Abstract
Background
High blood glucose and diabetes are amongst the conditions causing the greatest losses in years of healthy life worldwide. Therefore, numerous studies aim to identify reliable risk markers for development of impaired glucose metabolism and type 2 diabetes. However, the molecular basis of impaired glucose metabolism is so far insufficiently understood. The development of so called 'omics' approaches in the recent years promises to identify molecular markers and to further understand the molecular basis of impaired glucose metabolism and type 2 diabetes. Although univariate statistical approaches are often applied, we demonstrate here that the application of multivariate statistical approaches is highly recommended to fully capture the complexity of data gained using high-throughput methods.
Methods
We took blood plasma samples from 172 subjects who participated in the prospective Metabolic Syndrome Berlin Potsdam follow-up study (MESY-BEPO Follow-up). We analysed these samples using Gas Chromatography coupled with Mass Spectrometry (GC-MS), and measured 286 metabolites. Furthermore, fasting glucose levels were measured using standard methods at baseline, and after an average of six years. We did correlation analysis and built linear regression models as well as Random Forest regression models to identify metabolites that predict the development of fasting glucose in our cohort.
Results
We found a metabolic pattern consisting of nine metabolites that predicted fasting glucose development with an accuracy of 0.47 in tenfold cross-validation using Random Forest regression. We also showed that adding established risk markers did not improve the model accuracy. However, external validation is eventually desirable. Although not all metabolites belonging to the final pattern are identified yet, the pattern directs attention to amino acid metabolism, energy metabolism and redox homeostasis.
Conclusions
We demonstrate that metabolites identified using a high-throughput method (GC-MS) perform well in predicting the development of fasting plasma glucose over several years. Notably, not single, but a complex pattern of metabolites propels the prediction and therefore reflects the complexity of the underlying molecular mechanisms. This result could only be captured by application of multivariate statistical approaches. Therefore, we highly recommend the usage of statistical methods that seize the complexity of the information given by high-throughput methods.
Keywords: prediction, fasting glucose, type 2 diabetes, metabolomics, plasma, random forest, metabolite, regression, biomarker
Background
High blood glucose reduces life expectancy worldwide [1], and numerous studies have been performed to identify risk factors of impaired glucose metabolism and type 2 diabetes. Nevertheless, this is a topic that is subject to continuing discussion [2-5]. Established classical markers include: family history of diabetes, markers of adiposity, age and glycemic control itself. In recent years, high-throughput methods have been increasingly applied in clinical research [6-10]. In a recent article Wang et al. used a metabolomics approach for diabetes risk assessment [11]. They analysed baseline blood samples from 189 individuals that developed type 2 diabetes during a 12 year follow-up period as well as 189 matched control subjects. Using Liquid Chromatography coupled with Mass Spectrometry (LC-MS), they measured 61 metabolites. Applying paired t-test and McNemar's test, they identified isoleucine, leucine, valine, tyrosine and phenylalanine as being highly associated with future diabetes. We here show that multivariate statistical methods should be applied to account for dependencies within the metabolome. In doing so, we were able to define a complex pattern of metabolites that predicts future development of fasting plasma glucose levels with high accuracy. We also compare the quality of prediction between this metabolic pattern and established risk markers.
Methods
Fasting plasma samples were taken at baseline and at follow-up after an average of six years in subjects who participated in the prospective follow-up of the Metabolic Syndrome Berlin Potsdam (MESY-BEPO) study [12]. We took the samples under standardised conditions in the morning between 8 and 9 a.m. local time after an overnight fast. All patients gave written informed consent and the study was approved by the local ethical committee.
Fasting plasma glucose levels were measured applying a standard hexokinase assay. Furthermore, we analysed metabolic profiles of baseline fasting plasma samples in a random sub-cohort (n = 172; for characterisation see Table 1) using Gas Chromatography coupled with time-of-flight Mass Spectrometry (GC-MS) according to standard operating procedure [10]. We excluded subjects with type 1 or type 2 diabetes at baseline and subjects treated with oral anti-diabetics or insulin. We measured in total 286 metabolites, some of them are not yet identified. The measurements cover various biochemical classes, such as amino acids, carbohydrates, organic acids, fatty acids and steroids.
Table 1.
Characterisation of the investigated MESY-BEPO sub-cohort
| Clinical Characteristics | Baseline | Follow-up |
|---|---|---|
| Age [years] | 55.7 ± 11.7 | 61.5 ± 11.5 |
| Gender [% female] | 62.2 | |
| Waist circumference [cm] | 93.8 ± 13.8 | 94.6 ± 17.3 |
| Body mass index [kg/m2] | 28.6 ± 5.2 | 29.1 ± 5.3 |
| Fasting glucose [mg] | 92.1 ± 11.6 | 100.5 ± 13.6 |
| Δglucose [mg/(dl · a)] | 1.0 ± 2.3 | |
| Time between baseline and follow-up [years] | 5.6 ± 0.7 | |
Characterisation of the investigated MESY-BEPO sub-cohort (n = 172) at baseline and follow-up. Data are presented as mean ± standard deviation.
The chromatographic peaks were picked and identified using the Golm Metabolome Database (GMD) [13] and the R package TargetSearch [14]. Since missing values only occurred if metabolite concentration went below detection limit, these values were replaced by a value 0.7 times the minimum measured value. Log-transformation and normalisation of the measured relative intensities were performed according to Lisec et al. [15].
To quantify the development of fasting glucose levels we calculated the difference between glucose levels normalised by the elapsed time:
in (mg/dl)/a. We computed Spearman's rank correlation coefficient and p-values to identify significant correlation between Δglucose and single metabolites with a significance level of α = 0.05. Significantly correlating metabolites were used to build linear regression models using the R package stats as well as Random Forest regression models [16,17] using the R package randomForest. The correlation matrix was drawn using the R package corrplot. We also performed a nested feature selection based on the Random Forest importance measure as proposed by Svetnik et al. [18]. The importance measure is based on the decrease of accuracy, i.e. the increase of the mean square error (MSE), for the out-of-bag portion for each tree. The MSE was calculated for both the true as well as a permuted predictor variable. The difference between them was calculated for all trees, averaged and normalised and hence resulted in the importance measure. By removing the 50% of metabolites with the smallest importance measure, we iteratively bisected the number of metabolites used to predict Δglucose and thus removed irrelevant metabolites. During this reduction the average Random Forest model accuracy remained stable up to a pattern of nine metabolites. Thus, we employed this number to select metabolites for a Random Forest regression analysis to predict Δglucose. Furthermore, we added established risk markers to the Random Forest regression model. These risk markers were: gender, waist circumference, body mass index (BMI), age and baseline fasting glucose levels.
To evaluate the performance of all the regression models, we calculated the Pearson product-moment correlation coefficient between the real and calculated Δglucose. All feature selection and regression modeling was performed and validated on all samples as well as in the subsets of tenfold cross-validation [19]. To test for reliability the cross-validation including the metabolite selection and regression model building was repeated 100 times. Hence, the given cross-validation accuracies reflect the median of this replication.
