Abstract
We performed a multistage genome-wide association study of melanoma. In a discovery cohort of 1804 melanoma cases and 1026 controls, we identified loci at chromosomes 15q13.1 (HERC2/OCA2 region) and 16q24.3 (MC1R) regions that reached genome-wide significance within this study and also found strong evidence for genetic effects on susceptibility to melanoma from markers on chromosome 9p21.3 in the p16/ARF region and on chromosome 1q21.3 (ARNT/LASS2/ANXA9 region). The most significant single-nucleotide polymorphisms (SNPs) in the 15q13.1 locus (rs1129038 and rs12913832) lie within a genomic region that has profound effects on eye and skin color; notably, 50% of variability in eye color is associated with variation in the SNP rs12913832. Because eye and skin colors vary across European populations, we further evaluated the associations of the significant SNPs after carefully adjusting for European substructure. We also evaluated the top 10 most significant SNPs by using data from three other genome-wide scans. Additional in silico data provided replication of the findings from the most significant region on chromosome 1q21.3 rs7412746 (P = 6 × 10−10). Together, these data identified several candidate genes for additional studies to identify causal variants predisposing to increased risk for developing melanoma.
INTRODUCTION
Cutaneous melanoma (CM) is among the more common cancers, with more than 70 000 estimated new cases in the USA (excluding melanoma in situ) in 2011 (1). The risk for siblings of index cases who have CM is ∼2-fold higher than that for the general population (2). Linkage studies and genome-wide association studies (GWASs) have identified several loci influencing CM risk. Cyclin-dependent kinase inhibitor 2A (CDKN2A or p16), an alternate reading frame of CDKN2A (ARF or p14), and cyclin-dependent kinase 4 (CDK4) in region 9p21.3 have been identified as high-penetrance CM susceptibility genes (3), but mutations in these genes are rare and explain only a small fraction of familial CM risk. However, GWASs found evidence that common variation in the CDKN2A region also commonly influence risk for melanoma (4,5). Mutations in the melanocortin 1 receptor (MC1R) gene in the chromosomal region 16q24.3 influence skin pigmentation, and a candidate gene analysis followed by several confirmatory studies documented an ∼40% increased risk of CM in those who carried the variants conferring lighter skin pigmentation according to specific mutations (6–8).
In addition, GWASs have identified several other loci that may contribute to CM risk. GWAS analyses have revealed several additional genes associated with skin pigmentation, including TYR (tyrosinase, chromosome 11q21) (9), TYRP1 (tyrosinase-related protein 1, chromosome 9p23) (10), SLC45A2 (solute carrier family 45, member 2, chromosome 5p13.3), SLC24A4 (solute carrier family 24, member 4, chromosome 14q32.12) and IRF4 (interferon regulatory factor 4, chromosome 6p25.3). A case–control study (11) that used a pooling approach to identify common variants influencing CM risk found several genetic factors influencing risk across a broad region of chromosome 20q11.2 that encompasses the genetic loci, PIGU, EIF2S2, NCOA6 and MYH17B. Finally, the locus PLA2G6 (4) on chromosome 22q13 was also previously associated with CM risk in a GWAS. The goal of the current GWAS was to identify novel variants associated with CM risk by using dense single-nucleotide polymorphism (SNP) arrays on a large population of CM cases that were collected uniformly from a single cancer center in the USA and to then validate these findings with use of additional data from US, Australian and European populations.
RESULTS
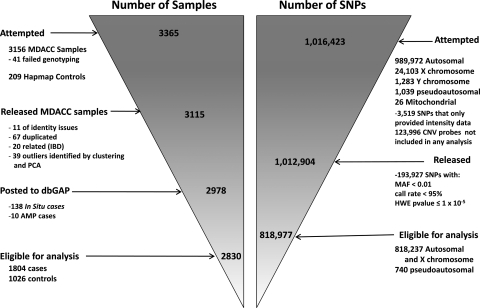
To identify risk variants for CM, we used an Illumina Omni1-Quad_v1-0_B array to genotype 1 016 423 SNPs in 3115 participants. After the application of quality control (QC) criteria (Supplementary Material, Tables S1 and S2 and Fig. 1), genotypes were available for 1804 Caucasian CM patients presenting to clinics (other than dermatology) at The University of Texas MD Anderson Cancer Center and 1026 Caucasian cancer-free controls who were friends or acquaintances accompanying patients at their clinical visits. We analyzed 1 012 904 probes with a mean sample call rate of 99.66%. After applying QC filters, we retained data for analysis from 818 977 SNPs (818 237 autosomal or X chromosome SNPs and 740 psuedoautosomal SNPs) that had a minor allele frequency (MAF) of >1%, were genotyped for 95% or more of retained participants and did not deviate from Hardy–Weinberg equilibrium (P> 1 × 10−5). Individual genotypes that had Illumina GenTrain version 1.0 quality scores of ≤0.15 were set to missing. Figure 1 and Supplementary Tables 1 and 2 present an overall description of the sample processing and in silico replication phases of this study.
Figure 1.
Description of the flow of samples and markers through the genotyping study reported for CM cases and controls from MD Anderson Cancer Center.
Table 1.
Associations of SNPs previously found in GWASs to be associated with CM risk, A1 is the minor allele in forward strand
| Chr. | SNP | Source | Gene | r2 | MAF | Reference | Position | P-value | Ref. All. | OR | 95% CI |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | rs16891982 | Imputed | SLC45A2 | 0.89 | 0.022 | 13 | 33 987 450 | 0.0026 | C | 0.54 | 0.37–0.81 |
| 5 | rs35391 | Typed | SLC45A2 | 0.014 | 13 | 33 991 430 | 0.020 | A | 0.58 | 0.37–0.91 | |
| 5 | rs28777 | Typed | SLC45A2 | 0.017 | 13 | 33 994 716 | 0.0048 | C | 0.56 | 0.37–0.84 | |
| 9 | rs1408799 | Typed | TYRP1 | 0.30 | 9,10, 13 | 12 662 097 | 0.70 | T | 0.98 | 0.87–1.10 | |
| 9 | rs2218220 | Imputed | near MTAP | 1.00 | 0.48 | 4 | 21 746 089 | 0.00014 | T | 0.81 | 0.73–0.90 |
| 9 | rs1335510 | Imputed | near MTAP | 1.00 | 0.39 | 4 | 21 747 803 | 5.01 × 10−5 | G | 0.79 | 0.71–0.89 |
| 9 | rs935053 | Imputed | near MTAP | 0.99 | 0.47 | 4 | 21 773 922 | 4.49 × 10−5 | A | 0.80 | 0.71–0.89 |
| 9 | rs10757257 | Imputed | MTAP | 0.99 | 0.38 | 4 | 21 796 564 | 1.96 × 10−5 | A | 0.78 | 0.70–0.88 |
| 9 | rs7023329 | Typed | MTAP | 0.47 | 4 | 21 806 528 | 1.86 × 10−5 | G | 0.79 | 0.71–0.88 | |
| 11 | rs1042602 | Typed | TYR | 0.36 | 4, 10 | 88 551 344 | 0.045 | A | 0.89 | 0.79–1.00 | |
| 11 | rs1393350 | Typed | TYR | 0.30 | 4, 10 | 88 650 694 | 0.0085 | A | 1.18 | 1.04–1.33 | |
| 11 | rs1847142 | Imputed | TYR | 0.99 | 0.34 | 4 | 88 661 222 | 0.0021 | A | 1.20 | 1.07–1.35 |
| 11 | rs10830253 | Imputed | TYR | 0.99 | 0.34 | 4 | 88 667 691 | 0.0022 | G | 1.20 | 1.07–1.34 |
| 11 | rs1806319 | Typed | TYR | 0.39 | 4 | 88 677 584 | 0.0050 | C | 1.17 | 1.05–1.31 | |
| 14 | rs12896399 | Typed | SLC24A4 | 0.46 | 10 | 91 843 416 | 0.33 | T | 1.06 | 0.95–1.18 | |
| 15 | rs1800407 | Typed | OCA2 | 0.08 | 13 | 25 903 913 | 0.67 | A | 0.95 | 0.78–1.17 | |
| 16 | rs1805006 | Typed | MC1R | 0.01 | 13 | 88 513 419 | 0.053 | A | 1.67 | 0.97–2.86 | |
| 16 | rs1805007 | Imputed | MC1R | 0.81 | 0.13 | 10, 13 | 88 513 618 | 4.1 × 10−9 | T | 1.74 | 1.44–2.11 |
| 16 | rs1805008 | Typed | MC1R | 0.10 | 10,13 | 88 513 645 | 0.001 | A | 1.37 | 1.13–1.67 | |
| 20 | rs4911414 | Typed | LOC729547 | 0.37 | 11 | 32 193 105 | 0.090 | T | 1.11 | 0.98–1.24 | |
| 20 | rs1015362 | Imputed | EIF2S2 | 0.96 | 0.27 | 11 | 32 202 273 | 0.31 | T | 0.94 | 0.83–1.06 |
| 20 | rs910873 | Typed | PIGU | 0.11 | 11 | 32 635 433 | 0.00017 | A | 1.42 | 1.18–1.71 | |
| 20 | rs17305573 | Typed | PIGU | 0.11 | 11 | 32 643 813 | 0.00017 | C | 1.42 | 1.18–1.71 | |
| 20 | rs4911442 | Imputed | NCOA6 | 0.98 | 0.15 | 11 | 32 818 707 | 0.0029 | G | 1.27 | 1.08–1.49 |
| 20 | rs1885120 | Typed | MYH7B | 0.11 | 11 | 33 040 650 | 0.00016 | C | 1.43 | 1.18–1.72 | |
| 22 | rs2284063 | Imputed | PLA2G6 | 0.97 | 0.35 | 4 | 36 874 244 | 0.0045 | G | 0.84 | 0.75–0.95 |
| 22 | rs6001027 | Typed | PLA2G6 | 0.34 | 4 | 36 875 565 | 0.0039 | G | 0.84 | 0.75–0.95 |
Table 2.
