Abstract
Plant height (PH), a crucial trait related to yield potential in crop plants, is known to be typically quantitatively inherited. However, its full expression can be inhibited by a limited water supply. In this study, the genetic basis of the developmental behaviour of PH was assessed in a 150-line wheat (Triticum aestivum L.) doubled haploid population (Hanxuan 10×Lumai 14) grown in 10 environments (year×site×water regime combinations) by unconditional and conditional quantitative trait locus (QTL) analyses in a mixed linear model. Genes that were expressed selectively during ontogeny were identified. No single QTL was continually active in all periods of PH growth, and QTLs with additive effects (A-QTLs) expressed in the period S1|S0 (the period from the original point to the jointing stage) formed a foundation for PH development. Additive main effects (a effects), which were mostly expressed in S1|S0, were more important than epistatic main effects (aa effects) or QTL×environment interaction (QE) effects, suggesting that S1|S0 was the most significant development period affecting PH growth. A few QTLs, such as QPh.cgb-6B.7, showed high adaptability for water-limited environments. Many QTLs, including four A-QTLs (QPh.cgb-2D.1, QPh.cgb-4B.1, QPh.cgb-4D.1, and QPh.cgb-5A.7) coincident with previously identified reduced height (Rht) genes (Rht8, Rht1, Rht2, and Rht9), interacted with more than one other QTL, indicating that the genetic architecture underlying PH development is a network of genes with additive and epistatic effects. Therefore, based on multilocus combinations in S1|S0, superior genotypes were predicted for guiding improvements in breeding for PH.
Keywords: Development, drought stress, epistasis, plant height, quantitative trait loci, Triticum aestivum L
Introduction
Plant height (PH), an important trait related to plant architecture and yield potential, is controlled polygenically (Cadalen et al., 1998; Peng et al., 1999; Sakamoto and Matsuoka, 2004). Twenty-one major genes influencing PH have been designated as reduced height (Rht) genes in wheat (Pestsova and Roder, 2002). Polygenes with quantitative effects on PH have been mapped on all 21 chromosomes (Cadalen et al., 1998; Korzun et al., 1998; Börner et al., 2002; Eriksen et al., 2003; Peng et al., 2003; Schnurbusch et al., 2003; McCartney et al., 2005; Liu et al., 2006; Pestsova et al., 2006; Quarrie et al., 2006). Segregation patterns in progeny of wheat crosses indicate that PH is under major gene control. Genetic components estimated from generation means (parental, F1, F2, F3, and backcross) showed that additive gene effects were the major components of variation in the majority of crosses, but in some crosses epistasis was the primary source of genetic variation (Fick and Qualset, 1973). Quantitative trait locus (QTL)×environment interaction (QE) effects were also important in determining terminal PH (Cao et al., 2001a; Yu et al., 2002; Li et al., 2003; Zhang et al., 2008).
Recently, there has been an increased interest in developmental genetics (Gray, 2004; Gaudet et al., 2004; Brady et al., 2006; Levesque et al., 2006; Gross et al., 2008). Unconditional analysis is a traditional method for studying developmental behaviour (Zhu, 1995). Depending upon the phenotype at various stages of development, the method can be used to reveal the static genetic control of traits at different growth stages. Sequential data reflect cumulative effects from the original to time t, rather than the real effects of gene expression during ontogeny. Zhu (1995) developed a mixed model approach, conditional analysis for analysing the net genetic effects in the period from time (t–1) to time t on trait development. The genetic effects revealed by conditional analysis, independently of the causal genetic effects at time (t–1), were influenced by the developmental status and external cropping environment of crops (Wu et al., 2002), revealing new genetic variation arising in specific periods during ontogeny. Thus, conditional analysis should be a valid method for identifying dynamic gene expression during the development of quantitative traits (Cao et al., 2001b). Conditional effects in early growth periods, as cumulative components, could affect later unconditional effects. The combination of unconditional and conditional analyses could more easily detect the expressional dynamics of PH QTLs, revealing the genetic bases for PH development, and perhaps uncovering a QTL expression pathway during plant growth. PH is easily measured throughout plant development, so it could serve as a suitable model trait for the study of developmental behaviour. The genetic control of PH development has been studied in rice, soybean, and mung bean by unconditional or conditional analysis (Yan et al., 1998a; Cao et al., 2001b; Khattak et al., 2002; Sun et al., 2006; Yang et al., 2006). These studies suggested that different QTLs/genes might control PH at different developmental stages, and a genetic model based on final phenotypes might not fully reflect the reality of morphological evolution. However, correlations among different QTLs across ontogeny, the changing genetic effects during ontogeny, and the vital developmental period for PH determination were not revealed in previous studies. Also, epistatic and QE effects were inadequately examined.
PH is a trait modified by environment. A limited water supply often inhibits PH development, consequently affecting yield (Sari-Gorla et al., 1999; Baum et al., 2003). QE is an important factor determining the stability of crop production in unfavourable environments (Lanceras et al., 2004; Maccaferri et al., 2008). Interactions among loci or between genes and environmental factors make substantial contributions to variation in complex traits (Carlborg and Haley, 2004). Mapping QTLs with genetic main effects and QE effects could help in understanding the nature of quantitative traits (Yan et al., 1998b; Cao et al., 2001b; Wu et al., 2002; Li et al., 2003; Ungerer et al., 2003; Yang et al., 2007). Therefore, epistasis and QE effects should not been ignored for studies of developmentally complex traits such as PH.
The objective of the present research was to map loci with genetic main effects and QE effects underlying the developmental behaviour of PH in doubled haploid lines (DHLs) of wheat (Triticum aestivum L.) by unconditional and conditional mapping, in order to uncover the genetic basis of PH development, even in water-limited environments. This approach was considered valuable for understanding the determination of final PH and for detecting PH markers that influence PH development during key periods and might be useful for marker-assisted selection (MAS).
Materials and methods
Experimental material
The 150 DHLs were the same as used in previous studies (Jing et al., 1999; Hao et al., 2003; Zhou et al., 2005; Yang et al., 2007). The population was derived from the cross of Hanxuan 10×Lumai 14. The two parents differed greatly in many morphological and physiological characters such as plant architecture, yield, and drought tolerance. Hanxuan 10, a tall drought-tolerant cultivar with a large number of roots, high root fresh weight and root dry weight, and a low maximum root length, ratio of root dry weight to shoot dry weight, and ratio of root fresh weight to shoot fresh weight (Zhou et al., 2005), released by Fenyang Station, Shanxi Academy of Agricultural Sciences in 1966, is still grown in arid and barren areas. Lumai 14, a short high-yielding cultivar, with a low number of roots, low root fresh weight and root dry weight, and a high maximum root length, ratio of root dry weight to shoot dry weight, and ratio of root fresh weight to shoot fresh weight (Zhou et al., 2005), adapted to abundant water and fertile conditions, was developed at the Yantai Institute of Agricultural Sciences, Shandong, and was widely grown in northern China during the 1990s. The Rht genes in both parents were detected by molecular markers (http://wheat.pw.usda.gov/cgi-bin/graingenes/). The marker Xgwm261 near Rht 8 was mapped on the short arm of chromosome 2D. Other Rht genes, such as Rht 1, Rht 2, and Rht 9 (Ellis et al., 2005), were also identified using the related markers, but could not be mapped to the linkage map because of no polymorphism between the two parents.
Field experiment
The 150 DHLs and their parents were grown at three sites over 3 years, The sites were Fenyang Station, Shanxi (111º47' E; 37º15' N) in 2001 (F01); Haidian, Beijing (116º28' E; 39º48' N) in 2005 (H05) and 2006 (H06); and Changping, Beijing (116º13' E; 40º13' N) in 2005 (Ch05) and 2006 (Ch06). The experimental fields at each site were managed under drought stress (DS) and well watered (WW) conditions. F01, H05, Ch05, H06, and Ch06 under DS were denoted as E1, E2, E3, E4, and E5, respectively, and those under WW management as E6, E7, E8, E9, and E10. DS treatments were represented by rain-fed conditions. The rainfall from July in the planting year to June (flowering stage) in the harvesting year was 296.7, 330.6, 395.4, 434.9, and 503.8 mm for each site. The total rainfall from mid November 2001 to mid June 2002 was only 35.4 mm in F01, a severe DS growth season. The WW treatments were irrigated with 900 m3 ha−1 at the pre-overwintering, jointing, flowering, and grain filling stages. A randomized complete block design was adopted to arrange the DHLs in each water regime, at one line one plot. The parental lines were planted alternatively at every 50th plot to evaluate the uniformity of the field. Each plot consisted of four 4 m rows at 30 cm spacing, with 180 plants per row in F01, Ch05, and Ch06, and two 2 m rows at 30 cm spacing, with 40 plants per row in H05 and H06.
At the jointing stage, PH (from the soil surface to the top of the plant canopy in each plot) was measured every 7 d until flowering when full PH was attained. A total of five measurements were taken during the growing period, and they were designated S1, S2, S3, S4, and S5. Fertility and cultivation regimes were consistent with wheat production in the relevant regions.
Data analysis
Based on the development theory proposed by Zhu (1995), the actual PH values at the different stages were defined as unconditional PHs, and the PH values obtained by the mixed model approach for conditional genetics of developmental quantitative traits as conditional PHs. The analyses for both unconditional and conditional PHs were used to detect dynamic genetic effects for PH; that is, unconditional analysis was used to detect the total accumulated genetic effects of genes expressed during the initial point to time t (0→t), and conditional analysis to reveal the net genetic effects from genes expressed in the period from time t−1 to t. When phenotypic values were first measured, the unconditional genetic effects were equivalent to the conditional genetic effects.
