Abstract
Transmembrane-4-L-six-family-1 (TM4SF1) is a tetraspanin-like membrane protein that is highly and selectively expressed by cultured endothelial cells (EC) and, in vivo, by EC lining angiogenic tumor blood vessels. TM4SF1 is necessary for the formation of unusually long (up to a 50 µm), thin (~ 100–300 nm wide), F-actin-poor EC cell projections that we term ‘nanopodia’. Immunostaining of nanopodia at both the light and electron microsopic levels localized TM4SF1 in a regularly spaced, banded pattern, forming TM4FS1-enriched domains (TMED). Live cell imaging of GFP-transduced HUVEC demonstrated that EC project nanopodia as they migrate and interact with neighboring cells. When TM4SF1 mRNA levels in EC were increased from the normal ~90 mRNA copies/cell to ~400 copies/cell through adenoviral transduction, EC projected more and longer nanopodia from the entire cell circumference but were unable to polarize or migrate effectively. When fibroblasts, which normally express TM4SF1 at ~5 copies/cell, were transduced to express TM4SF1 at EC-like levels, they formed typical TM4SF1-banded nanopodia, and broadened, EC-like lamellipodia. Mass-spectrophotometry demonstrated that TM4SF1 interacted with myosin-10 and β-actin, proteins involved in filopodia formation and cell migration. In summary, TM4SF1, like genuine tetraspanins, serves as a molecular organizer that interacts with membrane and cytoskeleton-associated proteins and uniquely initiates the formation of nanopodia and facilitates cell polarization and migration.
Keywords: TM4SF1, endothelial cell, nanopodia, myosin-10, β-actin
Introduction
Tetraspanins are integral membrane proteins that localize to microdomains where they associate with other tetraspanins, integrins, immunoglobulin superfamily members, growth factor receptors, and proteoglycans [1]; they are thought to stabilize cell signaling complexes that regulate cell adhesion, proliferation and migration, and are important players in many biological processes including angiogenesis [2]. In addition to the thirty-nine genuine tetraspanins, a group of six membrane proteins (the L6 family) has been described; L6 proteins resemble tetraspanins with respect to topology but not homology [3].
Transmembrane-4-L-six-family-1 (TM4SF1) is the founding member of the L6 family. It was originally described in tumor cells where it was found to share certain tetraspanin functions including roles in cell growth [4], motility [5] and metastasis [6]. Recently we demonstrated that TM4SF1 is not only highly expressed in tumor cells but also in vivo in the endothelial cells (EC) of angiogenic blood vessels supplying human cancers [7]. TM4SF1 is also highly expressed in cultured EC (~ 90 mRNA copies/cell) where it localizes to perinuclear vesicles, the plasma membrane, and thin cellular projections from the cell’s leading front and trailing rear; these projections either lack F-actin entirely or contain it proximally, within 10 µm of the cell body [7]. Here, we term these TM4SF1-banded cellular projections “nanopodia”, to signify their nano scale width and to distinguish them from F-actin-enriched structures such as filopodia and retraction fibers.
We now demonstrate that cells that express TM4SF1 at much lower levels, such as fibroblasts, do not project nanopodia but can be induced to do so when transduced to express TM4SF1 at EC-like levels. EC or fibroblasts that expressed TM4SF1 at much higher levels (~ 400 mRNA copies/cell) formed greatly increased numbers of nanopodia but experienced impaired cell polarization and migration. TM4SF1 was localized to TM4SF1-enriched domains (TMED) where it was found to interact with myosin-10, β-actin, and α5β1 integrin [7]. Thus, TM4SF1, like genuine tetraspanins, serves as a molecular organizer that is uniquely able to induce the formation of nanopodia and to establish the EC phenotype.
Materials and methods
Antibodies and reagents
Primary antibodies were: mouse anti-human TM4SF1 from Millipore (Billerica, MA) and from our own antibody production (paper in preparation), goat anti-human myosin-10 and CD9 (Santa Cruz Biotechnology, Santa Cruz, CA), and rabbit anti-human β-actin (Cell Signaling, Danvers, MA). Secondary antibodies were: Alexa fluor 488- or 594-labeled donkey-anti-mouse IgG (Invitrogen, Carlsbad, CA), and HRP-labeled goat anti-rabbit, goat anti-mouse, and rabbit anti-goat antibodies (Bio-Rad, Hercules, CA). Phalloidin-TRIC and mouse IgG were purchased from Sigma (Saint Louis, MO).
Cell culture and cell labeling
Human umbilical vein EC (HUVEC) from Lonza (Walkersville, MD) were grown in EGM-2-MV medium, and used at passage 5–6. Human dermal fibroblast (HDF) were acquired from the Cell Biology Core at our Center for Vascular Biology Research, cultured in DMEM/10%FBS, and used at passages 4–6. HUVEC at 60% confluence were labeled with CellMask red plasma membrane stain (Invitrogen) for 30 min according to manufacturer’s instructions and subcultured onto 8 mm collagen-1 coated glass discs (Fisher Scientific) for immunostaining.
Adenoviral constructs
Short hairpin RNA (shRNA) adenoviruses for TM4SF1 knockdown (KD) were described previously [7]; they reduce TM4SF1 mRNA and protein expression by ≥ 90% at day-3. For overexpression, full-length human TM4SF1 cDNA was cloned into pENT/SD/D-TOPO plasmids (Invitrogen). The empty pENT/SD/D-TOPO plasmid (control) and TM4SF1-inserted constructs were recombined with pAd/CMV/V5-DEST through LR recombination. Adenoviruses were purified using the Adenopure kit (PureSyn, Malvern, PA). Virus titer was determined by multiplicity of infection (moi) assays in 293A cells following manufacture’s instructions. HUVEC were treated with 15 or 50 moi of adenoviruses that were empty vector (control) or that contained TM4SF1 for 48h, or with 25 moi TM4SF1-KD constructs for 72h [7]. GFP-adenovirus construct were purchased from Vector Biolabs (Philadelphia, PA), and used at 15 moi to achieve GFP mRNA copy numbers of ~100 copies/cell. These adenoviruses achieve high transduction rates without overt cytotoxic effects at mois of 10–100, in most cultured cell lines, including the normal human fibroblasts and EC studied here [8].
