Abstract
The proteolysis of collagen triple-helical structure (collagenolysis) is a poorly understood yet critical physiological process. Presently, matrix metalloproteinase 1 (MMP-1) and collagen triple-helical peptide models have been utilized to characterize the events and calculate the energetics of collagenolysis via NMR spectroscopic analysis of 12 enzyme-substrate complexes. The triple-helix is bound initially by the MMP-1 hemopexin-like (HPX) domain via a four amino acid stretch (analogous to type I collagen residues 782–785). The triple-helix is then presented to the MMP-1 catalytic (CAT) domain in a distinct orientation. The HPX and CAT domains are rotated with respect to one another compared with the X-ray “closed” conformation of MMP-1. Back-rotation of the CAT and HPX domains to the X-ray closed conformation releases one chain out of the triple-helix, and this chain is properly positioned in the CAT domain active site for subsequent hydrolysis. The aforementioned steps provide a detailed, experimentally-derived, and energetically favorable collagenolytic mechanism, as well as significant insight into the roles of distinct domains in extracellular protease function.
Keywords: protease, matrix metalloproteinase, triple-helix, collagen, collagenolysis
Introduction
Collagens are the most abundant proteins in the human body and the main components of the extracellular matrix. The biophysical properties of collagens are related to their unique molecular structure that consists of three polyproline II-like chains, staggered by one residue and coiled to form a triple-helix1,2. Triple-helical structure provides collagens with exceptional mechanical strength, broad resistance to the proteolytic enzymes, and a distinct topology for protein-protein interactions3. The proteolysis of collagen is integral for numerous physiological functions including morphogenesis, tissue remodeling, and wound healing, and has been recognized as a contributing factor to multiple pathologies.
MMPs are a family of zinc-dependent proteolytic enzymes with the ability to catalyze the degradation of extracellular matrix components, including collagens. The collagenolytic MMPs are minimally composed of a CAT and an HPX domain connected by a linker of variable length4. Although mechanisms by which MMPs catalyze the hydrolysis of collagen have been postulated5–13, they are often contradictory and not validated on a molecular level. Understanding the roles of the individual domains and the molecular interactions that facilitate MMP catalyzed collagenolysis is still a scientific challenge.
In a broader context, a great variety of metallopeptidases are multidomain14, with little knowledge as to how domains cooperate to enhance proteolysis of macromolecular substrates. The present study has exploited advanced NMR approaches to document the interactions between MMP-1 and collagen-model triple-helical peptide (THP) substrates. The THPs encompass the MMP-1 cleavage site (residues 772–786) in the α1 chain of type I collagen (see Methods). To examine MMP-THP complexes, prodomain free full-length (FL) MMP-1 with a cadmium ion replacing the active site zinc and Glu219 mutated to Ala was utilized. This MMP-1 was unable to degrade the THP and therefore permitted examination of the interaction between the two species and any intermediates formed prior to catalysis. In addition, the interactions of the individual CAT and HPX domains with THPs were studied. Ultimately, the accumulated structural data was used to develop the first detailed mechanism for the collagenolytic process, and the energetics of this mechanism was evaluated explicitly.
Materials and Methods
Peptide synthesis and analysis
The α1(I)772–786 THP was assembled using Fmoc solid-phase chemistry by methods described previously15,16. 13C,15N-labeled Fmoc-amino acids were purchased from Cambridge Isotope Laboratories. Fmoc-Gln was prepared from 13C,15N-labeled Gln (Cambridge) as described17, and was coupled using 4-fold excesses of amino acid, N,N’-dicyclohexylcarbodiimide, and N-hydroxybenzotriazole. All other 13C,15N-labeled amino acids were coupled using 1.1-fold excesses of amino acid, 1-fold excess 2-(6-chloro-1H-benzotriazole-1-yl)-1,1,3,3-tetramethylaminium hexafluorophosphate (HCTU), and 2-fold excess N-methylmorpholine (NMM). All unlabeled amino acids were coupled using 4-fold excesses of amino acid, 3.9-fold excess HCTU, and 8-fold excess NMM. The couplings of 13C,15N-labeled amino acids were followed by double coupling of unlabeled amino acids. The sequence of α1(I)772–786 THP is as follows, with the labeled residues underlined and in bold:
Preparation of protein samples
The cDNA encoding for full-length MMP-1 (N106-N469) (FL-MMP-1 hereafter), the CAT domain of MMP-1 (N106-G261), and the HPX domain (T274-N469) was amplified from truclone c-dna (origene) by polymerase chain reaction (PCR) and cloned into the pET21 (Novagen) expression vector using NdeI and XhoI (New England BioLabs) as restriction enzymes. The recombinant vector was transformed into Escherichia coli strain BL21(DE3) Codon Plus RIPL, and colonies were selected for ampicillin and chloramphenicol resistance. The bacteria were grown in LB media. When a cell density corresponding to 0.6 A was reached, the expression of the protein was induced by adding 0.5 mM of isopropyl-β-d-thiogalactoside (IPTG) and the incubation at 37°C was continued for another 5 h. The full-length protein as well as the isolated domains precipitated in the inclusion bodies and these were solubilized, after lysis of the cells, in a solution of 8 M urea, 20 mM dithiotreithol (DTT), and 20 mM Tris–HCl (pH 8.2). The solubilized FL-MMP-1 and HPX were diluted with a buffer containing 6 M urea, 10 mM CaCl2, 0.1 mM ZnCl2, 20 mM cysteamine, and 20 mM Tris–HCl (pH 8) and refolded by using a multi-step dialysis against solutions containing 50 mM Tris–HCl (pH 7.4), 4 M urea, 10 mM CaCl2, 0.1 mM ZnCl2, 5 mM β-mercaptoethanol, and 1 mM 2-hydroxyethyl disulfide, then against a solution containing 50 mM Tris–HCl (pH 7.2), 2 M urea, 10 mM CaCl2, 0.1 mM ZnCl2, and 0.15 M NaCl, and then against the same solution without urea and with 0.3 mM CdCl2 instead of ZnCl2. The protein was purified by size exclusion chromatography on the HiLoad 26/60 Superdex 75 pg (Amersham). For the expression of 13C- and 15N-enriched FL-MMP-1 and HPX, bacteria were grown in minimal medium containing 15N-enriched (NH4)2SO4 and 13C-enriched glucose (Cambridge Isotope Laboratories)18. Samples of 2H,13C,15N-enriched FL-MMP-1 protein were obtained by adapting E. coli cells in medium with different percentages of deuterium, until 100% was reached, then growing the cells in OD2 Silantes media CDN. The inactive mutants E219A of FL-MMP-1 and CAT were produced using the Quick-Change site-directed mutagenesis kit (Qiagen), and the expression and purification of the protein and of its 15N- and 13C,15N-enriched versions completed using the same procedure described above. Samples of cadmium(II) substituted FL-MMP-1 and CAT proteins were prepared by exhaustive dialysis against a buffer containing 50 mM HEPES (pH 6.8), 10 mM CaCl2, 0.15 M NaCl, and 1 mM CdCl2.
NMR measurements
The experiments for assignment and mobility measurements of the THP were performed on peptide samples at concentrations ranging between 0.2 and 0.8 mM in the same buffer as FL-MMP-1. For FL-MMP-1, CAT, and HPX, all NMR experiments were performed on samples at a concentration of 0.2 mM. In all titrations with the THP, the THP was substoichiometric with respect to the protein to avoid excessive broadening in the NMR experiments. The binding of the THP to the labeled FL-MMP-1 was evaluated by adding increasing amounts of THP to 0.2 mM protein. The final concentration of the THP was 0.08 mM, providing a FL-MMP-1:THP ratio of 1:0.4. The Kd value for binding of the THP to FL-MMP-1 is 0.92 µM19 and under these experimental conditions the THP is 99% bound to FL-MMP-1. To monitor the effects of the THP on the HPX domain, a 0.2 mM solution of protein was titrated with increasing amounts of THP up to a final concentration of THP of 0.16 mM with a HPX:THP ratio of 1:0.8. The Kd value for binding of the THP to the HPX domain is 10.9 µM19 and under these experimental conditions the THP is 85% bound to the HPX domain.
NMR experiments were performed at 298 K and 310 K and acquired on Bruker AVANCE spectrometers operating at 500, 700, 800, and 900 MHz and equipped with triple resonance cryoprobes. All spectra were processed with the Bruker TOPSPIN software packages and analyzed by the program CARA (Computer Aided Resonance Assignment, ETH Zürich)20. The backbone resonance assignment was obtained by the analysis of HNCA21 and HNCACB22 spectra acquired at 900 MHz and by the analysis of HNCO23, HN(CA)CO24, and CBCA(CO)NH25 acquired at 500 MHz. Protonless experiments, CON26, CACO26, CBCACO26, and CC-flopsy26 were performed at 700 MHz. The assignment of the aliphatic side chain resonances was performed through the analysis of 3D (H)CCH-TOCSY27 spectra acquired at 500 MHz, together with 3D 13C- and 15N- NOESY-HSQC28 spectra at 900 MHz. The 1H, 13C, and 15N chemical shifts have been deposited in BMRB under the accession number 18083.