To investigate phenotypic variation in PH, correlation coefficients between PHs during development were analysed, and differences and relationships of PHs among environments were estimated by analysis of variance (ANOVA) and cluster methods individually. All phenotypic analyses were performed by SAS software (SAS Institute, 1996). Broadsense heritability of PH was estimated as:
Both unconditional and conditional PH during development were subjected to QTL analysis. A genetic linkage map, consisting of 395 marker loci, including 263 simple sequence repeats (SSRs) and 132 amplified fragment length polymorphisms (AFLPs), was available. The map was established from data on 150 DHLs and covered 3904 cM with an average distance of 9.9 cM between adjacent markers (Hao et al., 2003; Zhou et al., 2005; Yang et al., 2007). QTL analysis was implemented using mapping software QTLNetwork-2.0 based on the mixed linear model (Yang et al., 2007) to divide genetic effects into additive main effects (a effects), epistatic main effects (aa effects), and their environment interaction effects (QE, including ae and aae effects). QTLs with genetic main effects indicated that genes in these genomic regions were expressed in the same way across environments. QTLs with QE effects suggested that gene expression at these loci was environmentally dependent. An experiment-wise type I error of 0.05 was designated for candidate interval selection and putative QTL detection. The critical F-value to declare putative QTLs and to control genome-wise type I errors was accommodated by 1000 permutation tests. Both the testing window and filtration window were set at 10 cM, with a walk speed of 2 cM. QTLs were named according to the rule ‘QTL+trait+research department+chromosome’.
Results
Phenotypic variation
The female parent Hanxuan 10 was significantly taller than the male parent Lumai 14 at all five stages in all 10 environments (year×site×water regime combination) (P <0.05) (Table 1). The average PH of DHLs was between that of the two parents. High phenotypic variability was observed in the population, with coefficients of variation (CVs) ranging from 12.2% to 25.5%. The PHs of DHLs showed continuous variation, and transgressive segregation occurred at all stages in all environments. Most skew and kurt values were <1.0, suggestive of a quantitative trait.
Table 1.
Phenotypic values of PH for wheat DHLs and their parents at five growth stages in 10 environments
| Environment | Stage | H10 | L14 | DHLs |
||||
| Mean±SD | Range | CV (%) | Skew | Kurt | ||||
| E1 | S1 | 25.2 | 19.8 | 21.4±3.4 | 14.0–29.0 | 15.8 | –0.10 | –0.63 |
| S2 | 31.6 | 24.2 | 26.9±3.9 | 16.0–36.0 | 14.4 | –0.13 | –0.26 | |
| S3 | 40.8 | 29.8 | 32.9±4.8 | 20.0–44.0 | 14.6 | 0.11 | –0.19 | |
| S4 | 58.0 | 43.6 | 48.1±6.9 | 32.0–66.0 | 14.4 | 0.06 | –0.46 | |
| S5 | 75.8 | 56.0 | 61.1±7.9 | 40.0–81.0 | 13.0 | 0.00 | 0.04 | |
| E2 | S1 | 39.5 | 34.0 | 35.2±5.2 | 17.0–48.0 | 14.9 | 0.26 | 0.31 |
| S2 | 54.0 | 44.5 | 45.7±8.0 | 29.0–68.0 | 17.5 | 0.18 | –0.57 | |
| S3 | 65.0 | 52.0 | 55.0±9.5 | 32.0–79.0 | 17.2 | 0.09 | –0.47 | |
| S4 | 80.5 | 65.5 | 66.1±10.5 | 40.0–92.0 | 15.9 | 0.05 | –0.56 | |
| S5 | 102.0 | 72.0 | 81.5±13.0 | 49.0–109.0 | 15.9 | 0.11 | –0.62 | |
| E3 | S1 | 42.8 | 36.3 | 36.5±4.5 | 24.0–47.0 | 12.2 | 0.13 | –0.36 |
| S2 | 55.0 | 39.5 | 44.2±7.2 | 30.0–61.0 | 16.2 | –0.02 | –1.26 | |
| S3 | 69.3 | 43.0 | 54.0±12.6 | 32.0–77.0 | 23.4 | –0.01 | –1.50 | |
| S4 | 90.8 | 53.3 | 68.2±15.9 | 38.0–93.0 | 23.3 | –0.06 | –1.64 | |
| S5 | 101.3 | 69.5 | 81.8±14.3 | 47.0–105.0 | 17.5 | –0.14 | –1.34 | |
| E4 | S1 | 43.0 | 37.0 | 37.6±4.7 | 23.0–51.0 | 12.5 | 0.10 | 0.31 |
| S2 | 58.0 | 42.9 | 46.6±8.7 | 27.5–68.5 | 18.7 | 0.09 | –0.82 | |
| S3 | 78.0 | 50.6 | 61.8±14.2 | 35.0–86.0 | 22.9 | –0.05 | –1.59 | |
| S4 | 98.3 | 60.1 | 77.7±18.6 | 44.0–108.5 | 24.0 | –0.11 | –1.70 | |
| S5 | 125.0 | 70.9 | 94.7±21.2 | 53.0–125.0 | 22.4 | –0.11 | –1.62 | |
| E5 | S1 | 45.9 | 36.8 | 38.5±5.5 | 27.0–54.5 | 14.2 | 0.12 | –0.62 |
| S2 | 63.3 | 41.3 | 49.2±10.5 | 33.4–72.0 | 21.4 | 0.07 | –1.47 | |
| S3 | 91.0 | 52.8 | 66.5±16.9 | 40.0–95.0 | 25.5 | 0.00 | –1.71 | |
| S4 | 102.3 | 64.3 | 78.9±19.1 | 49.0–110.0 | 24.2 | –0.03 | –1.70 | |
| S5 | 111.3 | 70.0 | 87.7±16.3 | 54.0–117.0 | 18.6 | –0.11 | –1.49 | |
| E6 | S1 | 44.8 | 33.0 | 36.8±7.4 | 21.0–57.0 | 20.0 | 0.34 | –0.25 |
| S2 | 54.8 | 37.6 | 43.1±8.8 | 28.0–65.0 | 20.5 | 0.29 | –0.66 | |
| S3 | 71.0 | 57.6 | 59.9±10.6 | 41.0–82.0 | 17.7 | 0.09 | –1.06 | |
| S4 | 105.2 | 72.4 | 82.2±12.8 | 55.0–105.0 | 15.6 | 0.03 | –1.31 | |
| S5 | 107.6 | 79.8 | 87.2±13.3 | 60.0–116.0 | 15.3 | 0.08 | –1.14 | |
| E7 | S1 | 50.3 | 43.0 | 43.3±5.7 | 31.0–59.0 | 13.2 | 0.08 | –0.68 |
| S2 | 60.8 | 49.0 | 52.3±8.7 | 31.0–74.0 | 16.6 | 0.07 | –0.83 | |
| S3 | 80.5 | 59.8 | 67.2±11.7 | 40.0–92.0 | 17.4 | 0.00 | –1.04 | |
| S4 | 99.5 | 69.85 | 81.9±15.5 | 52.0–111.0 | 18.9 | 0.06 | –1.34 | |
| S5 | 120.5 | 78.0 | 96.4±18.5 | 63.0–132.0 | 19.2 | 0.07 | –1.32 | |
| E8 | S1 | 52.8 | 39.5 | 44.6±6.9 | 29.0–59.0 | 15.5 | 0.06 | –1.02 |
| S2 | 80.8 | 52.0 | 61.1±14.2 | 32.0–85.0 | 23.3 | –0.05 | –1.59 | |
| S3 | 101.8 | 63.0 | 78.3±19.8 | 43.0–105.0 | 25.3 | –0.08 | –1.77 | |
| S4 | 109.8 | 74.3 | 87.2±17.1 | 51.0–111.0 | 19.6 | –0.15 | –1.56 | |
| S5 | 113.3 | 79.3 | 93.1±13.7 | 54.0–117.0 | 14.8 | –0.25 | –1.01 | |
| E9 | S1 | 54.5 | 40.4 | 42.3±6.3 | 26.0–55.5 | 14.9 | –0.14 | –0.70 |
| S2 | 78.4 | 51.5 | 58.0±11.3 | 35.0–82.0 | 19.4 | 0.09 | –1.23 | |
| S3 | 99.6 | 64.0 | 78.1±15.9 | 44.8–102.3 | 20.4 | –0.07 | –1.62 | |
| S4 | 120.0 | 77.5 | 94.3±19.5 | 57.0–126.0 | 20.7 | –0.03 | –1.64 | |
| S5 | 132.8 | 84.5 | 106.4±19.5 | 60.0–141.0 | 18.3 | –0.07 | –1.34 | |
| E10 | S1 | 63.3 | 43.6 | 51.0±10.0 | 34.0–70.6 | 19.7 | 0.02 | –1.43 |
| S2 | 96.8 | 57.8 | 72.1±17.1 | 44.0–101.0 | 23.8 | –0.01 | –1.68 | |
| S3 | 100.5 | 66.8 | 82.4±18.2 | 49.0–115.0 | 22.1 | –0.04 | –1.69 | |
| S4 | 120.8 | 74.8 | 95.2±18.3 | 60.0–127.0 | 19.2 | –0.06 | –1.56 | |
| S5 | 123.1 | 78.1 | 99.2±16.1 | 62.0–133.1 | 16.3 | –0.06 | –1.25 | |
H10, Hanxuan 10; L14, Lumai 14; CV, coefficient of variation; E1, Fenyang, Shanxi province in 2001 (F01) under drought-stressed conditions (DS); E2 and E4, Haidian, Beijing in 2005 (H05) and 2006 (H06), DS; E3 and E5, Changping, Beijing, 2005 (Ch05) and 2006 (Ch06), DS; E6, Fenyang, Shanxi, 2001 (F01) under well-watered (WW); E7 and E9, Haidian, Beijing, 2005 (H05) and 2006 (H06), WW; E8 and E10, Changping, Beijing, 2005 conditions (Ch05) and 2006 (Ch06), WW; S1, S2, S3, S4 and S5 indicate the first, second, third, fourth and fifth measuring stage, respectively.
PH of the DHLs and their parents showed significant increases across all measuring stages in all environments (P <0.05). Positive correlations between PHs at different stages ranged from 0.63*** to 0.99*** (P <0.0001) in different environments (Supplementary Table S1 available at JXB online), indicating that PHs at different stages were closely correlated. Significant differences in PH were identified for genotypes, year×site combinations, water regimes, and all two-factor combinations except for the water regime×genotype combination which was not significant at S5 (Supplementary Table S2). The most significant difference occurred between water regimes. Highly significant differences in PH among environments were also detected at all measuring stages in the combined analysis over all 10 three-factor combination environments (P <0.0001) (Supplementary Table S3), indicating that the ontogeny of PH was influenced by environment. By cluster analysis, the 10 environments were further subdivided into three clusters: E1 stood alone as one cluster characterized as the severe DS cluster (mean PH among five stages, 38.1 cm), E8, E9, and E10 formed the second cluster, the WW group (mean PH, 76.2 cm), and the remainder formed a moderate DS cluster (mean, PH 61.8 cm). The broadsense heritabilities for unconditional PH were estimated as 34.1, 39.3, 45.4, 53.9, and 60.3%, respectively, for the different development stages.