Results
Fasting glucose development
We observed a median Δglucose of 0.8mg/(dl · a), within the range from -3.7mg/(dl · a) to 8.2mg/(dl · a). The standard deviation was 2.3mg/(dl · a). Compared to the baseline level, 61 subjects had a decreased fasting glucose level (Δglucose ≤ 0mg/(dl · a)), whereas 111 subjects showed an increased fasting glucose level (Δglucose >0mg/(dl · a)).
Correlation
To identify metabolites that are associated with the development of fasting plasma glucose levels, we calculated the Spearman's rank correlation coefficient between the single metabolites and Δglucose. We used the Spearman correlation since not all metabolites were normally distributed. All 30 metabolites that are significantly correlated with Δglucose are shown in Table 2. The observed correlation ranges from -0.27 to 0.40. Therefore, a single metabolite explains up to 16.2% of the observed variance in Δglucose (see Table 2). Consequently, the question arose whether the metabolites are not only capable of explaining but also predicting Δglucose. Thus, we applied a multivariate approach.
Table 2.
Spearman's rank correlation
| Spearman Correlation | p-value | % variance explained | |
|---|---|---|---|
| Hypoxanthine | 0.40 | <0.0001 | 16.20 |
| Aspartic acid | 0.30 | 0.0001 | 9.18 |
| Pyroglutamic acid | 0.29 | 0.0001 | 8.34 |
| 2-methyl-Malic acid | 0.27 | 0.0004 | 7.21 |
| NA 1033 (trisaccharide) | -0.27 | 0.0004 | 7.05 |
| NA 1034 | -0.23 | 0.0026 | 5.23 |
| NA 1052 (carbohydrate) | -0.22 | 0.0046 | 4.64 |
| Myo-inositol | 0.21 | 0.0052 | 4.51 |
| NA 1027 (sterol phosphate) | 0.21 | 0.0061 | 4.34 |
| Threonic acid | 0.21 | 0.0063 | 4.31 |
| NA 997 (Uridine-5'-monophosphate) | 0.20 | 0.0078 | 4.09 |
| Glutamic acid | 0.19 | 0.0104 | 3.80 |
| NA 831 | 0.19 | 0.0119 | 3.67 |
| NA 653 | 0.18 | 0.0179 | 3.25 |
| Ketopentose | 0.18 | 0.0181 | 3.24 |
| Fucose | 0.18 | 0.0192 | 3.18 |
| Uracil | 0.18 | 0.0213 | 3.08 |
| Fructose | -0.17 | 0.0233 | 2.99 |
| NA 631 (D-Glucopyranose) | -0.17 | 0.0235 | 2.98 |
| NA 613 | 0.17 | 0.0251 | 2.91 |
| NA 597 (pyranose) | -0.16 | 0.0312 | 2.70 |
| Isoleucine | 0.16 | 0.0358 | 2.57 |
| NA 275 | 0.16 | 0.0379 | 2.51 |
| NA 639 | 0.16 | 0.0388 | 2.49 |
| NA 442 | 0.16 | 0.0395 | 2.47 |
| NA 560 | 0.16 | 0.0402 | 2.45 |
| NA 854 (carbohydrate) | 0.16 | 0.0413 | 2.43 |
| Tartaric acid | 0.15 | 0.0429 | 2.39 |
| Fructose | -0.15 | 0.0466 | 2.31 |
| NA 632 | -0.15 | 0.0486 | 2.27 |
Spearman's rank correlation coefficient, p-values and % of explained variance for all metabolites significantly correlated with Δglucose (significance level α = 0.05). Not yet identified metabolites are marked with NA, putative biochemical structures are given in round brackets.
Linear Model
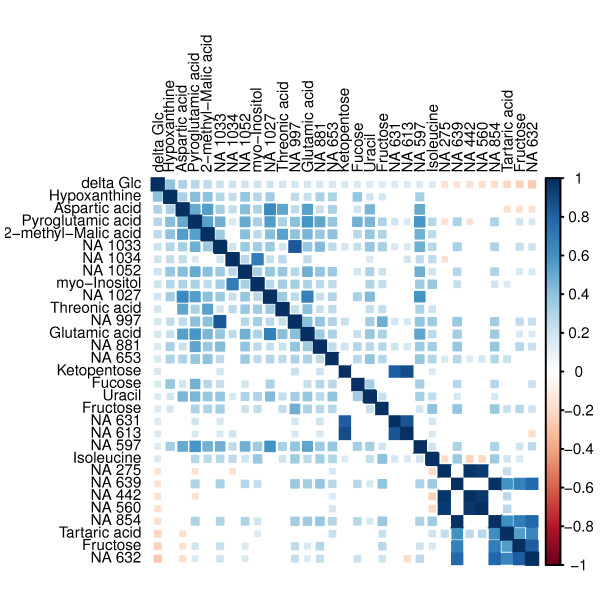
We used the Spearman correlation as a filter and included all metabolites significantly correlated with Δglucose to build a linear regression model. The accuracy (i.e. the Pearson correlation between true and estimated Δglucose) using all samples was 0.57, the median accuracy after tenfold cross-validation was 0.22. This accuracy is unsatisfactorily poor. Despite the weak accuracy of the linear model, it is known that multicollinearity often leads to an unstable model and affects the correct calculation of coefficients of linear regression models [20]. The calculated condition number of the correlation matrix K > >1000 indicated the strong collinearity within the chosen metabolites. However, high correlation within a metabolite matrix is not surprising, since the observed metabolites are not independent but connected via metabolism. The detected multicollinearity is illustrated with the correlation matrix in Figure 1. Hence, we had to find a regression model that is robust against multicollinearity. This characteristic is given for Random Forest models [21].
Figure 1.
Correlation matrix. This correlation matrix visualises not only the significant correlations between Δ glucose and the metabolites (first row/column) but also the correlation among the metabolites. The colour intensity and tile size indicate the strength of correlation. Positive correlation are marked blue, negative correlation are marked red.
Random Forest Regression Models
We built a Random Forest regression model using the metabolites selected by significant correlation to predict Δglucose. Compared to the linear model, the accuracy increase was noteworthy: using all samples the accuracy was 0.97, and after tenfold cross-validation the median accuracy was 0.41. Nevertheless, we investigated the scale of restriction due to using only metabolites that correlate with Δglucose.