The experiments for the determination of 15N longitudinal and transverse relaxation rates and 1H-15N NOE were recorded at 298 K and 700 MHz on 15N-enriched samples. The 15N longitudinal relaxation rates (R1) were measured using a sequence modified to remove cross correlation effects during the relaxation delay29. Inversion recovery times ranging between 2.5 and 2000 ms, with a recycle delay of 3.5 s, were used for the experiments. The 15N transverse relaxation rates (R2) were measured using a CPMG sequence30 with delays ranging between 8.5 and 1000 ms with a refocusing delay of 450 µs. The relaxation data are reported in Table S2.
THP structure calculation
Backbone dihedral angle constraints were obtained from 15N, 13C', 13Cα, 13Cβ, and Hα chemical shifts, using the TALOS prediction program31. Distance constraints for structure determination were obtained from 3D 15N- and 13C-NOESY-HSQC spectra. The crosspeak intensities were integrated using the integration routine implemented in CARA and converted into interatomic upper distance limits after calibration using the program CYANA. Automatic calibration was done by setting a distance of 4.6 Å for the median of the volumes. Then, CYANA32 was used to calculate a family of 600 structures of the central region of the THP starting from randomly generated conformers in 20,000 annealing steps. The 30 structures with the lowest Target Function values were selected to form a structure family. The solution structure statistics are reported in Table S9. Structure calculation statistics was provided by CYANA and the structural quality was evaluated using the program PROCHECK_NMR33. The structure was minimized using the program AMBER and deposited in PDB (PDB code: 2LLP).
MMP-1-THP structure calculations
The structure corresponding to the THP bound to the HPX domain was calculated using HADDOCK by imposing as “active residues” those experiencing in the THP the largest decrease in signal intensity and a water accessibility larger than 50% (1TV23, 1TV24, 1TG25, 1TL26, 2TV23, 2TV24, 2TG25, 2TL26, 3TV23, 3TV24, 3TG25, 3TL26) and HPX domain (R291, G292, E311, E313, N315, F316, I317, S318, V319, F320, Q323, N326). The solvent accessibility was calculated with the program NACCESS34. For calculating the structure of the FL-MMP-1 bound to the THP, active residues were those which experienced the largest decrease in signal intensity and water accessibility larger than 50% on the FL-MMP-1 and THP (N171, G178, N180, F207, R208, E209, H218, H222, H228, G233, Y237, S239, T241 for CAT; 1TP14, 1TQ15, 1TG16, 1TI17, 1TA18, 1TG19, 1TQ20, 1TR21, 1TV23, 1TV24, 1TG25, 1TL26, 2TV23, 2TV24, 2TG25, 2TL26, 3TV23, 3TV24, 3TG25, 3TL26 for THP; R291, G292, E311, E313, N315, F316, I317, S318, V319, F320, Q323, N326 for HPX).
Each calculation in HADDOCK consists of the following stages: randomization of orientations and rigid body minimization, semi-rigid simulated annealing in torsion angle space, and flexible solvent refinement where the structures obtained after the semi-rigid simulated annealing are refined with explicit solvent35. In each calculation the two MMP-1 domains were kept semi-rigid, allowing full flexibility of the linker. It should be noted that extensive docking calculations and simulations demonstrated that despite the linker being relatively long, only the 1T chain is accessible to the CAT domain once the HPX domain is bound to the 1T and 2T chains. Input files for docking calculations in HADDOCK were prepared from the PDB file of MMP-1 (PDB code: 2CLT) and from the member of the energy minimized family of 30 structures of THP closest to the average structure. Then, to generate the structural complexes reproducing the different stages of the THP unwinding, starting from the open/extended full length MMP-1, iterative docking calculations were performed by imposing a set of unambiguous restraints to selected residues (233, 241, 243, 247, 250, 271, 272, 300, 301, 316, 318, and 326) in order to induce a reorientation of the HPX and CAT domains toward the closed conformation of the X-ray structure.
The Crystallography & NMR System (CNS) potentials in HADDOCK were used to calculate the free energies of MMP-1-THP complexes. The first step was based on the experimentally determined, initial MMP-1-THP complex, and the free energy was set at 0 kcal/mol. The fourth step was based on the closed MMP-1 structure determined by X-ray crystallography36. The second and third steps were manually calculated. The energy of the four MMP-1-THP complexes were minimized as to draw a reasonable pathway based on the experimental restraints (the experimentally detected contacts of the THP with the HPX and the CAT domains) and to the reaction mechanism.
Results
Assignment and relaxation measurements of triple-helical structure
To assign residues in each individual THP chain, standard 3D spectra for backbone assignment were acquired on 13C,15N-enriched samples. A large number of signals corresponding to four distinct chains were identified and sequence-specific assigned (Figure 1). One of the four (ultimately designated 4M), accounting for about 30% of the total spectral intensity, displayed sharper signals and a distinct pattern of resonances with respect to the other three chains (designated 1T, 2T, and 3T). Carbon direct detection experiments37 permitted the full assignment of the labeled amino acids in each peptide chain despite overlapping signals.
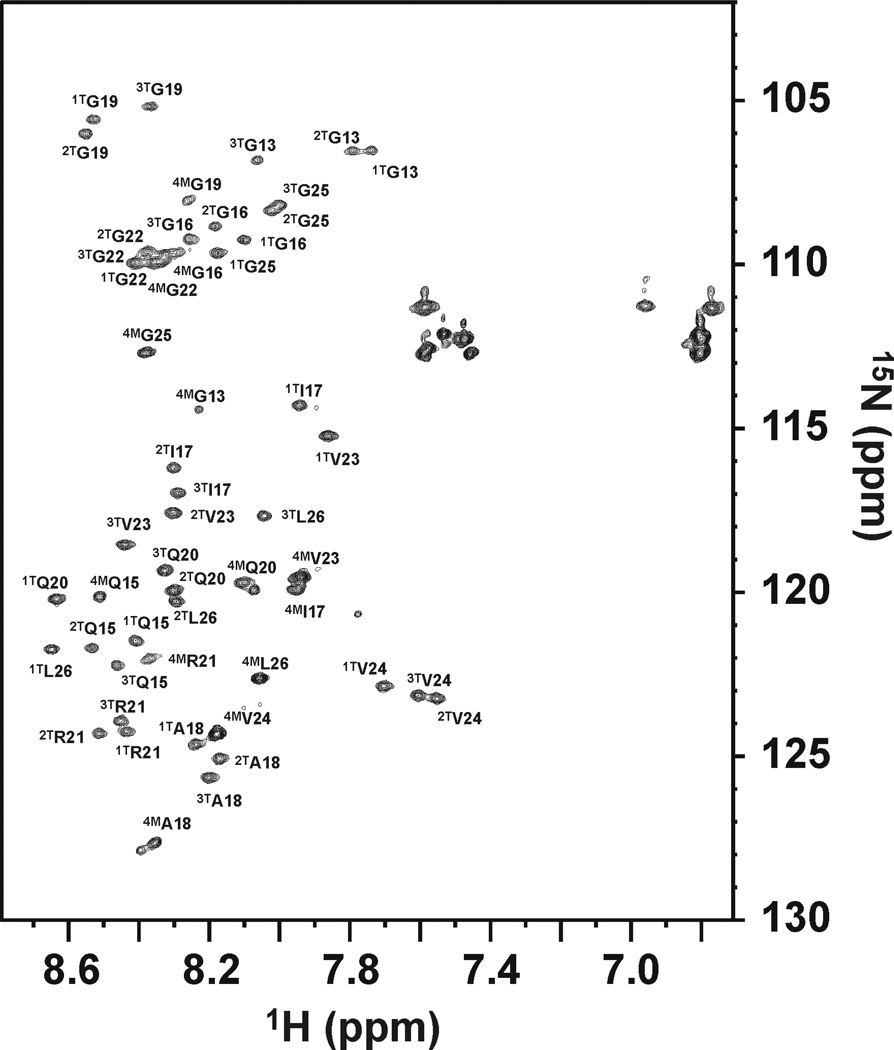
Figure 1.
1H-15N HSQC spectra of α1(I)772–786 THP at 298 K. All resonances belonging to the THP (1T-3T) and to the monomeric chain (4M) have been assigned. For each residue, the appearance of three cross peaks corresponding to triple-helical structure [since the three strands in a triple-helix are staggered by one residue (see Materials and Methods), the 15N-residue in each peptide chain of the THP would be in a different environment with respect to neighboring residues] and one cross peak corresponding to a monomeric state has been observed previously64–67.