Association of HERC2 rs12913832 with pigmentary phenotypes in the MD Anderson study
| GWAS phenotype | WT/VT | MAF | Estimate | SE | P-value |
|---|---|---|---|---|---|
| Hair color (blonde to black) | G/A | 0.2625 | 0.3924 | 0.0407 | 5.35 × 10−22 |
| Eye color (blue to brown) | G/A | 0.2620 | 0.9193 | 0.0283 | 1.79 × 10−231 |
| Tanning ability (none to deep) | G/A | 0.2589 | 0.2008 | 0.0489 | 4.07 × 10−5 |
WT, wild-type; VT, variant type; SE, standard error.
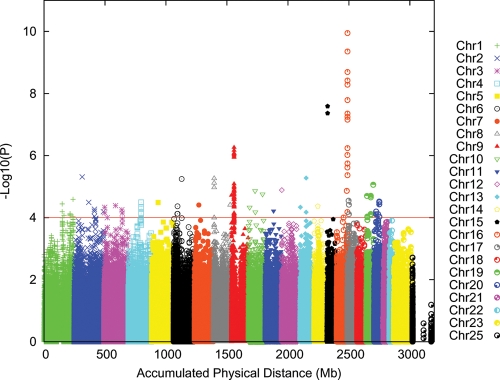
Initial analyses were performed using the additive logistic regression model implemented in PLINK (12). The Q–Q plots for the discovery sample are shown in Supplementary Material, Figures S1 and S2. The genomic inflation factor (λ) for the discovery sample was 1.020, and this was reduced when the first two principal components (PCs) were included as covariates (λ = 1.011). Association results across the genome after correcting for the two PCs are shown in Figure 2 and Supplementary Material, Table S3 (association tests with P < 1 × 10−4). The two regions reaching genome-wide significance (5 × 10−8) are located on 15q13.1 (HERC2/OCA2 region) and 16q24.3 (MC1R region) centered at or near the MC1R and HERC2/OCA2 genes, with a third highly significant region at chromosome 9p21.3 (near CDKN2A/ARF genes). In Table 1, we present the results for previously reported associations of CM risk with SNPs. Of the previously reported 27 risk-associated SNPs, 21 were significant in our study at P < 0.05. Individual SNPs that were previously associated with CM risk but did not reach significance in our study included rs1408799 in TYRP1, rs12896399 in SLC24A4, rs1800407 in OCA2, rs1805006 in MC1R, rs4911414 in LOC729547 and rs1015362 in EIF2S2, but associations for most previously reported SNPs were supported in our study, and except for the region around SLC24A4 on chromosome 14q32.12 there was at least one SNP in a region showing P < 1 × 10−4.
Figure 2.
Results from genome-wide association analysis of 818 977 (818 237 + 740 pseudo-autochromosomal) genotypes from 1804 CM cases and 1026 controls, after adjusting for two PCs.
To understand whether associations in regions that were previously reported to be associated with CM risk were likely to be due to effects from a single or multiple variants, we performed analyses in which the association of SNPs in a region was conditioned on the most significant SNP in that region. Conditioning on the most significant SNP in MC1R reduced the level of significance for all other SNPs, but some SNPs retained significance levels of 10−3 or higher (Supplementary Material, Fig. S3). Similarly, conditioning on the most significant SNP in the p16 region removed some, but not all, of the evidence for association in that region. These results suggest that multiple variants influence CM risk in these regions, or the SNP on which we conditioned may not be the causal variant, but was only in strong linkage disequilibrium (LD) with the causal variant, so that there remained some residual association. For 9p21.3 (CDKN2A region), associations were detected across a broad region that includes recombination hotspots even after conditioning on the most significant SNP, suggesting that multiple variants are involved.
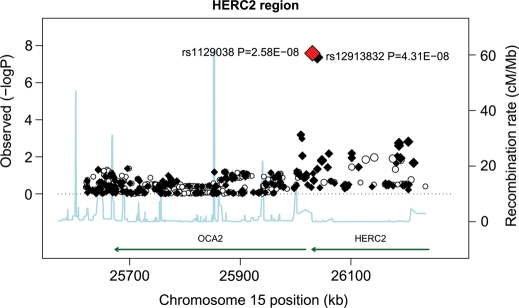
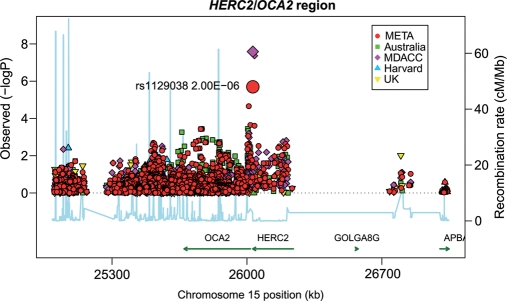
Figure 3 shows results from an association analysis of CM with SNPs in the HERC2/OCA2 region in samples from MD Anderson Cancer Center. The HERC2 SNP rs1129038 was the most significantly associated variant in the region [P = 2.58 × 10−8; odds ratio (OR) = 0.69; 95% confidence interval (CI) = 0.61–0.79]. The next most significant genotyped SNP in this region was rs12913832 (P = 4.31 × 10−8; OR = 0.69; 95% CI = 0.61–0.79). The variants rs1129038 and rs12913832 are in very high LD (r2 = 98.5%), but rs1129038 was not genotyped on other platforms used in subsequent studies; hence, further analyses were restricted to rs12913832.
Figure 3.
Association tests for the HERC2/OCA2 region on chromosome 15q13.1. Genotyped SNPs are indicated as diamonds and imputed SNPs as circles. The most significant SNP in the region is HERC2 rs1129038. The strength of the pairwise correlation between the surrounding markers and the most significant SNP is depicted by the size of the symbols: the larger the size, the stronger the LD. Genes in the region are annotated with location, range and orientation.
The SNP rs1800407 in the flanking region of 15q13.1 (HERC2/OCA2 region) showed moderate evidence (P = 0.004) for association with CM risk in a candidate gene study but was not associated with CM risk in this study (P = 0.62, OR = 0.95; 95% CI = 0.78–1.16). The most significant SNPs near the OCA2 gene yielded P-values (rs73377792, P = 6.3 × 10−4; rs4778138, P = 6.52 × 10−4) that were less extreme than those observed in the 15q13.1 (HERC2/OCA2) region in our study population. Further analyses conditioning on rs1129038 (Supplementary Material, Fig. S3) removed all remaining significant SNPs in the region.
