 48 h; barley glucan, 12–48 h) also small oligosaccharides were accumulated. In addition, no glucose was accumulated during the hydrolysis of CMC and barley glucan within incubation of 48 h. These results are also typical for an endo-wise action of the enzyme.
Fig. 4.
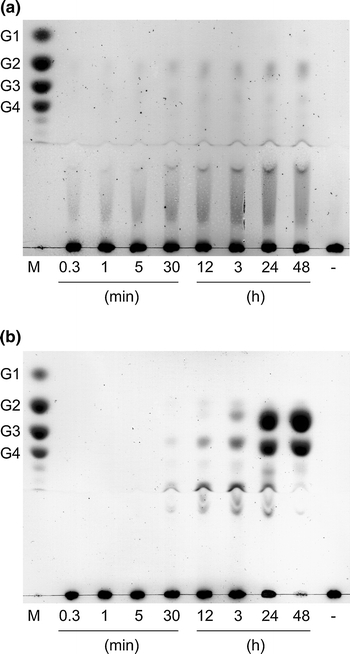
Thin layer chromatography analysis of hydrolysis products released by Cel01. Degradation of CMC (a) and barley glucan (b) was analyzed at the indicated time points. Lane M, mixed standard sugars: glucose (G1), cellobiose (G2), cellotriose (G3), and cellotetraose (G4); lane -, control containing substrate without enzyme
Purified Cel01 had significant activity between 30 and 50°C and exhibited optimal activity at 45 and 50°C (Fig. 5a). Cel01 was stable for 96 h below 50°C with more than 55% remaining activity (Supplementary Fig. 2). Incubation of Cel01 at 60°C caused complete inactivation of the enzyme. Interestingly, the optimal activities of other cellulases derived from soil metagenomic libraries ranged from 45 to 50°C and incubation at 60°C also resulted in rapid inactivation (Kim et al. 2008; Liu et al. 2011; Voget et al. 2006) (Table S3). Cel01 was highly active from pH 5 to pH 9 with optimal activity at pH 7 (Fig. 5b, Table S3). The cellulase activity was reduced by addition of MnCl2, ethanol, EDTA, and SDS to 62 ± 3.8%, 84 ± 5.5%, 12 ± 1.1%, and 18 ± 2.4%, respectively (Fig. 6). CoCl2 weakly stimulated the enzyme activity. When the enzyme was incubated in 3 M KCl or 4 M NaCl, it showed high halotolerance and retained more than 70% of its activity after 12 h incubation (Fig. 7).
Fig. 5.

Effect of temperature (a) and pH (b) on activity of Cel01. a Activity of Cel01 was determined using the standard assay at temperatures between 10 and 70°C. b Activity was measured at pH values between 4 and 10 using sodium acetate buffer (black circles), sodium phosphate buffer (white circles), Tris–HCl buffer (black triangles), and glycine-NaOH buffer (white triangles). The average of triplicate experiments is presented
Fig. 6.
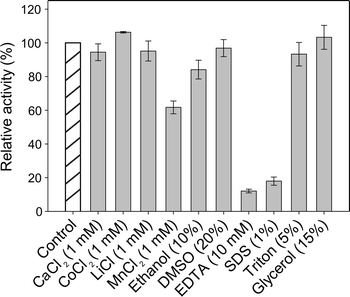
Effect of metal ions and chemical agents on activity of Cel01. Activity without addition of metal ions or chemical agents was defined as 100%. The average of triplicate experiments is presented
Fig. 7.
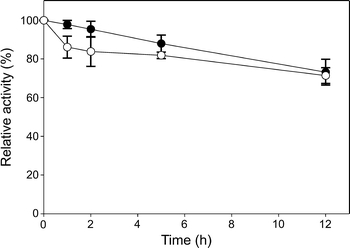
Halotolerance of Cel01. Halotolerance was investigated by measuring residual activity using the standard assay after incubation of Cel01 for 12 h in 3 M KCl (black circles) and 4 M NaCl (white circles). The average of triplicate experiments is presented
Conclusions
In this study, construction and screening of large-insert soil-derived metagenomic libraries has led to the discovery and characterization of two novel xylanases and one cellulase. The novelty of these enzymes arises from the enormous genetic diversity of uncultured soil microorganisms. The recovered enzymes have some interesting properties such as high activity of the xylanases over a wide range of temperatures and pH values. Especially the metagenome-derived cellulase has potential for industrial application, as the enzyme is highly active and stable over a wide pH range, and shows high halotolerance.