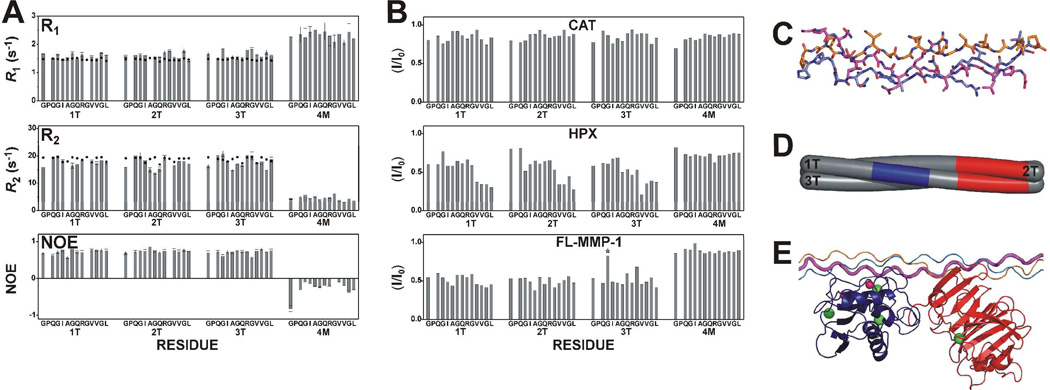
Longitudinal (R1) and transverse (R2) relaxation rates of the backbone amide nitrogens of the THP were consistent with trimeric (1T, 2T and 3T) and monomeric (4M) species (Figure 2A). The negative NOEs values observed for the 4M chain suggest that this monomeric peptide is characterized by fast motions and high flexibility and behaves as a random coil polypeptide in solution. The position of chains 1T, 2T, and 3T in the triple-helix were unequivocally identified by the analysis of the connectivities in the 3D 15N and 13C NOESY-HSQC spectra. The calculated THP structure (Figures 2C and S1) showed a left-handed superhelical arrangement with all dihedral angles falling in the polyPro II region of the Ramachandran plot. Up to 71 interchain NOE connectivities were observed (Table S1), uniformly spread along the labeled stretches in the THP. The distribution and number of interchain NOEs indicated a fully intact triple-helical structure. NMR experiments repeated at 310 K showed that uniform spreading of NOEs was maintained, indicating that the THP structure was still intact despite conditions being in the vicinity of the melting point [Tm for α1(I)772–786 THP = 313 K]. In addition, no relevant changes in chemical shifts, apart from the normal small drifts with temperature, were detected for any position in any of the three chains. Thus, it appeared that no region of the THP was prone to unwinding. The intactness of the triple-helical structure of the THP at physiological temperature (310 K) has been further verified by analyzing a set of 1H-15N HSQC spectra collected at 298, 310, and 316 K (Figure S2). Only at 316 K was the intensity of the THP cross-peaks drastically reduced with only the resonances corresponding to the 4M chain remaining visible and intense.
Figure 2.
(A) Experimental (bars) and theoretical (dots) R1, R2, and NOE values for rigid triple-helical (1T, 2T, and 3T) and non-triple-helical (4M) structure estimated from atomic coordinates of THPs using the program HYDRONMR and assuming for THP an S2 value of 0.80 at 298 K, typical for proteins. The THP structure was obtained by modeling the peptide on the crystal structure of an analogous THP (PDB code: 2d3h), and then refined with experimental NMR constraints. (B) Intensity changes per residue observed at 310 K in the 13C,15N THP upon the addition of unlabeled CAT (0.2 eq), HPX (0.2 eq), or FL-MMP-1 (0.2 eq). Residues showing signal overlap are indicated by stars. The only overlap is residue Gly16 from chain 3T with an impurity. (C) Solution structure of the THP. The three chains forming the THP are 1T (magenta), 2T (orange), and 3T (blue). (D) Schematic section of the THP with the stretch of chain 1T facing the active site of the CAT domain in blue and the HPX binding regions on chains 1T and 2T in red. (E) The adduct where the THP is still in a compact triple-helical conformation, with the CAT and HPX domains binding the cleavage site and the C-terminal region of the labeled stretch, respectively.
NMR spectroscopic evaluation of MMP-1 bound to a triple-helix
Structural data representative of interactions between MMP-1 and α1(I)772–786 THP in solution have been obtained by monitoring reciprocal effects on NMR spectra. The assignments of the FL-MMP-1 and its isolated domains were available elsewhere18. The FL-MMP-1, CAT, and HPX samples were found to be stable under the NMR experimental conditions for at least one month, and the α1(I)772–786 THP was stable in the presence of the inactivated proteins for more than two weeks.
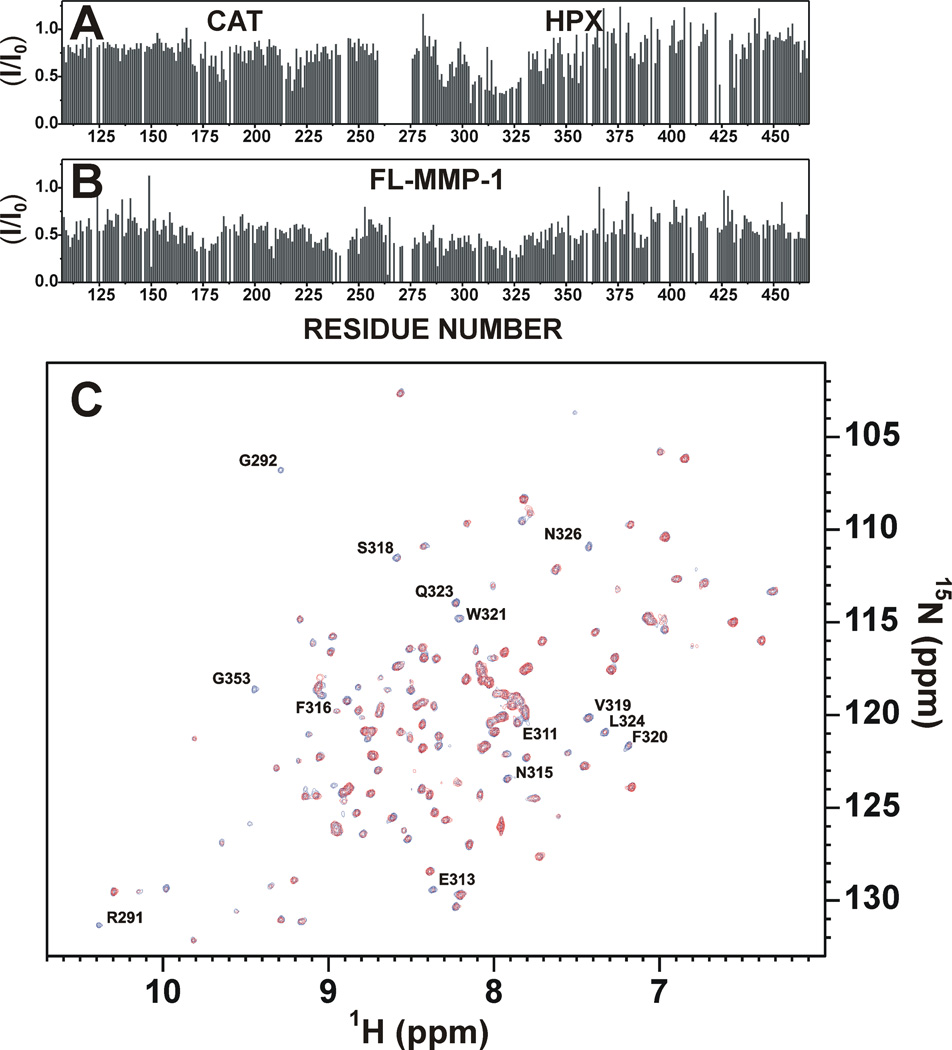
In the presence of unlabeled α1(I)772–786 THP, the 1H-15N TROSY-HSQC signals of FL-MMP-1 underwent intensity decrease both in the CAT and HPX domains without any sizable chemical shift perturbation (Figure 3A). After the addition of increasing concentrations of THP, the spectra of the isolated CAT and HPX domains also showed a decrease in height for several signals (Figure 3B). This can be explained by (a) an adduct of each of the two domains with the THP increasing the molecular weight, producing an increase in transverse relaxation rates and generalized decrease in signal intensity, and (b) the difference in chemical shift between the complex and the two interacting moieties generating a quasi-slow exchange regime which produces further broadening38. Comparison of the signal intensities (Figure 3A and 3B) indicated a larger overall decrease of the signal height in FL-MMP-1 with respect to the isolated domains. The pattern of the modulated residues on the isolated HPX domain clearly defined stretches 291–292 and 311–326 as the ones interacting with the THP (Figure 3C). Similar sites of interaction between α1(I)772–786 THP and FL-MMP-1 HPX domain were observed by hydrogen-deuterium exchange mass spectrometry39. At the same time, the pattern of NH crosspeaks experiencing height decrease in the isolated CAT domain showed that the affected nuclei belonged to the active site crevice (residues 160–199 and 216–22440). The intensity decrease was much more pronounced, at the same THP concentration, for HPX than for CAT, consistent with the higher affinity of the former for α1(I)772–786 THP (see also below)39.
Figure 3.
(A) Intensity changes per residue observed at 310 K for isolated 15N FL-MMP-1 domain upon the addition of unlabeled THP (0.4 eq). (B) Intensity changes per residue observed at 310 K in isolated 15N CAT and HPX domains upon the addition of unlabeled THP (0.8 eq). (C) 1H-15N HSQC spectra shows HPX domain before (blue) and after (red) the addition of THP (0.8 eq). Residues experiencing a large decrease in signal intensity are labeled in the spectra.