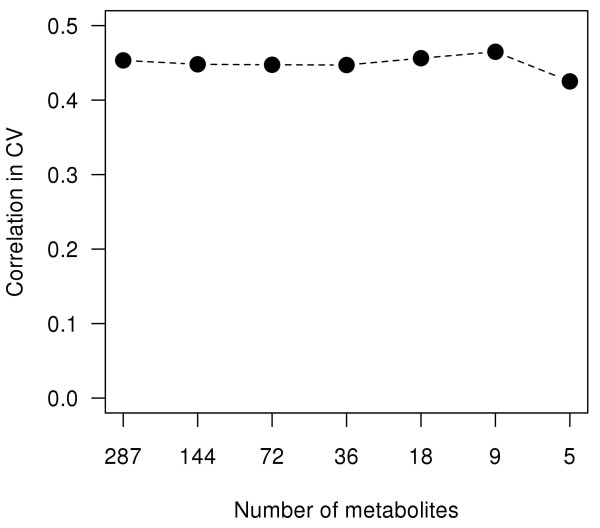
Thus, we performed a nested feature selection based on the Random Forest importance measure. To define a minimum number of metabolites necessary for accurate prediction of Δglucose, we stepwise bisected the number of metabolites. During this reduction the average model accuracy remained stable up to a pattern of nine metabolites (see Figure 2). Therefore, it is legitimate to use only the nine metabolites with the highest importance in the Random Forest model. These nine metabolites are shown in Table 3. The accuracy using these metabolites in a Random Forest regression model was 0.97. The median cross-validation accuracy was 0.47. Although the current selection of metabolites is smaller than the correlation based selection, the accuracy improved. Detailed examination of the two metabolite selections revealed an incomplete overlap. Metabolites highly correlated with Δglucose also showed a high Random Forest importance. However, some metabolites (e.g. the putative allantoin, citric acid and an unknown) showed high importance but no significant correlation with Δglucose. We assume that these metabolites are responsible for the increase in accuracy. Therefore, we conclude that not only linear but more complex relations may exist between metabolites and the fasting glucose development. Moreover, this assumption of complexity is underlined by the selected metabolites themselves and their location in biochemical pathways. The identified metabolites of the pattern are part of multiple metabolic pathways, e.g. purine degradation, energy metabolism and amino acid metabolism.
Figure 2.
Random Forest feature selection. Iterative bisection of the number of metabolites by removing the 50% of metabolites with the smallest importance measure. The remaining metabolites were used to build the Random Forest regression model. Shown is the median cross-validation accuracy. The accuracy remains stable up to a pattern of nine metabolites.
Table 3.
Random Forest Importance of the highest ranked metabolites
| Metabolite | Random Forest Importance |
|---|---|
| Hypoxanthine | 12.52 |
| Pyroglutamic acid | 9.37 |
| NA 1027 (sterol phosphate) | 8.13 |
| NA 611 (Allantoin) | 7.80 |
| NA 718 (carboxylic acid) | 7.56 |
| NA 1033 (trisaccharide) | 6.28 |
| Aspartic acid | 5.34 |
| NA 1034 | 5.26 |
| Citric acid | 4.60 |
Metabolites belonging to the pattern identified using Random Forests and their Importance. Not yet identified metabolites are marked with NA, putative biochemical structures are given in round brackets.
Furthermore, we added established risk markers (gender, waist circumference, BMI, age, baseline fasting glucose) to the metabolites and again performed feature selection based on Random Forest importance and built a Random Forest regression model. Surprisingly, these established risk markers did not improve the accuracy of the model. To further analyse this observation, we built a Random Forest regression model using only the established risk factors. This model had a cross-validation accuracy of 0.05, and therefore was not able to predict Δglucose.
All results of the different regression models can be found in Table 4. For identification of the so far unknown metabolites of our pattern, we revised the original spectra data using the Decision Tree tool provided by Hummel et al. [22] and added the information in Table 3.
Table 4.
Regression model accuracy
| Metabolite Selection | Model | all samples | CV |
|---|---|---|---|
| Spearman Correlation | linear Model | 0.57 | 0.22 |
| Spearman Correlation | Random Forest | 0.97 | 0.41 |
| RF importance | Random Forest | 0.97 | 0.47 |
| RF importance + Established markers | Random Forest | 0.97 | 0.46 |
| Established markers | Random Forest | 0.90 | 0.05 |
Accuracy of the models (median Pearson correlation between real and estimated Δglucose levels) based on metabolites and/or established risk markers was calculated using all samples of the training set and after tenfold cross-validation (CV). The established risk markers are: gender, waist circumference, BMI, age and baseline fasting glucose levels.
Discussion
In conclusion, our work demonstrates that metabolic profiles have a high performance in predicting fasting glucose level development (Table 4). However, this prediction ability was captured by using non-parametric Random Forest regression. This indicates that a rather complex non-linear pattern allows the prediction of Δglucose.
We also observed that metabolic profiles hold considerably more information than established markers. This may result from the different scopes of these variables: established markers describe a macroscopic phenotype that is underpinned by the molecular level. Although established markers may differ between healthy and diseased subjects, understanding diseases on their molecular level will support the development of individualised medicine and hence lead to better prevention and treatment in clinical practice.
We are aware that diurnal variation or other factors (e.g. physical activity or nutrition during the days before phenotyping) might have affected measurements of metabolite and glucose concentrations in our samples. We have tried to minimise this variation by sampling under standardised conditions. Due to the potentially remaining variance, important metabolites might not be included in the model, thus causing false negatives. Consequently, we might have underestimated the capacity of metabolites to predict glycemic control.
To put the metabolites belonging to the final pattern (Table 3) into a biochemical context, we reviewed existing literature.
Among all metabolites hypoxanthine showed the highest importance regarding fasting glucose development. It is a central intermediate in purine degradation and biosynthesis. Hypoxanthine is either enzymatically metabolised via xanthine to uric acid, or re-utilised to inosine monophosphate in the nucleotide salvage pathway. In the past decades several studies have shown an association between the purine degradation pathway and type 2 diabetes or the metabolic syndrome [23]. However, the causality of this association remains unclear. In their recent meta-analysis Pfister et al. did not observe a direct effect of serum uric acid or associated genetic variants on development of type 2 diabetes [24]. Nevertheless, other ways of interaction may exist [25,26]. As early as 30 years ago, Harkness et al. described that purine degradation is increased under ATP depleting conditions and small changes in hypoxanthine may reflect alterations of ATP turnover [27]. Whereas glucose uptake does not change hypoxanthine levels or purine degradation [28], fructose uptake leads to hepatic ATP degradation [25] and may play a major role in epidemic of metabolic syndrome and obesity [29]. The link between insulin resistance and increased serum urate might be mediated by the hexose phosphate shunt [26]. Elevated hypoxanthine levels might reflect an early stage of this mechanism.
The unknown NA611 is most likely allantoin. This metabolite is also closely related to purine degradation: Although it is not enzymatically produced in human body (due to knock-out mutation in urate oxidase gene [30]), it was referenced before in human blood, cerebrospinal fluid and urine. It is generated by spontaneous reaction of uric acid with radical oxygen species (ROS) [31]. Thus, allantoin levels reflect uric acid action as a ROS scavenger. The role of oxidative stress and ROS in diabetes development has been reviewed elsewhere [32].