Taking the relatively low number of screened library-containing E. coli clones screened into account the results presented here demonstrated that soil-based metagenomics in combination with use of a simple activity-based screening system is a method for the rapid isolation of diverse genes conferring the desired reactions.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The work has been funded by the DFG Priority Program 1374 “Infrastructure-Biodiversity-Exploratories” (DA 374/4-1 and DA 374/6-1). Field work permits were given by the responsible state environmental offices of Baden-Württemberg and Brandenburg (according to §72 BbgNatSchG). We thank the BEO (biodiversity exploratories office) for support and project administration.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Beg QK, Kapoor M, Mahajan L, Hoondal GS. Microbial xylanases and their industrial applications: a review. Appl Microbiol Biotechnol. 2001;56:326–338. doi: 10.1007/s002530100704. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Brennan Y, Callen WN, Christoffersen L, Dupree P, Goubet F, Healey S, Hernandez M, Keller M, et al. Unusual microbial xylanases from insect guts. Appl Environ Microbiol. 2004;70:3609–3617. doi: 10.1128/AEM.70.6.3609-3617.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazemier AE, Verdoes JC, van Ooyen AJ, Op den Camp HJ. Molecular and biochemical characterization of two xylanase-encoding genes from Cellulomonas pachnodae. Appl Environ Microbiol. 1999;65:4099–4107. doi: 10.1128/aem.65.9.4099-4107.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins T, Gerday C, Feller G. Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol Rev. 2005;29:3–23. doi: 10.1016/j.femsre.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Couto GH, Glogauer A, Faoro H, Chubatsu LS, Souza EM, Pedrosa FO. Isolation of a novel lipase from a metagenomic library derived from mangrove sediment from the south Brazilian coast. Genet Mol Res. 2010;9:514–523. doi: 10.4238/vol9-1gmr738. [DOI] [PubMed] [Google Scholar]
- Donato JJ, Moe LA, Converse BJ, Smart KD, Berklein FC, McManus PS, Handelsman J. Metagenomic analysis of apple orchard soil reveals antibiotic resistance genes encoding predicted bifunctional proteins. Appl Environ Microbiol. 2010;76:4396–4401. doi: 10.1128/AEM.01763-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan CJ, Feng JX. Mining metagenomes for novel cellulase genes. Biotechnol Lett. 2010;32:1765–1775. doi: 10.1007/s10529-010-0356-z. [DOI] [PubMed] [Google Scholar]
- Duan CJ, Xian L, Zhao GC, Feng Y, Pang H, Bai XL, Tang JL, Ma QS, et al. Isolation and partial characterization of novel genes encoding acidic cellulases from metagenomes of buffalo rumens. J Appl Microbiol. 2009;107:245–256. doi: 10.1111/j.1365-2672.2009.04202.x. [DOI] [PubMed] [Google Scholar]
- Ferrer M, Golyshina OV, Chernikova TN, Khachane AN, Reyes-Duarte D, Santos VA, Strompl C, Elborough K, et al. Novel hydrolase diversity retrieved from a metagenome library of bovine rumen microflora. Environ Microbiol. 2005;7:1996–2010. doi: 10.1111/j.1462-2920.2005.00920.x. [DOI] [PubMed] [Google Scholar]
- Fischer M, Bossdorf O, Gockel S, Hänsel F, Hemp A, Hessenmöller D, Korte G, Nieschulze J, et al. Implementing large-scale and long-term functional biodiversity research: the biodiversity exploratories. Basic Appl Ecol. 2010;11:473–485. doi: 10.1016/j.baae.2010.07.009. [DOI] [Google Scholar]
- Fontes CM, Gilbert HJ, Hazlewood GP, Clarke JH, Prates JA, McKie VA, Nagy T, Fernandes TH, et al. A novel Cellvibrio mixtus family 10 xylanase that is both intracellular and expressed under non-inducing conditions. Microbiology. 2000;146:1959–1967. doi: 10.1099/00221287-146-8-1959. [DOI] [PubMed] [Google Scholar]