To explore the binding sites for MMP-1 on the THP, the interaction of 13C,15N-labeled α1(I)772–786 THP with unlabeled samples of FL-MMP-1 and its isolated CAT and HPX domains was monitored through 1H-15N TROSY-HSQC at 298 and 310 K. By analyzing the signals of NH nuclei along the THP chains in the presence of the HPX domain, a decrease in signal heights without any sizable chemical shift perturbation was immediately apparent (Figure 2B). This decrease was more clearly pronounced for the nuclei belonging to the C-terminal region of the labeled stretch (Val23-Leu26). The monomeric chain 4M was only modestly affected, with a uniform distribution of the signal height decrease. These results demonstrated a specific localized interaction of HPX with the THP, and indicated that this region was relevant for collagen recognition and binding. A much more modest and relatively uniform decrease in signal intensity of the THP was observed in the presence of the isolated CAT domain (Figure 2B). A more substantial effect was observed for CAT domain interaction with the monomeric chain 4M, but only at 298 K (not shown). Most THP resonances were sizably affected by FL-MMP-1, whereas the monomeric chain was negligibly affected (Figure 2B). The decrease in height of the THP signals was more pronounced in the presence of FL-MMP-1 than in the presence of the isolated domains, analogously to what was previously observed for the protein resonances and consistent with prior studies of MMP-1 kinetics39. The above observations, taken together, indicated that (a) the isolated HPX domain had a relatively strong and localized interaction with the THP, while this domain did not show any significant affinity for the monomeric chain 4M; (b) the isolated CAT domain had preferential affinity for 4M compared with the THP (Figure S3); and (c) FL-MMP-1 interacted strongly with the THP and minimally bound 4M. Thus, the HPX domain of the full-length protein drives the interaction with the THP, and creates the conditions for the further interaction of the CAT domain with the THP, whereas this interaction is negligible for the isolated CAT domain. Such behavior mechanistically defines the critical function of the HPX domain in collagenolysis.
Upon addition of FL-MMP-1 to the THP a general decrease in the intensities of the THP interchain NOEs was observed, reflecting the decrease in intensity of all THP signals already discussed. However, a significant fraction of the 71 interchain NOEs (marked with asterisks in Table S1) showed a more pronounced decrease. Strikingly, the corresponding lengthening of interchain distances in the C-terminal region of the THP always involved chain 3T. Conversely, in the N-terminal region of the THP, it was chain 1T that weakened its contacts with either chain 2T or 3T. These data thus indicated that the THP was distorted towards unwinding when bound by FL-MMP-1. The addition of the isolated HPX domain to the THP performed under the same experimental conditions showed a less pronounced decrease in the intensities of the THP interchain NOEs, with fewer connectivities experiencing a sizable reduction in intensity with respect to that observed upon addition of the FL-MMP-1. In particular, only two interchain NOEs in the peptide stretch Val23-Leu26 (marked with triangles in Table S1) showed a more pronounced decrease in signal intensity versus six observed in the presence of FL-MMP-1. This experiment confirmed that the interaction mode of the HPX domain with THP is essentially the same as in the full-length enzyme, but that the interaction is reinforced in the latter case.
Structures of MMP-1-THP complexes
A series of structures were calculated41 to further examine molecular interactions in the FL-MMP-1-THP complex. First, the association of the isolated HPX domain with the THP was studied to determine the energetically most favorable binding conformations by using the decrease in height of the FL-MMP-1 and HPX domain signals as restraints (see Methods). To probe all of the possible binding conformations, calculations were performed with the active residues alternatively placed on one, two, or all three chains of the THP. The solutions were not energetically equivalent. The interaction with only one chain at a time was always unsatisfactory, while the interaction with all three chains simultaneously was practically impossible given the topology of the system. The only reasonable solutions were with two THP chains interacting with the protein at the same time. This result can be easily visualized if one recognizes that the MMP can interact with any of the three THP faces (chain pairs 1T-2T, 2T-3T, or 3T-1T) (Figure 3D). The adduct where the HPX domain binds mainly chains 1T-2T gave both the best combination of energy values and proper orientation of the CAT domain. Indeed, the HPX domain alone assumed somewhat different orientations when it interacted with chains 2T-3T and 3T-1T, and this caused a less plausible guidance of the CAT domain towards the Gly-Ile region in any of the three chains (see below). This result is in perfect agreement with the experimentally observed lengthening of the interaction of 3T with 1T and 2T (Table S1) in the very region of the THP indicated as facing the HPX domain.
The structure of the FL-MMP-1-THP adduct was calculated by imposing as active residues those already considered to dock the isolated HPX domain, and adding those around the THP amide bond to be cleaved (Gly16-Ile17) and those which experienced the largest decrease in signal intensity and water accessibility for the CAT domain in the FL-MMP-1-THP complex (see Methods). As noted above, the isolated CAT domain has negligible affinity for both THP and the isolated 4M at 310 K with a very little affinity for the latter only appearing at 298 K, while the CAT domain as part of the FL-MMP-1 interacted strongly with the THP and minimally bound 4M, and thus reductions in signal intensity due to 4M binding were negligible. Additional constraints were obtained by imposing the carbonyl oxygen of Gly16 to be close to the catalytic metal ion, and the hydrophobic Ile17 side-chain to point toward the S1’ pocket, as proposed for the enzyme-single polypeptide adduct42. In the FL-MMP-1-THP adduct (Figure 2E), the THP was still in a compact triple-helical conformation, with the CAT and HPX domains binding the cleavage site and the C-terminal region of the labeled stretch, respectively. The flexibility of the linker region was critical for this process, supporting the role of FL-MMP-1 Gly271 in collagenolysis43. In these experiments the HPX domain showed a marked preference for binding to chains 1T-2T, and allowed the CAT domain to face chain 1T (but not chain 2T or 3T) at the Gly-Ile region that is subsequently cleaved. Again, this is consistent with the perturbation of the interhelical NOEs (Table S1). In this respect, it was recently demonstrated that the Ile17 residue in one of the three chains at the site of hydrolysis has a distinct chemical shift, a higher J-coupling value, increased dynamics, and decreased local stability44. The present chain-specific assignment unambiguously establishes that the abnormal Ile17 residue belongs to chain 1T, the one that is facing the CAT domain in the present structure and is thus the one ready to undergo hydrolysis.
