Abstract
Maintenance of an intact mitochondrial genome is essential for oxidative phosphorylation in all eukaryotes. Depletion of mitochondrial genome copy number can have severe pathological consequences due to loss of respiratory capacity. In Saccharomyces cerevisiae, several bifunctional metabolic enzymes have been shown to be required for mitochondrial DNA (mtDNA) maintenance. For example, Ilv5 is required for branched chain amino acid biosynthesis and mtDNA stability. We have identified OXA1 and TIM17 as novel multicopy suppressors of mtDNA instability in ilv5 cells. In addition, overexpression of TIM17, but not OXA1, prevents the complete loss of mtDNA in cells lacking the TFAM homologue Abf2. Introduction of the disease-associated A3243G mutant mtDNA into human NT2 teratocarcinoma cells frequently causes mtDNA loss. Yet when human TIM17A is overexpressed in NT2 cybrids carrying A3243G mtDNA, the proportion of cybrid clones maintaining mtDNA increases significantly. TIM17A overexpression results in long-term mtDNA stabilization, since NT2 cybrids overexpressing TIM17A maintain mtDNA at levels similar to controls for several months. Tim17 is a conserved suppressor of mtDNA instability and is the first factor to be identified that can prevent mtDNA loss in a human cellular model of mitochondrial disease.
INTRODUCTION
The 13 proteins encoded by the human mitochondrial genome are essential for the maintenance of functional mitochondria and so for cell viability. Mutations in mitochondrial DNA (mtDNA) are responsible for a wide variety of inherited diseases (1,2) and have been recently linked to sporadic neurodegenerative diseases (3–6). The accumulation of mtDNA mutations may also have a role in the normal ageing process (3–5). In addition to mutations in the mtDNA, loss of the mitochondrial genome occurs in individuals with mtDNA depletion syndrome (MDDS) and related disorders such as progressive external opthalmoplegia (PEO), Alpers syndrome and ataxia-neuropathy syndrome. Symptoms of mtDNA depletion are heterogeneous and can include progressive muscle weakness, liver dysfunction and neurodegeneration (reviewed in 6). In recent years, molecular genetic studies of these disorders have found that mutations in several nuclear genes are linked to the depletion of mtDNA. The mtDNA polymerase catalytic subunit POLG and the mtDNA helicase TWINKLE have been linked to both PEO (7–9) and ataxia-neuropathy syndrome (10,11), whereas at least five different genes that function in supplying the mitochondrial deoxynucleotide triphosphate pool have been linked to MDDS (reviewed in 6).
mtDNA is packaged into nucleoprotein complexes, or nucleoids (12,13). Approximately 30 different protein species have been identified as potential components of the mtDNA nucleoid (14–17). These include proteins with well-defined DNA-binding properties, such as Abf2/TFAM, mtSSB in vertebrates and Rim1 in yeast. Abf2 and TFAM are high-mobility group (HMG) DNA-binding proteins of eukaryotic origin, which are evolutionarily conserved from yeast to humans (15). In yeast, the HMG protein Abf2 binds non-specifically to the mitochondrial genome and is essential for mtDNA maintenance in cells grown on fermentable carbon sources (18,19). abf2Δ cells lose mtDNA when grown on glucose, producing respiratory-deficient petite cells which lack mtDNA (ρ0) (18). In mammals, the Abf2 homologue, TFAM, binds the mitochondrial genome (20,21), and mouse models have shown that it is essential for mtDNA maintenance in a variety of tissues (22,23). In addition, TFAM has a C-terminal tail, not found in Abf2, which enables it to act as a transcription factor that facilitates assembly and promoter recognition of the mitochondrial transcription machinery (20,24,25).
In addition to the core HMG proteins, several other classes of proteins have been shown to be associated with the mitochondrial nucleoid. One class that occurs in both yeast and higher eukaryote nucleoids are metabolic proteins, such as aconitase and Ilv5 in yeast and 3-hydroxyacyl-CoA dehydrogenase in mammals (14,16,17). Genetic evidence also suggests a role for aconitase and Ilv5 in mtDNA stability in yeast. mtDNA is unstable in aco1Δ and ilv5Δ cells, and overexpression of either of these proteins suppresses mtDNA loss in abf2Δ cells (15,26). We previously identified point mutations in Ilv5 that cause loss of either the branched chain amino acid biosynthetic function or the mtDNA stability function of this enzyme (27). In the current study, we used the temperature-sensitive (ts) phenotype of one of these mutants (W327R) to screen for multicopy suppressors. This led to the identification of OXA1 and TIM17 as suppressors of the ts and mtDNA instability phenotypes in W327R and ilv5Δ cells. Overexpression of TIM17 also stabilizes mtDNA in abf2Δ cells, suggesting that Tim17 may be a general suppressor of mtDNA loss. To test this hypothesis, human TIM17A was overexpressed in human NT2 cybrids carrying A3243G mtDNA, which have been previously shown to lose mtDNA (28). Overexpression of TIM17A significantly reduced mtDNA loss in these cells, and overextended time periods mtDNA loss was completely prevented, suggesting that Tim17 is a conserved suppressor of mtDNA loss.
RESULTS
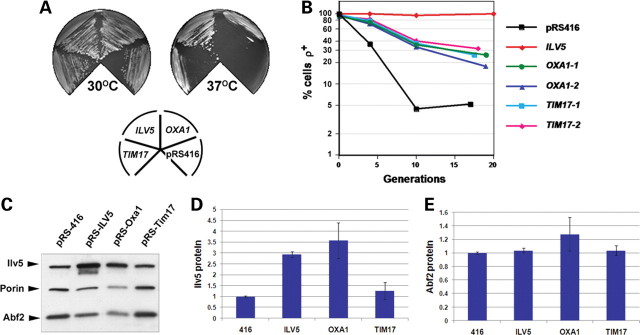
We previously identified point mutations in the bifunctional nucleoid protein Ilv5 of yeast that compromised either branched chain amino acid biosynthesis or mtDNA maintenance (27,29). Here, the ts phenotype of the most severe ilv5 DNA stability mutant (W327R; (27,29)) was used to screen for multicopy suppressors that enabled the W327R strain to grow on glycerol-containing medium at 37°C. Of 12 000 colonies screened, six suppressor clones (Supplementary Material, Table S1) were able to grow on YPGly at 37°C. Recovery of plasmid DNA and sequencing of genomic inserts identified that three genes—PET122, the mitochondrial inner-membrane insertase OXA1 (30) and mitochondrial inner-membrane presequence translocase TIM17 (31,32)—were potentially responsible for the suppression. Cloning into low copy number centromeric plasmids (two to three copies per cell) and re-introduction of each gene into W327R cells revealed that pRS-PET122 transformants remained ts (data not shown), whereas the ts phenotype was partially suppressed in W327R cells transformed with either pRS-Oxa1 or pRS-Tim17 (Fig. 1A; all pRS vectors contain the low copy number centromeric replication origin). Increased Oxa1 and Tim17 expression in these transformants was confirmed by immunoblot (Supplementary Material, Fig. S1). The suppression of the ts phenotype by OXA1 and TIM17 overexpression correlated with a 25% increase in mtDNA stability in pRS-Oxa1 transformants and a 32% increase in mtDNA stability in pRS-Tim17 transformants (mean percentage increase in ρ+ cells relative to pRS416 control at time course endpoint; Fig. 1B). Interestingly, although the suppressor screen was performed using a high copy number vector, a 2-fold increase in Tim17 expression was sufficient to suppress mtDNA loss in W327R cells (Fig. 1B and Supplementary Material, Fig. S1). Therefore, OXA1 and TIM17 are novel suppressors of the ts and mtDNA instability phenotypes of the ilv5 mutant W327R.