GFP-tagging of human TM4SF1 at either its N- or C-termini was conducted by cloning full-length cDNA into pAcGFP1-C1 and pAcGFP1-N1 vectors (Clontech, Mountain View, CA). The plasmids were then transfected to HUVEC through electroporation using the Amaxa HUVEC Nucleofector Kit according to the manufacturer’s protocol.
RNA isolation and Multi-Gene Transcriptional Profiling (MGTP)
Total RNA was isolated with the RNeasy kit following the manufacturer’s instructions (Qiagen, CA), and cDNA was prepared using reverse transcriptase III (Invitrogen) as described [7]. MGTP, a form of quantitative real-time PCR, was used to determine mRNA copy numbers per cell [9,10]. The number of mRNA copies per cell was calculated by normalization to 18S rRNA abundance, assuming that, on average, cells express ~106 18S-rRNA copies. Mean and standard error of the mean (mean ± SEM) were calculated from three cDNA samples prepared in three separate experiments. Real-time PCR primer sequences were as follows for β-actin (F: CTGGAACGGTGAAGGTGACA, R: AGTCCTCGGCCACATTGTG) and for myosin-10 (F: CTCAAGGGCACCGTAGAAGTG, R: AGTCCTATCGGCCATAATGATGTC).
Immunocytochemistry and electron microscopy
HUVEC plated on glass discs were fixed with 4% paraformaldehyde (PFA) in PBS at 25°C for 5 min and blocked with PBS containing 1% fetal bovine serum (FBS). Cells were stained with primary antibodies for 2h at 25°C. Secondary Alexa Fluor-488 or Alexa Fluor-494 antibodies were applied for 2h at 25°C, and rhodamine-conjugated phalloidin for 45 min at 37°C. ProLong Gold antifade reagent with DAPI (Invitrogen) was used for slide mounting.
Transmission electron-microscopy was performed on HUVEC fixed and immunostained as above, followed by a secondary goat anti-mouse Fab'-labeled with both Alexa Fluor-488 and nanogold (1.4 nm gold particles) from Nanoprobes (Yaphank, NY). Cells were postfixed in paraformaldehyde and glutaraldehyde for 15 minutes at 25°C, separated from the coverslip by cold fracture, and processed as previously described [11]. All immunocytochemistry and electron-microscopy images were representative selections from at least three separate experiments.
Wound healing assay
The wound healing assay was performed similarly to Rodriguez et. al. [12]. Briefly, a confluent monolayer was created by plating 8×104 HUVEC, pre-transduced with either empty vector control or TM4SF1 for 48h, in 24-well plates. Scratch wounds were generated 7h later with a p1000 pipette tip and cells were washed with fresh media. Three discs were immediately fixed in 4% PFA/PBS for use as reference points; the remaining three discs were cultured for an additional 18h and then fixed. We performed immunofluorescence anti-TM4SF1 staining and photographed three consecutive frames of images from the center of the scratched area. Twenty-seven total images were gathered from three independent experiments and measured the area of the wounded region that was cell-free at times 0 and 18h using Image-J [12]. We measured % mean recovery areas at 18h and divided by mean control areas at time 0h. Student t-test was used for statistical analysis.
Immunoprecipitation (IP), Western blotting and mass-spectrophotometry (MS)
HUVEC were grown to 80–90% confluency in 15-cm plate. Cells were trypsinized, and lysed in 2 ml Tris-buffered saline, pH 7.0, that contained 1% Brij99 and protease/phosphatase inhibitor cocktails (Sigma). Control mouse IgG was added to lysates for pre-IP before TM4SF1-IP. Protein bands for mass-spectrometry (MALDI-TOFF) were cut from SDS-PAGE, washed in 50% acetonitrile, and analyzed with a liquid chromatography-quadrupole mass spectrometer (MS/MS) in our proteomics core. Western blots were performed with indicated primary and secondary antibodies.
Live-cell imaging
Cells were plated on 30 mm plates (MatTek, Ashland, MA) coated with 1 µg/ml collagen (Sigma), and imaged over time in XY mode at 37°C, 5% CO2 using a time-lapse laser-scanning confocal microscope at 488 nm (LSM 510; Carl Zeiss, Inc.). Image analysis was performed with Volocity (PerkinElmer, Waltham, MA). Brightness and contrast were enhanced with Adobe Photoshop as indicated to improve fluorescence intensity. All live cell imaging movies (Supplemental Video-1 and -2) or frames from live cell imaging (Fig. 1D) were representative selections from at least five different movies
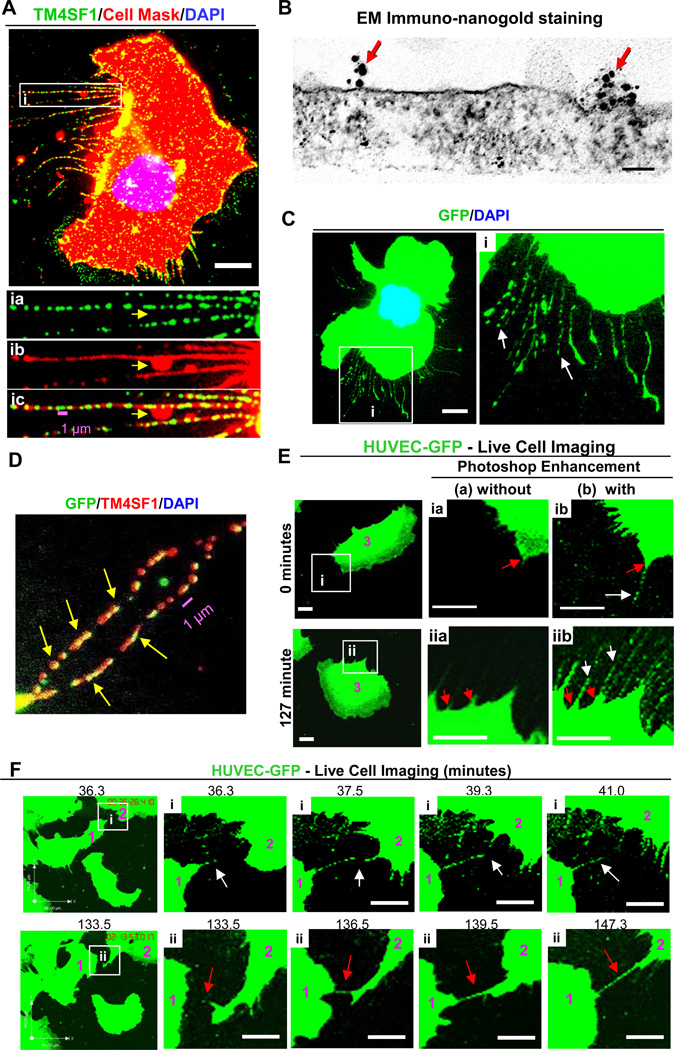
Fig. 1.