The mechanism of collagenolysis
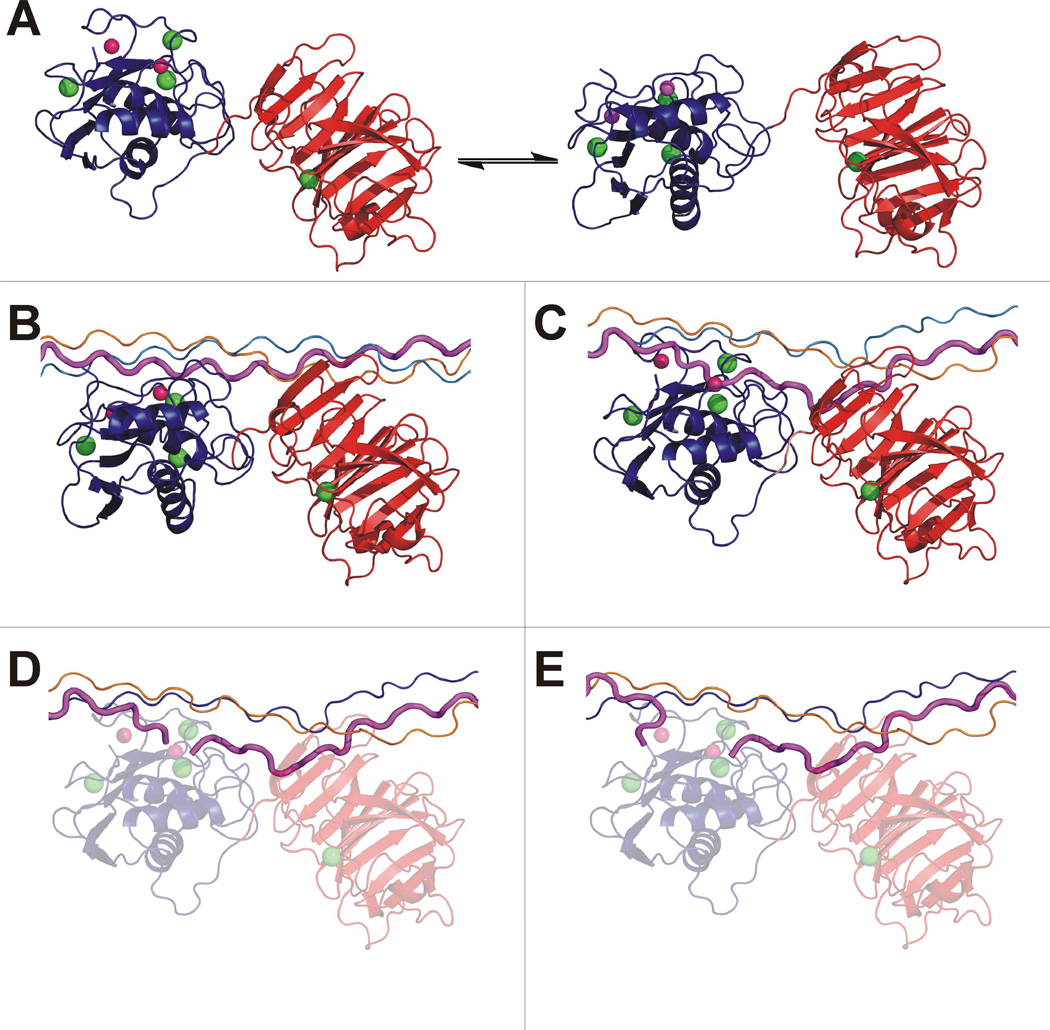
A detailed mechanism of collagenolysis was developed from examination of structures of MMPs and MMP-peptide complexes and docking experiments. MMP-1 is in equilibrium between open/extended and closed structures (Figure 4A). The HPX domain then binds chains 1T and 2T of the THP and, due to the flexibility of the linker, the CAT domain is guided towards the Gly16-Ile17 portion of chain 1T (Figure 2E). This structure (repeated in Figure 4B) would thus correspond to the first event of collagen recognition by MMP-1. Visual inspection of the complex at this point suggested that a back-rotation of the CAT and HPX domains would need to occur to achieve the X-ray crystallographic closed FL-MMP-1 conformation. To approximate this action, the residues at the interface between the HPX and CAT domains in the X-ray structure of FL-MMP-1 in the closed form36 were imposed as constraints in a docking calculation. In the resulting structure, with the CAT and HPX domains arranged in the X-ray crystallographic closed conformation, the THP was visibly unwound (Figure 4C). The domain movement drove chain 1T into the active site, allowing the polypeptide to establish a number of H-bonding interactions and the carbonyl oxygen of the cleavage site amide bond to coordinate the metal ion. This result is consistent with the experimentally observed weakening in NOEs for the interaction of chain 1T with chains 2T and 3T at the THP cleavage site (Table S1). Interestingly, the destabilization of the THP obtained in the present calculation, besides liberating the N-terminal part of chain 1T for hydrolysis, also caused a partial detachment of chain 3T near the C-terminus of the THP (Figure 4C), consistent with the observed lengthening in NOEs of chain 3T (Table S1). The position that the two peptide fragments would assume after cleavage in the present model (Figure 4D and 4E) was almost superimposable to the X-ray crystallographic structure of the complex between the MMP-12 CAT domain and the two fragments obtained by enzymatic cleavage of the α1(I) collagen model Pro-Gln-Gly-Ile-Ala-Gly hexapeptide at the Gly-Ile bond42. The active site is highly similar in all MMPs and it is reasonable to assume that the hydrolysis of the polypeptide chain proceeds with the same mechanism. Moreover, as MMP-12 catalyzes the hydrolysis of peptides at the same bonds (Gly-Ile) as MMP-145, basing the post-hydrolysis complex of MMP-1 on the structural data on MMP-12 is appropriate.
Figure 4.
The initial steps of collagenolysis. (A) Closed (left) and open/extended (right) forms of FL-MMP-1 in equilibrium. (B) The extended protein binds THP chains 1T-2T at Val23-Leu26 with the HPX domain and the residues around the cleavage site with the CAT domain. The THP is still in a compact conformation. (C) Closed FL-MMP-1 interacting with the released 1T chain (in magenta). (D) After hydrolysis, both peptide fragments (C- and N-terminal) are initially bound to the active site. (E) The C-terminal region of the N-terminal peptide fragment is released.
HADDOCK41 was then utilized to calculate the free energies of MMP-1-THP complexes to determine if the proposed mechanism of collagenolysis was sterically and energetically possible. To provide an accurate computational outcome, two steps were added between the initial MMP-1-THP complex (Figure 4B) and the X-ray closed structure complex (Figure 4C). The intermediate structures were generated in such way as to provide a smooth conformational transition between the initial and final states (as illustrated in Figure S4). At the initial MMP-1-THP complex (Figure 4B), the RMSD of FL-MMP-1 from the closed form observed in the X-ray crystallographic structure is 4.2 Å. As the RMSD of FL-MMP-1 approaches 3.9, 3.0, and 1.1 Å of the X-ray closed structure, the total free energy of the complex becomes −50, −100, and −300 kcal/mol, respectively. Thus, the calculated energy41 of the complex in Figure 4C is −300 kcal/mol compared with the initial complex in Figure 4B. Figure S4 is a four-frame movie which illustrates the structural and energetic changes simultaneously. Considering that the activation energies for FL-MMP-1 catalyzed hydrolysis of soluble type I collagen and type I collagen fibrils are 26.0–49.2 and 101 kcal/mol, respectively13,46, the free energy change associated with the collagenolysis mechanism described here is more than sufficient to account for catabolism of soluble and fibrillar collagen.
Discussion
There are a great variety of enzymes that utilize multiple domains to catalyze reactions. Extracellular metallopeptidases that act on macromolecular substrates are often multidomain, including members of the MMP, ADAM (a disintegrin and metalloproteinase), ADAMTS (a disintegrin-like and metalloproteinase domain with thrombospondin type 1 motifs) and meprin families, as well as procollagen C-endopeptidase/bone morphogenetic protein 114. It has long been recognized that selected members of the MMP family catalyze the hydrolysis of collagen, and that these MMPs utilize multiple domains to efficiently perform this function. The collagenolytic MMPs include MMP-1, MMP-2, MMP-8, MMP-9, MMP-13, MT1-MMP, and MT2-MMP47–50. In the cases of MMP-1, MMP-8, MMP-13, MT1-MMP, and MT2-MMP, efficient collagenolytic activity for the isolated enzyme requires both the CAT and HPX domains51–56. The linker region between these domains also participates in collagenolysis, either by direct binding of substrate57 or by allowing for the proper orientation of the CAT and HPX domains58–60. The gelatinase members of the MMP family (MMP-2 and MMP-9) possess three fibronectin type II (FN II) inserts within their CAT domains, and these inserts possess similar type I collagen binding sites61,62. On a mechanistic level, little is known as to how these domains participate in the collagenolytic process.
Several models have been proposed previously for collagenolysis. It has long been noted that the collagen triple-helix does not fit into the CAT domain active site cavity8. Models have generally accounted for the steric clash of the triple-helix with enzyme active sites by (a) requiring active unwinding of the triple-helix by an MMP8,9,63 and/or (b) considering that the site of hydrolysis within collagen has a distinct conformation, or conformational flexibility, rendering it more susceptible to proteolysis than other regions in collagen5. Active unwinding could be achieved via molecular tectonics, where each of the three collagen chains has a specific binding interaction with different domains6. Unwinding might also be accompanied by a conformational change in the active site of the MMP10. The “vulnerable” site hypothesis proposes that the distinct cleavage site region within collagen is alone responsible for collagenolysis12. Alternatively, there may be destabilization of the triple-helix and/or stabilization of an unwound triple-helix upon MMP binding13. As mentioned earlier, a recent study demonstrated that the Ile residue in one of the three chains at the site of hydrolysis has a distinct chemical shift, a higher J-coupling value, increased dynamics and decreased local stability44. This suggests that a single locally dynamic chain, rather than a labile region with three comparably dynamic chains, is the determining factor for collagen to be cleaved by MMPs44.
The present α1(I)772–786 THP has been reported to be hydrolyzed by FL-MMP-1 at the physiological cleavage site15,16, and therefore represented a suitable substrate for analyzing the collagenolytic process in solution. THPs have been shown, in numerous studies, to be highly accurate structural analogs of native collagen. Most importantly, results obtained from MMP studies with THPs have been recapitulated using collagen. For example, specific residues within the MMP-1 HPX domain were shown to be important for activity towards THPs39. Mutation of these residues significantly effected MMP-1 collagenolytic activity39. Similarly, demonstration that the CAT domain of collagenolytic MMPs had inherent (albeit low) activity towards triple-helices was initially observed with THPs15, followed by collagen12.
The 13C-15N enriched THP allowed us to assign the three chains around the cleavage site and to explore its structural features and dynamic properties. The presence of four different polypeptide chains, three of which form the homotrimer and the fourth isolated in solution, was apparent from the analysis of the NMR spectra. The signals of the isolated chain were much sharper and shifted with respect to those of the THP. The signals of the corresponding amino acids within the THP were not superimposable, with sizable differences for some residues. Thus, in the homotrimeric THP the corresponding amino acids in each of the three chains experience a different environment, in concert with prior studies64–67. The relaxation data were consistent with the rodlike structure of the THP as well as with the unfolded state of the isolated chain.
