Abstract
Alternative splicing is a fundamental feature in regulating the eukaryotic transcriptome, as ~95% of multi-exon human Pol II transcripts are subject to this process. Regulated splicing operates through the combinatorial interplay of positive and negative regulatory signals present in the pre-mRNA, which are recognized by trans-acting factors. All these RNA and protein components are assembled in a gigantic, 21 MDa, ribonucleoprotein splicing machine – the supraspliceosome. Because most alternatively spliced mRNA isoforms vary between different cell and tissue types, the ability to perform alternative splicing is expected to be an integral part of the supraspliceosome, which constitutes the splicing machine in vivo. Here we show that both the constitutively and alternatively spliced mRNAs of the endogenous human pol II transcripts: hnRNP A/B, survival of motor neuron (SMN) and ADAR2 are predominantly found in supraspliceosomes. This finding is consistent with our observations that the splicing regulators hnRNP G as well as all phosphorylated SR proteins are predominantly associated with supraspliceosomes. We further show that changes in alternative splicing of hnRNP A/B, affected by up regulation of SRSF5 (SRp40) or by treatment with C6-ceramide, occur within supraspliceosomes. These observations support the proposed role of the supraspliceosome in splicing regulation and alternative splicing.
Keywords: RNA splicing, ribonucleoprotein, macromolecular assembly, alternative splicing, SR proteins, supraspliceosome
1. Introduction
Alternative splicing is a process whereby different combinations of exons are spliced together to produce different mRNAs from a single pre-mRNA transcript. Alternative splicing is estimated to occur in ~95% of all human RNA polymerase II transcripts, thereby underlying a major source for the diversity of the human proteome (Brett et al., 2002; Garcia-Blanco et al., 2004; Modrek and Lee, 2002; Srebrow and Kornblihtt, 2006). The physiological significance of alternative splicing is manifested by the fact that the majority of the alternative exons are differentially expressed in various tissues and cell lines (Pan et al., 2008; Wang and Burge, 2008), and by the finding that mutations that affect alternative splicing are found in a large number of human diseases (Garcia-Blanco et al., 2004; Srebrow and Kornblihtt, 2006; Wang and Cooper, 2007).
Regulated splicing, as well as constitutive splicing, operates through the combinatorial interplay of positive and negative regulatory signals present in the pre-mRNA, which are recognized by trans-acting factors. The most studied of the latter are members of the hnRNP and SR protein family. SR proteins bind RNA through their RNA recognition motives (RRMs), while their SR domain appears to enable protein-protein and protein-RNA interactions during the splicing reaction. SR proteins show distinct RNA binding specificities for exonic splicing enhancers (ESEs), and multiple binding sites for several SR proteins are found in the same exon (Lin and Fu, 2007; Long and Caceres, 2009; Shepard and Hertel, 2009).
Alterations in alternative splicing associated with several human diseases, including cancer (Garcia-Blanco et al., 2004; Srebrow and Kornblihtt, 2006; Wang and Cooper, 2007), motivated search and identification of low molecular weight compounds that can affect alternative splicing, (reviewed in Sumanasekera et al., 2008). Among the substances identified were the ceramides. These compounds activate protein phosphatase 1 (PP1), by an unknown mechanism that probably involves dissociation of the phosphates from its inhibitors. PP1, which binds to an RRM of a number of splicing regulatory proteins, can thus affect splice site selection through dephosphorylation of its targets (Chalfant et al., 2001). Indeed, C6-ceramide was shown to affect alternative splicing of caspase 9 and Bcl-x (Chalfant et al., 2002).
Supraspliceosomes - isolated from mammalian cell nuclei are huge ribonucleoprotein (RNP) complexes having a molecular mass of ~21 MDa. They contain all five spliceosomal small nuclear RNP particles (snRNPs), as well as non-snRNP proteins. The entire repertoire of nuclear pre-mRNAs, independent of their length or the number of introns they contain, appear to be assembled in splicing-active supraspliceosomes (reviewed in Sperling et al., 2008). These complexes also harbor other components of pre-mRNA processing, such as the editing enzymes ADAR1 and ADAR2 (Raitskin et al., 2001), cap-binding proteins, and components of the 3’-end processing activity (Raitskin et al., 2002). Taken together, these observations support the view that the supraspliceosome is the nuclear pre-mRNA processing machine.
In addition to the constitutive splicing factors, a number of splicing regulatory factors were found to be predominantly associated with supraspliceosomes. These include all phosphorylated SR proteins (Yitzhaki et al., 1996); the splicing regulatory factor hnRNP G (Heinrich et al., 2009); and the alternative splicing factors RBM4 and WT1, which co-interact to influence alternative splicing (Markus et al., 2006).
Structural studies revealed that the supraspliceosome is composed of four apparently similar splicing active substructures – native spliceosomes – each resembling an in vitro assembled spliceosome, which are connected by the pre-mRNA (Azubel et al., 2004; Azubel et al., 2006; Cohen-Krausz et al., 2007; Medalia et al., 2002; Müller et al., 1998; Sperling et al., 1997). The native spliceosome, which is composed of a large and a small subunit, is arranged within the supraspliceosome such that the small subunits reside in the center of the supraspliceosome. This configuration allows communication between the native spliceosomes, which is a crucial element for regulated alternative splicing and for quality control of the resulting mRNAs (Azubel et al., 2006; Cohen-Krausz et al., 2007).
In this configuration the supraspliceosome can be regarded as a multiprocessor machine that can splice multi-intronic pre-mRNAs – four introns at a time – not necessarily in a consecutive manner (reviewed in Sperling et al., 2008). The observations cited above raised the question whether the regulation of alternative splicing occurs within the supraspliceosome. Here we show that endogenous alternatively spliced transcripts of hnRNP A/B, SMN and ADAR2 are predominantly found in the supraspliceosome. Furthermore, the regulation of alternative splicing of such endogenous transcripts, when affected by over expression of SR proteins or by treatment with C6-ceramide occurs within the supraspliceosome.
2. Material and methodss
2.1. Isolation of supraspliceosomes
Nuclear supernatants enriched in supraspliceosomes were prepared as described (Miriami et al., 1994; Sperling et al., 1985). Briefly, nuclear supernatants enriched for supraspliceosomes were prepared from purified nuclei of HeLa cells (CILBIOTECH, Mons, Belgium) by microsonication of the nuclei and precipitation of the chromatin in the presence of tRNA. The nuclear supernatants were fractionated in 10-45% linear glycerol gradients. The gradients were calibrated with 200S TMV particles run in a parallel gradient in fractions 9, 10. The bacterial 30S ribosomal subunit sedimented in fraction 18, and free tRNA in fraction 20. Aliquots from fractions corresponding to the 200S region of the glycerol gradient (27-32% glycerol) were taken for analysis and for visualization by EM. RNA was extracted from gradient fraction as described (Azubel et al., 2006).