Figure 1.
Overexpression of OXA1 and TIM17 suppresses the ts and mtDNA instability phenotypes of the ilv5 mutant W327R. (A) W327R cells transformed with either pRS416 (416), pRS-ILV5 (ILV5), pRS-Oxa1 (OXA1) or pRS-Tim17 (TIM17) were streaked onto YPG medium and grown at either 30°C or 37°C. Note the partial suppression of the ts phenotype conferred by OXA1 and TIM17. (B) The same transformants as in (A) were pre-grown in YNBG + cas at 30°C, then shifted to YNBD + cas and aliquots plated out at three time points and scored by TTC overlay for the fraction of petites in the population. Two independent pRS-Oxa1 (OXA1-1 and OXA1-2) or pRS-Tim17 (TIM17-1 and TIM17-2) transformant lines were scored. (C) Representative immunoblot of Ilv5 and Abf2 protein levels in W327R transformants. Quantification of Ilv5 and Abf2 protein levels are shown in (D) and (E), relative to the levels in pRS416 control transformants that was arbitrarily set as 1. (D) Ilv5 levels are increased 3.5-fold in W327R cells transformed with pRS-Oxa1, whereas Ilv5 levels in pRS-Tim17 transformants are similar to controls cells transformed with pRS416. As expected, Ilv5 protein levels were increased (2.7-fold) in pRS-ILV5 transformants. (E) Abf2 protein levels are similar to controls in pRS-Oxa1 and pRS-Tim17 transformants. All cultures were grown overnight in YNBG + cas prior to immunoblot analysis. Expression levels in (D) and (E) are calculated relative to porin-loading control and values shown are the average of two independent experiments (see Materials and Methods). Averages are shown ±1 SEM.
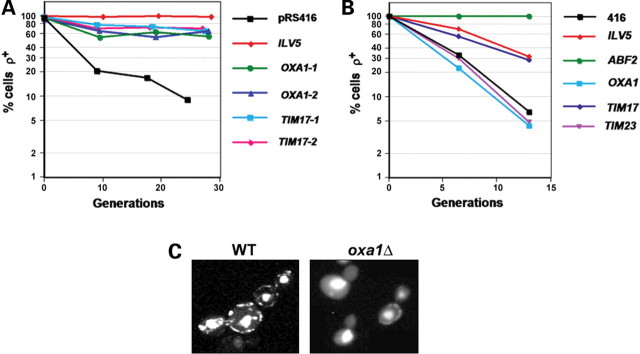
If elevated expression of Oxa1 and Tim17 attenuates mtDNA instability, then oxa1Δ or tim17Δ mutants might have a mtDNA instability phenotype. tim17Δ cells are not viable and so we could not address this question for Tim17. However, oxa1Δ cells are viable but respiratory deficient (30). Staining of oxa1Δ cells with DAPI indicated that they were largely devoid of mtDNA (Fig. 2C). Thus, Oxa1 is normally required to maintain mtDNA in Saccharomyces cerevisiae.
Figure 2.
TIM17, but not OXA1, is a general suppressor of mtDNA loss in yeast. (A) ilv5Δ cells transformed with pRS416, pRS-ILV5 (ILV5), pRS-Oxa1 (two independent transformant lines were used: OXA1-1 and OXA1-2) or pRS-Tim17 (two independent transformant lines were used: TIM17-1 and TIM17-2) were pre-grown in YNBG + cas, then shifted to YNBD + cas, aliquots removed at three time points and the fraction of petites in the population scored as in Figure 1B. (B) mtDNA loss in abf2Δ cells transformed with pRS416, pRS-ILV5 (ILV5), pRS-Oxa1 (OXA1), pRS-Tim17 (TIM17) or pRS-Tim23 (TIM23) was determined as in (A). (C) Wild-type and oxa1Δ cells were grown on YPD and stained with DAPI to identify mtDNA. The strong central staining in all cells is nuclear DNA, whereas the peripheral punctate staining is mtDNA (each punctate body contains one to two mtDNA genomes). oxa1Δ cells may be in the process of losing their mtDNA since, in some cells, a single punctate body can be seen in the mother cell and none in the daughter bud.
One possible explanation for the increase in mtDNA stability in W327R cells caused by increased Oxa1 or Tim17 could be that these proteins affect Ilv5 or Abf2 protein levels. Ilv5 levels increased 3.5-fold in cells overexpressing OXA1 (Fig. 1C and D), suggesting that OXA1 overexpression may stabilize mtDNA by increasing the abundance of this and possibly other nucleoid proteins, although not Abf2 (Fig. 1E). In cells overexpressing TIM17, Ilv5 and Abf2 levels were similar to the control (Fig. 1C, D and E), suggesting that the increase in mtDNA stability in these cells is not due to a change in Ilv5 or Abf2 protein levels.
To test whether OXA1 and TIM17 can suppress mtDNA instability in ilv5Δ cells as well as in the W327R hypermorph, ilv5Δ cells were transformed with pRS-Oxa1 and pRS-Tim17, and these transformants were assayed for mtDNA instability. Overexpression of OXA1 and TIM17 increased mtDNA stability in ilv5Δ cells by 65 and 76% respectively (Fig. 2A), demonstrating that OXA1 and TIM17 can increase mtDNA stability in the absence of Ilv5 as well as in the presence of mutant W327R protein.
The HMG group DNA-binding protein Abf2 is the most thoroughly characterized yeast mtDNA nucleoid component. abf2Δ cells rapidly lose mtDNA when grown on glucose, but unlike ilv5Δ cells (which produce ρ− petites), they produce ρ0 petites (33). The ability of Oxa1 and Tim17 overexpression to suppress ilv5 mtDNA instability phenotypes may be through a mechanism that is specific to Ilv5-mediated mtDNA stability or through a more general property of these proteins. To test this, we overexpressed Oxa1 and Tim17 in abf2Δ cells. Overexpression of OXA1 had no effect on mtDNA loss, whereas overexpression of TIM17 increased mtDNA stability in abf2Δ cells by 32% (Fig. 2B). ILV5 was originally identified as a suppressor of mtDNA loss in abf2Δ cells (26). Interestingly, overexpression of TIM17 suppressed the abf2Δ phenotype to a similar extent as overexpression of ILV5 (34% increase in mtDNA stability; Fig. 2B). These data suggest that TIM17 may be a general suppressor of mtDNA loss, whereas OXA1 is limited to the suppression of ilv5 phenotypes.
To address whether another component of the inner-membrane translocase (TIM) complex (32) can suppress mtDNA loss, abf2Δ cells were transformed with a centromeric plasmid containing TIM23 (pRS-Tim23). pRS-Tim23 transformants had a similar mtDNA instability phenotype to abf2Δ cells (Fig. 2B), suggesting that the ability of Tim17 to suppress mtDNA instability may be a specific property of this protein.