Reaction of the α1(I)772–786 THP with the CAT and HPX domains of MMP-1 and with the full-length protein was monitored by NMR spectroscopy. From the point of view of the substrate, we observed changes induced by the (a) isolated CAT domain, (b) isolated HPX domain, and (c) FL-MMP-1 on the THP and monomeric peptide. From the point of view of the enzyme, we observed changes induced by the THP and monomeric peptide on the (a) isolated CAT domain, (b) isolated HPX domain, and (c) FL-MMP-1. Thus, interactions of 12 complexes were monitored. Combined with the structure and assignment of the three chains for the THP based on experimental constraints, the present work resulted in the identification of MMP-collagen interactions that is only speculative at the actual cleavage step, which cannot occur here because the enzyme is inactive. The structure of the cleaved peptide bound to FL-MMP-1 was thus derived from a prior experimental result (see below)42.
The affinity of the THP for the HPX domain was higher than that observed for the CAT domain. An opposite behavior was observed for the unfolded peptide. Prior studies have demonstrated that the MMP-8 CAT domain digests a THP much slower than the analogous single-stranded peptide (t1/2 = 3 and ≫20 h, respectively)68. Conversely, full-length MMP-1 and MMP-8 preferred a THP compared with the analogous single-stranded peptide68. Deletion of the HPX domain from MMP-1 had little or no effect on activity towards SSPs, while activity towards THPs was reduced by as much as 99%15,39. Therefore, it appears that the HPX domain plays the pivotal role in collagenolysis by promoting the binding of MMP-1 to triple-helical structure. The interaction of the isolated CAT domain with the THP involved the residues around the active site crevice. However, the specificity for the THP observed for the HPX domain was not evident for the CAT domain. This suggested that the CAT domain alone cannot efficiently unwind the THP in order to properly bind the chain to be cleaved. More importantly, the experiments performed on the FL-MMP-1 showed an overall reinforcement of the interaction with the THP that is consistent with a cooperative behavior of the CAT and HPX domains. The HPX domain binds THP chains 1T and 2T downstream to the cleavage site, at Val23-Leu26. The CAT domain must bind the substrate with a specific orientation to hydrolyze the peptide bond. The interaction of the HPX domain with the Val23-Leu26 region of the THP facilitated the correct positioning of the CAT domain in front of the cleavage site. Our results indicate a mechanism by which distinct chains of the triple-helix are recognized by FL-MMP-1, access to the triple-helix is achieved by back rotation of the CAT and HPX domains, and a single locally dynamic chain allows Ile17 to enter the S1' pocket of the CAT domain.
During the proposed collagenolysis pathway the coordination of the three His residues to the active site Zn2+ is not perturbed upon binding of the THP carbonyl oxygen. The active site-coordinated water molecule has been removed in all steps for homogeneity, without implying its presence or absence. Prior studies with MMP-12 indicated that the water can remain coordinated in the presence of the carbonyl oxygen42.
The present study has presented a detailed mechanism for collagenolysis that, while aesthetically pleasing, needs to be considered in light of energetics. The rapid movement of the CAT and HPX domains between open and closed forms of full-length MMP18 (Figure 4A) indicates that all forms have essentially the same free energy, where the closed form is enthalpically-favored and the open forms equivalently entropically-favored. The initial binding of the HPX domain to the THP does not necessarily shift this equilibrium. The subsequent binding of the CAT domain to the THP in front of the Gly-Ile sequence of chain 1T to be cleaved (Figure 4B) is enthalpically-favored and entropically-disfavored. We experimentally observed that the isolated CAT domain has negligible affinity for the THP, gaining the ability to bind once held in place by the HPX domain in FL-MMP-1. The proposed back-rotation of FL-MMP-1 towards the X-ray closed structure and the associated liberation of chain 1T from the compact THP structure (Figure 4C) again implies a balance of enthalpic gains (the closing of FL-MMP-1 and the establishment of productive contacts between chain 1T and the active site of MMP) and losses (the breaking of the H-bonds between chain 1T and the other two THP chains). As discussed earlier, the free energy change associated with the collagenolysis mechanism can readily account for collagen catabolism. The final cleavage step yielding the complex in Figure 4D is energetically favorable46.
The open-closed equilibrium of MMP-1 (Figure 4A) was observed by NMR spectroscopy and small angle X-ray scattering18 and the formation of the initial FL-MMP-1-THP complex (Figure 4B) was demonstrated in the present work. The FL-MMP-1-THP complex in which one strand of the triple-helix is displaced from the other two (Figure 4C) was strongly supported by changes in intensity of the interhelical NOEs upon THP binding (Table S1) and is an energetically and mechanistically feasible route between the complexes in Figures 4B and 4D. The binding mode of the cleaved fragments (Figure 4D and 4E) had been revealed by X-ray crystallography42. Thus, Figure 4 represents an experimentally-based and energetically favorable description of the entire chain of events that lead to the initial cleavage of type I collagen by FL-MMP-1, and provides the basis for understanding catabolism of collagen fibrils69. The overall favorable energetics of the collagenolytic mechanism resolves the long-standing enigma as to why an external energy source (such as ATP) is not needed for collagen catabolism.
Our proposed mechanism does not explicitly account for the displacement of water present in collagen fibrils70. However, water associated with fibrillar collagen may ultimately aid collagenolysis. Collagen fibrils have a least three populations of water, the first tightly bound, the second found in the interstices of microfibrils, and the third localized in the interfibrillar space71,72. By accessing the edge of fibrils (see below), MMPs may have considerably less water to “strip” from the triple-helix. Individual triple-helices have two hydration shells, one of water directly hydrogen bonded to the polypeptide chain and one interacting only with other water molecules73,74. Hydration patterns in triple-helices are sequence dependent, and affect molecular packing of triple-helices73,74. Single water bridges between backbone atoms stabilize triple-helices in regions of low Pro/Hyp content, but are highly dynamic in nature and thus may facilitate unwinding in these regions including the one neighboring the MMP cleavage site75. In addition, it has been proposed that the release of the two water layers from the triple-helix promotes catalytic turnover76. We have shown that as the substrate is docking into the metalloproteinase active site, water movement in the environment slows down77. Water then helps trap the substrate in the active site.
MT1-MMP has been shown to cleave collagen efficiently without its HPX domain when it is cell surface bound78. In similar fashion, MMP-1 CAT can catalyze collagen hydrolysis when membrane tethered79. Cell surface collagenolysis may be facilitated by collagen binding integrins providing strain on the collagen63, protease binding partners80,81, and/or protease homodimerization82. Soluble MMP CAT domains can process triple-helical peptides and collagen, but less efficiently then full-length proteases12,39. The present model accounts for the need of the HPX domain for highly efficient collagenolysis without ancillary contributions from other biomolecules.
In vivo processing of collagen initially involves MMP interaction with fibrils. MMPs bind to multiple sites in collagen, but hydrolysis ultimately occurs only at a single site83. MMP-1 is a diffusion-based “Burnt Bridge” Brownian Ratchet capable of biased diffusion on the surface of collagen fibrils, where the bias is driven by proteolysis3. Surface-bound MT1-MMP movement is via a similar diffusion mechanism84. Hydrolysis of collagen proceeds at the outer edge of the fibril69. The exposure of the MMP cleavage site by removal of the collagen C-terminal telopeptide69 would permit interactions of the MMP-1 HPX and CAT domains described in the present work with triple-helices on the outer edge of the fibril. Once collagen undergoes initial processing, (a) gelatinolytic MMPs laterally diffuse on collagen, find the “tails” from the cleaved sites, denature the triple-helix, and further proteolyze the α chains85 and/or (b) the large collagen fragments undergo urokinase plasminogen activator receptor-associated protein (uPARAP)/Endo180-mediated endocytosis, lysosomal delivery, and cathepsin catalyzed degradation86–89. The present molecular mechanism is well integrated with current knowledge of fibrillar collagenolysis and is in concert with other real time, single molecule studies of collagen and collagenolytic enzymes3,63,77,83–85. In a broader context, the respective roles of the individual MMP-1 domains proposed here sheds light into the mode of action of numerous other extracellular metallopeptidases.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grant CA98799, NIH contract 268201000036C, the Multiple Sclerosis National Research Institute, and the Robert A. Welch Foundation to G.B.F. and grants from the European Commission (Projects MEST-CT-2004-504391, SFMET Number 201640, SPINE2-COMPLEXES Number 031220, and NanoDrugs Number LSHB-CT-2005-019102), the Ministero dell’Istruzione, dell’Universitá e della Ricerca (PRIN 2005, Protocollo Number 2005039878, Protocollo RBLA032ZM7, Protocollo RBIP06LSS2), and the Ente Cassa di Risparmio di Firenze.