2.2. Preparation and isolation of native spliceosomes
Native spliceosomes were prepared from supraspliceosomes as previously described (Azubel et al., 2004; Azubel et al., 2006). Briefly, purified supraspliceosomes were treated with 4 mM EDTA for 3 min and then incubated with a pool of oligodeoxynucleotides complementary to the 5’ splice site consensus sequence for additional 10 min. The sample was treated with RNase H (TAKARA; 70 units/ml), MgCl2 was added to a final concentration of 8 mM, and the mixture was incubated for 1 hr. The sample was centrifuged in a second linear 10-45% glycerol gradient, and the native spliceosomes were collected in aliquots from fractions corresponding to the 60-70S region of the gradient. All steps were performed at 4°C.
2.3. Reconstitution of supraspliceosomes
Reconstitution of supraspliceosomes from native spliceosomes and synthetic pre-mRNA was performed as described (Azubel et al., 2006). In vitro transcribed β-globin pre-mRNA (untagged or tagged with gold) was mixed to a final concentration of 10-50 ng/μl with native spliceosomes in a volume of 20 μl and incubated for 2 hr at 4° C.
2.4. Transfections, RNA isolation and RT-PCR analyses
These procedures were performed as described (Kamhi et al., 2006; Li et al., 2002). Human 293T or HeLa cells were grown to 20-40% confluency in tissue culture plates and transiently co-transfected with the appropriate constructs. Plasmids expressing SRSF5 (SRp40) (pCGT-SRp40), SRSF2 (SC35) and SRSF6 (SRp55) (Screaton et al., 1995) were a kind gift from Drs Adrian Krainer and Rotem Karni. Transfections with plasmids expressing SRSF5 were at 3-6 μg per 5×106 cells, and transfection with plasmids expressing SRSF6 or SRSF2 were at 3-8 μg per 5×106 cells. Treatment with C6-ceramide (Stratagene) of half confluent 293 cell cultures was at 20-40 mM. Cells were harvested at 24 h post transfection, or C6 ceramide treatment, and total cellular RNA was extracted with guanidinium thiocyanate as described (Li et al., 2002). For supraspliceosome analysis, nuclear supernatants, prepared from the treated cells, were fractionated in glycerol gradients as described above and RNA extracted from gradient fractions and analyzed by RT-PCR. For control, RNA was extracted from untreated cells. The following primer pairs were used: hnRNP A/B: ex3s (sense) 5’-AAATGGATCC CAACACTGGA-3’, and ex8as (antisense) 5’-TGGCTCTTGCCGTAGTTTGT-3’; ADAR2: ex3s 5’-CCTGGTCCTGGGTAAGTTTGGTG-3’, and ex7as 5’-GAATCGTCCCCTCACCAGACTCT-3’; SMN: ex5s 5’-CTATCATGCTGGCTGCCT 3’, and ex8a 5’-CTACAACACCCTTCTCACAG-3’; actin: s 5’-CAAGGCCAACCGCGAGAAGATGAC-3’, and as 5’-CAAGGCCAACCGCGAGAAGATGAC 3’; SRSF5: s 5’-CAAGAAGCAGGTCTCGATCCCG-3’, and as 5’-CCACGTTTCTGGCTCTTCTCAGG-3’.
2.5. Gold labeling of β-globin pre-mRNA
β-globin pre-mRNA was prepared as described (Azubel et al., 2006), by transcription in vitro of a BamHI-linearized pSP64HβΔ6 plasmid (Krainer et al., 1984). The sample was treated with DNase, and the RNA was recovered by extraction with phenol:chloroform:isoamyl alcohol (25:24:1) and precipitation in ethanol. In parallel, 50 pmole of 3’ thiolated oligodeoxynucleotide (5’-GGCCTTTTACCGG-SH-3’) was treated with polynucleotide kinase (10 Units), precipitated in ethanol and treated with tris(2-carboxyethyl)phosphine (TCEP) at 10 mM. β-globin pre-mRNA (4.15 μg, 25 pmole) was ligated to the oligo-SH using 22 units of T4 RNA ligase (Fermentas) as described by the manufacturer, and the ligated product was purified using a size-exclusion columns (G25 Sephadex, Roche) followed by precipitation in ethanol. The pellet was resuspended in 20 μl buffer (0.1 M NaPO4, pH 6.4, 1 mM EDTA, 2 mM vanadyl ribonucleoside) and incubated with 20 μl of Maleimido-Nanogold (41 pmole/μl, Nanoprobes) for 6 h at 4 °C. The gold-labeled β-globin pre-mRNA was fractionated on 2% agarose gel, blotted to Nitrocellulose membrane, and visualized using LI Silver enhancement kit (Nanoprobes).
2.6. Immunoprecipitation (IP) and Western blot analyses
IP experiments were performed as described (Raitskin et al., 2001), using immobilized anti-Sm monoclonal antibodies (MAb Y12). Anti-Sm-bound complexes were eluted with 0.2 M glycine-HCl, pH 2.0, RNA was extracted with phenol from the bound and unbound fractions and analyzed by RT-PCR. Proteins from the bound and unbound fractions were precipitated in acetone. As control we used immobilized normal rabbit serum (NRS). For Western blots, proteins were analyzed by SDS/PAGE, blotted, and probed with antibodies against the Sm proteins as described (Kamhi et al., 2006; Raitskin et al., 2001), and with antibodies against ribosomal protein S14, as described (Nadano et al., 2000).
2.7. Electron microscopy
Ten-microliter aliquots of the samples were absorbed on glow-discharged carbon-coated grids, washed with water, and negatively stained with 1% uranyl acetate. For visualizing Nanogold-tagged supraspliceosomes, aliquots were sampled on glow-discharged carbon-coated Nickel grids, washed, stained with gold-enhancement mix (Nanoprobes), washed again with water and stained with 1% uranyl acetate. A Tecnai 12 TEM (FEI), operating at acceleration voltage of 100 kV, equipped with a 1K X 1K TEMcam CCD camera was used.