There were significant correlations between conditional PHs in some periods and environments (Supplementary Table S4 at JXB online), implying the dynamic and environmentally influenced characteristics of PH ontogeny. As for unconditional PHs, significant differences were detected for conditional PHs among genotypes, year×site combinations, water regimes, and all two-factor combinations (P <0.05) except for water regime×genotype combinations with non-significant differences in S4|S3 and S5|S4 (Supplementary Table S2). The largest difference occurred between water regimes (P <0.0001). Similarly, highly significant differences (P <0.0001) were detected among 10 combination environments of year×site×water regime (Supplementary Table S3). Broadsense heritabilities for conditional PH were 34.1, 11.7, 6.1, 3.1, and 15.9%, respectively, for the five growth periods.
Unconditional QTL analysis for PH development
A total of 20 A-QTLs (Table 2) and 82 epistatic pairs (Supplementary Table S5 at JXB online) with significant genetic main effects and/or QE effects controlling unconditional PH at different development stages were detected. The A-QTLs were located on all chromosomes except 1A, 1D, 2A, 2B, 3D, 5D, 6D, and 7D, whereas epistasis involved contributions from all chromosomes except 6D, indicating that unconditional QTLs for PH were associated with nearly all chromosomes.
Table 2.
Unconditional QTLs affecting PH of wheat detected at five growth stages in 10 environments
| QTL | Flanking markers | Stage | a | ae | h2(a)% | h2(ae)% |
| QPh.cgb-1B.1 | Xgwm582–Xgwm273 | S1 | 0.93*** | 2.22 | ||
| QPh.cgb-1B.4 | Xwmc156–P3446.1 | S2 | 2.00*** | 2.88 | ||
| S3 | 1.12*** | 3.50 | ||||
| S4 | 5.08*** | 4.26 | ||||
| S5 | 4.50*** | 1.14* (ae7) | 4.55 | 0.32 | ||
| QPh.cgb-1B.19 | Xgwm259–Xwmc367 | S3 | –0.44* | 0.07 | ||
| S4 | –2.49*** | 0.07 | ||||
| S5 | –1.79*** | 0.08 | ||||
| QPh.cgb-2D.1 | Xwmc453.1–Xwmc18 | S1 | 1.85*** | –0.83** (ae1), 0.77* (ae10) | 3.57 | 0.53 |
| S2 | 3.89*** | –2.44*** (ae1), 1.87*** (ae8), 2.13*** (ae10) | 3.43 | 0.66 | ||
| S3 | 3.02*** | –1.92*** (ae1) | 3.18 | 0.64 | ||
| S4 | 4.70*** | –2.96*** (ae1) | 3.76 | 0.54 | ||
| S5 | 2.84*** | 4.05 | ||||
| QPh.cgb-2D.6 | P4233.2–P6411.4 | S4 | 1.83*** | 1.85 | ||
| QPh.cgb-2D.11 | P3176.1–P1123.1 | S1 | 0.92*** | 0.78 | ||
| S2 | 1.34*** | 0.84 | ||||
| S3 | 2.95*** | 0.81 | ||||
| S5 | 2.70*** | 1.39 | ||||
| QPh.cgb-3A.1 | Xcwm48.1–Xwmc532 | S2 | 0.72*** | 0.73 | ||
| S5 | 0.78*** | 1.42 | ||||
| QPh.cgb-3B.9 | P3622.4–P2076 | S1 | –1.07*** | –0.59* (ae10) | 0.96 | 0.33 |
| S2 | –1.74*** | –0.96* (ae10) | 1.68 | 0.35 | ||
| QPh.cgb-4A.5 | P6431.1–Xgwm160 | S1 | 0.62*** | 0.19 | ||
| S2 | 1.91*** | 0.24 | ||||
| S3 | 2.04*** | 0.07 | ||||
| S5 | 1.82*** | 0.36 | ||||
| QPh.cgb-4B.1 | Xgwm368–Xgwm107 | S3 | –2.12*** | 0.84 | ||
| S4 | –0.77*** | 0.79 | ||||
| QPh.cgb-4D.1 | Xgwm165.2–Xgwm192 | S1 | 1.32*** | –0.82* (ae1), 1.53*** (ae10) | 2.79 | 0.57 |
| S2 | 3.85*** | –2.66*** (ae1), 1.45* (ae8), 1.19* (ae9), 3.25*** (ae10) | 3.55 | 0.73 | ||
| S3 | 4.88*** | –2.83*** (ae1), –1.87** (ae2), 1.61* (ae8), 1.39* (ae9), 1.93*** (ae10) | 4.83 | 0.82 | ||
| S4 | 5.19*** | –2.66*** (ae1), –1.56* (ae2) | 5.74 | 0.63 | ||
| QPh.cgb-4D.2 | Xgwm192–Xwmc331 | S5 | 4.86*** | –2.36*** (ae1), 1.65* (ae4) | 6.81 | 0.66 |
| QPh.cgb-5A.6 | Xgwm595–Xwmc410 | S1 | –0.84*** | 0.32 | ||
| QPh.cgb-5A.7 | Xgwm291–Xgwm410 | S2 | –2.52*** | –1.10* (ae8), –1.35** (ae10) | 1.50 | 0.26 |
| S3 | –3.89*** | 1.21* (ae1), –1.26* (ae5) | 1.81 | 0.18 | ||
| S4 | –3.11*** | 1.42 | ||||
| QPh.cgb-5B.4 | Xgwm371–Xgwm335 | S1 | –0.96*** | 0.73 | ||
| QPh.cgb-6A.3 | P4232.4–Xcwm306 | S1 | –1.54*** | –0.74* (ae10) | 1.10 | 0.19 |
| QPh.cgb-6B.5 | Xgwm132–Xwmc104 | S1 | 1.58*** | –1.00*** (ae1), 0.99*** (ae10) | 0.67 | 0.20 |
| S2 | 3.06*** | –1.99*** (ae1), 1.55*** (ae10) | 0.65 | 0.12 | ||
| S3 | 3.16*** | –2.05*** (ae1) | 0.90 | 0.13 | ||
| S4 | 3.72*** | –1.67*** (ae1) | 0.66 | 0.09 | ||
| S5 | 3.95*** | –1.15* (ae1) | 0.43 | 0.06 | ||
| QPh.cgb-6B.7 | Xwmc269.3–P4232.1 | S1 | –1.83*** | 0.71* (ae1), 0.80* (ae4), –0.72* (ae6), –1.21*** (ae10) | 1.89 | 0.58 |
| S2 | –2.85*** | 1.81*** (ae1), –1.62*** (ae10) | 2.29 | 0.45 | ||
| S3 | –3.43*** | 1.99*** (ae1), 1.25* (ae2), –1.53** (ae10) | 3.94 | 0.67 | ||
| S4 | –6.39*** | 3.07*** (ae1), –1.45* (ae5), –1.84* (ae9) | 3.73 | 0.59 | ||
| S5 | –3.70*** | 2.04*** (ae1) | 3.39 | 0.56 | ||
| QPh.cgb-7A.3 | P3454.5–P3446.4 | S1 | 0.78*** | 1.40 | ||
| S2 | 2.27*** | –1.43*** (ae1), 1.67*** (ae10) | 1.61 | 0.35 | ||
| S3 | 3.07*** | –1.96*** (ae1), 1.31* (ae8), 1.13* (ae10) | 2.18 | 0.41 | ||
| S4 | 2.38*** | 2.85 | ||||
| S5 | 3.49*** | –1.58* (ae1) | 2.64 | 0.36 | ||
| QPh.cgb-7B.4 | Xpsp3033–Xgwm297 | S3 | –2.99*** | 1.74** (ae1), 1.34* (ae2), –1.55* (ae8) | 0.67 | 0.13 |
a, additive main effects; ae1, the additive QTL×environment interaction effects in E1, ae2, the additive QTL×environment interaction effects in E2, and so on; E1–E10 are as shown in Table 1; a positive value indicates that the Hanxuan 10 allele has a positive effects on PH, and a negative value that the Lumai 14 allele has a positive effect on PH; S1, S2, S3, S4, S5 are as shown in Table 1; h2(a)%, phenotypic variation explained (PVE) by a effects; h2(ae)%, PVE by ae effects.
* P=0.05, ** P=0.01, *** P=0.005. Only significant effects are listed.
Of the 20 A-QTLs, 13 were detected at 2–5 stages and seven at only one stage apart from S2 (Table 2), indicating that QTLs detected at one specific stage did not entirely represent those active at another stage, and that genes controlling PH might be selectively expressed during development. All 20 A-QTLs had significant additive main effects (a effects), and 11 of them had a effects (0.62***–5.19***) conferred by height-enhancing alleles from Hanxuan 10, implying that alleles for PH were dispersed between the two parents. Eleven QTLs were also detected with significant additive×environment interaction effects (ae effects) in 1–5 environments at 1–5 development stages (Table 2), indicating that their genetic sensitivities to environments changed during ontogeny. The reaction of QTLs to environments showed directional effects on ontogeny. QTLs such as QPh.cgb-2D.1, QPh.cgb-4D.1, QPh.cgb-6B.5, and QPh.cgb-7A.3, with negative ae effects (–2.96*** to –0.82*) in E1 (severe DS) and positive effects (0.77*–3.25***) in 1–3 environments of E8, E9, and E10 (WW) appeared to be up-regulated by WW environments at 2–3 stages, whereas QTLs such as QPh.cgb-5A.7, QPh.cgb-6B.7, and QPh.cgb-7B.4, having positive ae effects (0.71*–3.07***) in E1 and negative effects (–1.84* to –1.21***) in either E9 or E10, were up-regulated by severe DS environments at 1–5 stages (Table 2). This capacity for different QTLs to respond to environments varied. For the A-QTLs having both a effects and ae effects, the absolute values for the a effects were larger than those for ae effects in any one environment, and correspondingly the phenotypic variation explained (PVE) by a effects was larger than that explained by ae effects. Thus a effects predominated over ae effects at different growth stages. Additionally, of the QTLs detected at more than one stage, the a effects as well as the ae effects for the same environment were always in the same direction, but with unequal magnitudes at different stages. The absolute effects at later growth stages were usually greater than those at the first stage, indicating that the genetic effects of the first stage set a cumulative foundation for those at the later stages (Table 2). On the other hand, unequal magnitudes might result from net gene expression in different periods after the first stage.