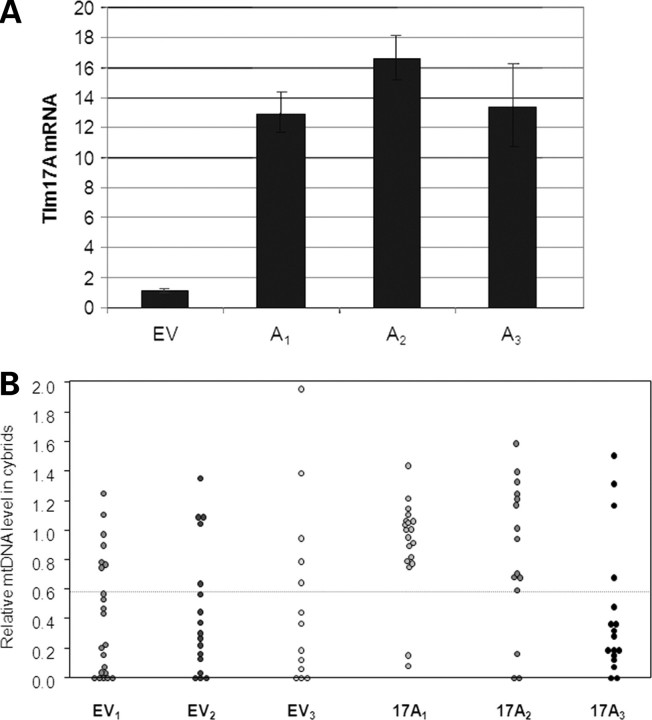
In a previous study, we showed that the introduction of A3243G mutant mtDNA into human NT2 teratocarcinoma cells was frequently associated with the loss of mtDNA (28). Such cells provided a means of testing the hypothesis that high levels of Tim17 could suppress the loss of mtDNA in mammalian cells. There are two isoforms of Tim17 in man, Tim17A and Tim17B, the former being most closely related to S. cerevisiae Tim17 (Supplementary Material, Fig. S2). Therefore, additional copies of human TIM17A were introduced into NT2 ρ0 cells carried on the plasmid pIRES.Hyg3, and the three clones (NT2.17A1, 17A2 and 17A3) that initially expressed TIM17A at levels 13–17-fold higher than sister clones containing empty vector (NT2.EV) were selected (Fig. 3A).
Figure 3.
Decreased frequency of mtDNA depletion in two out of three NT2 ρ0 cell lines carrying pIIRES.Tim17A. (A) TIM17A overexpression in NT2 ρ0 cells. NT2 ρ0 cells were transfected with pIRES.Hyg3 (empty vector—EV) or with pIRES.Hyg3 containing the human TIM17A gene (A1–A3). Hygromycin-resistant clones were expanded and RNA samples isolated for qPCR of TIM17A mRNA level relative to GAPDH. EV is the combined data from five control cell lines without the TIM17A gene and is arbitrarily set at 1. A1–A3 are the three clones that initially expressed TIM17A at a level >10-fold that of EV cell lines. Other clones showing more modest increases in TIM17A expression were discarded. qPCR and last-cycle PCR analysis failed to detect mtDNA in pIRES.EV and pIRES.17A clones, confirming that they remained devoid of mtDNA after transfection (data not shown). (B) Three NT2 ρ0 cell lines containing empty plasmid (EV1–EV3) and three NT2 ρ0 cell lines containing the TIM17A expressing plasmid (A1–A3) were fused with donor cytoplasts (75% A3243G mtDNA). Cybrids were grown for 5–7 weeks following fusion and mtDNA levels measured using qPCR. Each point represents a clonal cybrid cell line generated from these fusions. Levels of mtDNA are relative to the level of mtDNA in 143B cells that was arbitrarily set as 1.
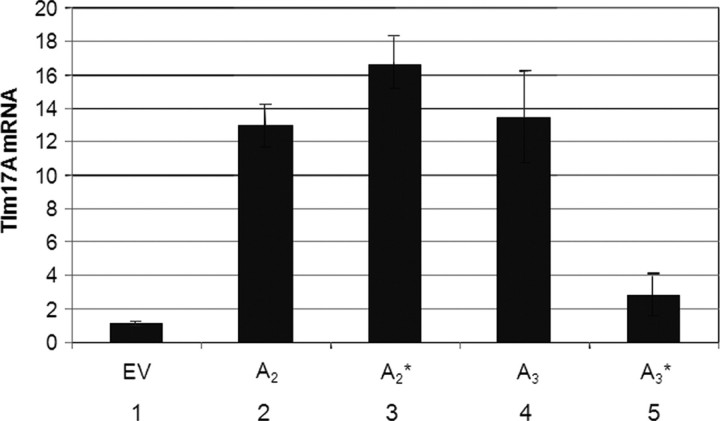
Three TIM17A-overexpressing and three control lines (NT2.EV1, EV2 and EV3) were fused with cytoplasts harbouring 75% A3243G mtDNA, and clonal cybrid uridine auxotrophic (28,34) cell lines were selected. mtDNA levels were analysed from clones isolated 35–42 days after fusion. These were scored as having suffered mtDNA loss if they had <60% of the level of mtDNA of a control cell line (143B osteosarcoma); clones with higher levels of mtDNA were categorized as falling within the normal range. By this measure, two-thirds of NT2.EV cybrids had reduced levels of mtDNA (Fig. 3B and Table 1), recapitulating earlier experiments with NT2 cybrids carrying A3243G mtDNA (28). In contrast, only 15% of cybrids derived from NT2.17A1 and NT2.17A2 fusion experiments lost mtDNA (Fig. 3B and Table 1). Overall, five of 33 NT2.17 A1 and A2 cybrids had reduced levels of mtDNA compared with 33 of 50 NT2.EV cybrids (P < 5 × 10−6). The increased frequency of mtDNA maintenance in sister cells (NT2.17A1 and A2) suggested that a high level of expression of TIM17A stabilizes A3243G mtDNA in the NT2 cell background. The data for cell line NT2.17A3 appeared at first to undermine the hypothesis, as three quarters of NT2.17A3 cybrids had low levels of mtDNA (Fig. 3B). In the case of cell lines NT2.17A2 and A3, there was an interval of 8 weeks between first quantifying TIM17A transcripts and cytoplast fusion, whereas NT2.17A1 cells were fused to A3243G containing cytoplasts within a few days of quantifying TIM17 mRNA. Retrospective analysis of TIM17A expression of the NT2.17A3 cell line revealed that overexpression decreased from 13-fold at first screening to 3-fold immediately prior to fusion, whereas NT2.17A2 maintained a much higher level of TIM17A expression (Fig. 4). Thus, a 3-fold increase in TIM17A transcript was insufficient to prevent mtDNA loss, whereas expression levels over 10-fold higher than controls favoured mtDNA maintenance.
Table 1.
Levels of mtDNA in NT2.EV and NT2.17A cybrids
| NT2 cell line | Cybrid clones (n) | Cybrids ↓mtDNA (n) | Cybrids ↓mtDNA (%) |
|---|---|---|---|
| EV1 | 22 | 15 | 68 |
| EV2 | 17 | 12 | 71 |
| EV3 | 11 | 6 | 55 |
| 17A1 | 18 | 2 | 11 |
| 17A2 | 15 | 3 | 20 |
| 17A3 | 17 | 13 | 76 |
The table contains the same data as in Figure 3B and shows the number and percentage of cybrids with 40% less mtDNA than the 143B cell control, which is indicated on the chart by a horizontal broken line.