Because the HERC2 gene was previously shown to be associated with pigmentary phenotypes (13), we further evaluated the association of the rs12913832 SNP with pigmentary phenotypes by using samples from MD Anderson (Supplementary Material, Table S4). Table 2 describes additional findings from the analysis of SNP rs12913832 in the MD Anderson participants. Results showed overwhelming evidence associating HERC2 rs12913832 with skin, eye and hair color phenotypes. Further analysis to evaluate the effect that this SNP had on these phenotypes showed that rs12913832 alone explained 50% of eye color (Supplementary Material, Table S5). In addition, we evaluated the association of HERC2 rs12913832 with pigmentary phenotypes in 10 183 participants in the Nurses' Health Study (NHS) and Health Professionals Follow-Up Study (HPFS). In these analyses, 567 cases and 7329 controls were genotyped using Illumina Bead arrays for HERC2 SNP rs12913832. We found extremely significant associations (Table 3). Stratifying by skin color, we observed strongest effects of the HERC2 SNP in lighter-skinned individuals (n = 1009, P = 0.0003, OR = 0.66, 95% CI = 0.53–0.83), a weaker effect in medium-skinned individuals (n = 828, P = 0.02, OR = 0.77, 95% CI = 0.61–0.96) and no effect in dark-skinned individuals (n = 117, P = 0.75, OR = 1.10, 95% CI = 0.61–2.00). In the Harvard cohort studies, the strongest effect was observed in light-skinned individuals (n = 2173, P = 0.08 OR = 0.80, 95% CI = 0.63–1.03), and no effect was observed in medium-skinned (n = 1916, P= 0.64, OR = 1.05, 95% CI = 0.85–1.32) or dark-skinned individuals (n = 1916, P= 0.72, OR = 0.94, 95% CI = 0.67–1.32).
Table 3.
Association of HERC2 rs12913832 with pigmentary phenotypes in the NHS and the HPFS
| GWAS phenotype | WT/VT | MAF | Effect | SE | P-value |
|---|---|---|---|---|---|
| Hair color (blonde to black) | G/A | 0.2497 | 0.3872 | 0.014 | 1.39 × 10−167 |
| Eye color (blue to brown) | G/A | 0.2647 | 0.8571 | 0.0175 | 7.68 × 10−523 |
| Tanning ability (none to deep) | G/A | 0.2534 | 0.1359 | 0.0139 | 1.44 × 10−22 |
WT, wild-type; VT, variant type; SE, standard error.
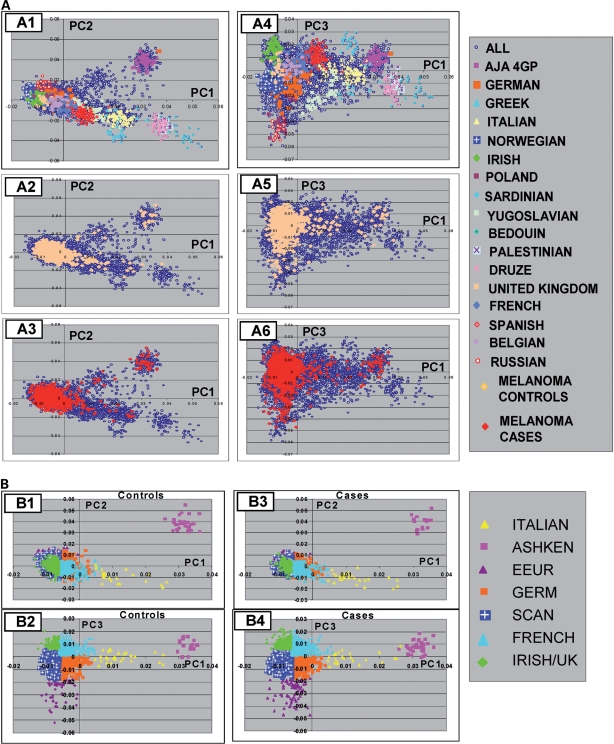
Because skin pigmentation and the HERC2 SNP frequency (Supplementary Material, Table S6) showed highly significant variability across European populations (14) and the CM risk is higher for northern European than for southern European populations, a potential concern can be raised that northern/southern European ancestry may be a confounder, causing a spurious association to be observed. We therefore derived the most likely European ancestry of the study participants (Fig. 4) by principal component analysis (PCA) with additional control samples from European groups. Results of conditioning on the most likely European ancestry showed no evidence for heterogeneity in the association of rs12913832 with CM risk by derived European populations, and stratified results were virtually identical to those obtained with allowing for European stratification (Supplementary Material, Table S7). In contrast, significant variation in the association between CM risk and SNPs in the p16 region did vary significantly among derived European populations, suggesting potential allelic heterogeneity in European populations for this SNP in CDKN2A (Supplementary Material, Table S7).
Figure 4.
PCA of CM cases and controls showing the relationship among populations of European and Middle-Eastern origin. (A) Analyses of 1804 CM cases and 1026 controls, with an additional 5001 Caucasians of known origin, are shown for PC1 and PC2 (A1, A2, A3), and for PC1 and PC3 (A4, A5, A6). The results are shown with either the known population groups color-coded (A1 and A4) or with controls color-coded (A2 and A5) or cases color-coded (A3 and A6). The population groups are indicated in the legend key. AJA 4GP are Ashkenazi Jewish Americans with four grandparents of Ashkenazi heritage. The group denoted ‘ALL’ are Americans of European ancestry. Results show similarity of PCA loadings for cases and controls in A2–A6. (B) The color code indicates the correspondence of controls or cases to seven different population groups with graphs for PC1 and PC2 (B1 and B3), and PC1 and PC3 (B2 and B4). The population groups were based on correspondence to PCA. The population groups corresponded to the range of the self-identified population group member defined by the first three PCs (see A). Where overlaps were present, the midpoints in the overlapped region were used to define the boundary between the population group assignment. The legend key shows the corresponding population groups with abbreviations for the following groups: Ashkenazi Jewish (ASHKEN), eastern European (EEUR), German (GER), Scandinavian (SCAN) and United Kingdom (UK). Analysis of CM case–control data alone yielded two PCs, whereas analysis including additional European controls yielded the additional third PC, shown in this figure.
We also sought in silico replication of the 10 most significant SNPs from the MD Anderson GWAS, excluding those loci that have been previously identified (in particular, we excluded the MC1R and CDKN2A regions, since these have been extensively resequenced in other studies), provided there was a second SNP with an r2 value of >0.8 and a P-value of <10−4. Results were provided to us by the GenoMEL consortium (based on European samples), an Australian collaboration, and from the Harvard cohorts (Supplementary Material, Table S8). To further evaluate effects for two most significant regions on chromosomes 1 and 15, we obtained data from the three collaborating groups for ∼1 Mb regions flanking the most significant SNP in our study. As shown in Figure 5, the most significant SNPs in the chromosome 15q region were rs1129038 in an intron of the HERC2 gene (P = 2.00 × 10−6, OR = 0.77, 95% CI = 0.69–0.85), but this SNP was genotyped only in the US and a subset of Australian samples and showed heterogeneity, with the Australian samples giving an OR of 0.97 (95% CI = 0.80–1.17). The next most significant SNP is rs4778138 in OCA2 (OR = 0.86, 95% CI = 0.80–0.92, P = 2.2 × 10−5). This SNP showed strong associations in the MD Anderson sample (P = 6.5 × 10−4, OR = 0.75, 95% CI = 0.64–0.89) and UK sample (P = 0.0064, OR = 0.88, 95% CI = 0.80–0.97) but weak association in the Australian sample (P = 0.14; OR = 0.89; 95% CI = 0.78–1.04) and was not genotyped in the UK sample. The next most significant SNP in the US study, rs12913832, was not associated with CM risk in the Australian samples (P = 0.63; OR = 0.98, 95% CI = 0.89–1.07) or UK samples (P = 0.44; OR = 0.96; 95% CI = 0.86–1.07), but showed a trend towards association in the Harvard cohort studies (P = 0.12; OR = 0.88, 95% CI = 0.76–1.03).
Figure 5.
Association tests from four studies and from fixed-effects meta-analysis for a region of chromosome 15 surrounding the HERC2/OCA2 region. The region telomeric to the HERC2 gene contains a region of many complex rearrangements including CHRFAM7A—a rearrangement of nicotinic acetylcholine receptor 7 and FAM7A. The gap in genotyping in the region between 25 168 374 and 25 271 522 base pairs overlays a region with some reported copy number variants in the GABRG3 gene.