- Gilad R, Rabinovich L, Yaron S, Bayer EA, Lamed R, Gilbert HJ, Shoham Y. CelI, a noncellulosomal family 9 enzyme from Clostridium thermocellum, is a processive endoglucanase that degrades crystalline cellulose. J Bacteriol. 2003;185:391–398. doi: 10.1128/JB.185.2.391-398.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant S, Sorokin DY, Grant WD, Jones BE, Heaphy S. A phylogenetic analysis of Wadi el Natrun soda lake cellulase enrichment cultures and identification of cellulase genes from these cultures. Extremophiles. 2004;8:421–429. doi: 10.1007/s00792-004-0402-7. [DOI] [PubMed] [Google Scholar]
- Hall A. Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Handelsman J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev. 2004;68:669–685. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hårdeman F, Sjöling S. Metagenomic approach for the isolation of a novel low-temperature-active lipase from uncultured bacteria of marine sediment. FEMS Microbiol Ecol. 2007;59:524–534. doi: 10.1111/j.1574-6941.2006.00206.x. [DOI] [PubMed] [Google Scholar]
- Heath C, Hu XP, Cary SC, Cowan D. Identification of a novel alkaliphilic esterase active at low temperatures by screening a metagenomic library from Antarctic desert soil. Appl Environ Microbiol. 2009;75:4657–4659. doi: 10.1128/AEM.02597-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong KS, Lim HK, Chung EJ, Park EJ, Lee MH, Kim JC, Choi GJ, Cho KY, et al. Selection and characterization of forest soil metagenome genes encoding lipolytic enzymes. J Microbiol Biotechnol. 2007;17:1655–1660. [PubMed] [Google Scholar]
- Jeon JH, Kim JT, Kang SG, Lee JH, Kim SJ. Characterization and its potential application of two esterases derived from the arctic sediment metagenome. Mar Biotechnol. 2009;11:307–316. doi: 10.1007/s10126-008-9145-2. [DOI] [PubMed] [Google Scholar]
- Jiang C, Hao ZY, Jin KE, Li SX, Che ZQ, Ma GF, Wu B. Identification of a metagenome-derived β-glucosidase from bioreactor contents. J Mol Catal B Enzym. 2010;63:11–16. doi: 10.1016/j.molcatb.2009.11.009. [DOI] [Google Scholar]
- Kim SJ, Lee CM, Kim MY, Yeo YS, Yoon SH, Kang HC, Koo BS. Screening and characterization of an enzyme with beta-glucosidase activity from environmental DNA. J Microbiol Biotechnol. 2007;17:905–912. [PubMed] [Google Scholar]
- Kim SJ, Lee CM, Han BR, Kim MY, Yeo YS, Yoon SH, Koo BS, Jun HK. Characterization of a gene encoding cellulase from uncultured soil bacteria. FEMS Microbiol Lett. 2008;282:44–51. doi: 10.1111/j.1574-6968.2008.01097.x. [DOI] [PubMed] [Google Scholar]
- Kulkarni N, Shendye A, Rao M. Molecular and biotechnological aspects of xylanases. FEMS Microbiol Rev. 1999;23:411–456. doi: 10.1111/j.1574-6976.1999.tb00407.x. [DOI] [PubMed] [Google Scholar]
- Lee ST, Lee JJ. Insoluble dye substrate for screening and assay of xylan-degrading enzymes. J Microbiol Meth. 1997;29:1–5. doi: 10.1016/S0167-7012(97)00986-X. [DOI] [Google Scholar]
- Lee CC, Kibblewhite-Accinelli RE, Wagschal K, Robertson GH, Wong DWS. Cloning and characterization of a cold-active xylanase from an environmental library. Extremophiles. 2006;10:295–300. doi: 10.1007/s00792-005-0499-3. [DOI] [PubMed] [Google Scholar]
- Li R, Kibblewhite R, Orts WJ, Lee CC. Molecular cloning and characterization of multidomain xylanase from manure library. World J Microbiol Biotechnol. 2009;25:2071–2078. doi: 10.1007/s11274-009-0111-6. [DOI] [Google Scholar]