Pyroglutamic acid (or 5-oxoproline) is a cyclised derivative of L-glutamic acid. In biological context it can be formed non-enzymatically from glutamate, glutamine and γ-glutamylated peptides or enzymatically by γ-glutamylcyclotransferase. The latter is part of the γ-glutamyl cycle, which synthesises and degrades glutathione. Glutathione is a main intracellular antioxidant [33] and thus plays an important role in maintaining redox homeostasis and eliminating ROS in the cell. S-glutathionylation of proteins as result of redox imbalance is thought to be a biological switch [34]. In our measurements pyroglutamic acid might also be derived from glutamic acid or glutamine during the derivatisation process. Both amino acids act not only in protein synthesis and degradation. Due to their potential to transfer amino groups, they occur in numerous pathways. The activity of several Glx consuming or producing enzymes (e.g. γ-glutamyltransferase [35-42], glutamine fructose-6-phosphate amidotransferase [43], glutamate pyruvate transaminase [40]) were reported to be associated with diabetes mellitus, impaired glucose tolerance and insulin resistance and are used as biomarkers to monitor liver functionality. Glutamate links the tricarboxylic acid (TCA) cycle with amino acid biosynthesis and degradation. Furthermore, several animal models were described that develop obesity and/or diabetes after injection of monosodium glutamate [44-48]. In such an animal model reduced nitric oxide and increased ROS production were also observed [49]. In addition, dietary monosodium glutamate increases weight gain, adiposity and reduces insulin sensitivity in an animal model [50]. According to Samocha-Bonett et al., oral glutamine reduced postprandial glycemia in type 2 diabetic subjects [51]. Finally, due to its role as neurotransmitter, glutamate is involved in numerous signaling processes [52,53] but is also potentially neurotoxic [54].
Aspartic acid is involved in protein synthesis and degradation and also acts in transamination reactions. It is directly linked to the TCA cycle via the malate shuttle. The aspartate aminotransferase was reported to be associated with glucose metabolism and insulin resistance [38]. Bousova et al. reported reduced aspartate amino transferase activity induced by fructose and the resulting glycation in vitro and observed beneficial effects of uric acid in that context [55]. Hageman et al. reported that feeding an aspartate rich protein blend decreased the postprandial glucose response in rats [56]. According to Arai et al. monosodium aspartate induces obesity, increased plasma insulin and increased acetyl-CoA carboxylase in animal models [57].
Citric acid is an intermediate of the TCA cycle, which is a central pathway in the organisms energy preservation. The TCA cycle oxidatively degrades acetyl-CoA, which originates from carbohydrate, amino acid or fatty acid degradation. An animal model for diabetes showed increased gluconeogenesis by increased TCA cycle substrate uptake and fluxes [58]. Citric acid is produced by citrate synthase. Ortenblad et al. [59] reported a reduced basal citrate synthase activity, reduced lipid oxidation and reduced insulin-mediated glucose oxidation in culture skeletal muscle cells from type 2 diabetic persons. They increased citrate synthase activity by incubation with insulin in cells from non-diabetic but not in cells from diabetic subjects. Co-incubation with palmitate abolished the stimulatory effect [59]. Furthermore, maximum velocity of citrate synthase was reduced in cultured pancreatic islet cells cultured with long-chain fatty acids or high dose glucose, which may play a role for glucotoxicity and lipotoxicity in β-cells [60]. Citrate also connects the TCA cycle with hepatic fatty acid and cholesterol synthesis by being exported via transport proteins from mitochondrion to cytosol where it is cleaved to acetyl-CoA by ATP-citrate lyase. The citrate carrier is the key component of citrate-malate-shuttle and it is located in the mitochondrial inner membrane. High levels of the citrate carrier can be found predominantly in liver, pancreas and kidney [61]. A reduction of citrate carrier activity and protein levels, both affected by insulin and glucose levels at different regulatory steps, was found in diabetic mice [62]. Knockout of gene SLC13A5, encoding for a sodium-coupled plasma membrane citrate transporter, protects mice from adiposity and insulin resistance [63]. The same group reported a 3-fold increased expression of SLC13A3, encoding another hepatic plasma membrane tri/dicarboxylate transporter, in the knockout mice. ATP-citrate lyase is the key enzyme in cellular lipid production. Its activity and mRNA levels are decreased in pancreatic islets of diabetic rats [64]. Administration of insulin increased ATP-citrate lyase activity and mRNA levels in liver of diabetic rats [65]. Chu et al. reported a suppression of ATP-citrate lyase expression and activity by palmitate and its critical role in pancreatic β-cell survival based on increased β-cell apoptosis in knockdown mutation [66]. A liver specific ATP-citrate lyase abrogation improved the systemic glucose metabolism in leptin receptor deficient mice [67]. Moreover, human platelets showed increased ATP-citrate lyase activity in diabetic subjects [68]. Furthermore, a decrease of adipose tissue and plasma insulin concentration were observed in diabetic mice fed with citric acid [69]. In addition, reduced urinary citrate excretion induced by insulin resistance may mediate the development of kidney stones in patients with the metabolic syndrome [70].
Taken together, the metabolites identified so far were reported to be associated with type 2 diabetes and fasting plasma glucose in numerous studies. The metabolites of our pattern seem to be representatives of an early stage of a perturbed energy metabolism. This is reflected by metabolites that are among other pathways closely linked to the TCA cycle as well as by metabolites of the purine degradation. The metabolites linked to the TCA cycle are also involved in many other pathways. Thus, they might be metabolic hubs that transfer the perturbation in energy metabolism to other pathways (e.g. amino acid metabolism) or vice versa. Subsequently, this perturbation transfer results in the complexity of fasting plasma glucose development. Furthermore, redox homeostasis seems to be involved in this development.
For a complete interpretation of our results the identification of the unknowns is desirable. However, the problem of a high number of unknown metabolites is still an intrinsic problem of metabolite profiling methods. In fact, many studies do not even consider the unknowns to circumvent this problem. However, our study demonstrates the potential importance of such unknowns, even if a complete identification is beyond the scope of this manuscript.
The identified metabolic pattern includes metabolites that are not part of a single pathway, but spread over several pathways. We conclude that glucose level development is comparable to a complex 'clockwork' with multiple key regulators. Small perturbations to single regulators can be buffered by the system, whereas bigger perturbation at several parts disturb the fine tuned balance and lead to elevated blood glucose levels. We interpret the metabolites that are part of the identified metabolic pattern as representatives for the regulatory parts of the 'clockwork'.
This complexity reflects the diabetes research results made on the genome level during the past years: no single gene, but rather a high number of genes are associated with type 2 diabetes [71-73] and fasting glucose development [74].
Finally, this complexity is the challenge for development of individualised medicine. For proper treatment, all disturbed metabolic regulators have to be identified and brought back to normal parameters.
We have mentioned above a recent study investigating the ability of single metabolites measured using LC-MS to predict type 2 diabetes and finally identifying five amino acids [11]. Although their approach is similar in style to ours, there are some differences that make a direct comparison delicate. These differences concern the technical platforms to measure metabolite levels, the number of metabolites measured, the number of patients, observation period, the prediction endpoints and the statistical approaches. Thus, further studies are needed to confirm the results.
Finally, to comment on the practical value of our work: We interpret our results as a first hint that complex metabolite pattern can predict clinical relevant endpoints and thus reveal new insights in molecular mechanisms at early stages of disease development. With respect to diabetes our results need to be confirmed in external cohorts before any application in a clinical setting.