Footnotes
Supporting Information Available: Interchain NOEs from 1H-15N NOESY-HSQC spectrum of the THP, 15N R1, R2, and NOEs for the THP, 1H and 15N chemical shifts for the THP, 13C resonance assignments for the THP, upper limit distances for structure calculation from 1H-15N NOESY- HSQC spectrum, torsion angles used for structure calculation, upper limit distances for N- and C-terminal residues, torsion angles used for N- and C-terminal residues, statistical analysis of the NMR structures of THP, solution structure of the THP, intensity changes observed in the THP upon addition of CAT domain, and the energetics calculation for collageonlysis. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Bella J, Eaton M, Brodsky B, Berman HM. Science. 1994;266:75–81. doi: 10.1126/science.7695699. [DOI] [PubMed] [Google Scholar]
- 2.Holmgren SK, Taylor KM, Bretscher LE, Raines RT. Nature. 1998;392:666–667. doi: 10.1038/33573. [DOI] [PubMed] [Google Scholar]
- 3.Saffarian S, Collier IE, Marmer BL, Elson EL, Goldberg G. Science. 2004;306:108–111. doi: 10.1126/science.1099179. [DOI] [PubMed] [Google Scholar]
- 4.Li J, Brick P, O'Hare MC, Skarzynski T, Lloyd LF, Curry VA, Clark IM, Bigg HF, Hazleman BL, Cawston TE, Blow DM. Structure. 1995;15:541–549. doi: 10.1016/s0969-2126(01)00188-5. [DOI] [PubMed] [Google Scholar]
- 5.Fields GB. J. Theor. Biol. 1991;153:585–602. doi: 10.1016/s0022-5193(05)80157-2. [DOI] [PubMed] [Google Scholar]
- 6.Overall CM. In: Methods in Molecular Biology 151: Matrix Metalloproteinase Protocols. Clark IM, editor. Totowa, NJ: Humana Press; 2001. pp. 79–120. [Google Scholar]
- 7.Lauer-Fields JL, Juska D, Fields GB. Biopolymers (Peptide Sci.) 2002;66:19–32. doi: 10.1002/bip.10201. [DOI] [PubMed] [Google Scholar]
- 8.Chung L, Dinakarpandian D, Yoshida N, Lauer-Fields JL, Fields GB, Visse R, Nagase H. EMBO J. 2004;23:3020–3030. doi: 10.1038/sj.emboj.7600318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tam EM, Moore TR, Butler GS, Overall CM. J. Biol. Chem. 2004;279:43336–43344. doi: 10.1074/jbc.M407186200. [DOI] [PubMed] [Google Scholar]
- 10.O'Farrell TJ, Guo R, Hasegawa H, Pourmotabbed T. Biochemistry. 2006;45:15411–15418. doi: 10.1021/bi060849d. [DOI] [PubMed] [Google Scholar]
- 11.Minond D, Lauer-Fields JL, Cudic M, Overall CM, Pei D, Brew K, Moss ML, Fields GB. Biochemistry. 2007;46:3724–3733. doi: 10.1021/bi062199j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salsas-Escat R, Nerenberg PS, Stultz CM. Biochemistry. 2010;49:4147–4158. doi: 10.1021/bi9021473. [DOI] [PubMed] [Google Scholar]
- 13.Han S, Makareeva E, Kuznetsova NV, DeRidder AM, Sutter MB, Losert W, Phillips CL, Visse R, Nagase H, Leikin S. J. Biol. Chem. 2010;285:22276–22281. doi: 10.1074/jbc.M110.102079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrett AJ, Rawlings ND, Woessner JF. Handbook of Proteolytic Enzymes, Second Edition. Vol. 1. Amsterdam: Elsevier/Academic Press; 2004. [Google Scholar]
- 15.Lauer-Fields JL, Tuzinski KA, Shimokawa K, Nagase H, Fields GB. J. Biol. Chem. 2000;275:13282–13290. doi: 10.1074/jbc.275.18.13282. [DOI] [PubMed] [Google Scholar]
- 16.Lauer-Fields JL, Nagase H, Fields GB. J. Chromatogr. A. 2000;890:117–125. doi: 10.1016/s0021-9673(00)00396-4. [DOI] [PubMed] [Google Scholar]
- 17.Myers AG, Gleason JL, Yoon T, Kung DW. J. Am. Chem. Soc. 1997;119:656–673. [Google Scholar]
- 18.Bertini I, Fragai M, Luchinat C, Melikian M, Mylonas E, Sarti N, Svergun DI. J. Biol. Chem. 2009;284:12821–12828. doi: 10.1074/jbc.M809627200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arnold LH, Butt L, Prior SH, Read C, Fields GB, Pickford AR. J. Biol. Chem. 2011 doi: 10.1074/jbc.M111.285213. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keller R. The Computer Aided Resonance Assignment Tutorial. Verlag: CANTINA; 2004. [Google Scholar]
- 21.Grzesiek S, Bax A. J. Magn. Reson. 1992;96:432–440. [Google Scholar]
- 22.Grzesiek S, Bax A. J. Magn. Reson. 1992;99:201–207. [Google Scholar]
- 23.Schleucher J, Sattler M, Griesinger C. Angew. Chem. Int. Ed. Eng. 1993;32:1489–1491. [Google Scholar]
- 24.Kay LE, Xu GY, Yamazaki T. J. Magn. Reson. Ser. A. 1994;109:129–133. [Google Scholar]
- 25.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 26.Bermel W, Bertini I, Duma L, Felli IC, Emsley L, Pierattelli R, Vasos PR. Angew. Chem. Int. Ed. Eng. 2005;44:3089–3092. doi: 10.1002/anie.200461794. [DOI] [PubMed] [Google Scholar]
- 27.Kay LE, Xu G-Y, Singer AU, Muhandiram DR, Forman-Kay JD. J. Magn. Reson. 1993;101 Series B:333–337. [Google Scholar]
- 28.Schleucher J, Schwendinger M, Sattler M, Schmidt P, Schedletzky O, Glaser SJ, Sørensen OW, Griesinger C. J. Biomol. NMR. 1994;4:301–306. doi: 10.1007/BF00175254. [DOI] [PubMed] [Google Scholar]
- 29.Kay LE, Nicholson LK, Delaglio F, Bax A, Torchia DA. J. Magn. Reson. 1992;97:359–375. [Google Scholar]
- 30.Peng JW, Wagner G. Meth. Enzymol. 1994;239:563–596. doi: 10.1016/s0076-6879(94)39022-3. [DOI] [PubMed] [Google Scholar]
- 31.Shen Y, Delaglio F, Cornilescu G, Bax A. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Güntert P, Mumenthaler C, Wüthrich K. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 33.Laskowski RA, Rullmann JAC, MacArthur MW, Kaptein R, Thornton JM. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 34.Hubbard SJ, Thornton JM. London: Department of Biochemistry and Molecular Biology, University College; 1993. [Google Scholar]
- 35.de Vries SJ, van Dijk AD, Krzeminski M, van Dijk M, Thureau A, Hsu V, Wassenaar T, Bonvin AM. Proteins: Struc. Funct. Bioinform. 2007;69:726–733. doi: 10.1002/prot.21723. [DOI] [PubMed] [Google Scholar]
- 36.Jozic D, Bourenkov G, Lim N-H, Visse R, Nagase H, Bode W, Maskos K. J. Biol. Chem. 2005;280:9578–9585. doi: 10.1074/jbc.M411084200. [DOI] [PubMed] [Google Scholar]
- 37.Bermel W, Bertini I, Felli IC, Peruzzini R, Pierattelli R. ChemPhysChem. 2010;11:689–695. doi: 10.1002/cphc.200900772. [DOI] [PubMed] [Google Scholar]
- 38.Erat MC, Slatter DA, Lowe ED, Millard CJ, Farndale RW, Campbell ID, Vakonakis I. Proc. Natl. Acad. Sci. USA. 2009;106:4195–4200. doi: 10.1073/pnas.0812516106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lauer-Fields JL, Chalmers MJ, Busby SA, Minond D, Griffin PR, Fields GB. J. Biol. Chem. 2009;284:24017–24024. doi: 10.1074/jbc.M109.016873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grams F, Reinemer P, Powers JC, Kleine T, Pieper M, Tschesche H, Huber R, Bode W. Eur. J. Biochem. 1995;228:830–841. doi: 10.1111/j.1432-1033.1995.tb20329.x. [DOI] [PubMed] [Google Scholar]
- 41.Dominguez C, Boelens R, Bonvin AM. J. Am. Chem. Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 42.Bertini I, Calderone V, Fragai M, Luchinat C, Maletta M, Yeo KJ. Angew. Chem. Int. Ed. Eng. 2006;45:7952–7955. doi: 10.1002/anie.200603100. [DOI] [PubMed] [Google Scholar]
- 43.Tsukada H, Pourmotabbed T. J. Biol. Chem. 2002;277:27378–27384. doi: 10.1074/jbc.M201367200. [DOI] [PubMed] [Google Scholar]
- 44.Xiao J, Addabbo RM, Lauer JL, Fields GB, Baum J. J. Biol. Chem. 2010;285:34181–34190. doi: 10.1074/jbc.M110.128355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taddese S, Jung MC, Ihling C, Heinz A, Neubert RHH, Schmelzer CEH. Biochim. Biophys. Acta. 2010;1804:731–739. doi: 10.1016/j.bbapap.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 46.Welgus HG, Jeffrey JJ, Eisen AZ. J. Biol. Chem. 1981;256:9516–9521. [PubMed] [Google Scholar]