- Lim HK, Chung EJ, Kim JC, Choi GJ, Jang KS, Chung YR, Cho KY, Lee SW. Characterization of a forest soil metagenome clone that confers indirubin and indigo production on Escherichia coli. Appl Environ Microbiol. 2005;71:7768–7777. doi: 10.1128/AEM.71.12.7768-7777.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Liu WD, Zhao XL, Shen WJ, Cao H, Cui ZL. Cloning and functional characterization of a novel endo-β-1, 4-glucanase gene from a soil-derived metagenomic library. Appl Microbiol Biotechnol. 2011;89:1083–1092. doi: 10.1007/s00253-010-2828-4. [DOI] [PubMed] [Google Scholar]
- Lynd LR, Weimer PJ, Willem HZ, Pretorius IS. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev. 2002;66:506–577. doi: 10.1128/MMBR.66.3.506-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynd LR, van Zyl WH, McBride JE, Laser M. Consolidated bioprocessing of cellulosic biomass: an update. Curr Opin Biotechnol. 2005;16:577–583. doi: 10.1016/j.copbio.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, et al. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- Morris DD, Gibbs MD, Chin CW, Koh MH, Wong KK, Allison RW, Nelson PJ, Bergquist PL. Cloning of the xynB gene from Dictyoglomus thermophilum Rt46B.1 and action of the gene product on kraft pulp. Appl Environ Microbiol. 1998;64:1759–1765. doi: 10.1128/aem.64.5.1759-1765.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacke H, Thürmer A, Wollherr A, Will C, Hodac L, Herold N, Schöning I, Schrumpf M, Daniel R. Pyrosequencing-based assessment of bacterial community structure along different management types in German forest and grassland soils. PLoS ONE. 2011;6:e17000. doi: 10.1371/journal.pone.0017000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacke H, Will C, Herzog S, Nowka B, Engelhaupt M, Daniel R. Identification of novel lipolytic genes and gene families by screening of metagenomic libraries derived from soil samples of the German Biodiversity Exploratories. FEMS Microbiol Ecol. 2011;78:188–201. doi: 10.1111/j.1574-6941.2011.01088.x. [DOI] [PubMed] [Google Scholar]
- Pang H, Zhang P, Duan CJ, Mo XC, Tang JL, Feng JX. Identification of cellulase genes from the metagenomes of compost soils and functional characterization of one novel endoglucanase. Curr Microbiol. 2009;58:404–408. doi: 10.1007/s00284-008-9346-y. [DOI] [PubMed] [Google Scholar]
- Parsiegla G, Belaïch A, Belaïch JP, Haser R. Crystal structure of the cellulase Cel9M enlightens structure/function relationships of the variable catalytic modules in glycoside hydrolases. Biochemistry. 2002;41:11134–11142. doi: 10.1021/bi025816m. [DOI] [PubMed] [Google Scholar]
- Polizeli ML, Rizzatti AC, Monti R, Terenzi HF, Jorge JA, Amorim DS. Xylanases from fungi: properties and industrial applications. Appl Microbiol Biotechnol. 2005;67:577–591. doi: 10.1007/s00253-005-1904-7. [DOI] [PubMed] [Google Scholar]
- Pottkämper J, Barthen P, Ilmberger N, Schwaneberg U, Schenk A, Schulte M, Ignatiev N, Streit WR. Applying metagenomics for the identification of bacterial cellulases that are stable in ionic liquids. Green Chem. 2009;11:957–965. doi: 10.1039/b820157a. [DOI] [Google Scholar]
- Pugsley AP. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosselló-Mora R, Amann R. The species concept for prokaryotes. FEMS Microbiol Rev. 2001;25:39–67. doi: 10.1016/S0168-6445(00)00040-1. [DOI] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Sanchez OJ, Cardona CA. Trends in biotechnological production of fuel ethanol from different feedstocks. Bioresour Technol. 2008;99:5270–5295. doi: 10.1016/j.biortech.2007.11.013. [DOI] [PubMed] [Google Scholar]
- Schneiker S, Perlova O, Kaiser O, Gerth K, Alici A, Altmeyer MO, Bartels D, Bekel T, et al. Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat Biotechnol. 2007;25:1281–1289. doi: 10.1038/nbt1354. [DOI] [PubMed] [Google Scholar]