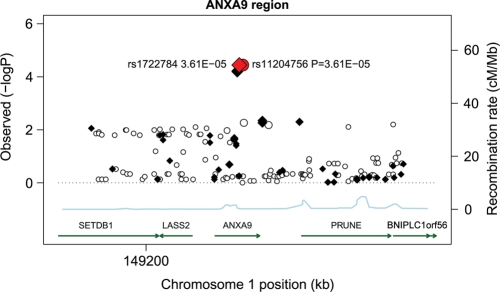
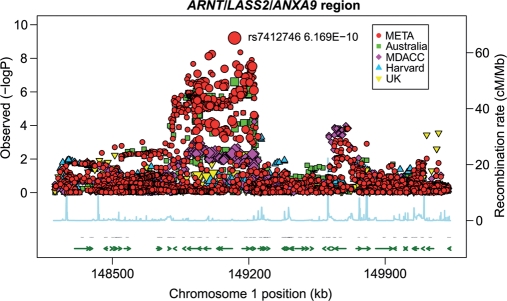
As shown in Figure 6, a region of chromosome 1q21.3 near the ARNT and LASS2 genes around 149.1 Mb was highly associated with CM risk in the samples from MD Anderson Cancer Center. The most significant SNP in the 1 Mb region including data from the other studies (Fig. 7) was rs7412746 (P = 6.17 × 10−10, OR = 0.88; 95% CI = 0.84–0.91), which significantly replicated in all of the studies but Harvard: MD Anderson Cancer Center (P = 0.003; OR = 0.85; 95% CI = 0.76–0.94), Australia (P = 2.52 × 10−7; OR = 0.82; 95% CI = 0.76–0.89), GenoMEL (P = 0.014; OR = 0.92; 95% CI = 0.86–0.98), but not in the smaller Harvard study (P = 0.12, OR = 1.11; 95% CI = 0.97–1.27). The most significant SNP in the MD Anderson Cancer Center study, rs11204756 (P = 3.61 × 10−5, OR = 1.26; 95% CI = 1.13–1.40) in the ANXA9 gene, had partially imputed data (r2 = 0.99) by the Australian study (P = 8 × 10−4; OR = 1.13; 95% CI = 1.05–1.22), yielding an overall association (P = 3.59 × 10−7, OR = 1.17, 95% CI = 1.10–1.24) and is, therefore, worthy of future investigations.
Figure 6.
Association tests for a region of chromosome 1 surrounding the LASS2/ANXA9 region. Genotyped SNPs are indicated as diamonds and imputed SNPs as circles. The most significant SNP in the region is rs1722784. The overall structure of the LD with SNPs in this region is reflected by estimated recombination rates. The strength of the pairwise correlation between the surrounding markers and the most significant SNP is depicted by the size of the symbols: the larger the size, the stronger the LD. Genes in the region are annotated with location, range and orientation.
Figure 7.
Association tests from four studies using fixed-effects meta-analysis for a region of chromosome 1 spanning the ARNT/LASS2/ANXA9 region.
DISCUSSION
This study provided confirmatory evidence of associations between genetic loci in several previously identified chromosome regions and CM risk, including the SLC45A2 region on chromosome 5p13.3, the 9p21.3 region encompassing CDKN2A, the region of TYR on chromosome 11q21, the region on chromosome 16q24.3 encompassing MC1R, a broad region on chromosome 20q11.2 and a region of 22q13 encompassing PLA2G6. Our study focused primarily on the discovery of novel variants influencing CM risk and we identified strong associations of SNPs in the HERC2/OCA2 region on chromosome 15q13.1 as well as a region of chromosome 1q21.3 that had not previously been well characterized. The HERC2 SNPs had previously been noted to associate strongly with pigmentation (5), but our finding is the first report that these SNPs influence CM risk in US populations. In the previous analysis of pigmentary phenotypes and SNPs in the HERC2/OCA2 region, the strongest association was found with SNPs rs12913832 and rs1129038 (13). These SNPs lie in a region that is in LD with both exons of HERC2 as well as the regulatory region of OCA2, so that further studies are needed to identify whether variation in HERC2 or OCA2 accounts for CM risk and pigmentary changes in this region. We also identified the SNP rs4778138 in OCA2, which showed a suggestive association with CM risk among the studies in which it was genotyped. These results implicate HERC2/OCA2 variation as a pigment-related gene that affects CM risk. Risk of CM could also be influenced by the effects of HERC2, which has recently been shown to play a role in ubiquitination after DNA damage (15) and to post-translationally regulate levels of XPA (16), a core protein involved in nucleotide-excision repair. HERC2 variants could reduce CM risk by increasing levels of XPA, a hypothesis that needs further mechanistic studies. Variation in the association of the HERC2 region SNPs among US, Australian and European regions may reflect varying patterns of sun exposure and thus further gene–environment studies are needed.
Pigmentation varies among European populations, as does the frequency of variants in rs12913832, but cases and controls from the two US studies were collected from the same institution and cohorts, respectively, limiting possible confounding effects. The GenoMEL study encompasses several countries within Europe. Differences in allelic background as well as variation in sun exposure may underlie the differences between US and Australian or GenoMEL study findings for the effect of HERC2/OCA2, chromosome 15q13.1 region SNPs on the risk of CM.
For the region on chromosome 1q21.3 that was highly associated with CM risk, the evidence of an association was the strongest for a region that includes ANXA9, LASS2, SETDB1 and ARNT, which are closely located. ANXA9 is an annexin that has previously been associated with TH2-related responses that are induced during pemphigus (17). LASS2 is a ceramide synthase that has previously been shown to influence apoptosis after ionizing irradiation (18) and to play a role in tumor suppression in hepatocellular carcinoma cell lines (19). SETDB1 plays a role in trimethylation required for proviral silencing, which would seem a less likely candidate for influencing CM risk. ARNT is the aryl hydrocarbon nuclear translocator and plays a key role in regulating response to exogenous compounds that are catabolized by P450 enzymes. In summary, this study identified signals in several chromosomal regions and suggests that candidate genes including OCA2, HERC2, ARNT, ANXA9 and LASS2 warrant additional investigations.
MATERIALS AND METHODS
MD Anderson Cancer Center Study
The study participants for the discovery analysis were from a hospital-based case–control study of CM, for which cases were recruited from non-Hispanic white patients and controls at MD Anderson between March 1998 and August 2008. Samples and data were available from 931 CM patients and 1026 cancer-free controls (friends or acquaintances of patients reporting to other clinics), who were frequency-matched on age and sex, completed a comprehensive skin lifestyle questionnaire and passed QC filters for genotyping. This questionnaire was administered by an interviewer to 70% of patients and controls and was self-administered for the remaining 30%. An additional case series comprising 873 individuals presenting for the treatment for CM at MD Anderson was also included, bringing the total number of CM patients to 1804. The study protocols were approved by the Institutional Review Board at MD Anderson, and informed consent was obtained from all participants.
Genotyping and data QC
Tissue samples were collected as whole blood, using various DNA extraction methods (including Gentra, Qiagen and phenol/chloroform). DNA samples for the first-stage GWAS were genotyped with use of the Illumina HumanOmni1-Quad_v1-0_B array and were called using the BeadStudio algorithm, at the John Hopkins University Center for Inherited Disease Research (CIDR). We were able to satisfactorily genotype 1 012 904 of the 1 016 423 SNPs attempted (99.6%) with a mean sample call rate of 99.86% (Supplementary Material, Table S1). Supplementary Material, Table S1 summarizes a series of SNP filters applied to the original 1 140 419 SNPs and CNV probes. Before data release, 3519 SNPs failed the CIDR QC process with a missing call rate of >5%. SNPs with an MAF of ≤0.01, call rate of <95% or Hardy–Weinberg equilibrium in controls with a P-value of ≤10−5 were excluded.
After the above criteria were applied, 818 237 genotyped autosomal or X chromosome SNPs and 740 pseudo-autosomal SNPs were available for the final association analysis. Imputation of ungenotyped SNPs in candidate regions (at least two genotyped SNPs in one region having P < 10−4) was performed for case and control samples with the use of the MACH program (20) using 1000 Genomes CEU (March 2010 release) as a reference panel. An additional 12 009 SNPs were imputed from those regions. In addition, genome-wide imputation using MACH and Hapmap2 Release 22 using the CEU population has been completed and data are available upon request for 2 373 692 SNPs with r2 > 0.8, with an average quality score of 99.35%.For the most significant SNP in the HERC2 region, rs12913832, we performed confirmatory genotyping of 528 cases and controls using TaqMan. One subject was misclassified between the two platforms, one subject was genotyped as G/G by TaqMan and A/G by Illumina and one subject could not be genotyped by TaqMan but was found to have the G/G genotype by Illumina.