Figure 4.
Declining TIM17A expression in NT2 ρ0 clone 17A3. Columns one, two and four: the mean level of expression of TIM17A in five clones carrying pIRES.Hyg3 (EV), expression of TIM17A transcript in clones NT2.17A2 (A2) and NT2.17A3 (A3), respectively (reproduced from Fig. 3A). Clones NT2.17A2 and NT2.17A3 were maintained in culture for a further 8 weeks before fusing to cytoplasts carrying A3243G mtDNA; at this time, RNA was again isolated and used to estimate TIM17A mRNA levels by qPCR (columns 3 and 5).
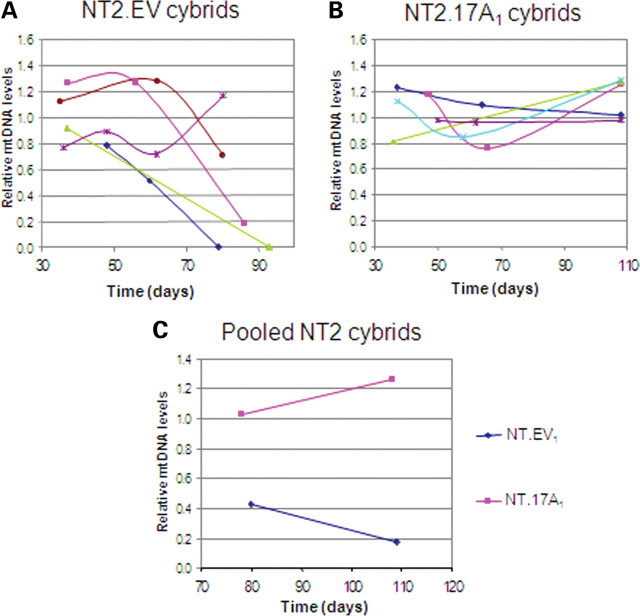
Several NT2.EV1 and NT2.17A1 cybrids were monitored for an extended period to determine whether there were subsequent changes in mtDNA copy number. Eighty days after fusion, the level of mtDNA fell below the arbitrary threshold (60% of the mtDNA level of 143B osteosarcoma cells) in three of five NT2.EV1 clones (Fig. 5A). Two clones had lost all mtDNAs and the third had dropped to 20%, whereas only one of the five clones stably maintained its mtDNA. There was no such decrease in mtDNA copy number in any of the five NT2.17A1 cybrids maintained up to 108 days after cybrid formation (Fig. 5B). Thus, the benefit conferred by increased TIM17A expression appeared to be permanent.
Figure 5.
Long-term maintenance of mtDNA in NT2 Tim17A cybrids. (A) and (B) show mtDNA levels in samples taken from NT2.3243 cybrids cultured for extended periods of time. Cybrids in (A) are derived from the empty vector containing ρ0 cell line EV1, and the cybrids in (B) are derived from the Tim17A-expressing ρ0 cell line A1. (C) shows pooled populations of cybrids from the same fusion experiments. Levels of mtDNA were measured by qPCR and are shown relative to the same control cell line (143B) that was arbitrarily set at 1.
In some cases, cybrids formed from the fusion of ρ0 cells with cytoplasts colonies were allowed to merge and form a polyclonal cell line consisting typically of over 100 clones. Polyclonal NT2.17A1 cybrids maintained their mtDNA, whereas mtDNA copy number fell to <20% of the control value in a polyclonal NT2.EV1 cybrid line (Fig. 5C). These data further confirm the potential for Tim17 overexpression to prevent mtDNA loss.
NT2 cybrids that retained their mtDNA had a similar mtDNA copy number, irrespective of whether the cells had been transfected with pIRES.hyg.Tim17A. Hence, there was no suggestion that a high level of Tim17A induced an increase in mtDNA copy number in NT2 cells. Likewise, there was no effect on mtDNA copy number from 4-fold or 29-fold overexpression of Tim 17A in 143B cells (Supplementary Material, Fig. S3). Surprisingly, overexpression of neither TFAM (35) nor Twinkle (36) is sufficient to produce an appreciable increase in copy number in cultured cells. This is in contrast to in vivo studies of TFAM, which have shown that mtDNA copy number is directly proportional to TFAM levels (37). Therefore the possibility that TIM17A overexpression may increase mtDNA copy number above wild-type levels requires further testing in vivo.
To begin to investigate the mechanism by which Tim17 prevents mtDNA loss, RNAi was used to determine the effect of Tim17 gene-silencing on mtDNA copy number and nucleoid morphology. If Tim17A interacts directly with the nucleoid and is required for mtDNA maintenance, then depletion of Tim17A would be expected to reduce mtDNA copy number and/or alter nucleoid morphology. TIM17A gene-silencing had only a slight effect on mtDNA copy number, similar to ATAD3 repression, yet unlike ATAD3 RNAi there was no effect on mitochondrial nucleoid structure, size or number (Supplementary Material, Fig. S4) (38). These data suggest that Tim17A is not normally required for mtDNA maintenance and is not a direct component of the mtDNA nucleoid.
DISCUSSION
Using a high copy number suppressor screen in budding yeast, we have identified OXA1 and TIM17 as novel suppressors of mtDNA instability in mutants of the bifunctional nucleoid gene ILV5. TIM17 is also able to suppress the severe mtDNA loss that occurs in cells lacking the TFAM homologue Abf2. In contrast, OXA1 overexpression has no effect in abf2Δ cells. In addition, TIM17A overexpression prevents short- and long-term mtDNA loss in human NT2 cybrids carrying A3243G mtDNA. Taken together, these data suggest that TIM17 may be a general suppressor of mtDNA loss.
The S. cerevisiae mitochondrial nucleoid contains at least 25 proteins, many of which have not been characterized with respect to their role in mtDNA inheritance (15,16). These proteins can be grouped into various classes according to function. One of the main classes is bifunctional metabolic enzymes that have presumably adapted to interact with mtDNA. Many of the bifunctional proteins identified as components of the yeast nucleoid have not been found in the mammalian equivalent. Some of these proteins, such as Ilv5, do not exist in mammals, but may be functionally replaced by metabolic proteins such as 3-hydroxyacyl-CoA dehydrogenase, an enzyme of fatty acid β-oxidation, which is found in human nucleoids (17). The best characterized proteins in this class are Ilv5 and Aco1, both of which have been shown to prevent mtDNA loss in abf2Δ cells when overexpressed (15,26,27). In addition, Aco1 and more recently Ilv5, have both been shown to bind mtDNA in vitro (39,40). In contrast, neither Oxa1 nor Tim17 has been identified in nucleoid preparations and so their ability to suppress mtDNA loss may be through an indirect mechanism.