Nanopodia in HUVEC. (A and inset i) HUVEC were plated at 60% confluency for 2h on collagen-I coated glass discs prior to staining with anti-TM4SF1 antibodies and CellMask. CellMask staining of nanopodia was largely continuous and formed occasional lateral membrane bulges (yellow arrow). (B) Transmission immuno-nanogold electron microscopic image of TM4SF1 distribution on apical plasma membrane of a nanopodium of a normal HUVEC. (Lower plasma membrane is not visualized because it remained attached to the glass matrix during processing) Red arrows indicate gold particle clusters (TMED). Scale bar, 100 nm. (C and inset i) GFP-transduced HUVEC were aldehyde-fixed 2h after plating. Intermittently GFP staining nanopodia (white arrows) project from a cell body. Photoshop was used to enhance green color. (D) GFP-transduced HUVEC as in (C) but additionally immunostained with TM4SF1. TM4SF1 commonly co-localized with GFP in nanopodia (yellow arrows). (E, F) Live cell imaging of GFP-transduced HUVEC (time-lapse image frames were selected from Supplemental Video 1). (E) Time-lapse images are shown without (a) or with (b) Photoshop enhancement. Nanopodia (white arrows) extend from short-spike-like projections (red arrows) at the cells’ leading front (0 minute) and trailing rear (127 minutes). (F) Nanopodia that projected from cells #1 and #2 formed transient (white arrows in inset i) or longer lasting contacts that in inset ii (red arrows) persisted for ≥ 12 min. These long lasting contacts may represent nanotubes of the type that have been reported in other types of cultured cells [27]. White scale bars, 10 µm.
Results
Nanopodia in cultured endothelial cells (EC)
We previously demonstrated that HUVEC extended lengthy (up to 50 µm), thin (~ 100–300 nm wide) projections that exhibited intermittent bands of TM4SF1 staining [7]. These structures, which we now call nanopodia, projected both from the cell’s leading front and trailing edge [7]. Whereas phalloidin staining is characteristic of filopodia and retraction fibers, phalloidin staining of nanopodia occurred only occassionally, and, when present, was confined proximally to within 10 µm of cell body [7]. Immunofluorescence staining demonstrated that TM4SF1 enriched microdomains (TMED) recurred with a periodicity of 1–3 per µm (Fig. 1A, Supplemental Figure 1A).
Using the CellMask stain, we now demonstrate that nanopodia are enveloped by plasma membrane throughout their length (Fig. 1A and insert). Transmission electron microscopy confirmed this finding and demonstrated that nanopodia were about as tall (237 ± 42 nm) as they were wide (Fig. 1B). Immunonanogold staining demonstrated TMED at intervals of 1.29 ± 0.10 per µm (Fig. 1B) along the course of nanopodia, within the range observed by immunofluorescence staining (Fig. 1A, Supplemental Figure 1A). In ~25% of TMED, label consisted of a single gold particle, but gold particles were commonly arranged in clusters (3.5 ± 0.2, mean ± SEM, ranging up to as many as 14 gold particles per TMED). Several other types of EC (coronary artery, pulmonary artery, dermal microvessel, and lymphatic) also formed TM4SF1-banded nanopodia and did so when plated on collagen-I (as shown here) or on fibronectin (not shown).
Nanopodia were highly susceptible to disruption. They were best preserved when aldehyde fixatives were added immediately after removing culture medium, and were generally lost when cells were cooled to room temperature before fixation. Methanol, acetone, and Triton X-100 (at concentrations >0.05%) were found to extract TM4SF1 and therefore rendered nanopodia invisible to TM4SF1 staining (data not shown).
GFP-transduced HUVEC
In order to follow the behaviour of nanopodia in living cells, we initially tagged TM4SF1 with GFP. However, TM4SF1 tagged with GFP at either its N- or C-termini lost its characteristic nanopodia staining pattern when transduced into HUVEC. Therefore, we transduced HUVEC with GFP alone. GFP-transduced HUVEC exhibited fluorescence staining not only of the cell cytoplasm but also intermittent staining of lengthy, nanopodia-like cell projections (Fig. 1C and inset). Further, GFP staining in these projections was found to co-localize with TM4SF1 (Fig. 1D), indicating that they were in fact nanopodia and therefore that nanopodia could be visualized directly in HUVEC with GFP staining. As in untransduced cells, F-actin, when present, was confined to portions of nanopodia closest to the cell body (Supplemental Figure 1C).
Live cell imaging (Supplemental Video 1) demonstrated that migrating HUVEC extended nanopodia from both their leading front and trailing rear (Fig. 1E). HUVEC changed direction frequently, typically once every 40–60 minutes. When the direction of migration changed, the leading front became the trailing edge, causing nanopodia that had been associated with the lamellipodium at the leading front to appear at the trailing edge. Nanopodia were commonly sloughed and left behind as matrix-adherent debris (Supplemental Figure 1B). Nanopodia at the cell’s leading front sometimes made contacts with nanopodia of neighboring cells (Fig. 1Fi, Fii). These contacts were often transient (Fig. 1Fi), but sometimes peristed for many minutes (Fig. Fii).