Conclusions
Taken together, our results indicate that specific metabolites are able to predict development of fasting glucose. However, this prediction is primarily driven by a complex pattern of metabolites rather than single metabolites, demonstrating the interrelation between different metabolic pathways in the regulation of circulating metabolite profiles and in the development of type 2 diabetes.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
MH and AL contributed equally to this manuscript and participated in study design, coordination, statistical analysis, interpretation of the results, discussion and preparation of the manuscript. FS participated in acquisition of the MESY-BEPO data and discussion. AFR participated in acquisition of the MESY-BEPO data and discussion. TB participated in acquisition of the MESY-BEPO data and discussion. AA participated in acquisition of the MESY-BEPO data and discussion. GSC participated in acquisition of GC-MS data and discussion. AFHP participated in acquisition of the MESY-BEPO data and discussion. LW participated in acquisition of GC-MS data and discussion. JSE participated in study design, interpretation of the results, discussion and helped to prepare the manuscript. JSP participated in study design, interpretation of the results, discussion and helped to prepare the manuscript. All authors read and approved the final manuscript.
Contributor Information
Manuela Hische, Email: manuela.hische@charite.de.
Abdelhalim Larhlimi, Email: Larhlimi@mpimp-golm.mpg.de.
Franziska Schwarz, Email: franziska.schwarz@charite.de.
Antje Fischer-Rosinský, Email: antje.fischer@charite.de.
Thomas Bobbert, Email: Thomas.Bobbert@charite.de.
Anke Assmann, Email: anke.assmann@charite.de.
Gareth S Catchpole, Email: gareth.catchpole@metanomics.de.
Andreas FH Pfeiffer, Email: andreas.pfeiffer@charite.de.
Lothar Willmitzer, Email: Willmitzer@mpimp-golm.mpg.de.
Joachim Selbig, Email: Selbig@mpimp-golm.mpg.de.
Joachim Spranger, Email: joachim.spranger@charite.de.
Acknowledgements
The authors like to thank Änne Eckhardt and Gudrun Wolter for their support on the GC-MS measurements, Nadine Huckauf and Katrin Sprengel for their assistance in the glucose measurements and Kristen Feher for revising the manuscript.
AFHP and JSP were supported by a Clinical Research Group (KFO218/1) of the Deutsche Forschungs-gemeinschaft (DFG). JSP was funded by a Heisenberg-Professorship (SP716/2-1) and a research group on molecular nutrition of the Bundesministerium für Bildung und Forschung (BMBF).
References
- Lopez AD, Mathers CD, Ezzati M, Jamison DT, Murray CJL, editor. Global Burden of Disease and Risk Factors. A copublication of The World Bank and Oxford University Press; 2006. http://files.dcp2.org/pdf/GBD/GBD.pdf [Google Scholar]
- von Eckardstein A, Schulte H, Assmann G. Risk for diabetes mellitus in middle-aged Caucasian male participants of the PROCAM study: implications for the definition of impaired fasting glucose by the American Diabetes Association. J Clin Endocrinol Metab. 2000;85(9):3101–8. doi: 10.1210/jc.85.9.3101. [DOI] [PubMed] [Google Scholar]
- Lindström J, Tuomilehto J. The diabetes risk score: a practical tool to predict type 2 diabetes risk. Diabetes Care. 2003;26(3):723–731. doi: 10.2337/diacare.26.3.725. [DOI] [PubMed] [Google Scholar]
- Lin X, Song K, Lim N, Yuan X, Johnson T, Abderrahmani A, Vollenweider P, Stirnadel H, Sundseth S, Lai E, Burns D, Middleton L, Roses A, Matthews P, Waeber G, Cardon L, Waterworth D, Mooser V. Risk prediction of prevalent diabetes in a Swiss population using a weighted genetic score - the CoLaus Study. Diabetologia. 2009;52(4):600–608. doi: 10.1007/s00125-008-1254-y. [DOI] [PubMed] [Google Scholar]
- Hische M, Luis-Dominguez O, Pfeiffer AF, Schwarz PE, Selbig J, Spranger J. Decision Trees as a simple-to-use and reliable tool to identify individuals with impaired glucose metabolism or type 2 diabetes mellitus. Eur J Endocrinol. 2010;163(4):565–571. doi: 10.1530/EJE-10-0649. [DOI] [PubMed] [Google Scholar]
- Denkert C, Budczies J, Kind T, Weichert W, Tablack P, Sehouli J, Niesporek S, Könsgen D, Dietel M, Fiehn O. Mass Spectrometry-Based Metabolic Profiling Reveals Different Metabolite Patterns in Invasive Ovarian Carcinomas and Ovarian Borderline Tumors. Cancer Res. 2006;66(22):10795–10804. doi: 10.1158/0008-5472.CAN-06-0755. http://cancerres.aacrjournals.org/content/66/22/10795.abstract [DOI] [PubMed] [Google Scholar]
- Denkert C, Budczies J, Weichert W, Wohlgemuth G, Scholz M, Kind T, Niesporek S, Noske A, Buckendahl A, Dietel M, Fiehn O. Metabolite profiling of human colon carcinoma - deregulation of TCA cycle and amino acid turnover. Mol Cancer. 2008;7:72. doi: 10.1186/1476-4598-7-72. http://www.molecular-cancer.com/content/7/1/72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukaetova-Ladinska EB, Monteith R, Perry EK. Cerebrospinal Fluid Biomarkers for Dementia with Lewy Bodies. Int J Alzheimers Dis. 2010;2010:17. doi: 10.4061/2010/536538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilvo M, Denkert C, Lehtinen L, Muller B, Brockmoller S, Seppanen-Laakso T, Budczies J, Bucher E, Yetukuri L, Castillo S, Berg E, Nygren H, Sysi-Aho M, Griffin JL, Fiehn O, Loibl S, Richter-Ehrenstein C, Radke C, Hyotylainen T, Kallioniemi O, Iljin K, Oresic M. Novel theranostic opportunities offered by characterization of altered membrane lipid metabolism in breast cancer progression. Cancer Res. 2011. http://cancerres.aacrjournals.org/content/early/2011/03/16/0008-5472.CAN-10-3894.abstract [DOI] [PubMed]
- Catchpole G, Platzer A, Weikert C, Kempkensteffen C, Johannsen M, Krause H, Jung K, Miller K, Willmitzer L, Selbig J, Weikert S. Metabolic profiling reveals key metabolic features of renal cell carcinoma. J Cell Mol Med. 2011;15:109–118. doi: 10.1111/j.1582-4934.2009.00939.x. http://dx.doi.org/10.1111/j.1582-4934.2009.00939.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C, O'Donnell CJ, Carr SA, Mootha VK, Florez JC, Souza A, Melander O, Clish CB, Gerszten RE. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011. http://www.nature.com/nm/journal/vaop/ncurrent/full/nm.2307.html [DOI] [PMC free article] [PubMed]