- 47.McCawley LJ, Matrisian LM. Curr. Opin. Cell Biol. 2001;13:534–540. doi: 10.1016/s0955-0674(00)00248-9. [DOI] [PubMed] [Google Scholar]
- 48.Overall CM. Mol. Biotech. 2002;22:51–86. doi: 10.1385/MB:22:1:051. [DOI] [PubMed] [Google Scholar]
- 49.Morrison CJ, Overall CM. J. Biol. Chem. 2006;281:26528–26539. doi: 10.1074/jbc.M603331200. [DOI] [PubMed] [Google Scholar]
- 50.Bigg HF, Rowan AD, Barker MD, Cawston TE. FEBS J. 2007;274:1246–1255. doi: 10.1111/j.1742-4658.2007.05669.x. [DOI] [PubMed] [Google Scholar]
- 51.Clark IN, Cawston TE. Biochem. J. 1989;263:201–206. doi: 10.1042/bj2630201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Knäuper V, Cowell S, Smith B, Lopez-Otin C, O'Shea M, Morris H, Zardi L, Murphy G. J. Biol. Chem. 1997;272:7608–7616. doi: 10.1074/jbc.272.12.7608. [DOI] [PubMed] [Google Scholar]
- 53.Knäuper V, Osthues A, DeClerk YA, Langley KE, Bläser J, Tschesche H. Biochem. J. 1993;291:847–854. doi: 10.1042/bj2910847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murphy G, Allan JA, Willenbrock F, Cockett MI, O'Connell JP, Docherty AJP. J. Biol. Chem. 1992;267:9612–9618. [PubMed] [Google Scholar]
- 55.Ohuchi E, Imai K, Fujii Y, Sato H, Seiki M, Okada Y. J. Biol. Chem. 1997;272:2446–2451. doi: 10.1074/jbc.272.4.2446. [DOI] [PubMed] [Google Scholar]
- 56.Hurst DR, Schwartz MA, Ghaffari MA, Jin Y, Tschesche H, Fields GB, Sang Q-XA. Biochem. J. 2004;377:775–779. doi: 10.1042/BJ20031067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Souza SJ, Pereira HM, Jacchieri S, Brentani RR. FASEB J. 1996;10:927–930. doi: 10.1096/fasebj.10.8.8666171. [DOI] [PubMed] [Google Scholar]
- 58.Hirose T, Patterson C, Pourmotabbed T, Mainardi CL, Hasty KA. Proc. Natl. Acad. Sci. USA. 1993;90:2569–2573. doi: 10.1073/pnas.90.7.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chung L, Shimokawa K, Dinakarpandian D, Grams F, Fields GB, Nagase H. J. Biol. Chem. 2000;275:29610–29617. doi: 10.1074/jbc.M004039200. [DOI] [PubMed] [Google Scholar]
- 60.Iyer S, Visse R, Nagase H, Acharya KR. J. Mol. Biol. 2006;362:78–88. doi: 10.1016/j.jmb.2006.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Steffensen B, Wallon UM, Overall CM. J. Biol. Chem. 1995;270:11555–11566. doi: 10.1074/jbc.270.19.11555. [DOI] [PubMed] [Google Scholar]
- 62.Xu X, Chen Z, Wang Y, Yamada Y, Steffensen B. Biochem. J. 2005;392:127–134. doi: 10.1042/BJ20050650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adhikari AS, Chai J, Dunn AR. J. Am. Chem. Soc. 2011;133:1686–1689. doi: 10.1021/ja109972p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fan P, Li MH, Brodsky B, Baum J. Biochemistry. 1993;32:13299–13309. doi: 10.1021/bi00211a043. [DOI] [PubMed] [Google Scholar]
- 65.Anachi RB, Siegel DL, Baum J, Brodsky B. FEBS Lett. 1995;368:551–555. doi: 10.1016/0014-5793(95)00738-u. [DOI] [PubMed] [Google Scholar]
- 66.Liu X, Siegel DL, Fan P, Brodsky B, Baum J. Biochemistry. 1996;35:4306–4313. doi: 10.1021/bi952270d. [DOI] [PubMed] [Google Scholar]
- 67.Yu Y-C, Roontga V, Daragan VA, Mayo KH, Tirrell M, Fields GB. Biochemistry. 1999;38:1659–1668. doi: 10.1021/bi982315l. [DOI] [PubMed] [Google Scholar]
- 68.Ottl J, Gabriel D, Murphy G, Knäuper V, Tominaga Y, Nagase H, Kröger M, Tschesche H, Bode W, Moroder L. Chem. Biol. 2000;7:119–132. doi: 10.1016/s1074-5521(00)00077-6. [DOI] [PubMed] [Google Scholar]
- 69.Perumal S, Antipova O, Orgel JPRO. Proc. Natl. Acad. Sci. USA. 2008;105:2824–2829. doi: 10.1073/pnas.0710588105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nagase H, Fushimi K. Connective Tissue Res. 2008;49:169–174. doi: 10.1080/03008200802151698. [DOI] [PubMed] [Google Scholar]
- 71.Knauss R, Fleischer G, Gründer W, Kärger J, Werner A. Magn. Reson. Med. 1996;36:241–248. doi: 10.1002/mrm.1910360211. [DOI] [PubMed] [Google Scholar]
- 72.Traore A, Foucat L, Renou JP. Biopolymers. 2000;53:476–483. doi: 10.1002/(SICI)1097-0282(200005)53:6<476::AID-BIP4>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 73.Kramer RZ, Bella J, Mayville P, Brodsky B, Berman HM. Nature Struct. Biol. 1999;6:454–457. doi: 10.1038/8259. [DOI] [PubMed] [Google Scholar]
- 74.Kramer RZ, Venugopal MG, Bella J, Mayville P, Brodsky B, Berman HM. J. Mol. Biol. 2000;301:1191–1205. doi: 10.1006/jmbi.2000.4017. [DOI] [PubMed] [Google Scholar]
- 75.Ravikumar KM, Hwang W. Proteins. 2008;72:1320–1332. doi: 10.1002/prot.22026. [DOI] [PubMed] [Google Scholar]
- 76.Eckhard U, Schönauer E, Nüss D, Brandstetter H. Nat. Struct. Mol. Biol. 2011;18:1109–1114. doi: 10.1038/nsmb.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grossman M, Born B, Heyden M, Tworowski D, Fields GB, Sagi I, Havenith M. Nat. Struct. Mol. Biol. 2011;18:1102–1108. doi: 10.1038/nsmb.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li X-Y, Ota I, Yana I, Sabeh F, Weiss SJ. Mol. Biol. Cell. 2008;19:3221–3233. doi: 10.1091/mbc.E08-01-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sabeh F, Li X-Y, Saunders TL, Rowe RG, Weiss SJ. J. Biol. Chem. 2009;284:23001–23011. doi: 10.1074/jbc.M109.002808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yañez-Mó M, Barreiro O, Gonzalo P, Batista A, Megías D, Genís L, Sachs N, Sala-Valdés M, Alonso MA, Montoya MC, Sonnenberg A, Arroyo AG, Sánchez-Madrid F. Blood. 2008;112:3217–3226. doi: 10.1182/blood-2008-02-139394. [DOI] [PubMed] [Google Scholar]
- 81.Lafleur MA, Xu D, Hemler ME. Mol. Biol. Cell. 2009;20:2030–2040. doi: 10.1091/mbc.E08-11-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Itoh Y, Ito N, Nagase H, Evans RD, Bird SA, Seiki M. Mol. Biol. Cell. 2006;17:5390–5399. doi: 10.1091/mbc.E06-08-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sun HB, Smith GN, Jr, Hasty KA, Yokota H. Anal. Biochem. 2000;283:153–158. doi: 10.1006/abio.2000.4629. [DOI] [PubMed] [Google Scholar]
- 84.Collier IE, Legant W, Marmer B, Lubman O, Saffarian S, Wakatsuki T, Elson E, Goldberg GI. PLoS One. 2011;6:e24029. doi: 10.1371/journal.pone.0024029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rosenblum G, Van den Steen PE, Cohen SR, Bitler A, Brand DD, Opdenakker G, Sagi I. PLoS One. 2010;5:e11043. doi: 10.1371/journal.pone.0011043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee H, Overall CM, McCulloch CA, Sodek J. Mol. Biol. Cell. 2006;17:4812–4826. doi: 10.1091/mbc.E06-06-0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wagenaar-Miller RA, Engelholm LH, Gavard J, Yamada SS, Gutkind JS, Behrendt N, Bugge TH, Holmbeck K. Mol. Cell. Biol. 2007;27:6309–6322. doi: 10.1128/MCB.00291-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Madsen DH, Engelholm LH, Ingvarsen S, Hillig T, Wagenaar-Miller RA, Kjoller L, Gardsvoll H, Hoyer-Hansen G, Holmbeck K, Bugge TH, Behrendt N. J. Biol. Chem. 2007;282:27037–27045. doi: 10.1074/jbc.M701088200. [DOI] [PubMed] [Google Scholar]
- 89.Madsen DH, Ingvarsen S, Jürgensen HJ, Melander MC, Kjoller L, Moyer A, Honoré C, Madsen CA, Garred P, Burgdorf S, Bugge TH, Behrendt N, Engelholm LH. J. Biol. Chem. 2011;286:26996–27010. doi: 10.1074/jbc.M110.208033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.