TM4SF1 overexpressing (TM4SF1-OE) HUVEC
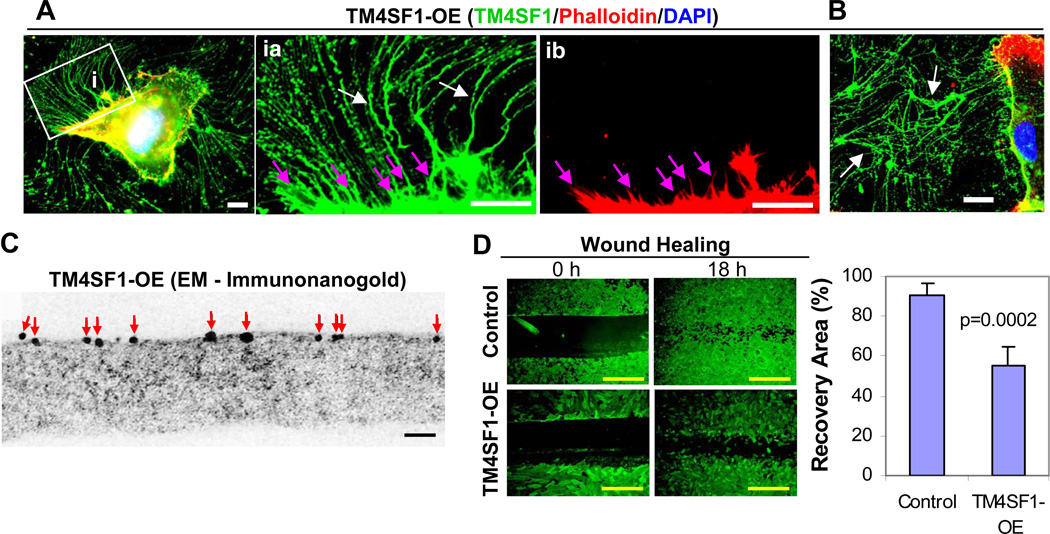
HUVEC normally express TM4SF1 at 91 ± 10 mRNA copies/cell, a level similar to that found in other types of cultured EC [7]. When transduced to overexpress TM4SF1 at 4.3-fold higher levels (386 ± 28 mRNA copies/cell), HUVEC extended many more and longer (80–100 µm) nanopodia from their entire perimeter (Fig. 2A and inset ia, white arrows). As they moved, TM4SF1-OE cells left behind large amounts of TM4SF1-stained, matrix-adherent debris (Fig. 2B, white arrows). In contrast to the clearly banded TMED pattern of nanopodia staining in control or mock-transfected HUVEC, TM4SF1 staining of nanopodia appeared almost continuous in TM4SF1-OE cells. However, the higher resolution provided by electron microscopic immunonanogold cytochemistry demonstrated greatly increased, though still discontinous, TM4SF1 labeling (Fig. 2C, frequency of 17.3 ± 5.0 gold particles/µm). F-actin, when detected, was confined to within 10 µm of the TM4SF1-OE cell body (Fig. 2Aib, pink arrows) as in the nanopodia of control [7] or empty vector-transduced HUVEC (Supplemental Figure 1A). Further, unlike control or GFP-transduced HUVEC (Supplemental Video 1), TM4SF1-OE HUVEC failed to form distinct lamellipodia, migrated poorly (Supplemental Video 2), and performed poorly in wound-healing scratch assays (Fig. 2D).
Fig. 2.
TM4SF1 overexpressing (OE) HUVEC. HUVEC were transduced with 50 moi adenovirus (empty vector control or full length TM4SF1) for 48h. OE HUVEC expressed TM4SF1 at ~400 mRNA copies/cell. Transduced cells were then subcultured at 60% confluence for 2h (A) or 24h (B) prior to immunostaining with TM4SF1 antibodies, phalloidin, and DAPI. TM4SF1-OE HUVEC projected greatly increased numbers of nanopodia that exhibited nearly continuous TM4SF1 staining (inset ia, white arrows), whereas phalloidin staining was confined to only the most proximal portions of nanopodia (inset ib, pink arrows). (B) By 24h in culture, TM4SF1-OE cells had shed extensive TM4SF1-positive debris that remained attached to substrate (white arrows). (C) Immuno-nanogold electron microscopic staining of TM4SF1 on a nanopodium of TM4SF1-OE HUVEC. Red arrows indicate gold particles. (D) TM4SF1-OE HUVEC performed poorly in an 18h wound healing scratch assay, compared with control, empty vector-transduced HUVEC. Black, white, and yellow scale bars are 0.1 µm, 10 µm and 100 µm, respectively.
Human dermal fibroblasts (HDF) expressing TM4SF1 at EC-like levels project typical nanopodia
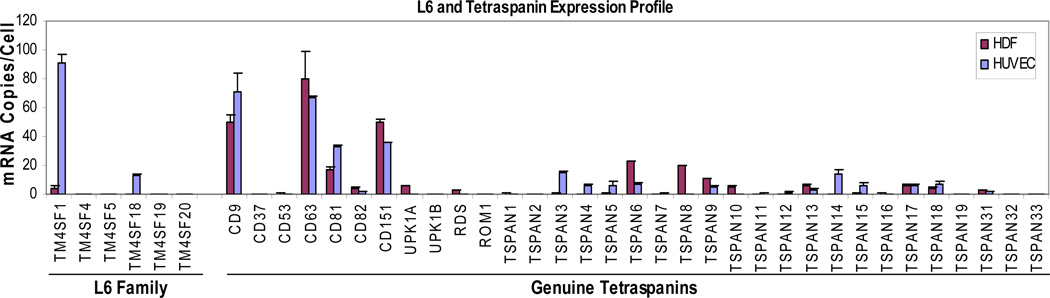
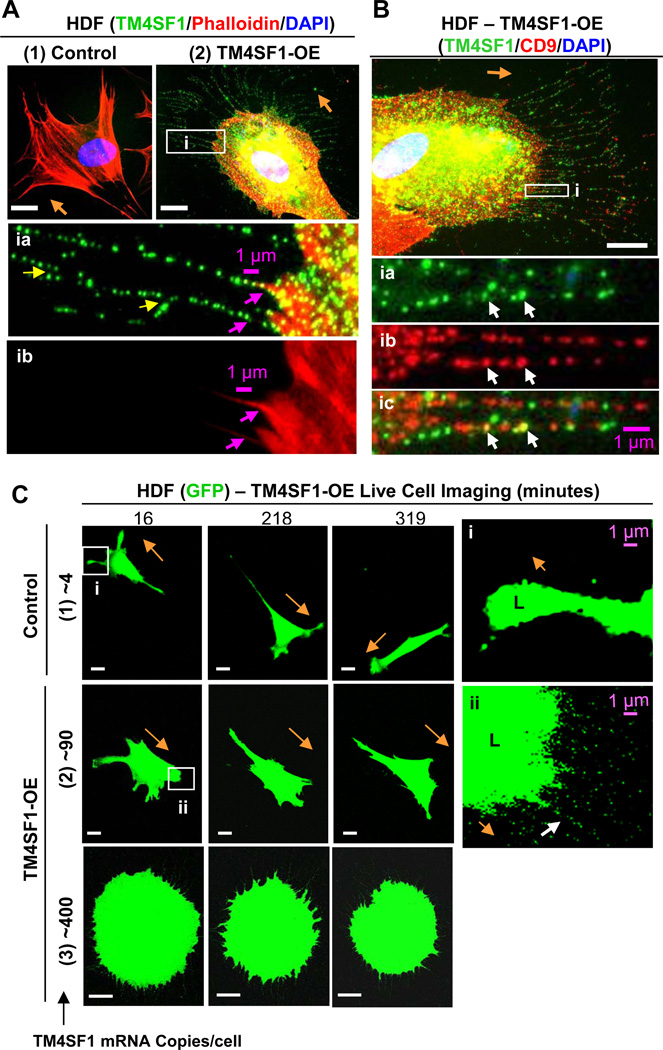
HDF and HUVEC express a variety of different tetraspanins (Fig. 3). However, whereas HUVEC expressed TM4SF1 at ~90 mRNA copies/cell, HDF, like many other types of normal cells in culture that we have tested other than EC, expressed TM4SF1 at only very low levels (Fig. 3, 4.42 ± 0.57 mRNA copies/cell, ~21-fold less than HUVEC), and below the detection range of immunofluorescence (Fig. 4A,1). HDF expressed other members of the L6 family at even lower levels (Fig. 3). When HDF were transduced with TM4SF1 at 15 moi of adenovirus, they expressed TM4SF1 at 92.4 ± 7.3 mRNA copies/cell, a level similar to that of normal HUVEC and other EC. TM4SF1-transduced HDF developed larger, broader and more rounded lamellipodia than control or mock-transduced HDF from which projected multiple long, TM4SF1-positive, branching nanopodia (Fig. 4A,2 and insets, yellow arrows). Like EC nanopodia, those in TM4SF1-transduced HDF contained F-actin only in their most proximal portions (Fig. 4A,2 and insets, pink arrows). Further, the periodicity of TMED in the nanopodia of TM4SF1-transduced HDF was similar to that in HUVEC (compare Fig. 4A,2 and insets with Fig. 1A and Supplemental Figure 1A).