Of the samples that were genotyped, 41 failed genotyping with a >10% missing rate across all SNPs; 11 samples had identity problems that could not be resolved. For this study, the identity-by-descent (IBD) coefficients were estimated using 116 002 autosomal SNPs in PLINK (12) and 5 unexpected duplicates and 15 related samples were removed. In total, 126 duplicated (67 expected duplicates), related (IBD) or outliers identified by PCA were excluded from the study. After these exclusions, 1952 cases and 1026 controls remained. Supplementary Material, Table S1 summarizes the whole-sample filters. From the total 2978 case and control subjects with data after QC, 138 in situ cases were removed from the study because they had indeterminate phenotypes; in addition, 10 patients with atypical melanocytic proliferation were excluded because they did not have invasive cancers. Data from 1804 cases and 1026 controls were analyzed for the association study of CM susceptibility.
PCA and population structure of the MD Anderson sample
We used an LD-pruned SNP set provided by the GENEVA (21) coordinating center for PCA adjustment to evaluate population structure. To select a set of SNPs for identifying population stratification, GENEVA thinned the initial SNPs to reduce LD to a set of 75 210 SNPs. SNPs were pruned using pairwise genotypic correlation in PLINK. The pruning procedure consisted of two stages. In the first stage, short-range LD was removed. We used a window size of 50 SNPs with a 5 SNP offset and r2 cut-off point of 0.2. For the second stage, we aimed to remove long-range LD. We used a window size of 150 SNPs with an offset of 5 Mb and r2 cut-off point of 0.2. We adjusted for the first two components from PCA in our GWAS. No other PCs varied significantly between cases and controls. Those two eigenvectors were treated as covariables to adjust for population structure among study subjects. The GENEVA coordinating center provided an initial quality assessment of data and helped to organize the data for submission to dbGAP.
Association analysis of the MD Anderson sample
Association analysis with CM of genotyped SNPs or most likely genotypes from the imputation study was performed using the PLINK –logistic and –covar options (12). A logistic regression model was built to measure the additive effect of each SNP on susceptibility to CM. A likelihood ratio test was performed under the null hypothesis of χ2 distribution with one degree of freedom. The first two PCs were included to adjust for population structure. Q–Q plots portraying the associations of markers with CM risk are shown in Supplementary Material, Figure S1, including all of the directly genotyped markers, and in Supplementary Material, Figure S2 after excluding previously identified genes strongly associated with CM risk (MC1R and p16/arf). Significant regions with multiple SNPs that each had a P-value of <10−4 were selected for further validation in other independent studies.
To evaluate associations with pigmentary phenotypes, we regressed ordinal coding for hair color (1 = blonde, 2 = red, 3 = brown and 4 = black), eye color (1 = blue/gray, 2 = hazel/green and 3 = brown/black) and frequency of burns on a 1–4 scale (1 = always or usually burn, 2 = moderately burn, 3 = minimally burn and 4 = rarely or never burn). We used linear regression to test the association between minor allele counts and pigmentary phenotypes, and the model was adjusted for age and sex.
Genomic imputation
MACH was used to impute ungenotyped SNPs in the candidate regions according to 1000 Genomes Project data. Based on the reference data with a denser set of markers, those untyped markers can be filled in by means of maximum-likelihood estimation. Imputation was run in one step since it was for small candidate regions. A total of 1682 SNPs with P-values of <10−4 were found among 16 regions on 12 chromosomes, excluding chromosome 9 near the centromere where p16/ARF/CDKN2A is located and chromosome 16 near MC1R, each of which contained at least two significant SNPs. Within a region of 50 kb from each side of these top SNPs, untyped genotypes were inferred with the use of MACH. Finally 12 009 SNPs were obtained with average r2> 0.875 for those candidate regions. The average posterior probability for the most likely genotype was 97.7%. As newer versions of the 1000 Genomes Project become available, better inference from it should be possible.
Further adjustment for population structure in the MD Anderson sample
Because skin pigmentation and CM risk show a North–South gradient across European populations, and allele frequencies in genes associated with skin pigmentation also show variation in allele frequencies, confounding between allele frequency and northern/southern European ancestry is a concern. We therefore performed additional studies to characterize the European ethnic background of study participants from MD Anderson and to allow for this inferred ancestry. Results, shown in Figure 4, demonstrate the inferred origins of study participants from MD Anderson according to the first two PCs. Supplementary Material, Table S7 displays results from analyses that were conditioned on the inferred most likely ancestral origin of each participant. The approach to selecting populations for characterizing European ancestry has been previously presented (22). Cases and controls that did not cluster within European ancestry groups were removed from analysis. In this analysis, 1795 cases and 931 controls were retained, whereas an additional 1587 controls genotyped on 610K platforms that had genotypes close to the existing cases according to the first six PCs were used as additional controls in the analysis.
Materials and methods from Harvard cohort studies
We have conducted several GWASs on different disease outcomes [NHS: breast cancer by Illumina 550, and pancreatic cancer by Illumina 550; HPFS: advanced prostate cancer by Illumina 610; both NHS (part of the CGEMS) (23–25) and HPFS: coronary heart disease by Affymetrix 6.0, type 2 diabetes by Affymetrix 6.0, kidney stone by Illumina 610 and glaucoma by Illumina 660]. We included only controls in each study, except for the kidney stone study, in which we used both cases and controls. We excluded those with personal history of squamous cell carcinoma and basal cell carcinoma. Participants with CM diagnosis were the CM cases in this study, and those without CM diagnosis were the controls. In addition, we genotyped the rest of the CM cases identified in both cohorts who were not in these previous GWASs by Illumina 610. Finally, we included 494 CM cases and 5628 controls.
Based on the genotyped SNPs and haplotype information in the NCBI Build 35 of phase II Hapmap CEU data, we imputed genotypes for >2.5 million SNPs using the program MACH (20). Only SNPs with imputation quality r2> 0.95 and with MAF > 2.5% in each study were included in the final analysis. A total of 1 579 307 (NHS: 1 518 067; HPFS: 1 533 499) SNPs were included in the final meta-analysis of the NHS set and HPFS set.
Information on natural hair color at age 20 years and on childhood and adolescent tanning tendency was collected in both the NHS and HPFS prospective questionnaires, and information on natural eye color was collected in the HPFS only. The summary of the basic information of the five-component cohort studies used in the GWAS of pigmentation is shown in Supplementary Material, Table S4. The imputed dosage data on chromosome 15 were used for the association study. We regressed ordinal coding for hair color (1 = blonde, 2 = light brown, 3 = dark brown and 4 = black; the participants with red hair color were excluded from the hair color GWAS), eye color (1 = blue/light, 2 = hazel/green/medium, 3 = brown/dark) and tanning ability (1 = practically none, 2 = light tan, 3 = average tan, 4 = deep tan in NHS; and 1 = pain: burn/peel, 2 = burn then tan, 3 = tan without burn in HPFS). We used linear regression to test the association between minor allele counts and pigmentary phenotypes, and the model was adjusted for the first four PCs separately for each SNP and trait; within-cohort association results were combined in an inverse variance weighted meta-analysis, and the software METAL (http://www.sph.umich.edu/csg/abecasis/Metal/index.html) was used for the meta-analysis across the different cohorts that were analyzed. Controlling for case–control status within each collection (e.g. type 2 diabetes in the HPFS) made no material difference to results (hence no adjustment was made). A summary of information from the five component studies used for SNP studies of pigmentation is shown in Supplementary Material, Table S4.