3. Results
3.1. Alternatively spliced mRNAs are predominantly found in supraspliceosomes
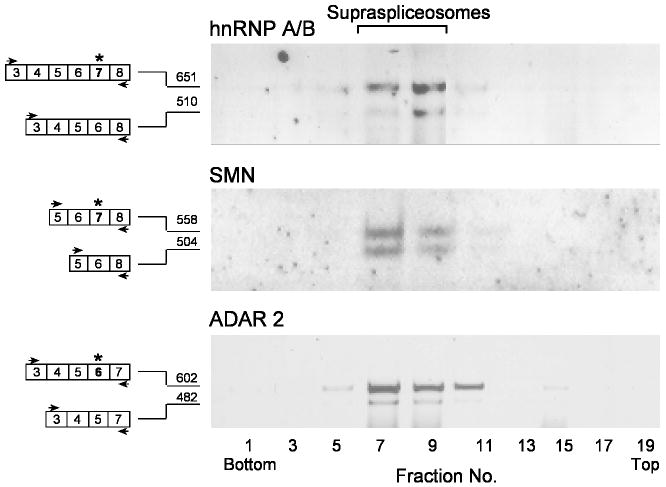
Because pre-mRNAs are assembled within supraspliceosomes during their entire lifetime in the nucleus, and because several regulatory splicing factors are found predominantly in supraspliceosomes (Sperling et al., 2008), we have asked whether specific endogenous alternatively spliced pol II transcripts are also packaged in supraspliceosomes. To this end we analyzed three human gene transcripts known to undergo alternative splicing: (i) hnRNP A/B, which is a member of the heterogeneous nuclear RNP proteins and has two alternatively spliced isoforms due to inclusion or skipping of exon 7 at its 3’ end, also known as apobec-1 binding protein (ABBP-1) (Khan et al., 1991; Lau et al., 1997); (ii) Survival motor neuron 1 (SMN1), which is involved in snRNP assembly, and the loss of its expression is the cause of the spinal muscular atrophy disease (SMA) (Lorson et al., 1999). A nearly identical copy gene called SMN2 produces low levels of the functional protein due to alternative splicing resulting in skipping of exon 7; and (iii) ADAR2, which is a member of the A-to-I RNA editing enzymes and a completely required gene in mammals. It edits the Q/R site of GluR-B subunit (Sommer et al., 1991), which is accompanied by dramatic decrease of Ca2+ permeability of the AMPA channel. ADAR2 is expressed ubiquitously and undergoes alternative splicing whereby its exon 6 is alternatively skipped (Gerber et al., 1997; Lai et al., 1997). Fig. 1 shows the distribution of these transcripts across a glycerol gradient in which a HeLa nuclear supernatant enriched for supraspliceosomes was fractionated (Azubel et al., 2006). It can be seen that all three transcripts and their respective alternatively spliced isoforms sedimented predominantly in the 200S region of the gradient, where the 21-MDa supraspliceosomes sediment. Their sedimentation patterns are similar to those of the phosphorylated SR-proteins (Heinrich et al., 2009; Raitskin et al., 2002; Yitzhaki et al., 1996) and hnRNP G (Heinrich et al., 2009), which were previously shown to be predominantly associated with supraspliceosomes in these fractions.
Fig. 1.
Constitutively and alternatively spliced mRNAs of endogenous human hnRNP A/B, SMN and ADAR2 transcripts are predominantly found in supraspliceosomes. Nuclear supernatants enriched in supraspliceosomes were prepared from HeLa cells, fractionated in a 10-45% glycerol gradient and RNA was extracted. Aliquots from odd fractions were analyzed by semi-quantitative RT-PCR, using primer pairs that flank the respective alternative exons. The constitutively and alternatively spliced forms of each of the three gene transcripts are schematically drawn on the left (the alternative exons are marked by asterisks), and the respective PCR primer pairs are indicated by arrowheads. Each part of the figure is representative of at least three different experiments. Supraspliceosomes peak in fractions 8-10. The gradients were calibrated with 200S TMV particles run in a parallel gradient in fractions 9, 10.
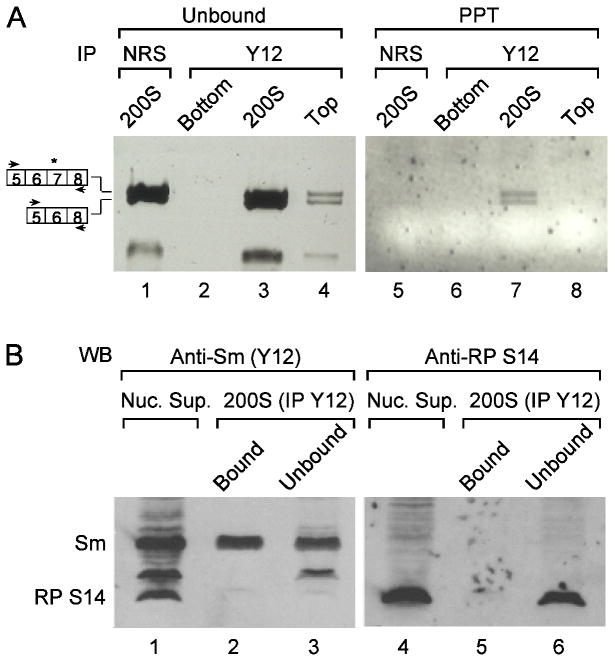
To further substantiate the association of both isoforms of alternatively spliced transcripts with supraspliceosomes we performed co-immunoprecipitation (co-IP) experiments using the Y12 monoclonal antibody (MAb), which is directed against the Sm proteins components of the supraspliceosome. For this experiment, the bottom fractions 1-4 (bottom), the supraspliceosomal fractions 8-11 (200S), and the top fractions 17-20 (top) of gradient-fractionated supraspliceosomes were each pooled together and immunoprecipitated with MAb Y12. RNA was extracted from each of the three unbound and precipitated pooled fractions, and RT-PCR analysis of the endogenous SMN transcript was performed. Fig. 2A shows that both alternative isoforms of SMN are found specifically associated with the supraspliceosome (200S) fraction (Fig. 2A, lane 7), whereas bottom and top fractions are devoid of these RNAs (Fig. 2A, lanes 5 and 8, respectively). The control experiment shows that normal rabbit serum (NRS) did not precipitate the SMN transcripts from the suprasplicesomal fraction (Fig. 2A, lane 5). Western blot (WB) analysis of the proteins extracted from the immunoprecipitated and unbound material of the supraspliceosomal fraction revealed that Sm proteins were efficiently and specifically precipitated (Fig. 2B, compare lane 2 to lane 3). Western blot analysis using antibodies against ribosomal protein (RP) S14 showed that this protein was not found associated with supraspliceosomes (Fig. 2B, compare lane 5 to lane 6), indicating that the supraspliceosome fraction was not contaminated with ribosomes. Together, these experiments suggest that the supraspliceosome is the machine where alternative splicing occurs.
Fig. 2.
Constitutively and alternatively spliced mRNAs of endogenous human SMN transcripts are associated with supraspliceosomes. (A) Immunoprecipitation (IP) of SMN splice isoforms. A nuclear supernatant enriched for splicing-active complexes was fractionated in a glycerol gradient as described (Azubel et al., 2006). Gradient fractions were pooled together: fractions 1-4 (bottom); 8-11 (200S); and 17-20 (top), and were each treated with immobilized anti-Sm monoclonal antibodies (Y12 MAb). The unbound and bound material (PPT, precipitate) was analyzed by RT-PCR, using SMN primer pairs. Immobilized normal rabbit serum (NRS) was used as a non-relevant control. The two isoforms of the alternatively spliced SMN transcripts are schematically drawn on the left (the alternative exon is marked). (B) Western blot (WB) analysis of precipitated supraspliceosomes, using anti-Sm MAbs (lanes 1-3) and reprobing of the same membrane with antibodies against the ribosomal protein S14 (lanes 4-6). Lane 1, Nuclear supernatant; lane 2, IP from the supraspliceosome fractions; lane 3, unbound material from the supraspliceosome fractions.
