Abstract
Cis-regulatory variation is considered to be an important determinant of human phenotypic variability, including susceptibility to complex disease. Recent studies have shown that the effects of cis-regulatory polymorphism on gene expression can differ widely between tissues. In the present study, we tested whether the effects of cis-regulatory variation can also differ between regions of the adult human brain. We used relative allelic expression to measure cis-effects on the RNA expression of five candidate genes for neuropsychiatric illness (ZNF804A, NOS1, RGS4, AKT1 and TCF4) across multiple discrete brain regions within individual subjects. For all five genes, we observed significant differences in allelic expression between brain regions in several individual subjects, suggesting regional differences in the effects of cis-regulatory polymorphism to be a common phenomenon. As well as highlighting an important caveat for studies of regulatory polymorphism in the brain, our findings indicate that it is possible to delineate brain areas in which cis-regulatory variants are active. This may provide important insights into the fundamental biology of neuropsychiatric phenotypes with which such variants are associated.
INTRODUCTION
Effects on gene expression can be classified according to whether they originate from the same DNA molecule as the regulated gene or from a remote source, the former said to be acting in cis, and the latter in trans. Variable cis-effects on gene expression can occur as a result of differences in DNA sequence or epigenetic modification at cis-regulatory elements (e.g. promoters, enhancers, locus control regions) distributed at the gene locus, as well as through DNA variation affecting the stability or splicing of the gene product. Such variation is common in the human genome (1–3) and likely to be an important determinant of phenotypic diversity, including susceptibility to complex disease. Trans-regulatory variation results from differences in the level or structure of trans-acting factors (e.g. transcription factors, hormones, microRNAs), which are usually transcribed at distinct gene loci and subject to similar regulation.
Whereas trans-acting influences operate on both chromosomal gene copies, cis-acting effects on gene expression operate in an allele-specific manner. This difference is exploited in assays of relative allelic expression, where the effects of cis-regulatory variants are exposed by comparing the level of mRNA transcribed from the two chromosomal gene copies in individual samples (1–4). The relative allelic expression method typically makes use of a single nucleotide polymorphism (SNP) within the mRNA sequence, allowing the RNA transcribed from each chromosome to be distinguished and quantified in heterozygous subjects. Significant departure from the genomic 1:1 ratio of the two alleles in cDNA is assumed to reflect heterozygosity for one or more cis-regulatory DNA variants or an allele-specific epigenetic process (e.g. imprinting) operating in the assayed sample.
Cis- and trans-regulatory variation may not always act independently. For example, the effect of a cis-regulatory variant in a binding site for a trans-regulatory factor will depend on the availability of that trans-factor. Cellular differences in the effects of transcription factor binding site polymorphism have been demonstrated in vitro (5,6) and provide a likely explanation for at least some of the tissue specificity of cis-effects detected through allelic expression (3,4,6–8) and expression quantitative trait loci mapping (9,10) studies. These findings suggest that multiple tissue sources may be required for adequate assessment of putative cis-regulatory variation.
In the present study, we tested whether the effects of cis-regulatory variation can differ between regions of the adult human brain, an organ in which there is a high degree of regional specialization. We selected for study five genes for which there is evidence of genetic association with neuropsychiatric illness [ZNF804A (11,12), NOS1 (11), RGS4 (13), AKT1 (14) and TCF4 (15)]. For each subject, we used relative allelic expression to measure cis-effects on the RNA expression of each gene across multiple discrete brain regions. One of the great advantages of this method is the ability to detect effects of cis-regulatory variation in individual samples (1–4). Moreover, by comparing cDNA allele ratios across brain regions, but within individual subjects, we were able to hold any (genetic) cis-regulatory variants constant, and could therefore expose any regional differences in their effects on gene expression.
RESULTS
Experiments were carried out on a total of 20 unrelated subjects for which postmortem tissue was available from at least 6 of 12 selected brain regions; namely dorsolateral prefrontal cortex [DLPFC, Brodmann area [BA] 9], occipital cortex (BA19), temporal cortex (BA21), parietal cortex (BA40), amygdala, hippocampus, thalamus, hypothalamus, nucleus accumbens, caudate, substantia nigra and cerebellum. Relative allelic expression was used to assay for regional differences in cis-regulatory effects in 8 of these subjects for ZNF804A, 11 for RGS4, 5 for AKT1, 5 for NOS1 and 7 for TCF4.
Of the eight subjects assayed for ZNF804A, six showed significant (P < 0.05) differences in cDNA allele ratios between brain regions (Table 1). Levels of significance survived correction for eight separate tests in three of these subjects. Allelic expression imbalance (AEI) was most pronounced in the hippocampus from two of these subjects and in the nucleus accumbens in the third. For all three subjects, there was at least one brain region where cDNA allele ratios were found to average within 5% of the average genomic DNA (1:1) allele ratio, suggesting little or no effect of the assumed cis-regulatory variant(s) in those regions. Repeat assays using freshly synthesized cDNA confirmed significant (P < 0.05) differences in allele ratios between the region showing the most pronounced AEI and the region showing the least AEI for all three subjects.
Table 1.