For the study on CM risk, the eligible cases consisted of participants in the NHS and the HPFS with pathologically confirmed CM, diagnosed any time after the baseline up to the 2006 follow-up cycle (for both cohorts), who had no previously diagnosed cancer. The controls were randomly selected from participants in the same cohorts who were free of cancer up to their participation and including the questionnaire cycle in which the case was diagnosed. In addition, after excluding the breast cancer cases in the NHS-BC set and the individuals diagnosed with skin cancer (CM, basal cell carcinoma, or squamous cell carcinoma), the participants in the five GWAS sets mentioned above were also included as controls. All subjects were US non-Hispanic Caucasians. We included 585 CM cases (mean age at diagnosis, 61.5 years) and 7363 controls. Laboratory personnel were blinded to case–control status, and blinded QC samples were inserted to validate genotyping procedures; concordance for the blinded samples was 100%. Primers, probes and conditions for genotyping assays are available upon request.
We regressed a binary coding for CM case and control (0 or 1) on each SNP (dosage file used) that passed QC filters. Among the 10 largest PCs, the first 5 were significantly associated with nevus count at a two-sided alpha of 0.05. Therefore, we adjusted for top five PCs of genetic variation in the regression model along with age. These PCs were calculated for all individuals on the basis of approximately 10 000 unlinked markers using the EIGENSTRAT software.
In silico replication studies
Confirmatory in silico replication was sought from GenoMEL, Harvard cohorts and the Australian consortium for the top 10 SNPs of significance for which another nearby SNP with LD r2> 0.8 also reached a significance level of <10−4.
Australian sample
DNA was extracted from peripheral blood or saliva samples. Australian twin and endometriosis sample controls were genotyped at deCODE Genetics (Reykjavik, Iceland) on the Illumina HumanHap610W Quad and Illumina HumanHap660 Quad Beadarrays, respectively. AMFS controls were genotyped by Illumina (San Diego) on Illumina Omni1-Quad arrays. Cases were genotyped by Illumina (San Diego) on Illumina Omni1-Quad (568 AMFS cases, 699 Q-MEGA cases) and HumanHap610W Quad arrays (998 Q-MEGA cases). All genotypes were called with the Illumina BeadStudio software. SNPs with a mean BeadStudio GenCall score <0.7 were excluded from the control data sets. All samples had successful genotypes for >95% of SNPs. SNPs with call rates either <0.95 (MAF > 0.05) or <0.99 (MAF > 0.01), Hardy–Weinberg equilibrium in controls P < 10−6 and/or MAF < 0.01 were excluded. Cryptic relatedness between individuals was assessed through the production of a full identity-by-state matrix. Ancestry outliers were identified by PC analysis, using data from 11 populations of the HapMap 3 Project and 5 northern European populations genotyped by the GenomeEUtwin consortium, using the EIGENSOFT package. Individuals lying ≥2 standard deviations from the mean PC1 and PC2 scores were excluded from subsequent analyses. Following these exclusions, there were 2168 case samples (1242 typed on the Omni1-Quad and 926 typed on the Hap610 arrays, respectively) and 4387 controls (431 typed on the Omni1-Quad and 3956 typed on the Hap610 arrays) retained for subsequent analyses (Table 1). Individuals typed on the Omni1-Quad array had genotypes for up to 816 169 SNPs, whereas individuals typed on the Hap610/670 arrays had genotypes for up to 544 483 SNPs. There were 299 394 SNPs passing QC and overlapping between these arrays (and hence directly genotyped on all Australian samples).
Imputation for the Australian samples was performed using MACH (20) with 1000 Genomes Project [June 2010 release] data obtained from people of northern and western European ancestry collected by the Centre d'Etude du Polymorphisme Humain. Imputation was based on a set of autosomal SNPs common to all CM case–control samples (n = 292 043). Imputation was run in two stages. First, data from a set of representative Australian sample individuals were compared with the phased haplotype data from the 1000 Genomes data to generate recombination and error maps. For the second stage, data were imputed for all individuals using the phased 1000 Genomes data as the reference panel and the recombination as well as error files generated in the first stage. In total, 5 480 804 1000 Genomes SNPs could be imputed with imputation r2 > 0.5.
Association analysis of genotyped SNPs was performed using the PLINK –assoc option (12). Analysis of dosage scores from the imputation analysis was done using mach2dat.
GenoMEL sample
The GenoMEL data incorporated in the in silico replication came from a GWAS of 2804 cases and 1835 controls for CM collected at 11 different centers across Europe and Israel. These were genotyped in two phases, phase 1 on the Illumina HumanHap300 BeadChip version 2 duo array (317K SNPs) and phase 2 on the Illumina Human610 quad array (610K SNPs). These are supplemented by 3878 controls from the WTCCC study [WTCCC (26)] genotyped on the Illumina HumanHap1.2 million array and 1905 French controls genotyped by Centre National de Genotypage on the Illumina Humancnv370k array. Individuals were excluded for low call rate (<97% on the array on which the sample was genotyped), non-European ethnicity (determined by PCA), sex discrepancy with recorded phenotype information, first-degree or closer relatedness with another sample or recommendation for exclusion by WTCCC (26). QC was applied to SNPs separately for each genotyping platform. SNPs were excluded for a low call rate (<97%), Hardy–Weinberg equilibrium P-value of <10−20 or recommendation for exclusion by WTCCC (usually on the basis of poor clustering). A trend test was applied to each SNP in turn stratified by broad geographic region (eight regions pre-specified).
WEB RESOURCES
EIGENSOFT: http://genepath.med.harvard.edu/~reich/Software.htm.
MACH: http://www.sph.umich.edu/csg/abecasis/MACH/index.html, release MACH1.016.
1000 Genomes: http://www.1000genomes.org.
The genotyping data and phenotypes that have been presented in this analysis are available online through dbGaP (study accession: phs000187.v1.p1).
Chromosomal region graphs were developed using the software developed by the Diabetes Genome Initiative (http://www.broadinstitute.org/science/projects/diabetes-genetics-initiative/plotting-genome-wide-association-results).
SUPPLEMENTARY MATERIAL
NOTE ADDED IN PROOF
Dr Houvras brought to our attention an elegant work that was performed showing that the SETDB1 gene (which lies within our region of association on chromosome 1q21.3) accelerates melanoma in cooperation with BRAF, increases invasiveness of the lesions and allows cells to bypass a senescence arrest. Therefore, this locus is also an excellent candidate for future studies to identify its role in melanoma susceptibility (27).
Conflict of Interest statement. None declared.
FUNDING
Research at MD Anderson Cancer Center was partially supported by NIH grants R01CA100264, 2P50CA093459, P30CA016672, R01CA133996 and by the Marit Peterson Fund for Melanoma Research.
The Center for Inherited Disease Research is supported by contract HHSN268200782096C. Geneva is supported by NIH grant HG004446.
The Australian Twin Registry is supported by an Australian National Health and Medical Research Council (NHMRC) Enabling Grant (2004–2009).
The QIMR Study was supported by grants from the Melanoma Research Alliance, the National Health and Medical Research Council (NHMRC) of Australia (241944, 339462, 389927,389875, 389891, 389892, 389938, 443036, 442915, 442981, 496610, 496739, 552485, 552498), the Cooperative Research Centre for Discovery of Genes for Common Human Diseases (CRC), Cerylid Biosciences (Melbourne), and donations from Neville and Shirley Hawkins.
S.M. is supported by NHMRC Career Development Awards (496674, 613705). G.W.M. is supported by the NHMRC Fellowships Scheme.
The AMFS study is funded by the National Health and Medical Research Council of Australia (NHMRC) (project grants 566946, 107359, 211172 and program grant number 402761 to G.J.M. and R.F.K.); the Cancer Council New South Wales (project grant 77/00, 06/10), the Cancer Council Victoria and the Cancer Council Queensland (project grant 371); and the US National Institutes of Health (via RO1 grant CA-83115-01A2 to the international Melanoma Genetics Consortium - GenoMEL). A.E.C. is the recipient of an NHMRC public health postdoctoral fellowship (520018) and a National Cancer Institute NSW Early Career Development Fellowship (10/ECF/2-06). B.K.A.'s research is supported by a University of Sydney Medical Foundation Program Grant, and J.L.H. is an Australia Fellow of the NHMRC.
The GenoMEL study was funded by the European Commission under the 6th Framework Programme, contract no: LSHC-CT-2006-018702; by Cancer Research UK Programme Awards, C588/A4994, and C588/A10589, and Cancer Research UK Project Grant C8216/A6129; and by US National Institutes of Health R01 CA83115. This research was also supported by the Intramural Research Program of the NIH, NCI and DCEG.
Emilia-Romagna: National Cancer Institute grants RO1 CA5558 to Maria Teresa Landi.