For unconditional epistatic effects on PH growth, one pair of QTLs, that is QPh.cgb-1B.4 and QPh.cgb-2B.6, had significant epistatic main effects (aa effects) at two stages (S4 and S5); another pair, QPh.cgb-2D.1 and QPh.cgb-4A.5 was found at four stages (S1, S2, S3, and S5), and all other pairs identified with significant aa effects and/or epistasis×environment interaction effects (aae effects) were detected at one specific stage (Supplementary Table S5 at JXB online). Thus epistatic effects were mainly short lived during PH development. All 82 epistatic pairs had significant aa effects; 43 of these, including two detected at two or more stages, had significant negative aa effects, implying that both parents contributed alleles for PH. Twelve pairs also had significant aae effects in 1–4 environments among E1, E2, E4, E6, E8, E9, and E10 at 1–5 stages, implying that most instances of epistasis were not affected by environment. For epistasis with aae effects, six pairs of QTLs, QPh.cgb-1B.11 and QPh.cgb-1B.16 at S1; QPh.cgb-1B.8 and QPh.cgb-3B.6, and QPh.cgb-2A.9 and QPh.cgb-6A.7 at S2; QPh.cgb-1B.19 and QPh.cgb-7B.4, QPh.cgb-2A.10 and QPh.cgb-7D.4, and QPh.cgb-2D.7 and QPh.cgb-7B.3 at S3, showed positive aae effects (0.45*–1.67***) in one or both environments of E8 and E10, or were negative (–2.14*** to –1.62***) in E1. In contrast, another six pairs, QPh.cgb-6A.4 and QPh.cgb-7B.4 at S1; QPh.cgb-4B.3 and QPh.cgb-5B.4, and QPh.cgb-5A.1 and QPh.cgb-7A.8 at S2, QPh.cgb-3B.14 and QPh.cgb-3D.1 at S4; and QPh.cgb-3A.1 and QPh.cgb-6A.6, and QPh.cgb-5A.2 and QPh.cgb-5A.4 at S5, displayed positive aae effects (1.05*–2.54***) in E1, or were negative (–1.57* to 1.14*) in either of E9 and E10. These epistatic pairs conferred adaptability for water-limited environments, consistent with some A-QTLs. For these instances of epistasis with aa effects and aae effects, similar to A-QTLs, the aa effects in absolute terms were larger than aae effects in each environment, and the PVE for aa effects was correspondingly larger than that of the aae effects; hence aa effects were more important in determining PH than aae effects.
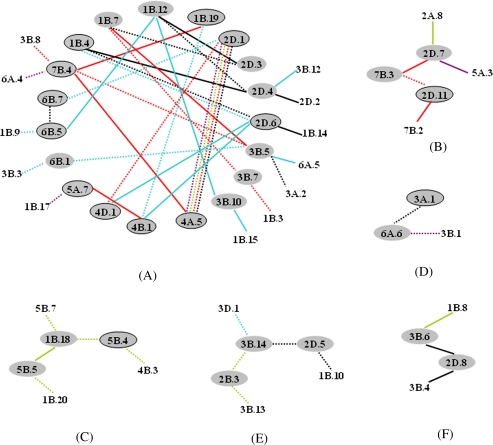
Interestingly, many A-QTLs (14 in total) also interacted with other QTLs, and thereby displayed modified functions (Supplementary Table S5). Moreover, 38.8% of QTLs participating in epistatic interactions (E-QTLs), including 13 A-QTLs and 27 non-individual E-QTLs (i.e. epistatic QTLs without additive effects), took part in two or more epistatic interactions at 1–5 stages, making up a QTL functional network associated with PH development (Fig. 1). For example, during PH development, through 11 A-QTLs and eight non-individual E-QTLs participating in two or more interactions, there was a large QTL network of 31 QTLs (Fig. 1A). Within this network, there were three kinds of interaction, that is 40.5% of interactions were between different A-QTLs, 21.6% were between A-QTLs and non-individual E-QTLs, and 37.8% were between different non-individual E-QTLs. Almost 60% of the interactions showed negative aa effects. QTLs interacting with more than one other QTL displayed both positive and negative aa effects. All of these interactions suggested that the genetic control of PH development was complex. Overall, the genetic effects involved in the QTL network of unconditional PH collectively accounted for 82.1% of the phenotypic variation occurring during PH development, whereas the remaining components of 15.4% and 2.5% of phenotypic variation, respectively, were attributed to those A-QTLs not involved in epistatic interactions and those non-individual QTLs not involved in the QTL network. Therefore, the genetic effects controlling PH growth were primarily expressed as part of the QTL network. Nevertheless, due to the additive and epistatic effects involved in the QTL network, unconditional PH QTLs contributed 55.3% and 26.8% of the phenotypic variation, and additive effects apparently constituted the major genetic basis of PH development.
Fig. 1.
Epistatic QTL network for unconditional PH at five stages in wheat DHLs.  indicates a QTL with additive effects (A-QTL);
indicates a QTL with additive effects (A-QTL);  indicates a QTL without individual effects but interacting with two more QTLs; other QTLs were detected only once. Solid lines indicate positive epistatic main (aa) effects to increase PH; dashed lines indicate negative aa effects to decrease PH.
indicates a QTL without individual effects but interacting with two more QTLs; other QTLs were detected only once. Solid lines indicate positive epistatic main (aa) effects to increase PH; dashed lines indicate negative aa effects to decrease PH.  ,
,  ,
,  ,
,  , and
, and  refer to positive aa effects at S1, S2, S3, S4, and S5, respectively.
refer to positive aa effects at S1, S2, S3, S4, and S5, respectively.  ,
,  ,
,  ,
,  , and
, and  refer to negative aa effects at S1, S2, S3, S4, and S5, respectively.
refer to negative aa effects at S1, S2, S3, S4, and S5, respectively.
Conditional QTL analysis for PH development
Conditional mapping was adopted to reveal the real gene expression during different developmental periods. By conditional mapping, a total of 20 A-QTLs and 23 epistatic pairs with genetic main effects and/or QE effects associated with PH growth were identified (Table 3, and Supplementary Table S6 at JXB online). A-QTLs occurred on all chromosomes except 1A, 2A, 2B, 3D, 4B, 5D, 6D, 7B, and 7D, and E-QTLs were associated with all chromosomes apart from 1D, 3A, 3D, 4D, 5D, and 6D. Some chromosomes with unconditional QTLs were not detected as conditional QTLs.
Table 3.
Conditional additive QTLs (A- QTLs) affecting PH of wheat in five periods and 10 environments
| QTL | Flanking markers | Period | a | ae | h2(a)% | h2(ae)% |
| QPh.cgb-1B.1 | Xgwm582–Xgwm273 | S1|S0 | 0.93*** | 2.22 | ||
| QPh.cgb-1B.5 | P3446.1–Xcwm65 | S5|S4 | 0.46*** | –0.83** (ae1), 0.91*** (ae2), 1.05*** (ae4), –0.86** (ae5), 1.63*** (ae7), –0.95*** (ae8), 0.84** (ae9), –0.68* (ae10) | 0.06 | 1.24 |
| QPh.cgb-1D.3 | Xwmc432–Xwmc222 | S5|S4 | –0.43*** | 0.99*** (ae4) | 0.42 | 0.18 |
| QPh.cgb-2D.1 | Xwmc453.1–Xwmc18 | S1|S0 | 1.85*** | –0.83** (ae1), 0.77* (ae10) | 3.57 | 0.53 |
| S3|S2 | –0.50*** | –0.74* (ae1), –0.86* (ae2), 0.74* (ae8) | 0.15 | 0.28 | ||
| QPh.cgb-2D.11 | P3176.1–P1123.1 | S1|S0 | 0.92*** | 0.78 | ||
| QPh.cgb-3A.1 | Xcwm48.1–Xwmc532 | S2|S1 | 0.49*** | –0.72** (ae1), 0.65* (ae5), –0.64* (ae6), –0.71** (ae7), 0.84*** (ae8) | 0.15 | 0.44 |
| QPh.cgb-3B.9 | P3622.4–P2076 | S1|S0 | –1.07*** | –0.59* (ae10) | 0.96 | 0.33 |
| QPh.cgb-3B.11 | Xwmc291–P3156.1 | S5|S4 | 0.34** | 0.97** (ae1), –1.62*** (ae4), 0.81* (ae8) | 0.36 | 0.53 |
| QPh.cgb-4A.5 | P6431.1–Xgwm160 | S1|S0 | 0.62*** | 0.19 | ||
| QPh.cgb-4D.1 | Xgwm165.2–Xgwm192 | S1|S0 | 1.32*** | –0.82* (ae1), 1.53*** (ae10) | 2.79 | 0.57 |
| S2|S1 | 0.36*** | –1.12*** (ae1), 0.63* (ae4), 1.09*** (ae5), –1.24*** (ae6), –0.87** (ae7), 0.66* (ae8) | 0.08 | 0.70 | ||
| QPh.cgb-4D.2 | Xgwm192–Xwmc331 | S3|S2 | –0.83** (ae2), –0.72* (ae6) | 0.43 | ||
| QPh.cgb-5A.6 | Xgwm595–Xwmc410 | S1|S0 | –0.84*** | 0.32 | ||
| QPh.cgb-5A.7 | Xgwm291–Xgwm410 | S2|S1 | –0.53*** | 0.67* (ae2), –0.61* (ae5), 0.80** (ae6), –1.05*** (ae8), –0.74* (ae10) | 0.23 | 0.49 |
| S3|S2 | –0.33*** | –0.68* (ae4), 1.27*** (ae10) | 0.10 | 0.36 | ||
| S4|S3 | 0.35*** | 0.83* (ae2), 1.05*** (ae5), –0.80* (ae6), –0.85* (ae7), –0.75* (ae9) | 0.06 | 0.34 | ||
| QPh.cgb-5B.4 | Xgwm371–Xgwm335 | S1|S0 | –0.96*** | 0.73 | ||
| QPh.cgb-6A.1 | Xgwm334–Xwmc297 | S3|S2 | 0.59*** | 0.27 | ||
| QPh.cgb-6A.3 | P4232.4–Xcwm306 | S1|S0 | –1.54*** | –0.74* (ae10) | 1.10 | 0.19 |
| S5|S4 | 1.60*** | 0.87 | ||||
| QPh.cgb-6B.5 | Xgwm132–Xwmc104 | S1|S0 | 1.58*** | –1.00*** (ae1), 0.99*** (ae10) | 0.67 | 0.20 |
| QPh.cgb-6B.7 | Xwmc269.3–P4232.1 | S1|S0 | –1.83*** | 0.71* (ae1), 0.80* (ae4), –0.72* (ae6), –1.21*** (ae10) | 1.89 | 0.58 |
| S5|S4 | –0.79** (ae2), –1.19*** (ae4), 0.90*** (ae5), 0.74* (ae8) | 0.65 | ||||
| QPh.cgb-6B.8 | Xgwm644.1–Xwmc417.2 | S5|S4 | 0.45*** | –0.61* (ae3) | 0.23 | 0.33 |
| QPh.cgb-7A.3 | P3454.5–P3446.4 | S1|S0 | 0.78*** | 1.40 |
In contrast to unconditional A-QTLs, conditional A-QTLs were rarely detected in two or more growth periods. Only one QTL, QPh.cgb-5A.7, was expressed in three periods, and four were expressed in two periods; these were QPh.cgb-2D.1, QPh.cgb-4D.1, QPh.cgb-6A.3, and QPh.cgb-6B.7. The remaining 75.0% of QTLs were detected in only one specific period. No QTL was expressed in all five periods, and most were expressed in the earliest period of PH development. This further indicated that genes for PH are expressed selectively during ontogeny (Table 3).