Average ZNF804A cDNA allele ratios at rs12476147 in 10 brain regions from eight subjects
| Subject no. | DLPFC | OCC | TEMP | PAR | AMYG | HIPP | CER | NAC | CAUD | SN | P |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1.00 (0.06) | 0.99 (0.09) | 0.86 (0.08) | 1.17 (0.22) | 1.71 (0.45) | 1.15 (0.05) | 1.30 (0.21) | 1.15 (0.05) | 1.15 (0.05) | 0.0032 | |
| 5 | 0.91 (0.17) | 0.89 (0.08) | 1.01 (0.03) | 1.01 (0.09) | 1.21 (0.10) | 1.05 (0.11) | 1.02 (0.09) | 1.35 (0.19) | 1.21 (0.14) | 0.88 (0.26) | 0.0031 |
| 10 | 0.98 (0.05) | 1.07 (0.04) | 0.99 (0.16) | 1.00 (0.09) | 1.14 (0.15) | 1.06 (0.13) | 0.81 (0.15) | 1.03 (0.09) | 1.19 (0.13) | 0.93 (0.33) | 0.058 |
| 15 | 1.09 (0.18) | 1.19 (0.28) | 1.12 (0.11) | 1.32 (0.33) | 1.17 (0.39) | 1.28 (0.16) | 1.07 (0.08) | 1.04 (0.11) | 0.605 | ||
| 22 | 1.01 (0.12) | 0.88 (0.05) | 1.01 (0.06) | 0.97 (0.06) | 1.06 (0.04) | 0.97 (0.05) | 0.044 | ||||
| 24 | 1.03 (0.03) | 1.07 (0.10) | 1.08 (0.03) | 1.08 (0.05) | 1.13 (0.06) | 1.34 (0.04) | 1.03 (0.08) | 0.021 | |||
| 30 | 1.00 (0.06) | 1.15 (0.13) | 1.19 (0.02) | 1.11 (0.07) | 1.24 (0.10) | 0.90 (0.06) | 0.0053 | ||||
| 34 | 1.18 (0.07) | 1.09 (0.03) | 1.13 (0.05) | 1.09 (0.04) | 1.22 (0.04) | 1.03 (0.05) | 1.15 (0.07) | 1.13 (0.05) | 1.02 (0.11) | 0.0096 |
Ratios are the average of four measures of each sample, corrected by the average allele ratio derived from concurrent assay of heterozygous genomic DNA. Standard deviations are shown in parentheses. Blank cells correspond to samples that were unavailable for assay. P-values are given for Kruskal–Wallis tests of differences in allelic expression across all assayed brain regions for each subject. P-values in bold are those that survived Bonferroni correction for eight tests.
DLPFC, dorsolateral prefrontal cortex; OCC, occipital cortex; TEMP, temporal cortex; PAR, parietal cortex; AMYG, amygdala; HIPP, hippocampus; CER, cerebellum; NAC, nucleus accumbens; CAUD, caudate; SN, substantia nigra.
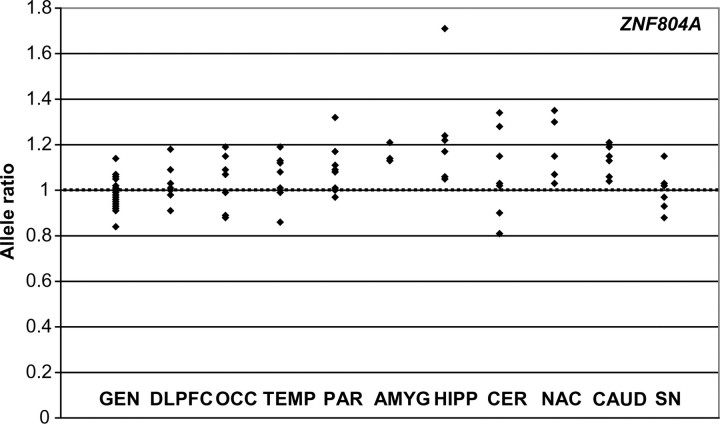
When ZNF804A allelic expression data from all regions and from all subjects were considered together, there was found to be a general over-expression of the A-allele of the assayed SNP rs12476147 relative to the T-allele (P < 0.0001), suggesting linkage disequilibrium with a functional regulatory variant. This is consistent with a recent study (12) that showed significant relative over-expression of the G-allele of another exonic ZNF804A SNP, rs4667001, with which the A-allele of rs12476147 is predicted to be consistently in phase (the two SNPs have an r2 of 1 in the HapMap CEU sample). We therefore performed a further analysis of ZNF804A allelic expression across brain regions in which we considered average allelic ratios from all eight assayed subjects together (data shown in Fig. 1). This analysis showed a general trend for a difference in ZNF804A allelic expression between assayed brain regions (Kruskal–Wallis χ2 = 16.5, d.f. = 9, P = 0.056). We therefore compared cDNA allele ratios from each brain region with genomic DNA allele ratios from all subjects using separate Mann–Whitney tests. Significant (P < 0.05) differences between cDNA and genomic DNA ratios were observed in the temporal cortex, parietal cortex, amygdala, hippocampus, nucleus accumbens and caudate, of which all but the temporal cortex survived Bonferroni correction for 10 tests. Differences between genomic DNA and cDNA allele ratios were most significant in the hippocampus (P = 0.00005, uncorrected), while cDNA allele ratios in the substantia nigra averaged very close to the 1:1 genomic ratio (average allele ratio = 0.996, P = 0.89 uncorrected).
Figure 1.
Allelic expression of ZNF804A at rs12476147 across 10 brain regions, compared with allele ratios in genomic DNA, in eight subjects. Data are represented as the average A/T allele ratio from four measures of each RNA sample and one measure of each genomic DNA sample, corrected by the average allele ratio observed in genomic DNA. The dotted horizontal line represents the average 1:1 genomic allele ratio. A general trend for a difference in allelic expression between brain regions was observed (P = 0.056). Significant (P < 0.05) differences between cDNA and genomic DNA ratios were observed in the temporal cortex (TEMP), parietal cortex (PAR), amygdala (AMYG), hippocampus (HIPP), nucleus accumbens (NAC) and caudate (CAUD). Allele ratios did not significantly differ between genomic DNA and cDNA from the DLPFC, occipital cortex (OCC), cerebellum (CER) or substantia nigra (SN).