3.2. A supraspliceosome is assembled on one pre-mRNA
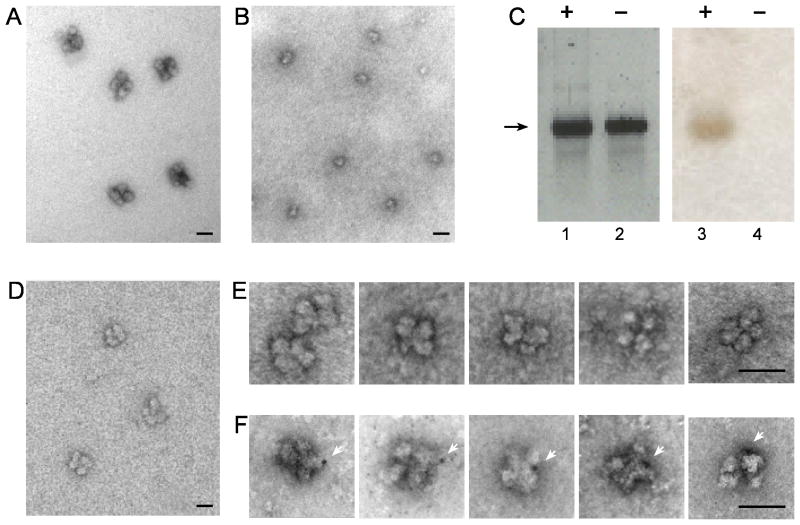
A prerequisite for the occurrence of efficient and precise alternative splicing within the supraspliceosome is that each complex should process a single RNA transcript at a time. This hypothesis has been implied in the supraspliceosome model (Azubel et al., 2006; Sperling et al., 2008) and is experimentally addressed here. To this end we used our previously published procedure for reconstituting tetrameric supraspliceosomes from monomeric native spliceosomes and an exogenously added synthetic pre-mRNA (Azubel et al., 2006). Visualization and distinction by EM of pre-mRNA molecules cannot be directly achieved because of the low-density weakly scattering atoms they contain. Tagging of the RNA with gold nanoclusters enables visualization by EM, as exemplified by gold-tagged synthetic mRNAs, labeled at multiple sites, and visualized by EM and AFM (Medalia et al., 1999). For the reconstitution experiment, we first prepared supraspliceosomes (Fig. 3A), which gave rise to native spliceosomes (Fig. 3B) upon specific cleavage of the endogenous pre-mRNAs as described (Azubel et al., 2004). Next, an in vitro transcribed human β-globin pre-mRNA was ligated to a thiolated 12-nt oligodeoxynucleotide. The RNA-oligo-SH was next tagged at its 3’ end with a single gold nanoparticle of 1.4 nm in diameter (Nanogold by Nanoprobes), and the gold-tagged and untagged RNAs were analyzed by gel electrophoresis (Fig. 3C). Staining the gel with ethidium bromide revealed both the Nanlgold-tagged and the untagged β-globin pre-mRNAs (Fig. 3C, lanes 1 and 2, respectively) whereas specifically staining the gold by silver enhancement, revealed only the Nanogold-tagged β-globin pre-mRNA (Fig. 3C, lane 3). Purified native spliceosomes were then incubated with the in vitro transcribed β-globin pre-mRNA (tagged or untagged with Nanogold) and the reconstituted supraspliceosomes were visualized by EM. Fig. 3D shows a field of supraspliceosomes reconstituted with untagged mRNA. Galleries of reconstituted supraspliceosomes are depicted in Figs 3E and 3F. To make the Nanogold tags visible by EM, the EM grids were gold-enhanced and stained with uranyl acetate. Fig. 4E shows a gallery of supraspliceosomes reconstituted with untagged RNA. Fig. 3F depicts supraspliceosomes reconstituted on Nanogold-tagged β-globin pre-mRNA showing only one enhanced cluster per reconstituted supraspliceosome. The images shown represent 75 images of supraspliceosomes tagged with one Nanogold particle out of 97 that had been checked (77%), the remaining were not tagged with gold.
Fig. 3.
Reconstitution of supraspliceosomes from native spliceosomes and Nanogold-tagged pre-mRNA. Native spliceosomes were prepared from supraspliceosomes by specific cleavage of the pre-mRNA as described (Azubel et al., 2004). For reconstitution of supraspliceosomes native spliceosomes were incubated with β-globin pre-mRNA (untagged or tagged with Nanogold) as described (Azubel et al., 2006). (A, B, D) Visualization by EM of a field of supraspliceosomes (A); Native spliceosomes (B); and reconstituted supraspliceosomes (D), negatively stained with 1% uranyl acetate. (C) Gel electrophoresis (2% agarose) analysis of untagged (lanes 2 and 4) and Nanogold-tagged β-globin pre-mRNA (lanes 1 and 3), used for the reconstruction experiment. Left panel, staining with ethidium bromide; right panel, staining of the blotted gel by silver enhancement. Galleries of images of reconstituted supraspliceosomes visualized by EM after staining with 1% uranyl acetate and gold enhancement. (E) Gallery of supraspliceosomes reconstituted from native spliceosomes and untagged β-globin pre-mRNA. (F) Gallery of supraspliceosomes reconstituted from native spliceosomes and Nanogold-tagged β-globin pre-mRNA. White arrowheads point to gold-enhanced Nanogold. Bar, 50 nm.
Fig. 4.
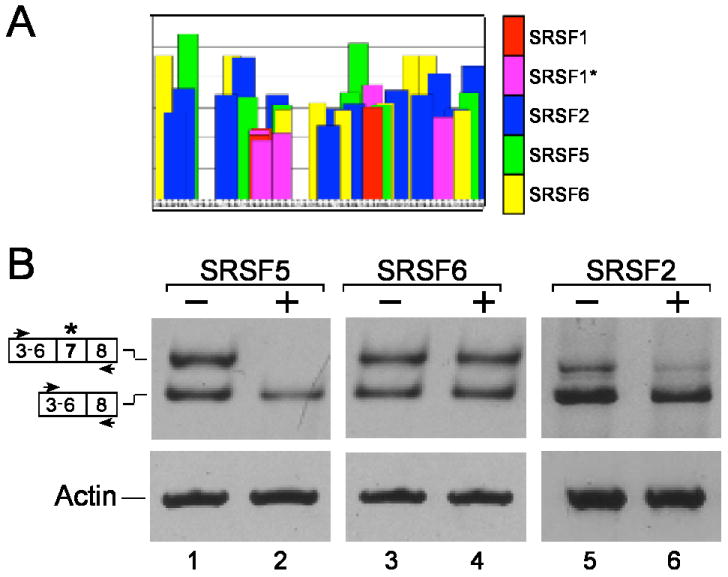
Upregulation of SRSF5 affects alternative splicing of human hnRNP A/B. (A) Potential ESEs in exon 7 of human hnRNP A/B, based on the ESEfinder program. (B) Effect of overexpression of SRSF5, SRSF6 and SRSF2 on alternative splicing of human hnRNP A/B. Total RNA was extracted from untreated HeLa cells (lanes 1, 3, and 5) and from cells transfected with the indicated plasmids (lanes 2, 4, and 6) and analyzed by RT-PCR using the indicated primer pairs. The two isoforms of hnRNPA/B are schematically drawn on the left. The results represent three independent experiments.