Genoa: IRCSS 2007 Italian Ministry of Health DGRST.4/4235-P1.9.A.B, Fondazione CARIGE, PRIN 2008 to G.B.-S.
Leeds: Cancer Research UK Programme grants Genetic Epidemiology of Cancer C588/A10589, C588/A4994 and Cancer Research UK Project grant C8216/A6129.
Lund: Swedish Cancer Society, Swedish Research Council, Region Skåne funds, Kamprad Foundation.
Norway: Grants from the Comprehensive Cancer Center, Oslo University Hospital and the University of Bergen.
Paris: Grants from Institut National du Cancer (INCa-PL016) and Ligue Nationale Contre Le Cancer (PRE05/FD and PRE 09/FD) to Florence Demenais, Programme Hospitalier de Recherche Clinique (PHRC 2007/AOM-07-195) to Marie-Françoise Avril and Florence Demenais, Institut National du Cancer (Melanoma Network RS Number 13), Association pour la Recherche sur le Cancer (ARC No. A09/5/5003), and Société Française de Dermatologie (SFD2009) to Brigitte Bressac-de Paillerets. Brigitte Bressac-de Paillerets has been awarded an INSERM Research Fellowship for hospital-based scientists.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge with appreciation all of the women who participated in the QIMR, OXEGENE and NHS studies. We thank Endometriosis Associations for supporting study recruitment as well as Sullivan Nicolaides and Queensland Medical Laboratory for pro bono collection and delivery of blood samples and other pathology services associated with blood collection. In addition, we thank Margaret J. Wright, Megan J. Campbell, Anthony Caracella, Margaret Lung, Zhensheng Liu, Yawei Qiao, Min Zhao, Dakai Zhu, B. Haddon, D. Smyth, H. Beeby, O. Zheng and B. Chapman for their input into project management, databases, sample processing and genotyping. We are grateful to the many research assistants and interviewers for assistance with the studies contributing to the QIMR collection.
APPENDIX
MEMBERS OF THE GENOMEL COLLABORATION
Australian Melanoma Family Study (AMFS): Graham J. Mann, John L. Hopper, Joanne F. Aitken, Bruce K. Armstrong, Graham G. Giles, Richard F. Kefford, Anne E. Cust, Mark A. Jenkins, Helen Schmid.
Barcelona: The participants of GenoMEL in Barcelona: Paula Aguilera, Celia Badenas, Cristina Carrera, Francisco Cuellar, Daniel Gabriel, Estefania Martinez, Melinda Gonzalez, Pablo Iglesias, Josep Malvehy, Rosa Marti-Laborda, Montse Mila, Zighe Ogbah, Joan-Anton Puig Butille, Susana Puig, and other members of the Melanoma Unit: Llúcia Alós, Ana Arance, Pedro Arguís, Antonio Campo, Teresa Castel, Carlos Conill, Jose Palou, Ramon Rull, Marcelo Sánchez, Sergi Vidal-Sicart, Antonio Vilalta and Ramon Vilella.
Brisbane: The Queensland study of Melanoma: Environmental and Genetic Associations (Q-MEGA) Principal Investigators: Nicholas G. Martin, Grant W. Montgomery, David L. Duffy, David C. Whiteman, Stuart MacGregor and Nicholas K. Hayward.
The Australian Cancer Study (ACS) Principal Investigators: David Whiteman, Penny Webb, Adele Green, Peter Parsons, David Purdie and Nicholas Hayward.
Emilia-Romagna: Maria Teresa Landi, Donato Calista, Giorgio Landi, Paola Minghetti, Fabio Arcangeli and Pier Alberto Bertazzi.
Genoa: Department of Internal Medicine (DIMI), University of Genoa: Giovanna Bianchi-Scarra, Paola Ghiorzo, Lorenza Pastorino, William Bruno, Linda Battistuzzi, Sara Gargiulo, Sabina Nasti, Sara Gliori, Paola Origone, Virginia Andreotti. Medical Oncology Unit, National Institute for Cancer Research: Paola Queirolo.
Glasgow: Rona Mackie, Julie Lang.
Leeds: Julia A. Newton Bishop, Paul Affleck, Jennifer H. Barrett, D. Timothy Bishop, Jane Harrison, Mark M. Iles, Juliette Randerson-Moor, Mark Harland, John C. Taylor, Linda Whittaker, Kairen Kukalizch, Susan Leake, Birute Karpavicius, Sue Haynes, Tricia Mack, May Chan, Yvonne Taylor, John Davies and Paul King.
Leiden: Department of Dermatology, Leiden University Medical Centre: Nelleke A. Gruis, Frans A. van Nieuwpoort, Coby Out, Clasine van der Drift, Wilma Bergman, Nicole Kukutsch and Jan Nico Bouwes Bavinck. Department of Clinical Genetics, Centre of Human and Clinical Genetics, Leiden University Medical Centre: Bert Bakker, Nienke van der Stoep, Jeanet ter Huurne. Department of Dermatology, HAGA Hospital, The Hague: Han van der Rhee. Department of Dermatology, Reinier de Graaf Groep, Delft: Marcel Bekkenk. Department of Dermatology, Sint Franciscus Gasthuis, Rotterdam: Dyon Snels, Marinus van Praag. Department of Dermatology, Ghent University Hospital, Ghent, Belgium: Lieve Brochez and colleagues. Department of Dermatology, St Radboud University Medical Centre, Nijmegen: Rianne Gerritsen and colleagues. Department of Dermatology, Rijnland Hospital, Leiderdorp: Marianne Crijns and colleagues. Dutch Patient Organization, Stichting Melanoom, Purmerend. The Netherlands Foundation for the Detection of Hereditary Tumors, Leiden: Hans Vasen.
Lund: Lund Melanoma Study Group: Håkan Olsson, Christian Ingvar, Göran Jönsson, Åke Borg, Anna Måsbäck, Lotta Lundgren, Katja Baeckenhorn, Kari Nielsen, Anita Schmidt Casslén.
Norway: Oslo University Hospital: Per Helsing, Per Arne Andresen, Helge Rootwelt.
University of Bergen: Lars A. Akslen, Anders Molven.
Paris: Marie-Françoise Avril, Brigitte Bressac-de Paillerets, Valérie Chaudru, Nicolas Chateigner, Eve Corda, Patricia Jeannin, Fabienne Lesueur, Mahaut de Lichy, Eve Maubec, Hamida Mohamdi, Florence Demenais, and the French Family Study Group including the following oncogeneticists and dermatologists: Pascale Andry-Benzaquen, Bertrand Bachollet, Frédéric Bérard, Pascaline Berthet, Françoise Boitier, Valérie Bonadona, Jean-Louis Bonafé, Jean-Marie Bonnetblanc, Frédéric Cambazard, Olivier Caron, Frédéric Caux, Jacqueline Chevrant-Breton, Agnès Chompret (deceased), Stéphane Dalle, Liliane Demange, Olivier Dereure, Martin-Xavier Doré, Marie-Sylvie Doutre, Catherine Dugast, Laurence Faivre, Florent Grange, Philippe Humbert, Pascal Joly, Delphine Kerob, Christine Lasset, Marie Thérèse Leccia, Gilbert Lenoir, Dominique Leroux, Julien Levang, Dan Lipsker, Sandrine Mansard, Ludovic Martin, Tanguy Martin-Denavit, Christine Mateus, Jean-Loïc Michel, Patrice Morel, Laurence Olivier-Faivre, Jean-Luc Perrot, Caroline Robert, Sandra Ronger-Savle, Bruno Sassolas, Pierre Souteyrand, Dominique Stoppa-Lyonnet, Luc Thomas, Pierre Vabres and Eva Wierzbicka.
Philadelphia: David Elder, Peter Kanetsky, Jillian Knorr, Michael Ming, Nandita Mitra, Althea Ruffin and Patricia Van Belle.
Poland: Tadeusz Dębniak, Jan Lubiński, Aneta Mirecka and Sławomir Ertmański.
Slovenia: Srdjan Novakovic, Marko Hocevar, Barbara Peric and Petra Cerkovnik.
Stockholm: Veronica Höiom and Johan Hansson.
Sydney: Graham J. Mann, Richard F. Kefford, Helen Schmid and Elizabeth A. Holland.