Of the 11 individuals assayed for RGS4, 4 showed significant (P < 0.05) differences in allelic expression between brain regions (Table 2). Levels of significance survived correction for 11 separate tests in two of these subjects (nos 2 and 18). The RGS4 allele that was over-expressed differed between the two subjects, suggesting multiple cis-regulatory variants and/or incomplete linkage disequilibrium between these variants and the exonic SNP used to measure relative allelic expression. Significant (P < 0.001) differences in cDNA allele ratios between the nucleus accumbens and parietal cortex were confirmed in subject 18 using freshly synthesized cDNA. Depletion of RNA precluded similar confirmatory assays for subject 2.
Table 2.
Average RGS4 cDNA allele ratios at rs10759 in 10 brain regions from 11 subjects
| Subject no. | DLPFC | OCC | TEMP | PAR | AMYG | HIPP | CER | NAC | CAUD | SN | P |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 0.97 (0.02) | 1.04 (0.03) | 1.11 (0.05) | 1.06 (0.07) | 0.95 (0.15) | 0.64 (0.21) | 0.58 (0.16) | 0.89 (0.10) | 0.00321 | ||
| 4 | 0.93 (0.02) | 1.02 (0.02) | 1.01 (0.03) | 0.99 (0.05) | 1.22 (0.41) | 1.08 (0.08) | 1.02 (0.05) | 0.138 | |||
| 5 | 0.99 (0.10) | 0.96 (0.04) | 1.12 (0.13) | 1.05 (0.08) | 0.84 (0.40) | 0.91 (0.12) | 0.303 | ||||
| 7 | 1.08 (0.08) | 1.13 (0.06) | 1.17 (0.03) | 1.15 (0.08) | 1.16 (0.06) | 1.14 (0.03) | 1.17 (0.16) | 1.01 (0.03) | 0.101 | ||
| 8 | 0.91 (0.02) | 0.99 (0.07) | 0.97 (0.05) | 0.70 (0.15) | 0.63 (0.40) | 1.01 (0.13) | 0.53 (0.36) | 0.064 | |||
| 9 | 1.06 (0.09) | 0.96 (0.09) | 1.11 (0.05) | 1.09 (0.06) | 0.71 (0.43) | 1.05 (0.10) | 0.76 (0.41) | 0.339 | |||
| 10 | 1.05 (0.02) | 1.06 (0.01) | 1.07 (0.01) | 1.05 (0.02) | 1.01 (0.06) | 1.14 (0.07) | 0.027 | ||||
| 11 | 1.06 (0.11) | 1.17 (0.07) | 1.12 (0.04) | 1.04 (0.03) | 1.28 (0.37) | 1.19 (0.31) | 1.00 (0.12) | 0.251 | |||
| 12 | 0.99 (0.04) | 0.97 (0.02) | 1.00 (0.07) | 0.98 (0.04) | 0.90 (0.05) | 0.99 (0.09) | 0.81 (0.10) | 1.01 (0.13) | 1.23 (0.22) | 0.00849 | |
| 13 | 1.07 (0.04) | 1.13 (0.04) | 1.16 (0.04) | 1.09 (0.04) | 0.99 (0.05) | 1.14 (0.09) | 1.14 (0.25) | 1.12 (0.08) | 1.21 (0.13) | 0.063 | |
| 18 | 1.01 (0.06) | 1.07 (0.04) | 1.12 (0.02) | 1.02 (0.09) | 0.90 (0.05) | 1.10 (0.05) | 1.79 (0.05) | 1.15 (0.08) | 1.15 (0.08) | 0.00116 |
Ratios are the average of four measures of each sample, corrected by the average allele ratio derived from concurrent assay of heterozygous genomic DNA. Standard deviations are shown in parentheses. Blank cells correspond to samples that were unavailable for assay. P-values are given for Kruskal–Wallis tests of differences in allelic expression across all assayed brain regions for each subject. P-values in bold are those that survived Bonferroni correction for 11 tests. See Table 1 legend for key to abbreviated brain regions.
Of the five subjects assayed for AKT1, three showed significant (P < 0.05) differences in cDNA allele ratios between brain regions (Table 3). The significance of all three observations survived correction for five separate tests. Only for one of these subjects (no. 4) did repeat assays using new cDNA confirm significant differences in allelic expression between the region showing the highest and the region showing the least AEI (in this case, cerebellum and hippocampus, respectively).
Table 3.