Nineteen of the 20 (95.0%) conditional A-QTLs were identified with significant a effects in 1–3 periods. Most of the favourable alleles came from Hanxuan 10, consistent with the unconditional results, implying that Hanxuan 10 carried most of the alleles for PH. Thirteen QTLs (65%) with ae effects were found in 1–8 environments in 1–3 periods. QPh.cgb-4D.2 was identified with clearly significant negative ae effects in E1 and E8, indicating that it was completely induced by environment.
Conditional ae effects across PH development had the following characteristics. First, conditional A-QTLs reacted differently to environments. Typically, QPh.cgb-2D.1, QPh.cgb-4D.1, and QPh.cgb-6B.5 expressed negative ae effects (–1.12*** to –0.74*) in E1 and were positive (0.66*–1.53***) in either E8 or E10 in 1–2 periods; QPh.cgb-6B.7 displayed positive ae effects in E1 (0.71*) and E4 (0.80*), and negative effects in E6 (–0.72*) and E10 (–1.21***) in S1|S0. The responses of these QTLs to environments were consistent with their unconditional behaviour. Secondly, conditional A-QTLs more readily responded to environments. On average each of these conditional QTLs had significant ae effects in three environments, compared with only two environments for the unconditional A-QTLs. This indicated that actual gene expression associated with PH was highly sensitive to environment. Finally, in contrast to A-QTLs, conditional A-QTLs displayed more ae effects in periods other than S1|S0. From 75% to 100% of A-QTLs were identified with ae effects in 2–5 environments in the periods after S1|S0, with each of their ae effects being larger in magnitude than the a effects. Consequently, ae effects contributed more to phenotypic variation than a effects. In contrast, only 50% of QTLs were detected with significant ae effects, and most were in E1 or E10 in S1|S0. Each ae effect for these QTLs was less than the corresponding a effect, hence a effects contributed more phenotypic variation than ae effects in S1|S0. Therefore, the real gene expression for PH in period S1|S0 was little affected by environments, whereas it was greatly affected thereafter.
For conditional A-QTLs identified in more than one period, the genetic effects varied greatly, in contrast to those of unconditional A-QTLs at more than one stage. For example, QPh.cgb-2D.1, QPh.cgb-5A.7, and QPh.cgb-6A.3 exhibited a effects with opposite signs in different periods, and their ae effects also differed between different periods. Another QTL, QPh.cgb-6B.7, was detected with significant a effects (–1.83***) in period S1|S0, but with only significant ae effects in periods S5|S4 in E2, E4, E5, and E8. This indicated that gene expression was developmentally dependent.
All conditional epistatic QTLs were detected in a specific period; no epistatic QTL pair was expressed in two or more periods. This further confirmed that epistatic effects were short lived. Over all five growth periods, the most (seven) epistatic QTL pairs were detected in S1|S0, whereas the least (three) pairs were in S4|S3 (Supplementary Table S6 at JXB online). Seventeen of 23 epistatic QTL pairs (73.9%) were identified with significant aa effects, 10 (58.8%) pairs of which expressed negative aa effects. There were 13 pairs (56.5%) of epistatic QTLs with significant aae effects in 1–8 environments, six of which had unique aae effects completely caused by environmental components. Among the 13 epistatic pairs with aae effects, on average one epistatic pair had aae effects in ∼3 environments compared with two environments for the unconditional epistasis. Additionally, two of seven (28.6%) epistatic pairs had significant aae effects in E10 in S1|S0, whereas 60.0–75.0% of epistatic pairs had significant aae effects in 1–6 environments (but excluding E3) in the other periods. Six epistatic pairs with unique aae effects in the later three periods were detected, with two such pairs in each period. Even for epistatic pairs with aa and aae effects in S1|S0, aa effects were clearly larger than aae effects, and the same trend existed in 66.7% of this kind of epistatic pair in S2|S1, but, in other periods, aae effects were always larger than aa effects. Therefore, for epistatic QTLs during PH development, aa effects were the important components in early periods (especially in S1|S0) of growth, whereas aae effects became predominant in the middle and later periods. This reality for epistatic QTLs was consistent with that for conditional A-QTLs, which were difficult to detect by unconditional analysis.
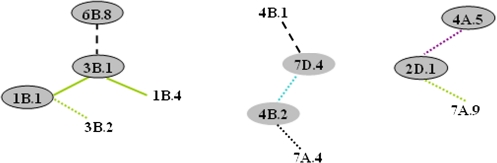
For conditional QTLs, 30% of A-QTLs (six QTLs) were involved in epistatic interactions. About 15% of QTLs involved in epistatic interaction, including three A-QTLs and three non-individual E-QTLs, interacted with 2–3 other QTLs in the same or different periods. Hence, QTL networks were detected across various periods of PH development (Fig. 2). The character was as revealed in the preceding unconditional analysis. Collectively, the QTL network-related effects explained 34.8% of the phenotypic variation, less than the 52.2% of phenotypic variation explained by A-QTLs which were not involved in the QTL networks. Additive effects involved in QTL networks accounted for up to 22.4% of the phenotypic variation (the remaining 12.5% was attributed to epistatic effects). Therefore, additive effects were the major contributors to gene expression during PH development, in accordance with the unconditional analysis.
Fig. 2.
QTL epistatic networks for conditional PH in five periods in wheat DHLs.  ,
,  , solid lines, and dashed lines are as shown in Fig. 1.
, solid lines, and dashed lines are as shown in Fig. 1.  ,
,  ,
,  ,
,  , and
, and  refer to positive aa effects in S1|S0, S2|S1, S3|S2, S4|S3, and S5|S4, respectively.
refer to positive aa effects in S1|S0, S2|S1, S3|S2, S4|S3, and S5|S4, respectively.  ,
,  ,
,  ,
,  , and
, and  refer to negative aa effects in S1|S0, S2|S1, S3|S2, S4|S3, and S5|S4, respectively.
refer to negative aa effects in S1|S0, S2|S1, S3|S2, S4|S3, and S5|S4, respectively.  ,
,  ,
,  ,
,  , and
, and  refer to epistasis only with aae effects in S1|S0, S2|S1, S3|S2, S4|S3, and S5|S4, respectively.
refer to epistasis only with aae effects in S1|S0, S2|S1, S3|S2, S4|S3, and S5|S4, respectively.
Common QTLs detected by unconditional and conditional analysis
Through unconditional and conditional analysis, 15 A-QTLs (Tables 2, 3) and seven epistatic pairs (Supplementary Tables S5, S6 at JXB online) were common between the two sets of results across the whole period of PH development. For common A-QTLs, the conditional a effects at least in one early period were consistent in direction with the late unconditional a effects at 1–5 stages; that is, 12 A-QTLs were revealed with a effects in S1|S0 in the same direction as detected at 1–5 stages later, three A-QTLs in S2|S1 with 2–3 other stages, and one QTL, QPh.cgb-5A.7, in S3|S2 to S3 and S4 (Tables 2, 3). The opposite effects were also observed for one QTL in different periods which might counteract each other, resulting in the detection of little or no cumulative effects of the locus at a later stage. For example, QPh.cgb-2D.1 had an a effect of –0.50*** in S3|S2, and was opposite in direction to 1.85*** in S1|S0, decreasing its unconditional a effect from 3.89*** at S2 to 3.02*** at S3. QPh.cgb-5A.7 had an a effect of 0.35*** in S4|S3, changing its unconditional a effect from –3.89*** at S3 to –3.11*** at S4. QPh.cgb-6A.3 was identified with an a effect of 1.60*** in S5|S4, but was reversed in direction to –1.54*** in S1|S0, leading to no a effect being identified at S5. As for the magnitude of a effects for a particular QTL, the absolute value of conditional a effects was always less than or equal to that of unconditional results. Hence, the real gene expression in the early period could explain gene action at a later stage. This relationship was equally applicable for common epistatic interactions. The conditional aa effects in S1|S0 could explain unconditional aa effects at 1–4 stages later.