- Fischer A, Fisher E, Möhlig M, Schulze M, Hoffmann K, Weickert MO, Schueler R, Osterhoff M, Pfeiffer AF, Boeing H, Spranger J. KCNJ11 E23K Affects Diabetes Risk and Is Associated With the Disposition Index. Diabetes Care. 2008;31:87–89. doi: 10.2337/dc07-1157. http://care.diabetesjournals.org/content/31/1/87.short [DOI] [PubMed] [Google Scholar]
- Hummel J, Selbig J, Walther D, Kopka J. In: Metabolomics, Volume 18 of Topics in Current Genetics. Nielsen J, Jewett M, editor. Springer Berlin/Heidelberg; 2007. The Golm Metabolome Database: a database for GC-MS based metabolite profiling; pp. 75–95.http://dx.doi.org/10.1007/4735_2007_0229 [Google Scholar]
- Cuadros-Inostroza A, Caldana C, Redestig H, Kusano M, Lisec J, Peña Cortés H, Willmitzer L, Hannah M. TargetSearch - a Bioconductor package for the efficient preprocessing of GC-MS metabolite profiling data. BMC Bioinformatics. 2009;10:1–12. doi: 10.1186/1471-2105-10-1. http://dx.doi.org/10.1186/1471-2105-10-428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc. 2006;1:387–396. doi: 10.1038/nprot.2006.59. [DOI] [PubMed] [Google Scholar]
- Breiman L. Random Forests. Mach Learn. 2001;45:5–32. doi: 10.1023/A:1010933404324. http://dx.doi.org/10.1023/A:1010933404324 [DOI] [Google Scholar]
- Liaw A, Wiener M. Classification and Regression by randomForest. R News. 2002;2(3):18–22. http://CRAN.R-project.org/doc/Rnews/Rnews_2002-3.pdf [Google Scholar]
- Svetnik V, Liaw A, Tong C, Wang T. In: Multiple classifier systems, proceedings, Volume 3077 of Lecture notes in computer science. Roli, Fabio, Kittler, Josef, Windeatt, editor. Terry; 2004. Application of Breiman's random forest to modeling structure-activity relationships of pharmaceutical molecules; pp. 334–343. 5th International Workshop on Multiple Classifier Systems, Cagliari, ITALY, JUN 09-SEP 11, 2004. [Google Scholar]
- Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning. Springer; 2009. [Google Scholar]
- Meloun M, Militky J, Hill M, Brereton RG. Crucial problems in regression modelling and their solutions. Analyst. 2002;127:433–450. doi: 10.1039/b110779h. http://dx.doi.org/10.1039/B110779H [DOI] [PubMed] [Google Scholar]
- Breiman L. Statistical modeling: The two cultures. Statistical Science. 2001;16(3):199–215. [Google Scholar]
- Hummel J, Strehmel N, Selbig J, Walther D, Kopka J. Decision tree supported substructure prediction of metabolites from GC-MS profiles. Metabolomics. 2010;6(2):322–333. doi: 10.1007/s11306-010-0198-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama S, Saito K, Yachi Y, Asumi M, Sugawara A, Totsuka K, Saito A, Sone H. Association Between Serum Uric Acid and Development of Type 2 Diabetes. Diabetes Care. 2009;32(9):1737–1742. doi: 10.2337/dc09-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister R, Barnes D, Luben R, Forouhi NG, Bochud M, Khaw KT, Wareham NJ, Langenberg C. No evidence for a causal link between uric acid and type 2 diabetes: a Mendelian randomisation approach. Diabetologia. 2011;54(10):2561–2569. doi: 10.1007/s00125-011-2235-0. [DOI] [PubMed] [Google Scholar]
- Fox I. Metabolic basis for disorders of purine nucleotide degradation. Metabolism-Clinical and Experimental. 1981;30(6):616–634. doi: 10.1016/0026-0495(81)90142-6. [DOI] [PubMed] [Google Scholar]
- Tsouli SG, Liberopoulos EN, Mikhailidis DP, Athyros VG, Elisaf MS. Elevated serum uric acid levels in metabolic syndrome: an active component or an innocent bystander? Metabolism-Clinical and Experimental. 2006;55(10):1293–1301. doi: 10.1016/j.metabol.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Harkness R. Hypoxanthine, Xanthine and Uridine in Body-Fluids, Indicators of ATP Depletion. Journal of Chromatography-Biomedical Applications. 1988;429:255–278. doi: 10.1016/s0378-4347(00)83873-6. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Moriwaki Y, Takahashi S, Tsutsumi Z, Yamakita J, Nakano T, Higashino K. Effect of glucose on the plasma concentration and urinary excretion of uridine and purine bases. Metabolism-Clinical and Experimental. 1999;48(3):338–341. doi: 10.1016/S0026-0495(99)90082-3. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Hu H, Zharikov S, Tuttle K, Short R, Glushakova O, Ouyang X, Feig D, Block E, Herrera-Acosta J, Patel J, Johnson R. A causal role for uric acid in fructose-induced metabolic syndrome. American Journal of Physiology-Renal Physiology. 2006;290(3):F625–F631. doi: 10.1152/ajprenal.00140.2005. [DOI] [PubMed] [Google Scholar]
- Yeldandi A, Wang X, Alvares K, Kumar S, Rao M, Reddy J. Human Urate Oxidase Gene - Cloning and Partial Sequence-Analysis Reveal a Stop Codon Within the 5th Exon. Biochemical and Biophysical Research Communications. 1990;171(2):641–646. doi: 10.1016/0006-291X(90)91194-W. [DOI] [PubMed] [Google Scholar]
- Kim KM, Henderson GN, Frye RF, Galloway CD, Brown NJ, Segal MS, Imaram W, Angerhofer A, Johnson RJ. Simultaneous determination of uric acid metabolites allantoin, 6-aminouracil, and triuret in human urine using liquid chromatography-mass spectrometry. Journal of Chromatography B-Analytical Technologies in the Biomedical and Life Sciences. 2009;877(1-2):65–70. doi: 10.1016/j.jchromb.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Jin X, Lam CWK, Yan SK. Oxidative stress and diabetes mellitus. Clinical Chemistry and Laboratory Medicine. 2011;49(11):1773–1782. doi: 10.1515/CCLM.2011.250. [DOI] [PubMed] [Google Scholar]
- Meister A. Glutathione - Metabolism and Function via gamma-Glutamyl Cycle. Life Sciences. 1974;15(2):177–190. doi: 10.1016/0024-3205(74)90206-9. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Uys JD, Tew KD, Townsend DM. S-Glutathionylation: From Molecular Mechanisms to Health Outcomes. Antioxidants & Redox Signaling. 2011;15(1):233–270. doi: 10.1089/ars.2010.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhailidis DP, Lioudaki E, Ganotakis ES. Liver enzymes: potential cardiovascular risk markers? Current pharmaceutical design. 2011;L17(33):3632–43. doi: 10.2174/138161211798220945. [DOI] [PubMed] [Google Scholar]