Average AKT1 cDNA allele ratios at rs1130233 in 10 brain regions from five subjects
| Subject no. | DLPFC | OCC | TEMP | PAR | AMYG | HIPP | CER | NAC | CAUD | SN | P |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 1.09 (0.03) | 1.09 (0.08) | 1.12 (0.07) | 1.11 (0.05) | 1.21 (0.01) | 0.99 (0.05) | 1.24 (0.03) | 1.11 (0.04) | 1.05 (0.03) | 0.00177 | |
| 7 | 1.11 (0.04) | 1.11 (0.04) | 1.17 (0.01) | 1.18 (0.04) | 1.10 (0.07) | 1.10 (0.11) | 1.16 (0.05) | 1.06 (0.06) | 1.09 (0.02) | 0.90 (0.11) | 0.00413 |
| 8 | 1.07 (0.03) | 0.95 (0.05) | 1.04 (0.02) | 1.34 (0.08) | 0.94 (0.05) | 1.26 (0.07) | 1.42 (0.06) | 1.37 (0.10) | 0.00025 | ||
| 9 | 0.87 (0.12) | 0.99 (0.11) | 0.93 (0.05) | 1.00 (0.05) | 0.92 (0.09) | 1.01 (0.09) | 0.92 (0.15) | 0.88 (0.21) | 0.652 | ||
| 11 | 0.93 (0.09) | 0.91 (0.05) | 0.93 (0.04) | 0.91 (0.02) | 0.83 (0.05) | 0.95 (0.03) | 0.88 (0.08) | 0.88 (0.03) | 0.92 (0.07) | 0.083 |
Ratios are the average of four measures of each sample, corrected by the average allele ratio derived from concurrent assay of heterozygous genomic DNA. Standard deviations are shown in parentheses. Blank cells correspond to samples that were unavailable for assay. P-values are given for Kruskal–Wallis tests of differences in allelic expression across all assayed brain regions for each subject. P-values in bold are those that survived Bonferroni correction for five tests. See Table 1 legend for key to abbreviated brain regions.
All five subjects assayed for NOS1 showed significant (P < 0.05) differences in allelic expression between brain regions (Table 4). Levels of significance survived correction for five separate tests in three of these individuals. Repeat assays using freshly synthesized cDNA confirmed significant (P < 0.05) differences in cDNA allele ratios between the parietal cortex and caudate in subject 2 and the temporal cortex and substantia nigra in subject 22.
Table 4.
Average NOS1 cDNA allele ratios at rs1047735 in 12 brain regions from five subjects
| Subject no. | DLPFC | OCC | TEMP | PAR | AMY | HIPP | CER | NAC | CAUD | SN | THAL | HYPO | P |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 1.15 (0.05) | 1.02 (0.04) | 1.18 (0.23) | 1.00 (0.12) | 1.16 (0.05) | 1.30 (0.13) | 1.66 (0.20) | 1.20 (0.25) | 0.98 (0.02) | 1.12 (0.03) | 0.0021 | ||
| 5 | 1.15 (0.18) | 1.11 (0.08) | 1.06 (0.16) | 1.01 (0.14) | 1.10 (0.09) | 1.04 (1.12) | 0.99 (0.09) | 0.88 (0.05) | 0.89 (0.16) | 1.01 (0.12) | 1.26 (0.11) | 1.36 (0.28) | 0.011 |
| 11 | 0.77 (0.05) | 0.83 (0.13) | 0.78 (0.07) | 0.71 (0.02) | 0.81 (0.04) | 0.77 (0.13) | 1.18 (0.13) | 0.76 (0.06) | 0.85 (0.05) | 0.81 (0.19) | 0.68 (0.12) | 0.75 (0.02) | 0.026 |
| 22 | 1.10 (0.10) | 1.03 (0.06) | 0.99 (0.08) | 0.95 (0.03) | 0.84 (0.03) | 0.58 (0.01) | 0.0009 | ||||||
| 34 | 0.97 (0.04) | 0.89 (0.01) | 0.91 (0.04) | 0.91 (0.03) | 0.85 (0.01) | 1.01 (0.03) | 0.93 (0.02) | 1.00 (0.04) | 1.03 (0.05) | 0.98 (0.03) | 0.0002 |
Ratios are the average of four measures of each sample, corrected by the average allele ratio derived from concurrent assay of heterozygous genomic DNA. Standard deviations are shown in parentheses. Blank cells correspond to samples that were unavailable for assay. P-values are given for Kruskal–Wallis tests of differences in allelic expression across all assayed brain regions for each subject. P-values in bold are those that survived Bonferroni correction for five tests. See Table 1 legend for key to abbreviated brain regions. Additional brain regions assayed for NOS1: THAL, thalamus; HYPO, hypothalamus.
All seven subjects assayed for TCF4 showed significant (P < 0.05) differences in allelic expression between brain regions, with levels of significance surviving Bonferroni correction in six cases (Table 5). Regions showing pronounced AEI largely differed between subjects. Repeat assays using freshly synthesized cDNA confirmed significant (P < 0.05) differences in allele ratios between the region showing the most pronounced AEI and the region showing the least AEI in four of the six subjects (nos 8, 11, 56 and 57). As for the assayed SNP in ZNF804A, when data from all subjects and regions were combined, there was a general trend for directional AEI at TCF4 exonic SNP rs8766 (P < 0.0001), suggesting linkage disequilibrium with a functional variant. However, in contrast to our assay of ZNF804A, there was little suggestion that the extent of TCF4 AEI followed a common regional pattern, as no regional differences in cDNA ratios were observed when all subjects were combined (Kruskal–Wallis χ2 = 11.5, d.f. = 8, P = 0.17). Instead, significant (P < 0.01) differences were seen between genomic DNA and cDNA allele ratios derived from every assayed brain region, with the G-allele of rs8766 relatively over-expressed in each case.
Table 5.