3.3. The SR protein SRSF5 affects alternative splicing in supraspliceosomes
We have previously shown that the phosphorylated forms of the SR proteins family, which play an important role in splicing and in regulated alternative splicing, are predominantly associated with supraspliceosomes (Yitzhaki et al., 1996). We therefore asked whether up-regulation of specific SR proteins would affect the ratio between the alternatively spliced hnRNP A/B RNA isoforms that were shown to reside in the supraspliceosome. SR proteins show binding specificities for multiple exonic splicing enhancer (ESE) sites within the same exon (Lin and Fu, 2007; Long and Caceres, 2009; Shepard and Hertel, 2009). We thus used the ESEfinder program (http://exon.cshl.edu/ESE) to look for ESEs in the alternative exon 7 of hnRNP A/B, and identified several potential SR proteins responsive elements for SRSF2 (SC35), SRSF5 and SRSF6 (SRp55) (Fig. 4A). HeLa cells were then transfected with plasmids over expressing either SRSF2, SRSF5 or SRSF6 (Screaton et al., 1995), total RNA was extracted and analyzed by RT-PCR to determine the effect of each of these plasmids on the alternative splicing pattern of the endogenous hnRNP A/B transcript (Fig. 4B). As can be seen only transfection with a plasmid encoding SRSF5 decreased significantly the inclusion of exon 7 of hnRNP A/B (Fig. 4B, lanes 1 and 2). Transfections with plasmids encoding for either SRSF6 or SRSF2 did not cause significant changes in the alternative splicing pattern of hnRNP A/B (Figure 4B, lanes 3,4 and 5,6, respectively), even when the amount of the transfected plasmid was doubled (data not shown).
Next we tested whether the effect of over expression of SRSF5 on shifting the alternative splicing of hnRNP A/B towards the skipping of exon 7 was also observed in hnRNP A/B transcripts assembled in supraspliceosomes. For this aim, we prepared supraspliceosomes from HeLa cells transfected with a plasmid encoding for SRSF5, and fractionated them on a glycerol gradient. RNA was prepared from even gradient fractions and analyzed by RT-PCR. For comparison, we analyzed parallel gradient fractions prepared from non-transfected HeLa cells. Fig. 5 (upper panel) shows that without transfection the exon 7-containing mRNA sedimented with 200S supraspliceosomes, while the analogous fractions from the SRSF5 transfected cells were devoid of that isoform Fig. 5 (lower panel). This experiment shows that SRSF5 increased skipping of exon 7 of hnRNP A/B in supraspliceosomes.
Fig. 5.
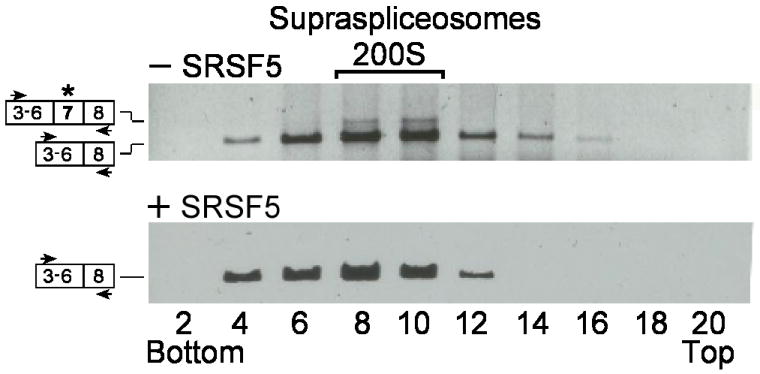
Up regulation of SRSF5 affects alternative splicing of hnRNP A/B in supraspliceosomes. Nuclear supernatants enriched in supraspliceosomes were prepared from HeLa cells, untreated (upper panel) or transfected with an SRSF5 encoding plasmid (lower panel) and fractionated on a 10-45 % glycerol gradient. RNA was extracted and analyzed by RT-PCR with primers from exons 3 (sense) and 8 (antisense) of hnRNP A/B. The constitutively and alternatively spliced isoforms of hnRNP A/B are schematically drawn on the left (the alternative exon is marked by asterisks), and the PCR primer pair is indicated by arrowheads. Each part of the figure is representative of at least three different experiments. Supraspliceosomes peak in fractions 8-10. The gradients were calibrated with 200S TMV particles running in fractions 9, 10.
3.4. C6-Ceramide affects alternative splicing in supraspliceosomes
Treatment of 293T cells with C6-ceramide, which had been shown to affect alternative splicing of a number of genes, resulted in an increased level of the exon 7-included isoform of the endogenous hnRNP A/B transcript (Fig. 6A). We next prepared supraspliceosomes from 293 cells treated with 30 mM ceramide or from untreated cells, and analyzed by RT-PCR the alternative splicing pattern of hnRNP A/B of RNA extracted from the supraspliceosome fractions (gradient fractions 7, 8, 9). Fig. 6B shows that C6-ceramide increased the inclusion of exon 7 of hnRNP A/B within supraspliceosomes.
Fig. 6.
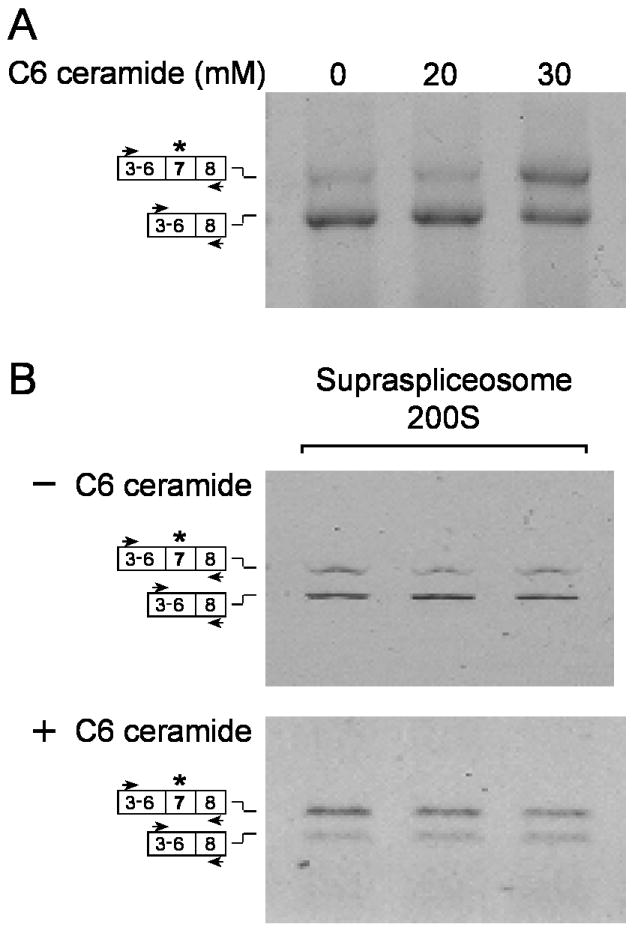
C6-Ceramide affects alternative splicing of hnRNP A/B in supraspliceosomes. (A) Total RNA was extracted from 293 cells, untreated or treated with C6-ceramide as indicated, and analyzed by RT-PCR using the indicated primer pair of hnRNP A/B. (B) Nuclear supernatants enriched in supraspliceosomes were prepared from 293 cells, untreated (upper panel) or treated with 30 mM C6-ceramide for 24 h (lower panel), and fractionated in a glycerol gradient. RNA was extracted from the supraspliceosome fractions (7-9) and analyzed by RT-PCR using the indicated primer pair. The two isoforms of hnRNP A/B are schematically drawn on the left.