OXA1 and TIM17 are the first multicopy suppressors of ilv5 mtDNA instability to be identified. Although Oxa1 and Tim17 are components of inner-membrane complexes that are involved in membrane insertion of mitochondrial proteins, our data suggest that the mechanism of Oxa1-mediated suppression differs significantly from that of Tim17. Oxa1 is a member of the conserved Oxa1/YidC/Alb3 protein family involved in membrane insertion of proteins (30,41–43). Oxa1 is the major component of the translocase machinery that inserts mitochondrial and nuclear encoded proteins into the inner membrane. Oxa1 has also been shown to interact with mitochondrial ribosomes and this association requires the matrix-exposed C-terminal region of the protein (44). Perhaps, the increased interaction of Oxa1 with ribosomes when Oxa1 is overexpressed indirectly stabilizes mtDNA through upregulation or stabilization of mitochondrial translation.
Tim17 is highly conserved throughout eukaryotes and is a component of the TIM23 presequence translocase of the mitochondrial inner membrane (45,46). The TIM23 complex accepts preproteins from the outer-membrane TOM complex and directs them either for insertion into the inner membrane or, in cooperation with the presequence translocase-associated motor (PAM), to the matrix (45,46). Tim17 is unlikely to form part of the translocase channel itself, since structural studies have shown that recombinant Tim23 is able to form channels in vitro (47). Rather than forming part of the channel, Tim17 is critical in conferring the dynamic properties of the TIM23 complex and in connecting TIM23 to the PAM complex. Patch clamp studies of reconstituted inner membranes recently showed that the TIM23 complex is composed of cooperatively gated twin pores (48). Tim17 is necessary for regulating pore structure and voltage gating of the Tim23 channel, such that depletion of Tim17 causes collapse into a single pore (48). Moreover, Tim17 has been shown to be required for the inner-membrane insertion function of the TIM23 complex and to act as a direct link to the PAM through interaction with Pam18 (31). Given the multiple concerted roles of Tim17, its expression level and stoichiometry within the TIM23 complex may be particularly important.
Although mtDNA copy number, size and individual nucleoid components differ significantly between yeast and mammals, the mode of inheritance of the genome and functional classes of the proteins composing the nucleoid are similar (12,13). The ability of Tim17 to prevent mtDNA loss in yeast and human cultured cells further suggests that there are conserved elements to mtDNA inheritance between the two systems. However, there are several phenotypic differences in Tim17-mediated prevention of mtDNA loss between yeast and human cells. Tim17 overexpression only partially suppresses mtDNA loss in abf2Δ cells. Similarly, overexpression of TIM17A in NT2 cybrids did not prevent mtDNA loss in all clones 5–7 weeks after fusion. However, in long-term experiments, mtDNA levels were similar to wild-type in all cybrid clones tested, as well as in a pooled population. Loss of mtDNA in yeast results in markedly different physiological changes compared with human cells. When grown on a fermentable carbon source, yeast cells can switch to fermentative metabolism and so the selective pressure to maintain a functional mitochondrial genome is reduced. This possibility does not exist in higher eukaryotes, and so the long-term stability of mtDNA in human cells overexpressing Tim17 may be a result of the strong selective pressure to maintain a functional mitochondrial genome.
The yeast and mammalian cell models of mtDNA loss used in this study, which are suppressed by overexpression of Tim17, are analogous to the mtDNA depletion seen in MDDS and related disorders (6). In recent years, these diseases have been shown to be linked to mutations in genes whose products function in mtDNA replication or supply mitochondria with the nucleotide precursors of replication (6). Given that Abf2 is a key regulator of mtDNA copy number (49), the ability of Tim17 to ameliorate mtDNA loss in abf2Δ and A3243G cells imply that increasing Tim17 activity may have the potential to be of therapeutic benefit in MDDS and related disorders. Since Abf2 is directly involved with mtDNA transactions, it seems more likely that such a strategy would be beneficial in diseases linked to mutations in proteins that function at the mtDNA replication fork, such as POLG and TWINKLE, rather than those which are involved in mitochondrial dNTP supply. The recently identified inner-membrane protein MPV17, mutated in infantile hepatic mtDNA depletion (50), may also be an interesting target.
To test the ability of Tim17 to prevent mtDNA loss in human cells, we used human NT2 teratocarcinoma cells containing A3243G mutant mtDNA. Since NT2 cells are tumour-derived, they may not be wholly representative of in vivo mtDNA loss. However, the failure to maintain A3243G mutant mtDNA in these cells does provide a valuable system for testing factors that potentially affect mtDNA loss. The A3243G mutation is often associated with the mitochondrial disease mitochondrial encephalo(myo)pathy, lactic acidosis and stroke-like episodes, or MELAS (51). A3243G is located in the leucine tRNA gene (51) and negatively affects the steady-state level, aminoacylation and extent of wobble base modification of tRNALeu(UUR) (52–54). The mechanism by which A3243G mtDNA in NT2 cybrids is lost has not been elucidated, but it may be analogous to the mtDNA instability in yeast caused by mutations in genes required for mitochondrial protein synthesis (55,56). It has been suggested that the inhibition of mitochondrial translation causes perturbation of the membrane potential (due to the lack of a component encoded by the mitochondrial genome), which prevents the import of specific proteins required for mtDNA stability (56). Overexpression of Tim17 may ameliorate this defect by increasing uptake of factors such as structural components of the nucleoid, or proteins involved in mtDNA replication or transmission (12,16,17). In accordance with this hypothesis, Iborra et al. (57) have suggested that the TIM23 complex is closely associated with mitochondrial ribosomes in order to coordinate the assembly of protein complexes encoded by both nuclear and mitochondrial genomes. It is unlikely that Tim17 is a physical component of the nucleoid since it has never been identified in yeast or animal mitochondrial nucleoprotein preparations (14,15,17,38). As such, Tim17 is unlikely to prevent mtDNA loss by binding to and physically stabilizing mtDNA in the manner of aconitase (39) or Ilv5 (40) in abf2Δ cells. Another argument against Tim17A interacting directly with nucleoids is the lack of effect of Tim17A RNAi on mitochondrial nucleoid structure, size or number.
Regardless of the mechanism, Tim17 has the ability to prevent mtDNA loss in both yeast and human cells. Moreover, this study is the first to identify a gene that can potentially prevent mtDNA loss in humans. Future studies will be aimed at determining the mechanism of Tim17-mediated prevention of mtDNA loss and testing Tim17A overexpression in other models of mitochondrial disease.
MATERIALS AND METHODS
Yeast strains and growth conditions
Yeast strains described were derivatives of 14CWW (MATa ade2-1 ura3-52 trp1 leu2-3,112 ρ+), 14CWWilv5Δ (MATa ade2-1 ura3-52 trp1 leu2-3,112 ilv5::URA3 ρ+), W327R (containing the strongest a+D− ilv5 allele integrated at the ILV5 locus, (27,29), 14CWWabf2Δ (26) or W303 (MATα leu2-3,112 his3-11,15 trp1-1 can1-100 ade2-1 ura3-1). The oxa1Δ strain was obtained by tetrad dissection of the commercially available strain diploid YER154W (Invitrogen).
Yeast cells were grown at 30°C in YP medium (1% yeast extract and 2% bactopeptone) containing either 2% dextrose (YPD), 3% glycerol (YPGly) or YNB medium (0.67% yeast nitrogen base without amino acids) containing 1% casamino acids and either 2% dextrose (YNBD + cas) or 3% glycerol (YNBG + cas). Additional amino acids were supplemented as required.