Fig. 3.
mRNA expression profiles of L6 family and genuine tetraspanins in HUVEC and HDF. TM4SF1 is strongly expressed in HUVEC but weakly in HDF.
Fig. 4.
HDF form nanopodia and alter their migration pattern following transduction with TM4SF1. HDF were transfected for 48h with empty vector (control) or TM4SF1 adenoviruses and subcultured at 60% confluence for 2h prior to immunostaining (A,B) or live cell imaging (C). (A1) Representative immunofluorescence image of HDF transduced with empty vector at 15 moi demonstrate lack of TM4SF1 staining. (A2) HDF transduced with 15 moi expressed TM4SF1 at ~90 mRNA copies/cell and acquired prominent, broadly rounded lamellipodia from which projected branching (yellow arrows) nanopodia that immunostained with TM4SF1 at a periodicity similar to that of EC nanopodia (inset ia). F-actin was confined proximally (insets ia and ib, pink arrows). (B) HDF expressing ~90 mRNA copies of TM4SF1/cell exhibited an intermittent staining pattern of both TM4SF1 and CD9 with only occasional co-localization (inset i, white arrows). (C) Representative time-lapse images of GFP-transduced HDF that were empty vector control- and TM4SF1-transduced at 15 or 50 moi to express TM4SF1 at ~90 or ~400 mRNA copies/cell. (1) Empty vector (control) transduced HDF exhibited a typical fibroblast migration pattern, projecting relatively narrow, splayed lamellipodia (L, inset i) and changing directions only every 3–5 h. (2) HUVEC transduced with both TM4SF1 (~90 mRNA copies/cell) and GFP projected prominent, larger, and more broadly rounded lamellipodia (L, inset ii) from the leading front and moved continuously in the direction of nanopodia projection (white arrow). (3) TM4SF1 (~400 mRNA copies/cell) and GFP co-transduced HDF projected numerous nanopodia from the entire cell perimeter and movement was impaired. Orange arrows indicate direction of cell migration. White scale bars, 10 µm.
Cultured HDF express high levels of CD9 (Fig. 3). CD9 is of interest because it is known to be enriched in microdomains [1] and to have a role in cell motility [13]. Staining of TM4SF1-transduced HDF with both TM4SF1 and CD9 demonstrated that CD9 membrane microdomains and TMED were for the most part distinct and only occasionally co-localized (Fig. 4B and insets, white arrows).
Live cell imaging was used to compare the migration patterns of GFP and GFP/TM4SF1-transduced HDF (Fig. 4C). As in fixed, stained preparations (Fig. 4B and inset), control HDF extended several narrow splayed lamellipodia (L) (Fig. 4C,1 and insect i), in contrast to EC which typically extended a large lamellipodium from the cell’s leading front (Supplemental Video 1). HDF changed direction much less frequently than HUVEC in culture, only about once every 3–5 hours (Fig. 4C,1). Also as in fixed-stained preparations (Fig. 4A,2), TM4SF1-transduced HDF that expressed TM4SF1 at EC-like levels (~90 mRNA copies/cell) formed broader, more rounded lamellipodia from which projected lengthy nanopodia in the direction of cell migration (Fig. 4C,2 and inset ii). When TM4SF1 expression levels were increased further to ~400 mRNA copies/cell, as in TM4SF1-OE HUVEC (Fig. 2, Supplemental Video 2), HDF projected nanopodia from the entire cell perimeter, failed to form distinct lamellipodia, and cell motility was greatly hindered (Fig. 4C,3).