4. Discussion
The entire repertoire of mammalian nuclear pre-mRNAs, independent of their length or number of introns, are individually found assembled in supraspliceosomes, which are composed of four native spliceosomes connected via the pre-mRNA (reviewed in Sperling et al., 2008). The supraspliceosome harbors all five spliceosomal snRNPs as well as non-snRNP proteins, including regulatory splicing factors such as the SR protein family and hnRNP G, all of which are predominantly found in supraspliceosomes. These observations, together with the findings that the native spliceosome as well as the supraspliceosome are active in splicing, make the supraspliceosome a suitable candidate to perform and regulate alternative splicing. In support of this notion we show here that both the constitutively and the alternatively spliced mRNAs of the alternatively spliced Pol II transcripts hnRNP A/B, SMN and ADAR2 are predominantly found in supraspliceosomes.
The supraspliceosome model presents a closed-structure composed of four native spliceosomes connected by one pre-mRNA molecule. A critical experiment in support of this arrangement showed that cleavage of the pre-mRNA yielded functional native spliceosomes that could be reconstituted into supraspliceosomes by incubation with exogenously added pre-mRNAs (Azubel et al., 2004; Azubel et al., 2006). Here we repeated this experiment to reconstitute supraspliceosomes from native spliceosomes and Nanogold-tagged pre-mRNA, showing by EM that only one transcript was assembled per supraspliceosome. Thus, the supraspliceosome can be portrayed as a multiprocessor machine that can simultaneously splice four introns of a pre-mRNA (Fig. 7). This model of the splicing machine accounts for the necessity to splice multiintronic pre-mRNAs in a coordinated manner and for alternative splicing.
Fig. 7.
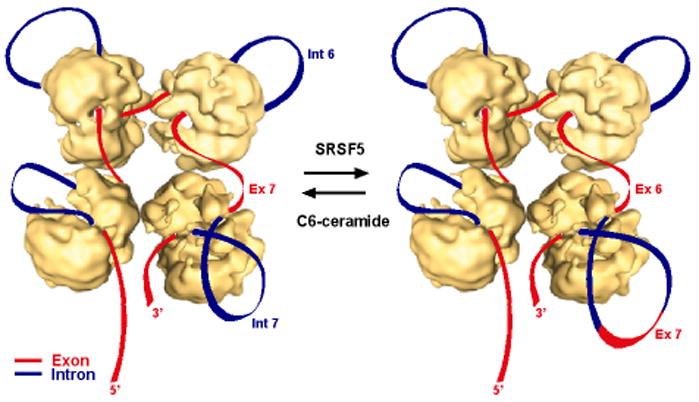
Alternative splicing within the supraspliceosome – a putative model. Schematic models of the supraspliceosome, in which the pre-mRNA (introns in blue, exons in red) is connecting four native spliceosomes. Transfection of cells with a plasmid expressing SRSF5 causes looping out of exon 7 (Ex 7; the alternative exon of hnRNP A/B) together with its flanking introns (depicted in the lower right corner), and thus facilitates skipping of exon 7. Treatment of cells with C6-ceramide enhances inclusion of exon 7 and looping out of introns 6 and 7 (Int 6 and Int 7, respectively). Adapted from Azubel et al. (2006).
The decision to include or skip an exon requires the selection of the appropriate splice sites through recognition of cis elements by trans acting factors. A well-characterized family of such trans factors that affect splicing and alternative splicing is the SR protein family (Lin and Fu, 2007; Long and Caceres, 2009; Shepard and Hertel, 2009). The family of SR proteins was originally shown to affect exon inclusion in regulated alternative splicing through binding to ESEs in the included exon (Ibrahim et al., 2005). However, several studies showed that exon skipping could also be affected by SR proteins. The mechanism underlying this effect is not yet well understood. In some cases, however, it was attributed to intronic binding sites for the SR proteins, or to the ESEs in other cases (Buratti et al., 2007; Gallego et al., 1997; Jiang et al., 1998; Lemaire et al., 1999; Solis et al., 2008; ten Dam et al., 2000). In a recent study it was shown that the exon skipping effect of SR proteins depends on their actions on a flanking constitutive exon and also requires joint effort of more than one SR protein (Han et al., 2011). We show here that SRSF5 affects exon skipping of exon 7 of hnRNP A/B. Inspection of the potential binding sites for SR proteins in the introns and the exons flanking exon 7 could not provide an explanation for the skipping of exon 7 of hnRNP A/B caused by SRSF5. Independent of the mechanism whereby SRSF5 affected skipping of exon 7 of hnRNP A/B, we show here that this skipping occurs in supraspliceosomes. We also tested the effect of C6-ceramide, which was previously shown to affect splice site selection. We have shown here that increased exon 7 inclusion of hnRNP A/B by C6 ceramide occurs in supraspliceosomes. Furthermore, our experiments show that both inclusion and skipping of exon 7 of hnRNP A/B are regulated in supraspliceosomes.
The selection of the right combination of 5’ and 3’ splice sites is a crucial stage in regulated alternative splicing. This process requires communication between spliceosomes for the choice of the right combination of splice sites before splicing commences. Being a tetrameric complex, the supraspliceosome can simultaneously splice four introns of a pre-mRNA, not necessarily in a consecutive manner. Splicing of a multi-intronic pre-mRNA can be facilitated by the translocation of the pre-mRNA through the complex in a ‘rolling model’ fashion. After processing of four introns the RNA roles in to place a new subset of introns in the correct position for their processing. For transcripts having less than four introns, the model predicts that some of its native spliceosome subunits are not occupied (reviewed in Sperling et al., 2008). Whether splicing occurs co-transcriptionally (de Almeida and Carmo-Fonseca, 2008; Kornblihtt, 2007; Pandit et al., 2008; Perales and Bentley, 2009) or not, the supraspliceosome is a stand-alone machine that can perform splicing, alternative splicing and the additional pre-mRNA processing activities required to process the pre-mRNA.