Five unconditional A-QTLs at 1–4 stages from S1 were not revealed to have conditional additive effects, whereas the same numbers of conditional A-QTLs in either periods S3|S2 or S5|S4 had no unconditional additive effects. In the same way, 91.5% of unconditional epistatic pairs were not detected with conditional epistatic effects, whereas 69.6% of conditional epistatic pairs were similarly not detected with unconditional epistatic effects. Therefore, by combining the conditional and unconditional QTL mapping of time-dependent measures, more loci were detected; consequently, it was possible to reveal dynamic gene expression for the development of a quantitative trait (Yan et al., 1998a).
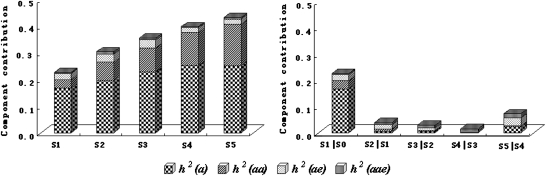
With regard to all genetic components, A-QTLs, E-QTLs, and their QE effects governed the development of PH of wheat, and, by unconditional mapping, a effects were identified as playing the most important roles at different development stages, accounting for 58.1–73.6% of the phenotypic variation; aa effects took second place, accounting for 14.1–36.0% of the phenotypic variation; and ae and aae effects contributed the least, accounting for 5.9–13.0% (Fig. 3). Conditional mapping showed that most of the phenotypic variation (73.6%) was due to a effects in S1|S0, whereas in periods after S1|S0, most phenotypic variation (64.8–94.0%) was attributable to ae and aae effects (Fig. 3). This implied that the net genetic effects varied greatly across ontogeny. Thus, as with the above analysis, genetic effects determined at different developmental stages after S1|S0 were basically the outcomes of real gene expression in S1|S0.
Fig. 3.
Contributions of genetic effects to PH development in wheat DHLs.
Superior genotype prediction based on mapped QTLs
In order to better utilize mapped QTLs in genetic improvement of wheat PH, the ideal multilocus combination of all putative QTLs (superior line, SL) was selected by comparing the total genetic effect of any individual with known QTL genotypes from the DHLs (Yang and Zhu, 2005). Because genetic main effects, particularly a effects expressed mostly in S1|S0, were the foremost genetic determinants of PH, SLs for PH in S1|S0 were predicted. With drought being a universal phenomenon, and increased PH being beneficial for exploiting soil moisture at depth under DS conditions, the predicted SLs were chosen with high values. The estimated total genetic effects for the taller parent (P1, Hanxuan 10) and SLs varied greatly in different environments; the predicted total genetic effects of SLs were far higher than that of P1 in each environment (Table 4), indicating the large potential for genetic improvement of PH. In E1, the severe DS environment, the predicted total genetic effect for SLs was 15.2 cm, whereas that for the taller parent P1 was 0.8 cm. Therefore, the SLs predicted in E1 adapted well to severe DS conditions.
Table 4.
Predicted genetic effects (G) (cm) for P1, P2, and a superior line (SL) on wheat PH in S1|S0
| Entry | G | G+GE1 | G+GE2 | G+GE3 | G+GE4 | G+GE5 | G+GE6 | G+GE7 | G+GE8 | G+GE9 | G+GE10 |
| P1 | 2.7 | 0.8 | 2.7 | 2.7 | 3.5 | 2.7 | 2.0 | 2.7 | 2.7 | 2.7 | 3.2 |
| P2 | –1.8 | –1.8 | –1.8 | –1.8 | –1.8 | –1.8 | –1.8 | –1.8 | –1.8 | –1.8 | –1.8 |
| SL | 18.6 | 15.2 | 18.6 | 18.6 | 17.8 | 18.6 | 19.3 | 18.6 | 18.6 | 18.6 | 25.5 |
G, general genetic effect; G+GE1 refers to the total genetic effects in E1, and so on; P1, Huanxuan 10; P2, Lumai 14; SL, superior line.
The same kind of QTL genotype was obtained for the predicted general superior line (GSL) and SLs in all 10 environments (Table 5), implying that the predicted SL genotype for PH in S1|S0 might be broadly adaptable; that is, the superior genotype could be adaptable to a range of environments. Compared with P1, the predicted QTL genotype for SL differed at eight loci, including six A-QTLs, QPh.cgb-3B.9, QPh.cgb-4A.5, QPh.cgb-5A.6, QPh.cgb-5B.4, QPh.cgb-6A.3, and QPh.cgb-6B.7, and two non-individual QTLs, QPh.cgb-3B.1 and QPh.cgb-6A.4, at which alleles increasing the PH were contributed by the lower value parent P2 (Lumai 14). Hence, the SL could be rapidly produced by substituting alleles of P1 by those of P2 by MAS.
Table 5.
QTL genotypes of the predicted general superior line (GSL) and SLs on wheat PH in S1|S0
| QTL | Flanking markers | GSL | SL |
|||||||||
| E1 | E2 | E3 | E4 | E5 | E6 | E7 | E8 | E9 | E10 | |||
| QPh.cgb-1B.1 | Xgwm582–Xgwm273 | |||||||||||
| QPh.cgb-2D.1 | Xwmc453.1–Xwmc18 | |||||||||||
| QPh.cgb-2D.11 | P3176.1–P1123.1 | |||||||||||
| QPh.cgb-3B.9 | P3622.4–P2076 | |||||||||||
| QPh.cgb-4A.5 | P6431.1–Xgwm160 | |||||||||||
| QPh.cgb-4D.1 | Xgwm165.2–Xgwm192 | |||||||||||
| QPh.cgb-5A.6 | Xgwm595–Xwmc410 | |||||||||||
| QPh.cgb-5B.4 | Xgwm371–Xgwm335 | |||||||||||
| QPh.cgb-6A.3 | P4232.4–Xcwm306 | |||||||||||
| QPh.cgb-6B.5 | Xgwm132–Xwmc104 | |||||||||||
| QPh.cgb-6B.7 | Xwmc269.3–P4232.1 | |||||||||||
| QPh.cgb-7A.3 | P3454.5–P3446.4 | |||||||||||
| QPh.cgb-1B.11 | P6934.3–P3446.6 | |||||||||||
| QPh.cgb-1B.16 | Xwmc269.2–Xcwm90 | |||||||||||
| QPh.cgb-2A.2 | Xwmc264.2–P8966.2 | |||||||||||
| QPh.cgb-7D.3 | Xwmc463–Xgwm295 | |||||||||||
| QPh.cgb-2B.1 | Xcwm529–Xwmc317 | |||||||||||
| QPh.cgb-7A.5 | Xgwm635.1–P2454.4 | |||||||||||
| QPh.cgb-2D.7 | Xwmc170–Xcwm96.2 | |||||||||||
| QPh.cgb-5A.3 | P2470–Xgwm154 | |||||||||||
| QPh.cgb-3B.1 | P2478.1–Xwmc505.1 | |||||||||||
| QPh.cgb-6A.6 | Xgwm617–Xcwm487 | |||||||||||
| QPh.cgb-6A.4 | Xpsp3071–Xgwm570 | |||||||||||
| QPh.cgb-7B.4 | Xpsp3033–Xgwm297 | |||||||||||
Discussion
Gene expression during ontogeny
Developmental quantitative genetics assume that development of complex morphological structures occurs through the actions and interactions of many genes acting differentially during ontogeny and whose expression is modified by interactions with other genes and by the cellular or organism environments (Atchley and Zhu, 1997; Wu et al., 2002). By unconditional and conditional analysis, a total of 129 QTLs were detected on 20 chromosomes (except 6D) associated with PH development. Of these, seven showed only individual additive effects, 104 only took part in epistatic interactions (i.e. modifying other QTLs), and the remaining 18 had both individual additive effects and modifying functions. Of these, 64.0% of the A-QTLs (16 of 25) and 23.2% of the epistatic pairs (23 of 99) also showed QE effects in 1–8 of the 10 environments. Expression of the A-QTLs and E-QTLs was developmental stage dependent; that is, most additive and interactive actions occurred in specific periods, with only a few A-QTLs being expressed in two or more periods, and no additive and interactive actions were continually active during the entire PH growth period. Consequently, most QTLs identified during the early periods were not detected in the later periods, suggesting that different genetic systems were responsible for PH development during different periods. This was also reported in previous studies (Yan et al., 1998a, b; Wu et al., 1999; Cao et al., 2001b; Ellis et al., 2004).
Several A-QTLs, such as QPh.cgb-2D.1, QPh.cgb-6B.5, QPh.cgb-6B.7, and QPh.cgb-7A.3, were expressed at all five stages. This was not ambivalent because QTLs detected at stage t were attributed to the cumulative gene expression from the initial time to stage t (Yan et al., 1998a), and not the real gene expression in the specific development period from stage t–1 to t. In reality, genetic behaviour measured at stage t is the confounded result of genes expressed before stage (t–1) and within the period from (t–1) to t (Zhu, 1995). QTL behaviour at stage t suggested the sequential and hierarchical character of development; that is, an event at stage t can have significant consequences on subsequent phenotypes later in ontogeny (Atchley and Zhu, 1997). Thus, QTLs significantly expressed in one or two periods simultaneously had significant genetic effects at all developmental stages. For example, QPh.cgb-2D.1, detected with significant a effects (1.85***) in S1|S0 and S3|S2 (–0.50***), also exhibited significant a effects (1.85*** to 4.70***) at all five stages. Such properties of QPh.cgb-4D.1 was proven in a recombinant inbred line (RIL) derived from the same cross Hanxuan 10 × Lumai 14 (Wang. et al., 2010). Therefore, genetic information disclosed by both conditional mapping in a specific growth period from t–1 to t, and by unconditional mapping at stage t is necessary for revealing the genetic expression of the developmental behaviour of a quantitative trait. Parts of the genetic actions and interactions were observed to have only unconditional effects, and not conditional effects, and vice versa. The absence of conditional effects might result from the effects being too small to be detected at the statistical level, whereas the absence of unconditional effects might be associated with the counteraction of conditional effects for the same QTL during different growth periods, as shown in other studies (Yan et al., 1998a, b; Wu et al., 1999; Cao et al., 2001b).