To measure petite formation in the various strains, mid-log-phase cultures of strains grown in YNBG+cas medium were transferred to YNBD + cas medium and grown for the required number of generations. At various time points, cells were plated onto YNBD + cas plates and grown for 3 days at 30°C. ρ+ and petite colonies were distinguished by 2,3,5-triphenyltetrazolium chloride (TTC) overlay (58), using 0.2% TTC (Sigma) in 0.8% agarose. Between 50 and 300 colonies were counted for each time point.
Multicopy suppressor screen
The W327R strain was transformed with a Yep24 URA3 yeast genomic library (59) and grown on YNBD + cas plates. About 12 000 URA+ transformants were recovered. These colonies were then replica-plated into YPGly and YNBD + cas. The YPGly plates were incubated for 3–4 days at 37°C, to identify cells able to grow at the non-permissive temperature. Suppressor colonies were re-streaked on YPGly at the non-permisive temperature and plasmid DNA recovered from these clones. Genomic inserts in each plasmid were sequenced at both ends and compared with the S. cerevisiae genome using NCBI BLAST search.
Plasmid construction
pRS416-ILV5 was described previously (26). The PET122 coding sequence (PET122 has a complex promoter) was cloned by PCR using primers: 5′-CATAAGAACTCTAGACGTCATGGTGACTATCACG-3′ and 5′-GCTCCACGTCGACTTATGTGTTGATTTCAAATCC-3′. The PCR product was then digested with XbaI and SalI and ligated in to p416-TEF (60). TIM17 was cloned using primers 5′-GATGAAAACACTAGTCGATACGGCC-3′ and 5′-TAAAAGGCTCGAGAATACATCTGGC-3′, which amplify the coding sequence and promoter region. The PCR product was then digested with SpeI and XhoI and ligated into pRS416 (61). The OXA1 coding sequence (OXA1 has a complex promoter) was amplified using primers 5′-CCACAGAATAAACTAGTCGACGATCAAACTC-3′ and 5′-CAGAGAGATATAGAGTCGACATTCATTTTTTG-3′, then digested with SpeI and SalI and cloned into p416-TEF. The Tim23 coding sequence and promoter region were amplified using primers 5′-GATACTAGTCGCAGTCGTGGTGACTCTGG-3′ and 5′-CGGTGCACTTGTCTCGAGTTTTTTCAAGC-3′, digested with SpeI and XhoI and cloned into pRS416. Restriction enzyme sites are in boldface. All constructs were confirmed by DNA sequencing.
Immunoblotting
Immunoblotting was performed as in Bateman et al. (29). Antibodies used were rabbit anti-Ilv5 (16) and mouse anti-Porin (Molecular Probes). Rabbit anti-Tim17, anti-Tim23 (62) and anti-Oxa1 (63) were kindly provided by Thomas Langer. Anti-rabbit or anti-mouse immunoglobulin G-coupled horseradish peroxidase (Bio-Rad) were used as secondary antibodies and were visualized using the ECL system (Amersham Biosciences). To determine fold changes in protein levels in transformants, immunoblot bands were quantified using ImageJ. The integrated density of each band of interest was divided by the integrated density of the porin band in the same lane. The relative increase in the protein was calculated by comparison with the level of same protein in control cells transformed with empty vector (pRS416). The average increase from two independent transformants was used to obtain the final value.
Human cell culture and transformation
All cells were grown in DMEM supplemented with 1 mm sodium pyruvate and 10% fetal bovine serum. In the case of ρ0 cells that lack mtDNA, 50 µm uridine was also included. The Tim17A gene was purchased from the Mammalian Genome Collection, amplified using primers 5′-TCCCCTTAAGATGGAGGAGTACGCGCGAG-3′ and 5′-TCCCCCCGGGCTACTGATATTGTCGATAGTC-3′ and cloned into plasmid pIRES hyg3 (Clontech) after digestion with AflII and XmaI. A total of 5 × 106 NT2 ρ0 cells were transformed with 5 µg plasmid with and without the TIM17A gene by electroporation in 0.5 ml PBS, 25 mm glucose, 1.25% DMSO. Electroporation conditions were 300 mV and 950 µFD (Gene Pulser II, BioRad), with time constants of 18. An amount of 300 µg/ml hygromycin B was applied 48 h after electroporation colonies formed after 2–3 weeks.
Cybrid formation and analysis
Cell cybrids were generated by fusing NT2 ρ0 cells and enucleated osteosarcoma cells (cytoplasts) carrying A3243G mutant mtDNA, as described previously (28), except that the mitochondrial recipient NT2 ρ0 cells carried pIRES hyg3 with or without the TIM17A gene. Monoclonal and polyclonal cybrid cell lines were expanded after cell–cytoplast fusion and DNA and RNA harvested at intervals. mtDNA copy number and transcript levels were determined by quantitative PCR (qPCR) as described (64).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Muscular Dystrophy Campaign (MRC); the European Union (Mitocombat integrated programme) (I.J.H.); King’s College London; The Royal Society (J.M.B.).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Philip S. Perlman for his support and advice on the manuscript. This manuscript is dedicated to the memory of Ronald A. Butow.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Dimauro S., Davidzon G. Mitochondrial DNA and disease. Ann. Med. 2005;37:222–232. doi: 10.1080/07853890510007368. [DOI] [PubMed] [Google Scholar]
- 2.Taylor R.W., Turnbull D.M. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krishnan K.J., Greaves L.C., Reeve A.K., Turnbull D.M. Mitochondrial DNA mutations and aging. Ann. NY Acad. Sci. 2007;1100:227–240. doi: 10.1196/annals.1395.024. [DOI] [PubMed] [Google Scholar]
- 4.Kujoth G.C., Bradshaw P.C., Haroon S., Prolla T.A. The role of mitochondrial DNA mutations in mammalian aging. PLoS Genet. 2007;3:e24. doi: 10.1371/journal.pgen.0030024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wallace D.C. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Copeland W.C. Inherited mitochondrial diseases of DNA replication. Annu. Rev. Med. 2008;59:131–146. doi: 10.1146/annurev.med.59.053006.104646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirano M., Marti R., Ferreiro-Barros C., Vila M.R., Tadesse S., Nishigaki Y., Nishino I., Vu T.H. Defects of intergenomic communication: autosomal disorders that cause multiple deletions and depletion of mitochondrial DNA. Semin. Cell Dev. Biol. 2001;12:417–427. doi: 10.1006/scdb.2001.0279. [DOI] [PubMed] [Google Scholar]
- 8.Van Goethem G., Dermaut B., Lofgren A., Martin J.J., Van Broeckhoven C. Mutation of POLG is associated with progressive external ophthalmoplegia characterized by mtDNA deletions. Nat. Genet. 2001;28:211–212. doi: 10.1038/90034. [DOI] [PubMed] [Google Scholar]
- 9.Spelbrink J.N., Li F.Y., Tiranti V., Nikali K., Yuan Q.P., Tariq M., Wanrooij S., Garrido N., Comi G., Morandi L., et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat. Genet. 2001;28:223–231. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- 10.Hakonen A.H., Heiskanen S., Juvonen V., Lappalainen I., Luoma P.T., Rantamaki M., Goethem G.V., Lofgren A., Hackman P., Paetau A., et al. Mitochondrial DNA polymerase W748S mutation: a common cause of autosomal recessive ataxia with ancient European origin. Am. J. Hum. Genet. 2005;77:430–441. doi: 10.1086/444548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Goethem G., Luoma P., Rantamaki M., Al Memar A., Kaakkola S., Hackman P., Krahe R., Lofgren A., Martin J.J., De Jonghe P., et al. POLG mutations in neurodegenerative disorders with ataxia but no muscle involvement. Neurology. 2004;63:1251–1257. doi: 10.1212/01.wnl.0000140494.58732.83. [DOI] [PubMed] [Google Scholar]