The supraspliceosome model presents a platform on which splice junctions could be checked prior to introns excision (Fig. 7). According to the supraspliceosome model (Sperling et al., 2008), the pre-mRNA that is not being processed is folded and protected within the cavities of the native spliceosome. However, when a staining protocol that allows visualization of nucleic acids was used, RNA strands and loops were seen emanating from the supraspliceosomes (Müller et al., 1998). Under these conditions the RNA kept in the cavity is proposed to unfold and loop-out. Our structural studies have shown that the small subunit of each native spliceosome, proposed to harbor non-snRNP components such as SR proteins and hnRNP proteins, is placed at the center of the supraspliceosome (Cohen-Krausz et al., 2007). This configuration allows communication between the native spliceosomes, which is a crucial element for regulated alternative splicing and for quality control of the resulting mRNAs. This setting places the large subunit of each native spliceosome, where catalysis by the U-snRNPs presumably takes place, in the periphery of the supraspliceosome. An attractive feature of such a machine is that it allows rearrangement of splice junction combinations to select the appropriate ones. This way it comprises an important tool to ensure the fidelity of splicing and alternative splicing. For the particular example addressed here we assume that SRSF5 facilitates the looping out of exon 7 with its flanking introns (Fig. 7, right). In this configuration, the splice sites required for the skipping of exon 7 can be juxtaposed for the formation of the alternative splice junction. C6-ceramide appears to assist in the inclusion of exon 7 and looping out of introns 6 and 7 (Fig. 7, left). Focusing on alternative splicing, we have shown here that alternative splicing of endogenous transcripts (both inclusion and exclusion of an alternative exon) occur within the supraspliceosome. This study offers novel insight into the functioning of alternative splicing.
Acknowledgments
We thank Aviva Pecho for excellent technical assistance. We are grateful to Dr. Meir Wilchek for useful advice on affinity purification, Drs Adrian Krainer and Rotem Karni for plasmids. We thank the US NIH, grant GM079549 to R. S. and J.S. and the Helen and Milton Kimmelman Center for Biomolecular Structure and Assembly at the Weizmann Institute of Science (J.S.) for financial support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Azubel M, Wolf SG, Sperling J, Sperling R. Three-dimensional structure of the native spliceosome by cryo-electron microscopy. Mol Cell. 2004;15:833–9. doi: 10.1016/j.molcel.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Azubel M, Habib N, Sperling J, Sperling R. Native spliceosomes assemble with pre-mRNA to form supraspliceosomes. J Mol Biol. 2006;356:955–966. doi: 10.1016/j.jmb.2005.11.078. [DOI] [PubMed] [Google Scholar]
- Brett D, Pospisil H, Valcárcel J, Reich J, Bork P. Alternative splicing and genome complexity. Nat Genet. 2002;30:29–30. doi: 10.1038/ng803. [DOI] [PubMed] [Google Scholar]
- Buratti E, Stuani C, De Prato G, Baralle FE. SR protein-mediated inhibition of CFTR exon 9 inclusion: molecular characterization of the intronic splicing silencer. Nucleic Acids Res. 2007;35:4359–68. doi: 10.1093/nar/gkm444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfant CE, Ogretmen B, Galadari S, Kroesen BJ, Pettus BJ, Hannun YA. FAS activation induces dephosphorylation of SR proteins; dependence on the de novo generation of ceramide and activation of protein phosphatase 1. J Biol Chem. 2001;276:44848–55. doi: 10.1074/jbc.M106291200. [DOI] [PubMed] [Google Scholar]
- Chalfant CE, Rathman K, Pinkerman RL, Wood RE, Obeid LM, Ogretmen B, Hannun YA. De novo ceramide regulates the alternative splicing of caspase 9 and Bcl-x in A549 lung adenocarcinoma cells. Dependence on protein phosphatase-1. J Biol Chem. 2002;277:12587–95. doi: 10.1074/jbc.M112010200. [DOI] [PubMed] [Google Scholar]
- Cohen-Krausz S, Sperling R, Sperling J. Exploring the architecture of the intact supraspliceosome using electron microscopy. J Mol Biol. 2007;368:319–27. doi: 10.1016/j.jmb.2007.01.090. [DOI] [PubMed] [Google Scholar]
- de Almeida SF, Carmo-Fonseca M. The CTD role in cotranscriptional RNA processing and surveillance. FEBS Lett. 2008;582:1971–6. doi: 10.1016/j.febslet.2008.04.019. [DOI] [PubMed] [Google Scholar]
- Gallego ME, Gattoni R, Stevenin J, Marie J, Expert-Bezancon A. The SR splicing factors ASF/SF2 and SC35 have antagonistic effects on intronic enhancer-dependent splicing of the beta-tropomyosin alternative exon 6A. EMBO J. 1997;16:1772–84. doi: 10.1093/emboj/16.7.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Blanco MA, Baraniak AP, Lasda EL. Alternative splicing in disease and therapy. Nat Biotechnol. 2004;22:535–46. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- Gerber A, O’Connell MA, Keller W. Two forms of human double-stranded RNA-specific editase 1 (hRED1) generated by the insertion of an Alu cassette. RNA. 1997;3:453–463. [PMC free article] [PubMed] [Google Scholar]
- Han J, Ding JH, Byeon CW, Kim JH, Hertel KJ, Jeong S, Fu XD. SR proteins induce alternative exon skipping through their activities on the flanking constitutive exons. Mol Cell Biol. 2011;31:793–802. doi: 10.1128/MCB.01117-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich B, Zhang Z, Raitskin O, Hiller M, Benderska N, Hartmann AM, Bracco L, Elliott D, Ben-Ari S, Soreq H, Sperling J, Sperling R, Stamm S. Heterogeneous Nuclear Ribonucleoprotein G Regulates Splice Site Selection by Binding to CC(A/C)-rich Regions in Pre-mRNA. J Biol Chem. 2009;284:14303–15. doi: 10.1074/jbc.M901026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim EC, Schaal TD, Hertel KJ, Reed R, Maniatis T. Serine/arginine-rich protein-dependent suppression of exon skipping by exonic splicing enhancers. Proc Natl Acad Sci USA. 2005;102:5002–7. doi: 10.1073/pnas.0500543102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang ZH, Zhang WJ, Rao Y, Wu JY. Regulation of Ich-1 pre-mRNA alternative splicing and apoptosis by mammalian splicing factors. Proc Natl Acad Sci USA. 1998;95:9155–60. doi: 10.1073/pnas.95.16.9155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamhi E, Yahalom G, Kass G, Hacham Y, Sperling R, Sperling J. AUG sequences are required to sustain nonsense-codon-mediated suppression of splicing. Nucleic Acids Res. 2006;34:3421–3433. doi: 10.1093/nar/gkl390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan FA, Jaiswal AK, Szer W. Cloning and sequence analysis of a human type A/B hnRNP protein. FEBS Lett. 1991;290:159–61. doi: 10.1016/0014-5793(91)81249-8. [DOI] [PubMed] [Google Scholar]
- Kornblihtt AR. Coupling transcription and alternative splicing. Adv Exp Med Biol. 2007;623:175–89. doi: 10.1007/978-0-387-77374-2_11. [DOI] [PubMed] [Google Scholar]