Tel Aviv: Esther Azizi, Gilli Galore-Haskel, Eitan Friedman, Orna Baron-Epel, Alon Scope, Felix Pavlotsky, Emanuel Yakobson, Irit Cohen-Manheim, Yael Laitman, Roni Milgrom, Iris Shimoni and Evgeniya Kozlovaa.
See also: www.GenoMEL.org.
REFERENCES
- 1.Siegel R., Ward E., Brawley O., Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J. Clin. 2011;61:212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Cust A.E., Schmid H., Maskiell J.A., Jetann J., Ferguson M., Holland E.A., Agha-Hamilton C., Jenkins M.A., Kelly J., Kefford R.F., et al. Population-based, case-control-family design to investigate genetic and environmental influences on melanoma risk: Australian Melanoma Family Study. Am. J. Epidemiol. 2009;170:1541–1554. doi: 10.1093/aje/kwp307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyle K.D., Guldberg P. Genetic risk factors for melanoma. Hum. Genet. 2009;126:499–510. doi: 10.1007/s00439-009-0715-9. [DOI] [PubMed] [Google Scholar]
- 4.Bishop D.T., Demenais F., Iles M.M., Harland M., Taylor J.C., Corda E., Randerson-Moor J., Aitken J.F., Avril M.F., Azizi E., et al. Genome-wide association study identifies three loci associated with melanoma risk. Nat. Genet. 2009;41:920–925. doi: 10.1038/ng.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sturm R.A., Duffy D.L., Zhao Z.Z., Leite F.P., Stark M.S., Hayward N.K., Martin N.G., Montgomery G.W. A single SNP in an evolutionary conserved region within intron 86 of the HERC2 gene determines human blue-brown eye color. Am. J. Hum. Genet. 2008;82:424–431. doi: 10.1016/j.ajhg.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fargnoli M.C., Argenziano G., Zalaudek I., Peris K. High- and low-penetrance cutaneous melanoma susceptibility genes. Expert Rev. Anticancer Ther. 2006;6:657–670. doi: 10.1586/14737140.6.5.657. [DOI] [PubMed] [Google Scholar]
- 7.Kanetsky P.A., Rebbeck T.R., Hummer A.J., Panossian S., Armstrong B.K., Kricker A., Marrett L.D., Millikan R.C., Gruber S.B., Culver H.A., et al. Population-based study of natural variation in the melanocortin-1 receptor gene and melanoma. Cancer Res. 2006;66:9330–9337. doi: 10.1158/0008-5472.CAN-06-1634. [DOI] [PubMed] [Google Scholar]
- 8.Pho L., Grossman D., Leachman S.A. Melanoma genetics: a review of genetic factors and clinical phenotypes in familial melanoma. Curr. Opin. Oncol. 2006;18:173–179. doi: 10.1097/01.cco.0000208791.22442.09. [DOI] [PubMed] [Google Scholar]
- 9.Gudbjartsson D.F., Sulem P., Stacey S.N., Goldstein A.M., Rafnar T., Sigurgeirsson B., Benediktsdottir K.R., Thorisdottir K., Ragnarsson R., Sveinsdottir S.G., et al. ASIP and TYR pigmentation variants associate with cutaneous melanoma and basal cell carcinoma (vol 40, pg 886, 2008) Nat. Genet. 2008;40:1029. doi: 10.1038/ng.161. [DOI] [PubMed] [Google Scholar]
- 10.Sulem P., Gudbjartsson D.F., Stacey S.N., Helgason A., Rafnar T., Magnusson K.P., Manolescu A., Karason A., Palsson A., Thorleifsson G., et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat. Genet. 2007;39:1443–1452. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- 11.Brown K.M., Macgregor S., Montgomery G.W., Craig D.W., Zhao Z.Z., Iyadurai K., Henders A.K., Homer N., Campbell M.J., Stark M., et al. Common sequence variants on 20q11.22 confer melanoma susceptibility. Nat. Genet. 2008;40:838–840. doi: 10.1038/ng.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: A tool set for whole-genome association and population-based linkage analyses 1. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duffy D.L., Zhao Z.Z., Sturm R.A., Hayward N.K., Martin N.G., Montgomery G.W. Multiple pigmentation gene polymorphisms account for a substantial proportion of risk of cutaneous malignant melanoma. J. Invest. Dermatol. 2010;130:520–528. doi: 10.1038/jid.2009.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cann H.M., de Toma C., Cazes L., Legrand M.F., Morel V., Piouffre L., Bodmer J., Bodmer W.F., Bonne-Tamir B., Cambon-Thomsen A., et al. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- 15.Bekker-Jensen S., Rendtlew D.J., Fugger K., Gromova I., Nerstedt A., Lukas C., Bartek J., Lukas J., Mailand N. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat. Cell Biol. 2010;12:80–86. doi: 10.1038/ncb2008. [DOI] [PubMed] [Google Scholar]
- 16.Kang T.H., Lindsey-Boltz L.A., Reardon J.T., Sancar A. Circadian control of XPA and excision repair of cisplatin-DNA damage by cryptochrome and HERC2 ubiquitin ligase. Proc. Natl Acad. Sci. USA. 2010;107:4890–4895. doi: 10.1073/pnas.0915085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caproni M., Giomi B., Cardinali C., Salvatore E., Pestelli E., D'Agata A., Bianchi B., Toto P., Feliciani C., Fabbri P. Further support for a role for Th2-like cytokines in blister formation of pemphigus. Clin. Immunol. 2001;98:264–271. doi: 10.1006/clim.2000.4974. [DOI] [PubMed] [Google Scholar]
- 18.Mesicek J., Lee H., Feldman T., Jiang X., Skobeleva A., Berdyshev E.V., Haimovitz-Friedman A., Fuks Z., Kolesnick R. Ceramide synthases 2, 5, and 6 confer distinct roles in radiation-induced apoptosis in HeLa cells. Cell. Signal. 2010;22:1300–1307. doi: 10.1016/j.cellsig.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang N., Jin J., Deng Y., Ke R.H., Shen Q.J., Fan S.H., Qin W.X. LASS2 interacts with V-ATPase and inhibits cell growth of hepatocellular carcinoma. Sheng Li Xue Bao. 2010;62:196–202. [PubMed] [Google Scholar]
- 20.Li Y., Abecasis G. Mach 1.0: rapid haplotype reconstruction and missing genotype inference. Am. J. Hum. Genet. 2006;S79:2290. [Google Scholar]
- 21.Cornelis M.C., Agrawal A., Cole J.W., Hansel N.N., Barnes K.C., Beaty T.H., Bennett S.N., Bierut L.J., Boerwinkle E., Doheny K.F., et al. The Gene, environment association studies consortium (GENEVA): maximizing the knowledge obtained from GWAS by collaboration across studies of multiple conditions. Genet. Epidemiol. 2010;34:364–372. doi: 10.1002/gepi.20492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian C., Kosoy R., Nassir R., Lee A., Villoslada P., Klareskog L., Hammarstrom L., Garchon H.J., Pulver A.E., Ransom M., et al. European population genetic substructure: further definition of ancestry informative markers for distinguishing among diverse European ethnic groups. Mol. Med. 2009;15:371–383. doi: 10.2119/molmed.2009.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han J., Kraft P., Nan H., Guo Q., Chen C., Qureshi A., Hankinson S.E., Hu F.B., Duffy D.L., Zhao Z.Z., et al. A genome-wide association study identifies novel alleles associated with hair color and skin pigmentation. PLoS Genet. 2008;4:e1000074. doi: 10.1371/journal.pgen.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nan H., Kraft P., Qureshi A.A., Guo Q., Chen C., Hankinson S.E., Hu F.B., Thomas G., Hoover R.N., Chanock S., et al. Genome-wide association study of tanning phenotype in a population of European ancestry. J. Invest. Dermatol. 2009;129:2250–2257. doi: 10.1038/jid.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunter D.J., Kraft P., Jacobs K.B., Cox D.G., Yeager M., Hankinson S.E., Wacholder S., Wang Z., Welch R., Hutchinson A., et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.WTCCC. Genome-wide association study of 14 000 cases of seven common diseases and 3000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ceol C.J., Houvras Y., Jane-Valbuena J., Bilodeau S., Orlando D.A., Battisti V., Fritsch L., Lin W.M., Hollmann T.J., Ferré F., et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471:513–517. doi: 10.1038/nature09806. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.