Inductive interactions are common in sequential and hierarchical systems. One component (e.g. a gene, protein, cell, or tissue) may ‘induce’ activities of other components and alter the eventual phenotype in a unidirectional or ‘cause and effect’ fashion (Atchley and Zhu, 1997). Epistatic QTL mapping studies in model organisms have shown that epistasis makes large contributions to the genetic regulation of complex traits (Carlborg and Haley, 2004). The effect of a gene on a phenotype is a collective property of a network of genes, rather than a property of a single gene (Wade, 2002; Malmberg et al., 2005). In the current study, a number of QTLs, including A-QTLs and E-QTLs without individual effects, participated in more than one epistatic interaction during ontogeny. Interestingly, some QTLs mapping at known Rht loci and reported in previous research, such as QPh.cgb-2D.1 mapping to Rht8 (Korzun et al., 1998), QPh.cgb-4B.1 to Rht1 (Cadalen et al., 1998; Liu et al., 2006) or Rht3 (Cadalen et al., 1998; Liu et al., 2006), QPh.cgb-4D.1 to Rht2 (Cadalen et al., 1998; Ellis et al., 2002; McCartney et al., 2005) or Rht10 (Cadalen et al., 1998; Ellis et al., 2002; McCartney et al., 2005), QPh.cgb-5A.7 to Rht9 (Ellis et al., 2005), and QPh.cgb-5A.6 to Rht12 (Ellis et al., 2005), participated in both additive and epistatic effects, and all of them, except QPh.cgb-5A.6, participated in more than one epistatic interaction, suggesting that the major Rht genes for PH acted as a network during ontogeny; that is, they display effects that depend upon genotypes at other loci. This kind of action for the major dwarfing genes was also reported for PH in rice (Jiang et al., 1994). Additionally, four of five A-QTLs detected in the present study were coincident with previously reported PH QTLs (Cadalen et al., 1998; Huang et al., 2003; Schnurbusch et al., 2003; Sourdille et al., 2003), including QPh.cgb-1B.1, QPh.cgb-1B.4, QPh.cgb-5B.4, and QPh.cgb-7B.4, and also participated in two or more epistatic interactions. Close to QPh.cgb-1B.4 (0.3 cM), QPh.cgb-1B.5, identified in S5|S4, also controlled the interval length from the flag leaf ligule to the spike base. It was detected in another study on the same population (unpublished data). This provided direct evidence for gene expression of PH during development. The remaining A-QTL, QPh.cgb-6A.3, acted individually, rather than being involved in interactions with other QTLs. Additionally, among three new QTLs, QPh.cgb-6B.5 and QPh.cgb-6B.7 had significant a effects with absolute values ranging from 1.58*** to 6.39*** at all five growth stages, and were also involved in three and two epistatic interactions, respectively. The third new QTL, QPh.cgb-7A.3, with significant a effects (0.78***–3.49***) at five stages, did not participate in epistatic interaction with other QTLs. Thus there is enough evidence to infer that the genetic architecture underlying PH development was represented as a network of epistatic and additive effects, and gene expression in ontogeny was regulatory and interactive.
Genetic components of PH development
Additive gene effects are the major components of PH variation (Fick and Qualset, 1973). As in the present study, PH was attributed to the major gene control model based on segregation patterns (Fick and Qualset, 1973). It was determined here that additive main effects (a effects), epistatic main effects (aa effects), and their QE effects control PH development. Among these, a effects were the major genetic determinants of PH, whereas aa effects and QE effects contributed much less. Although aa effects contributed less to phenotypic variation of PH compared with a effects, they were an important genetic component in that most loci participated in epistatic interactions, rather than functioning alone. This also occurred in other studies (Fick and Qualset, 1973; Jiang et al., 1994; Yu et al., 2002; Malmberg et al., 2005; Zhang et al, 2008).
Genetic components governing PH were time dependent; a effects were much more important than aa effects and QE effects at all five growth stages. Moreover, a effects were the predominant genetic components compared with other effects in S1|S0, whereas QE effects became major contributors to phenotypic variation after S1|S0; that is, the genetic effects of phenotypic variability become modified during ontogeny. A similar trend occurred with aa effects, which played a secondary role at all five growth stages, and also were more important than QE effects in S1|S0, whereas they became less important than QE effects after S1|S0. Developmental quantitative genetics showing that age-specific genetic variation at time t was conditioned by causal genetic effects at time t–1 imply the generation of significant episodes of new genetic variation arising during the period t–1 to t (Atchley and Zhu, 1997; Wu et al., 2002), and traits at different developmental stages are due to the accumulated results of genetic main effects and QE effects at all previous stages (Cao et al., 2001b). Therefore, genetic effects emerging at different developmental periods were more variable. Thus, our conclusion that the predominance of a effects at stages after S1|S0 was mostly attributable to the additive genetic variation generated in S1|S0 was reasonable; that is, a effects expressed in S1|S0 play the most important role for genetic control of PH, allowing for efficient early selection for PH improvement.
Environmental effects on QTL expression
The development of a complex trait reflects the adaptation of an organism to a particular environment and represents the impacts of genetic and environmental interactions (Wu et al., 2002). GE is a very important factor determining the stability of crop varieties, and has received considerable attention in plant breeding programmes (Xing et al., 2002). Loci conditioned by environmental signals (environmentally dependent genes) provide plants with large amounts of flexibility in responding to environmental changes; that is, these environmentally dependent loci might be of great importance in some environments, although less important in other environments (Lark et al., 1995). In the present study, 64.0% of A-QTLs and 23.5% of epistatic pairs had significant interaction effects in one to eight environments. Among them, QTLs highly responsive to water-limited environments might have practical consequences for breeding programmes. QTLs such as QPh.cgb-2D.1, QPh.cgb-4D.1, QPh.cgb-6B.5, and QPh.cgb-7A.3 were distinctly more adaptable to WW conditions (E10) at growth stages 2–3 (before S3); whereas QTLs such as QPh.cgb-5A.7, QPh.cgb-6B.7, and QPh.cgb-7B.4 were more responsive under DS conditions (E1) at stages 1–5. Drought tolerance is always a major goal in wheat improvement for water-limited areas. Under drought conditions increased PH is beneficial for exploiting soil moisture at depth, and for enabling mechanical harvesting, therefore leading to increased biological as well as grain yield (Baum et al., 2003). Hence QTLs such as QPh.cgb-5A.7, QPh.cgb-7B.4, and, especially, QPh.cgb-6B.7 might be useful for improving wheat PH by MAS under DS conditions.
Superior genotypes predicted from QTLs
In the present study, additive effects were the primary genetic component for PH growth, but E-QTLs should not been ignored in order to attain the greatest PH improvement. Since many QTLs were dependent on other QTLs rather than acting independently, predicted superior genotypes will be unreliable if epistasis is ignored (Jiang et al., 1994). Therefore, the best multilocus combination of all the putative QTLs could be used to predict and select SLs (Yang and Zhu, 2005).
QTLs detected with the conditional genetic model are associated with the main genes affecting the developmental trajectory of a morphological trait during ontogeny. A MAS programme for biologically real conditional QTLs can be designed to alter growth trajectories at particular stages and hence to achieve maximum final growth, whereas conventional MAS incorporating QTLs for final growth will be less efficient in effective early selection (Wu et al., 2002). With the knowledge that genetic effects in S1|S0, with minimal QE effects, played decisive roles in determining final PH, predicted SLs can be based on QTLs identified in S1|S0.
Our predicted SL was far superior to the better parent P1, implying a large potential among the progeny for further improvement of PH. Furthermore, the estimated total genetic effects for the SL varied in different environments (15.20–25.53 cm), indicating that PH improvement might be obtained differently in different environments. Furthermore, the predicted QTL genotype for SLs in S1|S0 was stable across environments. Thus, the predicted SL should be broadly adaptable to enable the optimal PH improvement in multitude environments using the same SL. To evaluate the predicted SL, further experiments are needed to substitute P1 alleles by P2 alleles using the appropriate molecular markers, then to reveal the effects of PH on grain yields of these artificial lines, which will be rather valuable for practical breeders.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Correlation coefficients between stages for plant height of wheat grown in 10 environments
Table S2. ANOVA for developmental behaviour of plant height among different environmental components in wheat DHLs
Table S3. ANOVA for developmental behaviour of plant height among different environmental combinations (year×site×water regime) in wheat DHLs
Table S4. Correlation coefficients between periods for plant height of wheat grown in 10 environments
Table S5. Unconditional epistatic QTLs affecting plant height of wheat at five growth stages in 10 environments
Table S6. Conditional epistatic QTLs affecting plant height of wheat in five periods in 10 environments
Acknowledgments
We thank Professor Jun Zhu (College of Science, Zhejiang University) for kind advice regarding the use of mapping software and data analysis. We thank Professor Robert A McIntosh (Plant Breeding Institute, University of Sydney, NSW, Australia) for revising the manuscript. This work was supported by the National High Technology Research and Development Program of China (2006AA100201), Generation Challenge Program (G4007.06), and the National Basic Research Program of China (2010CB125905).