TM4SF1 interaction with myosin-10 and β-actin
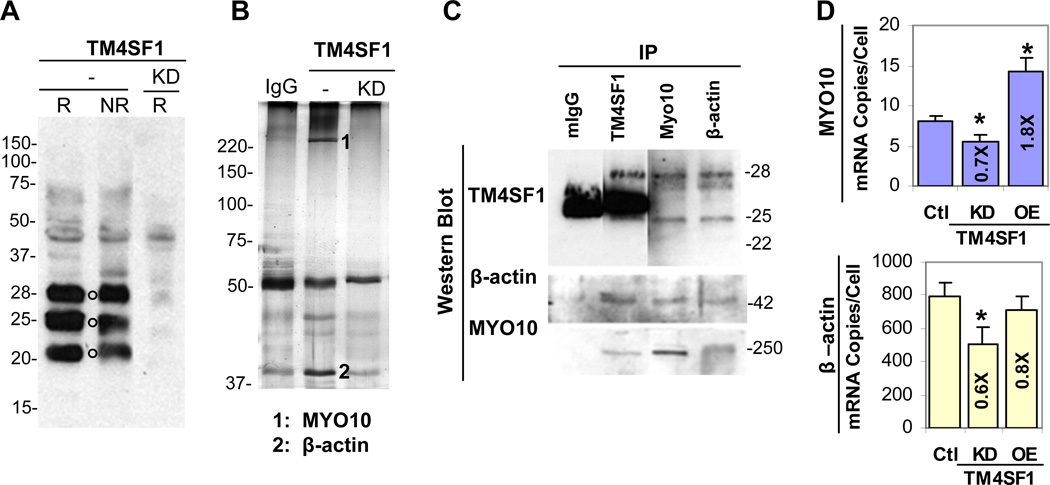
Based on its amino acid sequence, TM4SF1 is predicted to have a MW of 21.6-kD. However, due to glycosylation and other possible modifications, TM4SF1 appeared as multiple bands in Western blots of tumor cell lysates [14–16]. In HUVEC, Western blots demonstrated three major bands with MW of ~ 22-, 25-, and 28-kD under both reducing and non-reducing conditions, along with several larger minor bands of unclear significance. The intensities of all major and most minor bands, except for a minor band at ~ 45kD, were greatly diminished in knock down (KD) cells in which TM4SF1 mRNA levels were reduced by >95% (Fig. 5A).
Fig. 5.
TM4SF1 interaction with MYO10 and β-actin in HUVEC. HUVEC were transduced with adenoviruses expressing TM4SF1 (same experimental conditions as in Fig. 2), TM4SF1 knockdown (KD) shRNA, or empty vector (control) [7]. (A) Western blot under reducing (R) or non-reducing (N) conditions stained with TM4SF1 antibody demonstrates three major bands with molecular weights of ~22-, 25-, and 28-kD (black circles). In TM4SF1-KD HUVEC, all three bands were greatly reduced. (B) Cell lysates prepared from 3 day control and TM4SF1-KD HUVEC were pre-absorbed with mouse IgG, immunoprecipitated with anti-TM4SF1 antibodies, and silver stained following SDS-PAGE. Several prominent bands were diminished in TM4SF1-KD lysates. Mass spectrometry (MALDI-TOFF) revealed that band #1 was MYO10 and band #2 was β-actin; sixteen and fourteen peptide bands matched with MYO10 and β–actin, respectively. (C) IP of control HUVEC lysates with TM4SF1 (mouse anti-human), β–actin (rabbit anti-human), or MYO10 (goat anti-human) antibodies, followed by Western blots with anti-TM4SF1 antibodies, consistently demonstrated 28-kD and 25-kD bands. TM4SF1 immunoprecipitates also demonstrated β-actin- and MYO10-reactive bands at their expected molecular weights, 42- and 250-kD, respectively. (D) MYO10 mRNA expression increased 1.8-fold (p=0.004) in TM4SF1-OE HUVEC (~400 TM4SF1 mRNA copies/cell) and decreased by ~30% (p=0.015) in TM4SF1-KD cells. β-actin mRNA levels were not significantly (p=0.17) affected by TM4SF1-OE, but decreased by ~40% (p=0.022) in TM4SF1-KD cells. Student t-test was used to obtain p-values.
To identify proteins that might interact with TM4SF1 and affect nanopodia projection and cell polarization, we prepared 1% Brij-99 lysates of control and TM4SF1-KD shRNA HUVEC, and performed immunoprecipitation (IP) with antibodies against TM4SF1. SDS-PAGE revealed greatly diminished bands in IPs of TM4SF1-KD lysates (Fig. 5B). Further, mass-spectrometry (MALDI-TOFF) identified two of the bands missing in KD lysates to be myosin-10 (MYO10) and β-actin. Western blots demonstrated that MYO10 and β-actin proteins immunoprecipitated with antibodies against TM4SF1 (Fig. 5C). Also, antibodies against MYO10 and β-actin immunoprecipitated the ~28-kD and ~25-kD TM4SF1 bands, but not the 22-kD band. mRNA expression levels of MYO10, and to a lesser extent those of β-actin, correlated with TM4SF1 expression levels (Fig. 5D).
Discussion
The data presented here, coupled with those from our earlier report [7], demonstrate nanopodia to be a newly recognized type of cell projection that has important roles in EC polarization and migration (Fig. 1). Nanopodia formation is dependent on the expression of TM4SF1, a member of the tetraspanin-related L6 family. The tetraspanin family has thirty-nine members [1,2], many of which are highly expressed in both EC and fibroblasts (Fig. 3). In contrast, TM4SF1 expression is highly restricted to EC (among normal cells thus far tested) and to tumor cells [4–7]. Nanopodia are greatly reduced following TM4SF1 knockdown [7], whereas many more and longer nanopodia formed when TM4SF1 was overexpressed (Fig. 2). The singular importance of TM4SF1 for generating nanopodia was further demonstrated when TM4SF1 was transduced into fibroblasts, cells that normally express TM4SF1 at extremely low levels and that do not normally form nanopodia. Fibroblasts expressing TM4SF1 at EC-like levels underwent fundamental cytoskeletal re-organization, forming larger, more rounded lamellipodia, extending lengthy EC-like nanopodia with typical TMED banding, and adapting an EC-like pattern of migration (Fig. 4). Similar changes were observed when TM4SF1 was transduced into melanocytes and mouse 3T3 cells, cells that, like HDF, normally express TM4SF1 at extremely low levels (< 5 mRNA copies/cell, data not shown). However, when TM4SF1 was expressed at much higher levels (~400 mRNA copies/cell) in HUVEC (Fig. 2 and Supplemental Video 1), in HDF (Fig. 4C,3), and in the other cell types (data not shown), excessive numbers of nanopodia formed and cell polarization and migration were impaired.