- 12.Chen X.J., Butow R.A. The organization and inheritance of the mitochondrial genome. Nat. Rev. Genet. 2005;6:815–825. doi: 10.1038/nrg1708. [DOI] [PubMed] [Google Scholar]
- 13.Kucej M., Butow R.A. Evolutionary tinkering with mitochondrial nucleoids. Trends Cell. Biol. 2007;17:586–592. doi: 10.1016/j.tcb.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 14.Bogenhagen D.F., Wang Y., Shen E.L., Kobayashi R. Protein components of mitochondrial DNA nucleoids in higher eukaryotes. Mol. Cell Proteomics. 2003;2:1205–1216. doi: 10.1074/mcp.M300035-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Chen X.J., Wang X., Kaufman B.A., Butow R.A. Aconitase couples metabolic regulation to mitochondrial DNA maintenance. Science. 2005;307:714–717. doi: 10.1126/science.1106391. [DOI] [PubMed] [Google Scholar]
- 16.Kaufman B.A., Newman S.M., Hallberg R.L., Slaughter C.A., Perlman P.S., Butow R.A. In organello formaldehyde crosslinking of proteins to mtDNA: identification of bifunctional proteins. Proc. Natl Acad. Sci. USA. 2000;97:7772–7777. doi: 10.1073/pnas.140063197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y., Bogenhagen D.F. Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J. Biol. Chem. 2006;281:25791–25802. doi: 10.1074/jbc.M604501200. [DOI] [PubMed] [Google Scholar]
- 18.Diffley J.F., Stillman B. A close relative of the nuclear, chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc. Natl Acad. Sci. USA. 1991;88:7864–7868. doi: 10.1073/pnas.88.17.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diffley J.F., Stillman B. DNA binding properties of an HMG1-related protein from yeast mitochondria. J. Biol. Chem. 1992;267:3368–3374. [PubMed] [Google Scholar]
- 20.Fisher R.P., Clayton D.A. Purification and characterization of human mitochondrial transcription factor 1. Mol. Cell Biol. 1988;8:3496–3509. doi: 10.1128/mcb.8.8.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaufman B.A., Durisic N., Mativetsky J.M., Costantino S., Hancock M.A., Grutter P., Shoubridge E.A. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol. Biol. Cell. 2007;18:3225–3236. doi: 10.1091/mbc.E07-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larsson N.G., Wang J., Wilhelmsson H., Oldfors A., Rustin P., Lewandoski M., Barsh G.S., Clayton D.A. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 23.Silva J.P., Kohler M., Graff C., Oldfors A., Magnuson M.A., Berggren P.O., Larsson N.G. Impaired insulin secretion and beta-cell loss in tissue-specific knockout mice with mitochondrial diabetes. Nat. Genet. 2000;26:336–340. doi: 10.1038/81649. [DOI] [PubMed] [Google Scholar]
- 24.McCulloch V., Shadel G.S. Human mitochondrial transcription factor B1 interacts with the C-terminal activation region of h-mtTFA and stimulates transcription independently of its RNA methyltransferase activity. Mol. Cell Biol. 2003;23:5816–5824. doi: 10.1128/MCB.23.16.5816-5824.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parisi M.A., Clayton D.A. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science. 1991;252:965–969. doi: 10.1126/science.2035027. [DOI] [PubMed] [Google Scholar]
- 26.Zelenaya-Troitskaya O., Perlman P.S., Butow R.A. An enzyme in yeast mitochondria that catalyzes a step in branched-chain amino acid biosynthesis also functions in mitochondrial DNA stability. EMBO J. 1995;14:3268–3276. doi: 10.1002/j.1460-2075.1995.tb07330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bateman J.M., Perlman P.S., Butow R.A. Mutational bisection of the mitochondrial DNA stability and amino acid biosynthetic functions of ilv5p of budding yeast. Genetics. 2002;161:1043–1052. doi: 10.1093/genetics/161.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turner C.J., Granycome C., Hurst R., Pohler E., Juhola M.K., Juhola M.I., Jacobs H.T., Sutherland L., Holt I.J. Systematic segregation to mutant mitochondrial DNA and accompanying loss of mitochondrial DNA in human NT2 teratocarcinoma cybrids. Genetics. 2005;170:1879–1885. doi: 10.1534/genetics.105.043653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bateman J.M., Iacovino M., Perlman P.S., Butow R.A. Mitochondrial DNA instability mutants of the bifunctional protein Ilv5p have altered organization in mitochondria and are targeted for degradation by Hsp78 and the Pim1p protease. J. Biol. Chem. 2002;277:47946–47953. doi: 10.1074/jbc.M209071200. [DOI] [PubMed] [Google Scholar]
- 30.Bonnefoy N., Chalvet F., Hamel P., Slonimski P.P., Dujardin G. OXA1, a Saccharomyces cerevisiae nuclear gene whose sequence is conserved from prokaryotes to eukaryotes controls cytochrome oxidase biogenesis. J. Mol. Biol. 1994;239:201–212. doi: 10.1006/jmbi.1994.1363. [DOI] [PubMed] [Google Scholar]
- 31.Chacinska A., Lind M., Frazier A.E., Dudek J., Meisinger C., Geissler A., Sickmann A., Meyer H.E., Truscott K.N., Guiard B., et al. Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell. 2005;120:817–829. doi: 10.1016/j.cell.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 32.Kutik S., Guiard B., Meyer H.E., Wiedemann N., Pfanner N. Cooperation of translocase complexes in mitochondrial protein import. J. Cell Biol. 2007;179:585–591. doi: 10.1083/jcb.200708199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Megraw T.L., Chae C.B. Functional complementarity between the HMG1-like yeast mitochondrial histone HM and the bacterial histone-like protein HU. J. Biol. Chem. 1993;268:12758–12763. [PubMed] [Google Scholar]
- 34.King M.P., Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 35.Pohjoismaki J.L., Wanrooij S., Hyvarinen A.K., Goffart S., Holt I.J., Spelbrink J.N., Jacobs H.T. Alterations to the expression level of mitochondrial transcription factor A, TFAM, modify the mode of mitochondrial DNA replication in cultured human cells. Nucleic Acids Res. 2006;34:5815–5828. doi: 10.1093/nar/gkl703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wanrooij S., Goffart S., Pohjoismaki J.L., Yasukawa T., Spelbrink J.N. Expression of catalytic mutants of the mtDNA helicase Twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic Acids Res. 2007;35:3238–3251. doi: 10.1093/nar/gkm215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ekstrand M.I., Falkenberg M., Rantanen A., Park C.B., Gaspari M., Hultenby K., Rustin P., Gustafsson C.M., Larsson N.G. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum. Mol. Genet. 2004;13:935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- 38.He J., Mao C.C., Reyes A., Sembongi H., Di Re M., Granycome C., Clippingdale A.B., Fearnley I.M., Harbour M., Robinson A.J., et al. The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J. Cell Biol. 2007;176:141–146. doi: 10.1083/jcb.200609158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen X.J., Wang X., Butow R.A. Yeast aconitase binds and provides metabolically coupled protection to mitochondrial DNA. Proc. Natl Acad. Sci. USA. 2007;104:13738–13743. doi: 10.1073/pnas.0703078104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Macierzanka M., Plotka M., Pryputniewicz-Drobinska D., Lewandowska A., Lightowlers R., Marszalek J. Maintenance and stabilization of mtDNA can be facilitated by the DNA-binding activity of Ilv5p. Biochim. Biophys. Acta. 2008;1783:107–117. doi: 10.1016/j.bbamcr.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 41.Bauer M., Behrens M., Esser K., Michaelis G., Pratje E. PET1402, a nuclear gene required for proteolytic processing of cytochrome oxidase subunit 2 in yeast. Mol. Gen. Genet. 1994;245:272–278. doi: 10.1007/BF00290106. [DOI] [PubMed] [Google Scholar]