Average TCF4 cDNA allele ratios at rs8766 in 10 brain regions from seven subjects
| Subject no. | DLPFC | OCC | TEMP | PAR | AMYG | HIPP | CER | NAC | CAUD | SN | P |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 1.26 (0.03) | 1.31 (0.05) | 1.35 (0.04) | 1.27 (0.03) | 1.31 (0.02) | 1.19 (0.02) | 1.34 (0.04) | 1.20 (0.01) | 0.00074 | ||
| 8 | 1.18 (0.02) | 1.24 (0.01) | 1.10 (0.03) | 1.28 (0.04) | 1.23 (0.03) | 1.07 (0.02) | 1.24 (0.03) | 1.15 (0.01) | 0.00022 | ||
| 11 | 1.15 (0.03) | 1.21 (0.04) | 1.17 (0.02) | 1.06 (0.02) | 1.05 (0.02) | 1.14 (0.02) | 1.09 (0.02) | 1.09 (0.06) | 1.09 (0.02) | 1.21 (0.14) | 0.00107 |
| 13 | 1.14 (0.02) | 1.18 (0.02) | 1.18 (0.01) | 1.09 (0.05) | 1.11 (0.03) | 1.12 (0.04) | 1.08 (0.03) | 1.16 (0.04) | 1.13 (0.03) | 1.14 (0.02) | 0.019 |
| 56 | 1.13 (0.05) | 1.22 (0.02) | 1.33 (0.03) | 1.15 (0.02) | 1.15 (0.05) | 1.07 (0.02) | 1.24 (0.05) | 0.00069 | |||
| 57 | 1.10 (0.03) | 1.05 (0.03) | 1.09 (0.03) | 1.07 (0.03) | 1.13 (0.03) | 1.25 (0.03) | 1.08 (0.02) | 1.18 (0.04) | 0.0012 | ||
| 58 | 1.19 (0.02) | 1.27 (0.02) | 1.19 (0.01) | 1.13 (0.02) | 1.13 (0.02) | 1.37 (0.03) | 1.21 (0.04) | 0.00056 |
Ratios are the average of four measures of each sample, corrected by the average allele ratio derived from concurrent assay of heterozygous genomic DNA. Standard deviations are shown in parentheses. Blank cells correspond to samples that were unavailable for assay. P-values are given for Kruskal–Wallis tests of differences in allelic expression across all assayed brain regions for each subject. P-values in bold are those that survived Bonferroni correction for seven tests. See Table 1 legend for key to abbreviated brain regions.
The subjects showing significant regional differences in allelic expression (surviving correction for multiple tests) differed between gene assays, with the exception of one subject showing regional differences in both RGS4 and NOS1 and two subjects showing such differences in both AKT1 and TCF4. Regional differences were therefore seen in a large proportion of the assayed subjects (14/20), rather than arising from a few particular samples. None of the DNase-treated RNA samples gave rise to a visible product on agarose in the absence of reverse transcription (RT), indicating that differences in allelic expression were not the result of residual genomic DNA contamination (which would artificially bias cDNA ratios towards the 1:1 genomic ratio).
DISCUSSION
Using measures of relative allelic expression across multiple brain regions, within individual subjects, we have shown that the effects of cis-variation on gene expression can significantly differ between sampled areas of the adult brain. Only by testing within individual subjects could we be certain that the genetic cis-regulatory variants were constant between brain regions. For all five assayed genes, we observed significant regional differences in allelic expression in multiple subjects, suggesting this to be a common phenomenon. In the case of ZNF804A, a robustly supported susceptibility gene for schizophrenia (11,12,16), we observed a general regional pattern of allelic expression at the assayed SNP. Our data therefore extend previous observations of tissue specificity of allelic expression e.g. (4,6–8) in showing that this can also differ between regions of the adult human brain.
All five assayed genes have been reported to show genetic association with neuropsychiatric illness (11–16). Previous evidence exists for common genotypic effects on the expression of all of these genes (12,14,16–18), with the exception of TCF4 [a susceptibility gene for schizophrenia (15) which is mutated in Pitt–Hopkins syndrome (19,20)]. We therefore biased our screen in favor of genes that would contain common (genetic) cis-regulatory variants. However, for none of these genes was there any prior evidence that effects of cis-regulatory variation could differ depending on brain region.
We have not attempted to relate our findings to DNA variants for which phenotypic associations have been reported (as exemplified in 21) because our subject numbers are too small for meaningful statistical comparison between genotypes. The precise mechanisms underlying AEI of these genes, and its variation between brain regions, therefore remain open to speculation. AEI can have a genetic or epigenetic basis, the former explained by heterozygosity for DNA sequence variants affecting cis-regulatory elements, and the latter by allele-specific modifications of DNA or chromatin. A plausible (genetic) explanation for our findings is that the effects of DNA variation in cis-regulatory elements (which might include DNA variants associated with disease) vary depending on regional differences in the level of interacting trans-regulators (e.g. transcription factors, hormones, microRNA). This explanation is consistent with in vitro findings of cellular differences in the effect of DNA variants in transcription factor binding sites on gene expression (5,6). An alternative (epigenetic) explanation is that the assayed genes are subject to an imprinting mechanism occurring in only a proportion of cells, which are then differentially sampled between brain regions. However, imprinting appears to affect only a small proportion of human genes (22), and there is no previous evidence that any of the five genes assayed in this study are subject to such regulation. Recent evidence suggests that a significant proportion of human genes are subject to random epigenetic allele silencing that can persist in clonal cell lines (23). However, this also appears an unlikely explanation for our findings, since the number of sampled cells from each region would presumably result in any purely random bias for either allele being canceled out. Within subjects, we observed a general consistency in which allele was over-expressed, again arguing against a purely random mechanism. Recent studies have indicated that common allelic differences in DNA methylation are usually associated with DNA sequence variation (24,25). Moreover, there is evidence that such allele-specific methylation can differ between tissues (25). Differences between brain regions in the extent of methylation at variable DNA sites may therefore provide an alternative explanation for our findings.