- Shedova EN, Berezina OV, Lunina NA, Zverlov VV, Schwarz WH, Velikodvorskaya GA. Cloning and characterization of a large metagenomic DNA fragment containing glycosyl-hydrolase genes. Mol Gen Microbiol Virol. 2009;24:12–16. doi: 10.3103/S0891416809010030. [DOI] [PubMed] [Google Scholar]
- Simon C, Daniel R. Achievements and new knowledge unraveled by metagenomic approaches. Appl Microbiol Biotechnol. 2009;85:265–276. doi: 10.1007/s00253-009-2233-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon C, Daniel R. Metagenomic analyses: past and future trends. Appl Environ Microbiol. 2011;77:1153–1161. doi: 10.1128/AEM.02345-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele HL, Jaeger KE, Daniel R, Streit WR. Advances in recovery of novel biocatalysts from metagenomes. J Mol Microbiol Biotechnol. 2009;16:25–37. doi: 10.1159/000142892. [DOI] [PubMed] [Google Scholar]
- Strohl WR. The role of natural products in a modern drug discovery program. Drug Discov Today. 2000;5:39–41. doi: 10.1016/S1359-6446(99)01443-9. [DOI] [PubMed] [Google Scholar]
- Sunna A, Antranikian G. Xylanolytic enzymes from fungi and bacteria. Crit Rev Biotechnol. 1997;17:39–67. doi: 10.3109/07388559709146606. [DOI] [PubMed] [Google Scholar]
- Sunna A, Bergquist PL. A gene encoding a novel extremely thermostable 1, 4-beta-xylanase isolated directly from an environmental DNA sample. Extremophiles. 2003;7:63–70. doi: 10.1007/s00792-002-0296-1. [DOI] [PubMed] [Google Scholar]
- Ten LN, Im W-T, Kim M-K, Lee S-T. A plate assay for simultaneous screening of polysaccharide- and protein-degrading microorganisms. Lett Appl Microbiol. 2005;40:92–98. doi: 10.1111/j.1472-765X.2004.01637.x. [DOI] [PubMed] [Google Scholar]
- Thompson J, Higgins D, Gibson T. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voget S, Steele HL, Streit WR. Characterization of a metagenome-derived halotolerant cellulase. J Biotechnol. 2006;126:26–36. doi: 10.1016/j.jbiotec.2006.02.011. [DOI] [PubMed] [Google Scholar]
- Wang F, Li F, Chen G, Liu W. Isolation and characterization of novel cellulase genes from uncultured microorganisms in different environmental niches. Microbiol Res. 2009;164:650–657. doi: 10.1016/j.micres.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Warnecke F, Luginbühl P, Ivanova N, Ghassemian M, Richardson TH, Stege JT, Cayouette M, Mc Hardy AC, et al. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature. 2007;450:560–565. doi: 10.1038/nature06269. [DOI] [PubMed] [Google Scholar]
- Waschkowitz T, Rockstroh S, Daniel R. Isolation and characterization of metalloproteases with a novel domain structure by construction and screening of metagenomic libraries. Appl Environ Microbiol. 2009;75:2506–2516. doi: 10.1128/AEM.02136-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu EY, Kwon MA, Lee M, Oh JY, Choi JE, Lee JY, Song BK, Hahm DH, Song JK. Isolation and characterization of cold-active family VIII esterases from an arctic soil metagenome. Appl Microbiol Biotechnol. 2011;90:573–581. doi: 10.1007/s00253-011-3132-7. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhao J, Zeng R. Expression and characterization of a novel mesophilic protease from metagenomic library derived from Antarctic coastal sediment. Extremophiles. 2011;15:23–29. doi: 10.1007/s00792-010-0332-5. [DOI] [PubMed] [Google Scholar]
- Zhao S, Wang J, Bu D, Liu K, Zhu Y, Dong Z, Yu Z. Novel glycoside hydrolases identified by screening a Chinese Holstein dairy cow rumen-derived metagenome library. Appl Environ Microbiol. 2010;76:6701–6705. doi: 10.1128/AEM.00361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