- 42.Bonnefoy N., Fiumera H.L., Dujardin G., Fox T.D. Roles of Oxa1-related inner-membrane translocases in assembly of respiratory chain complexes. Biochim. Biophys. Acta. 2008 doi: 10.1016/j.bbamcr.2008.05.004. in press. doi:10.1016/j.bbamcr.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hell K., Herrmann J.M., Pratje E., Neupert W., Stuart R.A. Oxa1p, an essential component of the N-tail protein export machinery in mitochondria. Proc. Natl Acad. Sci. USA. 1998;95:2250–2255. doi: 10.1073/pnas.95.5.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jia L., Dienhart M., Schramp M., McCauley M., Hell K., Stuart R.A. Yeast Oxa1 interacts with mitochondrial ribosomes: the importance of the C-terminal region of Oxa1. EMBO J. 2003;22:6438–6447. doi: 10.1093/emboj/cdg624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baker M.J., Frazier A.E., Gulbis J.M., Ryan M.T. Mitochondrial protein-import machinery: correlating structure with function. Trends Cell Biol. 2007;17:456–464. doi: 10.1016/j.tcb.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 46.Bolender N., Sickmann A., Wagner R., Meisinger C., Pfanner N. Multiple pathways for sorting mitochondrial precursor proteins. EMBO Rep. 2008;9:42–49. doi: 10.1038/sj.embor.7401126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Truscott K.N., Kovermann P., Geissler A., Merlin A., Meijer M., Driessen A.J., Rassow J., Pfanner N., Wagner R. A presequence- and voltage-sensitive channel of the mitochondrial preprotein translocase formed by Tim23. Nat. Struct. Biol. 2001;8:1074–1082. doi: 10.1038/nsb726. [DOI] [PubMed] [Google Scholar]
- 48.Martinez-Caballero S., Grigoriev S.M., Herrmann J.M., Campo M.L., Kinnally K.W. Tim17p regulates the twin pore structure and voltage gating of the mitochondrial protein import complex TIM23. J. Biol. Chem. 2007;282:3584–3593. doi: 10.1074/jbc.M607551200. [DOI] [PubMed] [Google Scholar]
- 49.Zelenaya-Troitskaya O., Newman S.M., Okamoto K., Perlman P.S., Butow R.A. Functions of the high mobility group protein, Abf2p, in mitochondrial DNA segregation, recombination and copy number in Saccharomyces cerevisiae. Genetics. 1998;148:1763–1776. doi: 10.1093/genetics/148.4.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spinazzola A., Viscomi C., Fernandez-Vizarra E., Carrara F., D’Adamo P., Calvo S., Marsano R.M., Donnini C., Weiher H., Strisciuglio P., et al. MPV17 encodes an inner mitochondrial membrane protein and is mutated in infantile hepatic mitochondrial DNA depletion. Nat. Genet. 2006;38:570–575. doi: 10.1038/ng1765. [DOI] [PubMed] [Google Scholar]
- 51.Goto Y., Nonaka I., Horai S. A mutation in the tRNA(Leu)(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature. 1990;348:651–653. doi: 10.1038/348651a0. [DOI] [PubMed] [Google Scholar]
- 52.Chomyn A., Martinuzzi A., Yoneda M., Daga A., Hurko O., Johns D., Lai S.T., Nonaka I., Angelini C., Attardi G. MELAS mutation in mtDNA binding site for transcription termination factor causes defects in protein synthesis and in respiration but no change in levels of upstream and downstream mature transcripts. Proc. Natl Acad. Sci. USA. 1992;89:4221–4225. doi: 10.1073/pnas.89.10.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.King M.P., Koga Y., Davidson M., Schon E.A. Defects in mitochondrial protein synthesis and respiratory chain activity segregate with the tRNA(Leu(UUR)) mutation associated with mitochondrial myopathy, encephalopathy, lactic acidosis, and strokelike episodes. Mol. Cell Biol. 1992;12:480–490. doi: 10.1128/mcb.12.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yasukawa T., Suzuki T., Ishii N., Ohta S., Watanabe K. Wobble modification defect in tRNA disturbs codon–anticodon interaction in a mitochondrial disease. EMBO J. 2001;20:4794–4802. doi: 10.1093/emboj/20.17.4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Contamine V., Picard M. Maintenance and integrity of the mitochondrial genome: a plethora of nuclear genes in the budding yeast. Microbiol. Mol. Biol. Rev. 2000;64:281–315. doi: 10.1128/mmbr.64.2.281-315.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Myers A.M., Pape L.K., Tzagoloff A. Mitochondrial protein synthesis is required for maintenance of intact mitochondrial genomes in Saccharomyces cerevisiae. EMBO J. 1985;4:2087–2092. doi: 10.1002/j.1460-2075.1985.tb03896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iborra F.J., Kimura H., Cook P.R. The functional organization of mitochondrial genomes in human cells. BMC Biol. 2004;2:9. doi: 10.1186/1741-7007-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ogur M., St John R., Nagai S. Tetrazolium overlay technique for populations studies of respiration deficiency in yeast genetics. Science. 1957;125:928–929. doi: 10.1126/science.125.3254.928. [DOI] [PubMed] [Google Scholar]
- 59.Carlson M., Botstein D. Two differentially regulated mRNAs with different 5′ ends encode secreted with intracellular forms of yeast invertase. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- 60.Mumberg D., Muller R., Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 61.Sikorski R.S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tatsuta T., Model K., Langer T. Formation of membrane-bound ring complexes by prohibitins in mitochondria. Mol. Biol. Cell. 2005;16:248–259. doi: 10.1091/mbc.E04-09-0807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kaser M., Kambacheld M., Kisters-Woike B., Langer T. Oma1, a novel membrane-bound metallopeptidase in mitochondria with activities overlapping with the m-AAA protease. J. Biol. Chem. 2003;278:46414–46423. doi: 10.1074/jbc.M305584200. [DOI] [PubMed] [Google Scholar]
- 64.Tyynismaa H., Sembongi H., Bokori-Brown M., Granycome C., Ashley N., Poulton J., Jalanko A., Spelbrink J.N., Holt I.J., Suomalainen A. Twinkle helicase is essential for mtDNA maintenance and regulates mtDNA copy number. Hum. Mol. Genet. 2004;13:3219–3227. doi: 10.1093/hmg/ddh342. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.