The profound effects of TM4SF1 on cell shape and motility implied corresponding changes in the cell cytoskeleton and prompted a search for cytoskeletal partners with which TM4SF1 might interact. We demonstrated here that TM4SF1 interacts with MYO10 and β-actin (Fig. 5). Further, MYO10 expression closely correlated with that of TM4SF1 in HUVEC (Fig. 5D). MYO10 has been shown to interact with β1-integrin via its tail domain, and to bind β-actin through head/neck domains [17–19]. Thus, TM4SF1’s interactions with MYO10 and β-actin may be indirectly mediated by integrins as TM4SF1 also interacts with integrin-α5β1 as we showed previously [7]. Studies to determine which domains of TM4SF1 interact with integrins, MYO10, and β-actin are currently underway.
CD9, a ubiquitously expressed tetraspanin, has been shown to recruit TM4SF1 to CD9-enriched microdomains in tumor cells [16]. However, CD9 only occasionally co-localized with TMED in TM4SF1-transduced HDF (Fig. 4B), suggesting that in HDF TM4SF1 and CD9 function independently. TM4SF1 behaved in a dominant fashion in HDF, as in HUVEC; i.e., HDF transduced to express TM4SF1 at EC-like levels projected typical EC-like nanopodia and lamellipodia. CD9 was also expressed in HUVEC at ~71±12 mRNA copies/cell (Fig. 3), a level similar to that in HDF, and its expression was not altered by TM4SF1 knockdown under circumstances where HUVEC were unable to project nanopodia (data not shown).
A curious feature of nanopodia, in contrast to filopodia and other cell projections, is their relative lack of F-actin content. This raises questions as to the mechanisms by which nanopodia form. Rho family GTPases such as CDC42 are thought to have essential roles in filopodia formation, acting to nucleate and elongate actin filaments [23–26]. Membrane projections of different lengths can also be generated by exogenous expression of different Rho GTPases such as Rif, TC10 and RhoT [23,22,21]; generally, though not invariably, these projections include F-actin [24–26]. However, filopodia are also reported to form in cells lacking CDC42, as well as in cells lacking its effector, the Wiskott-Aldrich syndrome protein (WASP). Recent work has shown that filopodia-like membrane projections can be generated independently of actin by BAR superfamily domains, which deform membranes into tubular structures [24–26]. These data provide a potential mechanism for the formation of nanopodia that express F-actin, if at all, only in their most proximal parts.
Migrating HUVEC change direction every 45 min or so (Supplemental Video 1). As they changed directions, nanopodia that were once at the advancing front reverted to the cell’s trailing edge. In some instances, nanopodia interacted with nanopodia from neighboring cells to form prolonged, long distance contacts. These intercellular connections resemble nanotubes, fine intercellular structures (~50–300 nm in width) that were initially described in PC12 cells and which play a role in intercellular communication [27]. Nanotubes have now been found in a variety of cultured cells [28], including EC [29]. Studies to determine whether nanopodia participate in EC nanotube formation and therefore in intercellular molecular and organelle trafficking are ongoing.
In summary, we have shown that TM4SF1, a member of the L6 protein family, functions, like genuine tetraspanins, as a ‘molecular organizer’, modulating cell motility and directional migration [1]. Through a unique ability to generate nanopodia, possibly by partnering with molecules specific to TM4SF1, and interacting with cytoskeletal and membrane molecules that are ubiquitously expressed, TM4SF1 plays a dominant role in cytoskeletal organization, cell polarization, and migration. The functions of nanopodia are not as yet entirely clear but may include sensing the microenvironment and directing cell movement at longer range than filopodia [30,31].
Supplementary Material
Acknowledgement
We thank Dr. Youichiro Wada for helpful discussions. This work was supported by NIH grant P01 CA92644 and by a contract from the National Foundation for Cancer Research.