- Krainer AR, Maniatis T, Ruskin B, Green MR. Normal and mutant human β-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984;36:993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- Lai F, Chen C-X, Carter KC, Nishikura K. Editing of glutamate receptor B subunit ion channel RNAs by four alternatively spliced DRADA2 double-stranded RNA adenosine deaminases. Mol Cell Biol. 1997;17:2413–2424. doi: 10.1128/mcb.17.5.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau PP, Zhu HJ, Nakamuta M, Chan L. Cloning of an Apobec-1-binding protein that also interacts with apolipoprotein B mRNA and evidence for its involvement in RNA editing. J Biol Chem. 1997;272:1452–1455. doi: 10.1074/jbc.272.3.1452. [DOI] [PubMed] [Google Scholar]
- Lemaire R, Winne A, Sarkissian M, Lafyatis R. SF2 and SRp55 regulation of CD45 exon 4 skipping during T cell activation. Eur J Immunol. 1999;29:823–37. doi: 10.1002/(SICI)1521-4141(199903)29:03<823::AID-IMMU823>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Li B, Wachtel C, Miriami E, Yahalom G, Friedlander G, Sharon G, Sperling R, Sperling J. Stop codons affect 5’ splice site selection by surveillance of splicing. Proc Natl Acad Sci USA. 2002;99:5277–5282. doi: 10.1073/pnas.082095299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Fu XD. SR proteins and related factors in alternative splicing. Adv Exp Med Biol. 2007;623:107–22. doi: 10.1007/978-0-387-77374-2_7. [DOI] [PubMed] [Google Scholar]
- Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci USA. 1999;96:6307–11. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus MA, Heinrich B, Raitskin O, Adams DJ, Mangs H, Goy C, Ladomery M, Sperling R, Stamm S, Morris BJ. WT1 interacts with the splicing protein RBM4 and regulates its ability to modulate alternative splicing in vivo. Exp Cell Res. 2006;312:3379–88. doi: 10.1016/j.yexcr.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Medalia O, Heim M, Guckenberger R, Sperling R, Sperling J. Gold-Tagged RNA-A Probe for Macromolecular Assemblies. J Struct Biol. 1999;127:113–119. doi: 10.1006/jsbi.1999.4134. [DOI] [PubMed] [Google Scholar]
- Medalia O, Typke D, Hegerl R, Angenitzki M, Sperling J, Sperling R. Cryoelectron microscopy and cryoelectron tomography of the nuclear pre-mRNA processing machine. J Struct Biol. 2002;138:74–84. doi: 10.1016/s1047-8477(02)00027-8. [DOI] [PubMed] [Google Scholar]
- Miriami E, Sperling J, Sperling R. Heat shock affects 5’ splice site selection, cleavage and ligation of CAD pre-mRNA in hamster cells, but not its packaging in lnRNP particles. Nucleic Acids Res. 1994;22:3084–3091. doi: 10.1093/nar/22.15.3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek B, Lee C. A genomic view of alternative splicing. Nat Genet. 2002;30:13–9. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- Müller S, Wolpensinger B, Angenitzki M, Engel A, Sperling J, Sperling R. A supraspliceosome model for large nuclear ribonucleoprotein particles based on mass determinations by scanning transmission electron microscopy. J Mol Biol. 1998;283:383–394. doi: 10.1006/jmbi.1998.2078. [DOI] [PubMed] [Google Scholar]
- Nadano D, Ishihara G, Aoki C, Yoshinaka T, Irie S, Sato TA. Preparation and characterization of antibodies against human ribosomal proteins: heterogeneous expression of S11 and S30 in a panel of human cancer cell lines. Jpn J Cancer Res. 2000;91:802–10. doi: 10.1111/j.1349-7006.2000.tb01017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–5. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Pandit S, Wang D, Fu XD. Functional integration of transcriptional and RNA processing machineries. Curr Opin Cell Biol. 2008;20:260–5. doi: 10.1016/j.ceb.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales R, Bentley D. “Cotranscriptionality”: the transcription elongation complex as a nexus for nuclear transactions. Mol Cell. 2009;36:178–91. doi: 10.1016/j.molcel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raitskin O, Angenitzki M, Sperling J, Sperling R. Large nuclear RNP particles-the nuclear pre-mRNA processing machine. J Struct Biol. 2002;140:123–30. doi: 10.1016/s1047-8477(02)00541-5. [DOI] [PubMed] [Google Scholar]
- Raitskin O, Cho DS, Sperling J, Nishikura K, Sperling R. RNA editing activity is associated with splicing factors in lnRNP particles: The nuclear pre-mRNA processing machinery. Proc Natl Acad Sci USA. 2001;98:6571–6. doi: 10.1073/pnas.111153798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screaton GR, Caceres JF, Mayeda A, Bell MV, Plebanski M, Jackson DG, Bell JI, Krainer AR. Identification and characterization of three members of the human SR family of pre-mRNA splicing factors. EMBO J. 1995;14:4336–49. doi: 10.1002/j.1460-2075.1995.tb00108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard PJ, Hertel KJ. The SR protein family. Genome Biol. 2009;10:242. doi: 10.1186/gb-2009-10-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis AS, Peng R, Crawford JB, Phillips JA, 3rd, Patton JG. Growth hormone deficiency and splicing fidelity: two serine/arginine-rich proteins, ASF/SF2 and SC35, act antagonistically. J Biol Chem. 2008;283:23619–26. doi: 10.1074/jbc.M710175200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer B, Köhler M, Sprengel R, Seeburg PH. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67:11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- Sperling J, Azubel M, Sperling R. Structure and Function of the Pre-mRNA Splicing Machine. Structure. 2008;16:1605–1615. doi: 10.1016/j.str.2008.08.011. [DOI] [PubMed] [Google Scholar]
- Sperling R, Sperling J, Levine AD, Spann P, Stark GR, Kornberg RD. Abundant nuclear ribonucleoprotein form of CAD RNA. Mol Cell Biol. 1985;5:569–575. doi: 10.1128/mcb.5.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling R, Koster AJ, Melamed-Bessudo C, Rubinstein A, Angenitzki M, Berkovitch-Yellin Z, Sperling J. Three-dimensional image reconstruction of large nuclear RNP (lnRNP) particles by automated electron tomography. J Mol Biol. 1997;267:570–583. doi: 10.1006/jmbi.1997.0898. [DOI] [PubMed] [Google Scholar]
- Srebrow A, Kornblihtt AR. The connection between splicing and cancer. J Cell Sci. 2006;119:2635–41. doi: 10.1242/jcs.03053. [DOI] [PubMed] [Google Scholar]
- Sumanasekera C, Watt DS, Stamm S. Substances that can change alternative splice-site selection. Biochem Soc Trans. 2008;36:483–90. doi: 10.1042/BST0360483. [DOI] [PubMed] [Google Scholar]
- ten Dam GB, Zilch CF, Wallace D, Wieringa B, Beverley PC, Poels LG, Screaton GR. Regulation of alternative splicing of CD45 by antagonistic effects of SR protein splicing factors. J Immunol. 2000;164:5287–95. doi: 10.4049/jimmunol.164.10.5287. [DOI] [PubMed] [Google Scholar]
- Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8:749–61. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–13. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yitzhaki S, Miriami E, Sperling J, Sperling R. Phosphorylated Ser/Arg-rich proteins: Limiting factors in the assembly of 200S large nuclear ribonucleoprotein particles. Proc Natl Acad Sci USA. 1996;93:8830–8835. doi: 10.1073/pnas.93.17.8830. [DOI] [PMC free article] [PubMed] [Google Scholar]