In the case of ZNF804A and TCF4, we observed a general consistency across subjects in which allele was relatively over-expressed, suggesting either that the assayed SNPs have direct effects on gene expression or (perhaps more likely) that they are in linkage disequilibrium with DNA variants of that effect. General AEI of ZNF804A in human brain tissue has recently been reported at an exonic SNP with which ours is likely to be in perfect linkage disequilibrium (r2 = 1 in the HapMap CEU sample), and which is itself in strong linkage disequilibrium with the intronic SNP showing strongest association with schizophrenia (12). It should be noted, however, that in a comparison between heterozygotes and homozygotes for the risk SNP, the authors found no difference in (cortical) allelic expression, leading to the conclusion that the risk SNP was not in itself responsible for AEI. In the present study, we have found that allelic expression of ZNF804A is generally pronounced in some brain regions and very little in others. In contrast, in our assay of TCF4, we observed similar levels of general AEI across all assayed brain regions. However, on the individual subject level, we found that, while there was directional AEI of TCF4 in all brain regions, the extent of this imbalance would frequently differ significantly (and reproducibly) between regions. Given that there appears to be little consistency in the region(s) showing pronounced AEI across subjects, it is possible that there are multiple regionally sensitive regulatory variants in TCF4, leading to individual differences. Alternatively, there may be a single cis-regulatory variant with differential effects on the many TCF4 transcripts known to exist, which are then differentially expressed/sampled between regions and subjects. Transcript-specific assays of allelic expression are likely to be informative for this and other genes assayed in the present study.
The human brain serves a wide range of functions, from physiological homeostasis to higher cognition. While multiple brain regions are typically recruited for any given function, it is clear that there exists a high degree of regional specialization. The effects of cis-regulatory variation are therefore likely to have distinct phenotypic consequences depending on where in the brain they are manifest. Our data show that such effects are often far more restricted than expression of the regulated gene per se.
Our demonstration of differential allelic expression across brain regions has important implications for studies of cis-regulatory variation in the brain. First, it highlights the caveat that it may not be possible to generalize findings to brain areas that have not been assayed. Second, it suggests that power to detect association between particular gene variants and gene expression may be improved by careful matching of tissue samples. Third, it shows that specific regions of the brain where the effects of cis-regulatory variants are manifest (or pronounced) can be delineated. Using such an approach, it should therefore be possible to define brain areas, and perhaps even cell populations, in which cis-regulatory variants conferring susceptibility to neuropsychiatric disease are active.
MATERIALS AND METHODS
Brain samples
Experiments were carried out using postmortem brain tissue from 20 unrelated Caucasian subjects obtained from the MRC London Neurodegenerative Diseases Brain Bank. Subjects comprised 12 males and 8 females with a mean age of death of 66 years (SD = 18 years). All subjects were free from psychiatric or neurological diagnosis at the time of death. Subjects were selected from an initial sample of 64 control individuals on the basis of heterozygosity for an expressed SNP in one or more of the five assayed genes and availability of tissue from at least six of the selected brain regions. Twelve discrete brain regions were selected for study: DLPFC (BA9), occipital cortex (BA19), temporal cortex (BA21), parietal cortex (BA40), amygdala, hippocampus, thalamus, hypothalamus, nucleus accumbens, caudate, substantia nigra and cerebellum. Where available, ∼100 mg tissue from each of the 12 brain areas was dissected from one hemisphere of the frozen postmortem brain from each subject by a trained neuropathologist (T.H.). Each tissue sample was placed in a separate 2 ml Lysing Matrix D tube (MP Biomedicals), homogenized in 1 ml Tri-Reagent® solution (Ambion) using a FastPrep®-24 (MP Biomedicals) and RNA extracted following the standard Tri-Reagent protocol. Total RNA was treated with Turbo DNase™ (Ambion) prior to RT. Absence of residual genomic DNA was confirmed by PCR, using amplification primers designed for the allelic expression assays, and UV visualization on agarose. Two RT reactions were performed for each sample using ∼1 µg total RNA, random decamers (Ambion) and M-MLV reverse transcriptase (Ambion). For those subjects showing significant regional differences in allelic expression (surviving correction for multiple testing), a further two RT reactions were performed for both the region showing the largest AEI and the region showing the least AEI. All cDNA samples were diluted 1/7 prior to assay of allelic expression.