References
- 1.Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol. 2005;6(10):801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 2.Hemler ME. Targeting of tetraspanin proteins--potential benefits and strategies. Nat Rev Drug Discov. 2008;7(9):747–758. doi: 10.1038/nrd2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wright MD, Ni J, Rudy GB. The L6 membrane proteins--a new four-transmembrane superfamily. Protein Sci. 2000;9(8):1594–1600. doi: 10.1110/ps.9.8.1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hellstrom I, Horn D, Linsley P, Brown JP, Brankovan V, Hellstrom KE. Monoclonal mouse antibodies raised against human lung carcinoma. Cancer Res. 1986;46(8):3917–3923. [PubMed] [Google Scholar]
- 5.Chang YW, Chen SC, Cheng EC, Ko YP, Lin YC, Kao YR, Tsay YG, Yang PC, Wu CW, Roffler SR. CD13 (aminopeptidase N) can associate with tumor-associated antigen L6 and enhance the motility of human lung cancer cells. Int J Cancer. 2005;116(2):243–252. doi: 10.1002/ijc.21089. [DOI] [PubMed] [Google Scholar]
- 6.Kao YR, Shih JY, Wen WC, Ko YP, Chen BM, Chan YL, Chu YW, Yang PC, Wu CW, Roffler SR. Tumor-associated antigen L6 and the invasion of human lung cancer cells. Clin Cancer Res. 2003;9(7):2807–2816. [PubMed] [Google Scholar]
- 7.Shih SC, Zukauskas A, Li D, Liu G, Ang LH, Nagy JA, Brown LF, Dvorak HF. The L6 protein TM4SF1 is critical for endothelial cell function and tumor angiogenesis. Cancer Res. 2009;69(8):3272–3277. doi: 10.1158/0008-5472.CAN-08-4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi T, Takahashi K, Daniel TO. High-Efficiency and Low-Toxicity Adenovirus-Assisted Endothelial Transfection. In: Baker AH, editor. Methods in Molecular Medicine, Vascular Disease: Molecular Biology and Gene Therapy Protocols. Totowa, NJ: © Humana Press Inc.; 1999. pp. 307–314. [DOI] [PubMed] [Google Scholar]
- 9.Shih SC, Smith LE. Quantitative multi-gene transcriptional profiling using real-time PCR with a master template. Exp Mol Pathol. 2005;79(1):14–22. doi: 10.1016/j.yexmp.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Wada Y, Li D, Merley A, Zukauskas A, Aird WC, Dvorak HF, Shih SC. A multi-gene transcriptional profiling approach to the discovery of cell signature markers. Cytotechnology. 2010 doi: 10.1007/s10616-010-9315-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown LF, Lanir N, McDonagh J, Tognazzi K, Dvorak AM, Dvorak HF. Fibroblast migration in fibrin gel matrices. Am J Pathol. 1993;142(1):273–283. [PMC free article] [PubMed] [Google Scholar]
- 12.Rodriguez LG, Wu X, Guan JL. Wound-healing assay. Methods Mol Biol. 2005;294:23–29. doi: 10.1385/1-59259-860-9:023. [DOI] [PubMed] [Google Scholar]
- 13.Deissler H, Kuhn EM, Lang GE, Deissler H. Tetraspanin CD9 is involved in the migration of retinal microvascular endothelial cells. Int J Mol Med. 2007;20(5):643–652. [PubMed] [Google Scholar]
- 14.Marken JS, Schieven GL, Hellstrom I, Hellstrom KE, Aruffo A. Cloning and expression of the tumor-associated antigen L6. Proc Natl Acad Sci U S A. 1992;89(8):3503–3507. doi: 10.1073/pnas.89.8.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marken JS, Bajorath J, Edwards CP, Farr AG, Schieven GL, Hellstrom I, Hellstrom KE, Aruffo A. Membrane topology of the L6 antigen and identification of the protein epitope recognized by the L6 monoclonal antibody. J Biol Chem. 1994;269(10):7397–7401. [PubMed] [Google Scholar]
- 16.Lekishvili T, Fromm E, Mujoomdar M, Berditchevski F. The tumour-associated antigen L6 (L6-Ag) is recruited to the tetraspanin-enriched microdomains: implication for tumour cell motility. J Cell Sci. 2008;121(Pt 5):685–694. doi: 10.1242/jcs.020347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berg JS, Derfler BH, Pennisi CM, Corey DP, Cheney RE. Myosin-X, a novel myosin with pleckstrin homology domains, associates with regions of dynamic actin. J Cell Sci. 2000;113(Pt 19):3439–3451. doi: 10.1242/jcs.113.19.3439. [DOI] [PubMed] [Google Scholar]
- 18.Zhang H, Berg JS, Li Z, Wang Y, Lang P, Sousa AD, Bhaskar A, Cheney RE, Stromblad S. Myosin-X provides a motor-based link between integrins and the cytoskeleton. Nat Cell Biol. 2004;6(6):523–531. doi: 10.1038/ncb1136. [DOI] [PubMed] [Google Scholar]
- 19.Weber KL, Sokac AM, Berg JS, Cheney RE, Bement WM. A microtubule-binding myosin required for nuclear anchoring and spindle assembly. Nature. 2004;431(7006):325–329. doi: 10.1038/nature02834. [DOI] [PubMed] [Google Scholar]
- 20.Pi X, Ren R, Kelley R, Zhang C, Moser M, Bohil AB, Divito M, Cheney RE, Patterson C. Sequential roles for myosin-X in BMP6-dependent filopodial extension, migration, and activation of BMP receptors. J Cell Biol. 2007;179(7):1569–1582. doi: 10.1083/jcb.200704010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupton SL, Gertler FB. Filopodia: the fingers that do the walking. Sci STKE. 2007;2007(400):re5. doi: 10.1126/stke.4002007re5. [DOI] [PubMed] [Google Scholar]
- 22.Mattila PK, Lappalainen P. Filopodia: molecular architecture and cellular functions. Nat Rev Mol Cell Biol. 2008;9(6):446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- 23.Ohta Y, Suzuki N, Nakamura S, Hartwig JH, Stossel TP. The small GTPase RalA targets filamin to induce filopodia. Proc Natl Acad Sci U S A. 1999;96(5):2122–2128. doi: 10.1073/pnas.96.5.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Itoh T, De Camilli P. BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature. Biochim Biophys Acta. 2006;1761(8):897–912. doi: 10.1016/j.bbalip.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 25.Ahmed S, Goh WI, Bu W. I-BAR domains, IRSp53 and filopodium formation. Semin Cell Dev Biol. 2009 doi: 10.1016/j.semcdb.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 26.Saarikangas J, Zhao H, Pykalainen A, Laurinmaki P, Mattila PK, Kinnunen PK, Butcher SJ, Lappalainen P. Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr Biol. 2009;19(2):95–107. doi: 10.1016/j.cub.2008.12.029. [DOI] [PubMed] [Google Scholar]
- 27.Rustom A, Saffrich R, Markovic I, Walther P, Gerdes HH. Nanotubular highways for intercellular organelle transport. Science. 2004;303(5660):1007–1010. doi: 10.1126/science.1093133. [DOI] [PubMed] [Google Scholar]
- 28.Gerdes HH, Carvalho RN. Intercellular transfer mediated by tunneling nanotubes. Curr Opin Cell Biol. 2008;20(4):470–475. doi: 10.1016/j.ceb.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 29.Wang X, Veruki ML, Bukoreshtliev NV, Hartveit E, Gerdes HH. Animal cells connected by nanotubes can be electrically coupled through interposed gap-junction channels. Proc Natl Acad Sci U S A. 2010;107(40):17194–17199. doi: 10.1073/pnas.1006785107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozma R, Ahmed S, Best A, Lim L. The Ras-related protein Cdc42Hs and bradykinin promote formation of peripheral actin microspikes and filopodia in Swiss 3T3 fibroblasts. Mol Cell Biol. 1995;15(4):1942–1952. doi: 10.1128/mcb.15.4.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81(1):53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.