- Wu J, Yan Wh, Qiu L, Chen Xq, Guo Xz, Wu W, Xia Ly, Qin Xz, Liu Yh, Ding Ht, Han Sm, Xu Cl, Zhu Gj. High prevalence of coexisting prehypertension and prediabetes among healthy adults in northern and northeastern China. BMC Public Health. 2011;11 doi: 10.1186/1471-2458-11-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Xu M, Huang Y, Wang T, Li M, Wu Y, Song A, Li X, Bi Y, Ning G. Elevated serum gamma-glutamyltransferase predicts the development of impaired glucose metabolism in middle-aged and elderly Chinese. Endocrine. 2011;40(2):265–272. doi: 10.1007/s12020-011-9468-z. [DOI] [PubMed] [Google Scholar]
- Succurro E, Arturi F, Grembiale A, Iorio F, Fiorentino TV, Andreozzi F, Sciacqua A, Hribal ML, Perticone F, Sesti G. One-hour post-load plasma glucose levels are associated with elevated liver enzymes. Nutrition Metabolism and Cardiovascular Diseases. 2011;21(9):713–718. doi: 10.1016/j.numecd.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Sun J, Ren J, Pang ZC, Gao WG, Nan HR, Wang SJ, Zhang L, Qian Q. The association of gamma-glutamyltransferase and C-reactive protein with IFG/IGT in Chinese adults in Qingdao, China. Clinica Chimica Acta. 2011;412(17-18):1658–1661. doi: 10.1016/j.cca.2011.05.019. [DOI] [PubMed] [Google Scholar]
- Rueckert IM, Heier M, Rathmann W, Baumeister SE, Doering A, Meisinger C. Association between Markers of Fatty Liver Disease and Impaired Glucose Regulation in Men and Women from the General Population: The KORA-F4-Study. Plos One. 2011;6(8) doi: 10.1371/journal.pone.0022932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evliyaoglu O, Kibrisli E, Yildirim Y, Gokalp O, Colpan L. Routine enzymes in the monitoring of type 2 diabetes mellitus. Cell Biochemistry and Function. 2011;29(6):506–512. doi: 10.1002/cbf.1779. [DOI] [PubMed] [Google Scholar]
- Bonnet F, Ducluzeau PH, Gastaldelli A, Laville M, Anderwald CH, Konrad T, Mari A, Balkan B, Grp RS. Liver Enzymes Are Associated With Hepatic Insulin Resistance, Insulin Secretion, and Glucagon Concentration in Healthy Men and Women. Diabetes. 2011;60(6):1660–1667. doi: 10.2337/db10-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y, Ahmad M, Chen S, Gillespie P, Le N, Mennona F, Mischke S, So SS, Wang H, Burghardt C, Tannu S, Conde-Knape K, Kochan J, Bolin D. Discovery of 1-arylcarbonyl-6,7-dimethoxyisoquinoline derivatives as glutamine fructose-6-phosphate amidotransferase (GFAT) inhibitors. Bioorganic & Medicinal Chemistry Letters. 2011;21(21):6264–6269. doi: 10.1016/j.bmcl.2011.09.009. [DOI] [PubMed] [Google Scholar]
- Nagata M, Suzuki W, Iizuka S, Tabuchi M, Maruyama H, Takeda S, Aburada M, Miyamo K. Type 2 diabetes mellitus in obese mouse model induced by monosodium glutamate. Experimental Animals. 2006;55(2):109–115. doi: 10.1538/expanim.55.109. [DOI] [PubMed] [Google Scholar]
- Cameron D, Poon T, Smith G. Effects of Monosodium Glutamate Administration in Neonatal-Period on Diabetic Syndrome in KK Mice. Diabetologia. 1976;12(6):621–626. doi: 10.1007/BF01220641. [DOI] [PubMed] [Google Scholar]
- Iwase M, Yamamoto M, Iino K, Ichikawa K, Shinohara N, Yoshinari M, Fujishima M. Obesity induced by neonatal monosodium glutamate treatment in spontaneously hypertensive rats: An animal model of multiple risk factors. Hypertension Research-Clinical and Experimental. 1998;21(1):1–6. doi: 10.1291/hypres.21.1. [DOI] [PubMed] [Google Scholar]
- Komeda K, Yokote M, Oki Y. Diabetic Syndrome in the Chinese-Hamster Induced with Monosodium Glutamate. Experientia. 1980;36(2):232–234. doi: 10.1007/BF01953751. [DOI] [PubMed] [Google Scholar]
- Nakajima H, Tochino Y, Fujinokurihara H, Yamada K, Gomi M, Tajima K, Kanaya T, Miyazaki A, Miyagawa J, Hanafusa T, Mashita K, Kono N, Moriwaki K, Nonaka K, Tarui S. Decreased Incidence of Diabetes-Mellitus by Monosodium Glutamate in the non-Obese Diabetic (NOD) Mouse. Research Communications in Chemical Pathology and Pharmacology. 1985;50(2):251–257. [PubMed] [Google Scholar]
- Lobato NS, Filgueira FP, Akamine EH, Davel APC, Rossoni LV, Tostes RC, Carvalho MHC, Fortes ZB. Obesity induced by neonatal treatment with monosodium glutamate impairs microvascular reactivity in adult rats: Role of NO and prostanoids. Nutrition Metabolism and Cardiovascular Diseases. 2011;21(10):808–816. doi: 10.1016/j.numecd.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Collison KS, Zaidi MZ, Saleh SM, Inglis A, Mondreal R, Makhoul NJ, Bakheet R, Burrows J, Milgram NW, Al-Mohanna FA. Effect of trans-fat, fructose and monosodium glutamate feeding on feline weight gain, adiposity, insulin sensitivity, adipokine and lipid profile. British Journal of Nutrition. 2011;106(2):218–226. doi: 10.1017/S000711451000588X. [DOI] [PubMed] [Google Scholar]
- Samocha-Bonet D, Wong O, Synnott EL, Piyaratna N, Douglas A, Gribble FM, Holst JJ, Chisholm DJ, Greenfield JR. Glutamine Reduces Postprandial Glycemia and Augments the Glucagon-Like Peptide-1 Response in Type 2 Diabetes Patients. Journal of Nutrition. 2011;141(7):1233–1238. doi: 10.3945/jn.111.139824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkman HO. Circumstantial evidence for a role of glutamine-synthetase in suicide. Medical Hypotheses. 2011;76(6):905–907. doi: 10.1016/j.mehy.2011.03.005. [DOI] [PubMed] [Google Scholar]
- Nakazawa T, Shimura M, Ryu M, Nishida K, Pages G, Pouyssegur J, Endo S. ERK1 plays a critical protective role against N-methyl-D-aspartate-induced retinal injury. Journal of Neuroscience Research. 2008;86(1):136–144. doi: 10.1002/jnr.21472. [DOI] [PubMed] [Google Scholar]
- Abu Fanne R, Nassar T, Heyman SN, Hijazi N, Higazi AAR. Insulin and glucagon share the same mechanism of neuroprotection in diabetic rats: role of glutamate. American Journal of Physiology-Regulatory Integrative and Comparative Physiology. 2011;301(3):R668–R673. doi: 10.1152/ajpregu.00058.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousova I, Bakala H, Chudacek R, Palicka V, Drsata J. Glycation-induced inactivation of aspartate aminotransferase, effect of uric acid. Molecular and Cellular Biochemistry. 2005;278(1-2):85–92. doi: 10.1007/s11010-005-6933-0. [DOI] [PubMed] [Google Scholar]