Allelic expression assays
Assays of relative allelic expression required that subjects were heterozygous for an SNP in exonic sequence, in order to distinguish and quantify RNA transcribed from each parental chromosome. Exonic SNP rs12476147 was used to assay ZNF804A, rs10759 to assay RGS4, rs1130233 to assay AKT1, rs1047735 to assay NOS1 and rs8766 to assay TCF4. Assayable heterozygotes were initially identified by genotyping genomic DNA from 64 control individuals for each of these polymorphisms. Assays of relative allelic expression were performed for each gene on a per-subject basis, with cDNA from all available brain regions from each subject assayed concurrently, alongside genomic DNA from heterozygous subjects and H2O negative controls, to ensure no contamination which could bias allele ratios. Sequence containing the exonic SNP was first PCR-amplified from two RT reactions for each brain region, two heterozygous genomic DNA samples and two negative controls. PCR primers were based on a single exon sequence, designed to produce the same amplicon from both cDNA and genomic DNA (primer sequences available on request). PCR was carried out in a total reaction volume of 12 µl, containing 1× HotStarTaq® Plus Master Mix (Qiagen), 0.3 µm primers and 6 µl of either genomic DNA (at 4 ng/µl), cDNA preparation or H2O, with 35 cycles and annealing temperatures of between 50°C and 60°C. After completion, 4 µl of each PCR reaction was electrophoresed on agarose and visualized by UV illumination to confirm amplification and no PCR contamination. PCR reactions were then incubated with 1 U shrimp alkaline phosphatase (GE Healthcare) and 2 U Exonuclease I (New England Biolabs) at 37°C for 45 min and then at 85°C for 15 min prior to primer extension reactions. As for PCR, primer extension reactions were carried out on the PCR products from all brain regions from each individual concurrently, alongside those from heterozygous genomic DNA. Primer extension reactions were performed in a total volume of 10 µl, containing 2 µl treated PCR product, 1.25 µl SNaPshot® reagent (Applied Biosystems), 5.75 µl H2O and 1 pM extension primer (sequences available on request). Primer extension thermocycling conditions consisted of an initial step of 95°C for 2 min, followed by 30 cycles of 95°C for 5 s, 43°C for 5 s and 60°C for 5 s. Aliquots of 2 µl SNaPshot reaction product were combined with 8 µl Hi-Di formamide (Applied Biosystems) and electrophoresed on a 3130 Genetic Analyzer (Applied Biosystems). Peak heights of allele-specific extended primers were determined using GeneMarker® software (SoftGenetics) and the ratio of the two peak heights calculated for each reaction. For each plate, the average ratio of allelic peak heights for genomic DNA was used as a correction factor for all allele ratios, since this can be assumed to reflect a perfect 1:1 ratio of the two alleles and can therefore be used to correct for any inequalities in allelic representation specific to each assay (2). The above procedure was performed twice for each sample, giving a total of four corrected cDNA allele ratios for each brain region. For the additional two RT reactions per region from subjects showing significant regional differences in allelic expression in the first analysis, the above procedure was performed four times, giving a total of eight corrected cDNA allele ratios for each repeated brain region.
Statistical analyses
Statistical tests were performed using SPSS 15.0. cDNA allele ratios were initially compared between all assayed brain regions within each subject using Kruskal–Wallis tests. Derived P-values for each subject assay were corrected by the number of individual tests for each gene (i.e. subjects assayed) using Bonferroni correction. In subjects showing significant differences in cDNA ratios between brain regions (surviving Bonferroni correction), allele ratios from the region originally showing the greatest AEI were compared with those from the region originally showing the least AEI using fresh preparations of cDNA and Mann–Whitney tests. Differences were considered to be significantly confirmed when the direction of allelic imbalance was the same as in the initial assay and when P < 0.05. All tests were two-tailed.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by the Medical Research Council, UK (Grant ID: 89465).
REFERENCES
- 1.Yan H., Yuan W., Velculescu V.E., Vogelstein B., Kinzler K.W. Allelic variation in human gene expression. Science. 2002;297:1143. doi: 10.1126/science.1072545. doi:10.1093/humrep/deh509. [DOI] [PubMed] [Google Scholar]
- 2.Bray N.J., Buckland P.R., Owen M.J., O'Donovan M.C. Cis-acting variation in the expression of a high proportion of genes in human brain. Hum. Genet. 2003;113:149–153. doi: 10.1007/s00439-003-0956-y. doi:10.1001/jama.273.11.854. [DOI] [PubMed] [Google Scholar]
- 3.Lo H.S., Wang Z., Hu Y., Yang H.H., Gere S., Buetow K.H., Lee M.P. Allelic variation in gene expression is common in the human genome. Genome Res. 2003;13:1855–1862. doi: 10.1101/gr.1006603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cowles C.R., Hirschhorn J.N., Altshuler D., Lander E.S. Detection of regulatory variation in mouse genes. Nat. Genet. 2002;32:432–437. doi: 10.1038/ng992. doi:10.1093/humrep/13.2.500. [DOI] [PubMed] [Google Scholar]
- 5.Koch O., Kwiatkowski D.P., Udalova I.A. Context-specific functional effects of IFNGR1 promoter polymorphism. Hum. Mol. Genet. 2006;15:1475–1481. doi: 10.1093/hmg/ddl071. doi:10.1001/jama.257.8.1079. [DOI] [PubMed] [Google Scholar]
- 6.Sun C., Southard C., Witonsky D.B., Olopade O.I., Di Rienzo A. Allelic imbalance (AI) identifies novel tissue-specific cis-regulatory variation for human UGT2B15. Hum. Mutat. 2010;31:99–107. doi: 10.1002/humu.21145. doi:10.1097/00002030-200403260-00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilkins J.M., Southam L., Price A.J., Mustafa Z., Carr A., Loughlin J. Extreme context specificity in differential allelic expression. Hum. Mol. Genet. 2007;16:537–546. doi: 10.1093/hmg/ddl488. doi:10.1016/S0264-410X(00)00460-6. [DOI] [PubMed] [Google Scholar]
- 8.Zhang K., Li J.B., Gao Y., Egli D., Xie B., Deng J., Li Z., Lee J.H., Aach J., Leproust E.M., et al. Digital RNA allelotyping reveals tissue-specific and allele-specific gene expression in human. Nat. Methods. 2009;6:613–618. doi: 10.1038/nmeth.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emilsson V., Thorleifsson G., Zhang B., Leonardson A.S., Zink F., Zhu J., Carlson S., Helgason A., Walters G.B., Gunnarsdottir S., et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. doi:10.1097/QAD.0b013e3282703879. [DOI] [PubMed] [Google Scholar]
- 10.Dimas A.S., Deutsch S., Stranger B.E., Montgomery S.B., Borel C., Attar-Cohen H., Ingle C., Beazley C., Gutierrez Arcelus M., Sekowska M., et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. doi:10.1093/humrep/17.12.3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Donovan M.C., Craddock N., Norton N., Williams H., Peirce T., Moskvina V., Nikolov I., Hamshere M., Carroll L., Georgieva L., et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat. Genet. 2008;40:1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- 12.Williams H.J., Norton N., Dwyer S., Moskvina V., Nikolov I., Carroll L., Georgieva L., Williams N.M., Morris D.W., Quinn E.M., et al. Fine mapping of ZNF804A and genome-wide significant evidence for its involvement in schizophrenia and bipolar disorder. Mol. Psychiatry. 2010 doi: 10.1038/mp.2010.36. Epub ahead of print]. doi:10.1038/mp.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chowdari K.V., Mirnics K., Semwal P., Wood J., Lawrence E., Bhatia T., Deshpande S.N., B K.T., Ferrell R.E., Middleton F.A., et al. Association and linkage analyses of RGS4 polymorphisms in schizophrenia. Hum. Mol. Genet. 2002;11:1373–1380. doi: 10.1093/hmg/11.12.1373. [DOI] [PubMed] [Google Scholar]
- 14.Emamian E.S., Hall D., Birnbaum M.J., Karayiorgou M., Gogos J.A. Convergent evidence for impaired AKT1-GSK3beta signaling in schizophrenia. Nat. Genet. 2002;36:131–137. doi: 10.1038/ng1296. doi:10.1093/humrep/deg431. [DOI] [PubMed] [Google Scholar]
- 15.Stefansson H., Ophoff R.A., Steinberg S., Andreassen O.A., Cichon S., Rujescu D., Werge T., Pietiläinen O.P., Mors O., Mortensen P.B., et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. doi:10.1093/humrep/17.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riley B., Thiselton D., Maher B.S., Bigdeli T., Wormley B., McMichael G.O., Fanous A.H., Vladimirov V., O'Neill F.A., Walsh D., Kendler K.S. Replication of association between schizophrenia and ZNF804A in the Irish case–control study of schizophrenia sample. Mol. Psychiatry. 2010;15:29–37. doi: 10.1038/mp.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reif A., Jacob C.P., Rujescu D., Herterich S., Lang S., Gutknecht L., Baehne C.G., Strobel A., Freitag C.M., Giegling I., et al. Influence of functional variant of neuronal nitric oxide synthase on impulsive behaviors in humans. Arch. Gen. Psychiatry. 2009;66:41–50. doi: 10.1001/archgenpsychiatry.2008.510. doi:10.1046/j.1537-2995.1997.37897424411.x. [DOI] [PubMed] [Google Scholar]
- 18.Emilsson L., Saetre P., Jazin E. Low mRNA levels of RGS4 splice variants in Alzheimer's disease: association between a rare haplotype and decreased mRNA expression. Synapse. 2006;59:173–176. doi: 10.1002/syn.20226. [DOI] [PubMed] [Google Scholar]
- 19.Amiel J., Rio M., de Pontual L., Redon R., Malan V., Boddaert N., Plouin P., Carter N.P., Lyonnet S., Munnich A., Colleaux L. Mutations in TCF4, encoding a class I basic helix-loop-helix transcription factor, are responsible for Pitt–Hopkins syndrome, a severe epileptic encephalopathy associated with autonomic dysfunction. Am. J. Hum. Genet. 2007;80:988–993. doi: 10.1086/515582. doi:10.1093/humrep/deh460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brockschmidt A., Todt U., Ryu S., Hoischen A., Landwehr C., Birnbaum S., Frenck W., Radlwimmer B., Lichter P., Engels H., et al. Severe mental retardation with breathing abnormalities (Pitt–Hopkins syndrome) is caused by haploinsufficiency of the neuronal bHLH transcription factor TCF4. Hum. Mol. Genet. 2007;16:1488–1494. doi: 10.1093/hmg/ddm099. doi:10.1080/1464770312331369343. [DOI] [PubMed] [Google Scholar]
- 21.Bray N.J., Preece A., Williams N.M., Moskvina V., Buckland P.R., Owen M.J., O'Donovan M.C. Haplotypes at the dystrobrevin binding protein 1 (DTNBP1) gene locus mediate risk for schizophrenia through reduced DTNBP1 expression. Hum. Mol. Genet. 2005;14:1947–1954. doi: 10.1093/hmg/ddi199. doi:10.1093/humrep/deh828. [DOI] [PubMed] [Google Scholar]
- 22.Bartolomei M.S. Genomic imprinting: employing and avoiding epigenetic processes. Genes Dev. 2009;23:2124–2133. doi: 10.1101/gad.1841409. doi:10.1097/01.aids.0000200532.56490.fe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gimelbrant A., Hutchinson J.N., Thompson B.R., Chess A. Widespread monoallelic expression on human autosomes. Science. 2007;318:1136–1140. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- 24.Schilling E., El Chartouni C., Rehli M. Allele-specific DNA methylation in mouse strains is mainly determined by cis-acting sequences. Genome Res. 2009;19:2028–2035. doi: 10.1101/gr.095562.109. doi:10.1093/humrep/dei459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schalkwyk L.C., Meaburn E.L., Smith R., Dempster E.L., Jeffries A.R., Davies M.N., Plomin R., Mill J. Allelic skewing of DNA methylation is widespread across the genome. Am. J. Hum. Genet. 2010;86:196–212. doi: 10.1016/j.ajhg.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]