- Hageman R, Severijnen C, van de Heijning BJM, Bouritius H, van Wijk N, van Laere K, van der Beek EM. A specific blend of intact protein rich in aspartate has strong postprandial glucose attenuating properties in rats. Journal of Nutrition. 2008;138(9):1634–1640. doi: 10.1093/jn/138.9.1634. [DOI] [PubMed] [Google Scholar]
- Arai T, Sasaki M, Shiomi M, Nonaka T, Ochiai K, Oki Y. Alterations in Acetyl Coenzyme-A Carboxylase Activities in Voles and Mice Treated with Monosodium Aspartate. Journal of Veterinary Medical Science. 1992;54(1):131–135. doi: 10.1292/jvms.54.131. [DOI] [PubMed] [Google Scholar]
- Large V, Beylot M. Modifications of citric acid cycle activity and gluconeogenesis in streptozotocin-induced diabetes and effects of metformin. Diabetes. 1999;48(6):1251–1257. doi: 10.2337/diabetes.48.6.1251. [DOI] [PubMed] [Google Scholar]
- Ortenblad N, Mogensen M, Petersen I, Hojlund K, Levin K, Sahlin K, Beck-Nielsen H, Gaster M. Reduced insulin-mediated citrate synthase activity in cultured skeletal muscle cells from patients with type 2 diabetes: Evidence for an intrinsic oxidative enzyme defect. Biochimica et Biophysica Acta-Molecular Basis of Disease. 2005;1741(1-2):206–214. doi: 10.1016/j.bbadis.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Liu Y, Tornheim K, Leahy J. Shared biochemical properties of glucotoxicity and lipotoxicity in islets decrease citrate synthase activity and increase phosphofructokinase activity. Diabetes. 1998;47(12):1889–1893. doi: 10.2337/diabetes.47.12.1889. [DOI] [PubMed] [Google Scholar]
- Gnoni GV, Priore P, Geelen MJH, Siculella L. The Mitochondrial Citrate Carrier: Metabolic Role and Regulation of its Activity and Expression. IUBMB Life. 2009;61(10):987–994. doi: 10.1002/iub.249. [DOI] [PubMed] [Google Scholar]
- Gnoni GV, Giudetti AM, Mercuri E, Damiano F, Stanca E, Priore P, Siculella L. Reduced Activity and Expression of Mitochondrial Citrate Carrier in Streptozotocin-Induced Diabetic Rats. Endocrinology. 2010;151(4):1551–1559. doi: 10.1210/en.2009-1352. [DOI] [PubMed] [Google Scholar]
- Birkenfeld AL, Lee HY, Guebre-Egziabher F, Alves TC, Jurczak MJ, Jornayvaz FR, Zhang D, Hsiao JJ, Martin-Montalvo A, Fischer-Rosinsky A, Spranger J, Pfeiffer AF, Jordan J, Fromm MF, Koenig J, Lieske S, Carmean CM, Frederick DW, Weismann D, Knauf F, Irusta PM, De Cabo R, Helfand SL, Samuel VT, Shulman GI. Deletion of the Mammalian INDY Homo log Mimics Aspects of Dietary Restriction and Protects against Adiposity and Insulin Resistance in Mice. Cell Metabolism. 2011;14(2):184–195. doi: 10.1016/j.cmet.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan NM, Longacre MJ, Ahmed MS, Kendrick MA, Gu H, Ostenson CG, Fukao T, MacDonald MJ. Lower succinyl-CoA:3-ketoacid-CoA transferase (SCOT) and ATP citrate lyase in pancreatic islets of a rat model of type 2 diabetes: Knockdown of SCOT inhibits insulin release in rat insulinoma cells. Archives of Biochemistry and Biophysics. 2010;499(1-2):62–68. doi: 10.1016/j.abb.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SW, Kim KS, Whang SK, Kim JS, Kim YS. Induction of hepatic ATP-citrate lyase by insulin in diabetic rat-effects of insulin on the contents of enzyme and its mRNA in cytosol, and the transcriptional activity in nuclei. Yonsei medical journal. 1994;35(1):25–33. doi: 10.3349/ymj.1994.35.1.25. [DOI] [PubMed] [Google Scholar]
- Chu KY, Lin Y, Hendel A, Kulpa JE, Brownsey RW, Johnson JD. ATP-Citrate Lyase Reduction Mediates Palmitate-induced Apoptosis in Pancreatic Beta Cells. Journal of Biological Chemistry. 2010;285(42):32606–32615. doi: 10.1074/jbc.M110.157172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Jiang L, Wang J, Li S, Yu Y, You J, Zeng R, Gao X, Rui L, Li W, Liu Y. Abrogation of Hepatic ATP-Citrate Lyase Protects Against Fatty Liver and Ameliorates Hyperglycemia in Leptin Receptor-Deficient Mice. Hepatology. 2009;49(4):1166–1175. doi: 10.1002/hep.22774. [DOI] [PubMed] [Google Scholar]
- Michno A, Skibowska A, Raszeja-Specht A, Cwikowska J, Szutowicz A. The role of adenosine triphosphate-citrate lyase in the metabolism of acetyl-coenzyme A and function of blood platelets in diabetes mellitus. Metabolism-Clinical and Experimental. 2004;53(1):66–72. doi: 10.1016/j.metabol.2003.07.012. [DOI] [PubMed] [Google Scholar]
- Muroyama K, Murosaki S, Yamamoto Y, Odaka H, Chung H, Miyoshi M. Anti-obesity effects of a mixture of thiamin, arginine, caffeine, and citric acid in non-insulin dependent diabetic KK mice. Journal of Nutritional Science and Vitaminology. 2003;49(1):56–63. doi: 10.3177/jnsv.49.56. [DOI] [PubMed] [Google Scholar]
- Cupisti A, Meola M, D'Alessandro C, Bernabini G, Pasquali E, Carpi A, Barsotti G. Insulin resistance and low urinary citrate excretion in calcium stone formers. Biomedicine & Pharmacotherapy. 2007;61(1):86–90. doi: 10.1016/j.biopha.2006.09.012. [DOI] [PubMed] [Google Scholar]
- McCarthy MI. Genomic Medicine Genomics, Type 2 Diabetes, and Obesity. New England Journal of Medicine. 2010;363(24):2339–2350. doi: 10.1056/NEJMra0906948. [DOI] [PubMed] [Google Scholar]
- Froguel P Genetics of obesity and type 2 diabetes Diabetes Obesity & Metabolism 2010121522384427 [Google Scholar]
- Lyssenko V, Jonsson A, Almgren P, Pulizzi N, Isomaa B, Tuomi T, Berglund G, Altshuler D, Nilsson P, Groop L. Clinical Risk Factors, DNA Variants, and the Development of Type 2 Diabetes. New England Journal of Medicine. 2008;359(21):2220–2232. doi: 10.1056/NEJMoa0801869. [DOI] [PubMed] [Google Scholar]
- Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL, Lindgren CM, Magi R, Morris AP, Randall J, Johnson T, Elliott P, Rybin D, Thorleifsson G, Steinthorsdottir V, Henneman P, Grallert H, Dehghan A, Hottenga JJ, Franklin CS, Navarro P, Song K, Goel A, Perry JRB, Egan JM, Lajunen T, Grarup N. et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nature Genetics. 2010;42(2):105–U32. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]