Glossary
Abbreviations
- a effect
additive main effect
- aa effect
additive×additive epistatic main effect
- ae effect
additive QTL×environment interaction effect
- aae effect
epistasis×environment interaction effect
- A-QTL efffect
QTL with individual additive effect
- Ch05 and Ch06
Changping Beijing in 2005 and 2006, respectively
- DHLs
doubled haploid lines
- DS
drought stress
- E1
F01 under DS
- E2
H05 under DS
- E3
Ch05 under DS
- E4
H06 under DS
- E5
Ch06 under DS
- E6
F01 under WW conditions
- E7
H05 under WW
- E8
Ch05 under WW
- E9
H06 under WW
- E10
Ch06 under WW
- E-QTL
QTL participating in epistatic effects
- F01
Fenyang Shanxi in 2001
- GSL
general superior line
- H05 and H06
Haidian Beijing in 2005 and 2006, respectively
- MAS
marker-assisted selection
- PH
plant height
- PVE
phenotypic variation explained
- QE
QTL×environment interaction, including ae and aae
- QTL
quantitative trait locus
- S1, S2, S3, S4 and S5
the first, second, third, fourth and fifth measuring stage, respectively
- S1|S0, S2|S1, S3|S2, S4|S3 and S5|S4
the first, second, third, fourth and fifth measuring period, respectively
- SL
superior line
- WW
well watered
References
- Atchley WR, Zhu J. Developmental quantitative genetics, conditional epigenetic variability and growth in mice. Genetics. 1997;147:765–776. doi: 10.1093/genetics/147.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum M, Grando S, Backes G, Jahoor A, Sabbagh A, Ceccarelli S. QTLs for agronomic traits in the Mediterranean environment identified in recombinant inbred lines of the cross ‘Arta’ × H. spontaneum 41-1. Theoretical and Applied Genetics. 2003;107:1215–1225. doi: 10.1007/s00122-003-1357-2. [DOI] [PubMed] [Google Scholar]
- Börner A, Schumann E, Fürste A, Cöster H, Leithold B, Röder MS, Weber WE. Mapping of quantitative trait loci determining agronomic important characters in hexaploid wheat (Triticum aestivum L.) Theoretical and Applied Genetics. 2002;105:921–936. doi: 10.1007/s00122-002-0994-1. [DOI] [PubMed] [Google Scholar]
- Brady SM, Long TA, Benfey PN. Unraveling the dynamic transcriptome. The Plant Cell. 2006;18:2101–2111. doi: 10.1105/tpc.105.037572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadalen T, Sourdille P, Charmet G, Tixier MH, Gay G, Boeuf C, Bernard S, Leroy P, Bernard M. Molecular markers linked to genes affecting plant height in wheat using a doubled-haploid population. Theoretical and Applied Genetics. 1998;96:933–940. [Google Scholar]
- Cao G, Zhu J, He C, Gao Y, Wu P. QTL analysis for epistatic effects and QTL × environment interaction effects on final height of rice (Oryza sativa L.) Yi Chuan Xue Bao. 2001a;28:135–143. [PubMed] [Google Scholar]
- Cao G, Zhu J, He C, Gao Y, Yan J, Wu P. Impact of epistasis and QTL × environment interaction on the developmental behavior of plant height in rice (Oryza sativa L.) Theoretical and Applied Genetics. 2001b;103:153–160. [Google Scholar]
- Carlborg Ö, Haley CS. Epistasis: too often neglected in complex trait studies? Nature Reviews Genetics. 2004;5:618–625. doi: 10.1038/nrg1407. [DOI] [PubMed] [Google Scholar]
- Ellis MH, Rebetzke GJ, Azanza F, Richards RA, Spielmeyer W. Molecular mapping of gibberellin-responsive dwarfing genes in bread wheat. Theoretical and Applied Genetics. 2005;111:423–430. doi: 10.1007/s00122-005-2008-6. [DOI] [PubMed] [Google Scholar]
- Ellis MH, Rebetzke GJ, Chandler P, Bonnett D, Spielmeyer W, Richards RA. The effect of different height reducing genes on the early growth of wheat. Functional Plant Biology. 2004;31:583–589. doi: 10.1071/FP03207. [DOI] [PubMed] [Google Scholar]
- Ellis MH, Spielmeyer W, Gale KR, Rebetzke GJ, Richards RA. ‘Perfect’ markers for the Rht-B1b and Rht-D1b dwarfing genes in wheat. Theoretical and Applied Genetics. 2002;105:1038–1042. doi: 10.1007/s00122-002-1048-4. [DOI] [PubMed] [Google Scholar]
- Eriksen L, Borum F, Jahoor A. Inheritance and localisation of resistance to Mycosphaerella graminicola causing septoria tritici blotch and plant height in the wheat (Triticum aestivum L.) genome with DNA markers. Theoretical and Applied Genetics. 2003;107:515–527. doi: 10.1007/s00122-003-1276-2. [DOI] [PubMed] [Google Scholar]
- Fick GN, Qualset CO. Genes for dwarfness in wheat, Triticum aestivum L. Genetics. 1973;75:531–539. doi: 10.1093/genetics/75.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet J, Muttumu S, Horner M, Mango SE. Whole genome analysis of temporal gene expression during foregut development. PLoS Biology. 2004;2:1828–1842. doi: 10.1371/journal.pbio.0020352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM. Hormonal regulation of plant growth and development. PLoS Biology. 2004;2:1270–1273. doi: 10.1371/journal.pbio.0020311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J, Protas M, Conrad M, Scheid P, Vidal O, Jeffery W, Borowsky R, Tabin C. Synteny and candidate gene prediction using an anchored linkage map of Astyanax mexicanus. Proceedings of the National Academy of Sciences, USA. 2008;105:20106–20111. doi: 10.1073/pnas.0806238105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Z, Chang X, Guo X, Jing R, Li R, Jia J. QTL mapping for drought tolerance at stages of germination and seedling in wheat (Triticum aestivum L.) using a DH population. Agricultural Sciences in China. 2003;2:943–949. [Google Scholar]
- Huang XQ, Coster H, Ganal MW, Roder MS. Advanced backcross QTL analysis for the identification of quantitative trait loci alleles from wild relatives of wheat (Triticum aestivum L.) Theoretical and Applied Genetics. 2003;106:1379–1389. doi: 10.1007/s00122-002-1179-7. [DOI] [PubMed] [Google Scholar]
- Jiang C, Pan X, Gu M. The use of mixture models to detect effects of major genes on quantitative characters in a plant breeding experiment. Genetics. 1994;136:383–394. doi: 10.1093/genetics/136.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing R, Chang X, Jia J, Hu R. Establishing wheat doubled haploid population for genetic mapping by anther culture. Biotechnology. 1999;9:4–8. [Google Scholar]
- Khattak GSS, Ashraf M, Haq MA, McNeilly T, Rha ES. Genetic basis of plant height and its degree of indetermination in mungbean (Vigna radiata (L.) Wilczek) Hereditas. 2002;137:52–56. doi: 10.1034/j.1601-5223.2002.1370107.x. [DOI] [PubMed] [Google Scholar]
- Korzun V, Roder MS, Ganal MW, Worland AJ, Law CN. Genetic analysis of the dwarfing gene (Rht8) in wheat. Part I. Molecular mapping of Rht8 on the short arm of chromosome 2D of bread wheat (Triticum aestivum L) Theoretical and Applied Genetics. 1998;96:1104–1109. [Google Scholar]
- Lanceras JC, Pantuwan G, Jongdee B, Toojinda T. Quantitative trait loci associated with drought tolerance at reproductive stage in rice. Plant Physiology. 2004;135:384–399. doi: 10.1104/pp.103.035527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lark K, Chase K, Adler F, Mansur L, Orf J. Interactions between quantitative trait loci in soybean in which trait variation at one locus is conditional upon a specific allele at another. Proceedings of the National Academy of Sciences, USA. 1995;92:4656–4660. doi: 10.1073/pnas.92.10.4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque MP, Vernoux T, Busch W, et al. Whole genome analysis of the SHORT–ROOT developmental pathway in Arabidopsis. PLoS Biology. 2006;4:739–752. doi: 10.1371/journal.pbio.0040143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZK, Yu SB, Lafitte HR, et al. QTL × environment interactions in rice. I. Heading date and plant height. Theoretical and Applied Genetics. 2003;108:141–153. doi: 10.1007/s00122-003-1401-2. [DOI] [PubMed] [Google Scholar]
- Liu S, Zhou R, Dong Y, Li P, Jia J. Development, utilization of introgression lines using a synthetic wheat as donor. Theoretical and Applied Genetics. 2006;112:1360–1373. doi: 10.1007/s00122-006-0238-x. [DOI] [PubMed] [Google Scholar]
- Maccaferri M, Sanguineti MC, Corneti S, et al. Quantitative trait loci for grain yield and adaptation of durum wheat (Triticum durum Desf.) across a wide range of water availability. Genetics. 2008;178:489–511. doi: 10.1534/genetics.107.077297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmberg RL, Held S, Waits A, Mauricio R. Epistasis for fitness-related quantitative traits in Arabidopsis thaliana grown in the field and in the greenhouse. Genetics. 2005;171:2013–2027. doi: 10.1534/genetics.105.046078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney CA, Somers DJ, Humphreys DG, Lukow O, Ames N, Noll J, Cloutier S, McCallum BD. Mapping quantitative trait loci controlling agronomic traits in the spring wheat cross RL4452 × ‘AC Domain’. Genome. 2005;48:870–883. doi: 10.1139/g05-055. [DOI] [PubMed] [Google Scholar]
- Peng J, Richards DE, Hartley NM, et al. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature. 1999;400:256–261. doi: 10.1038/22307. [DOI] [PubMed] [Google Scholar]
- Peng J, Ronin Y, Fahima T, Röder MS, Li Y, Nevo E, Korol A. Domestication quantitative trait loci in Triticum dicoccoides, the progenitor of wheat. Proceedings of the National Academy of Sciences, USA. 2003;100:2489–2494. doi: 10.1073/pnas.252763199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestsova EG, Börner A, Röder MS. Development and QTL assessment of Triticum aestivum–Aegilops tauschii introgression lines. Theoretical and Applied Genetics. 2006;112:634–647. doi: 10.1007/s00122-005-0166-1. [DOI] [PubMed] [Google Scholar]
- Pestsova E, Roder M. Microsatellite analysis of wheat chromosome 2D allows the reconstruction of chromosomal inheritance in pedigrees of breeding programmes. Theoretical and Applied Genetics. 2002;106:84–91. doi: 10.1007/s00122-002-0998-x. [DOI] [PubMed] [Google Scholar]
- Quarrie SA, Quarrie SP, Radosevic R, Rancic D, Kaminska A, Barnes JD, Leverington M, Ceoloni C, Dodig D. Dissecting a wheat QTL for yield present in a range of environments: from the QTL to candidate genes. Journal of Experimental Botany. 2006;57:2627–2637. doi: 10.1093/jxb/erl026. [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Matsuoka M. Generating high-yielding varieties by genetic manipulation of plant architecture. Current Opinion in Biotechnology. 2004;15:144–147. doi: 10.1016/j.copbio.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Sari-Gorla M, Krajewski P, Fonzo ND, Villa M, Frova C. Genetic analysis of drought tolerance in maize by molecular markers. II. Plant height and flowering. Theoretical and Applied Genetics. 1999;99:289–295. [Google Scholar]
- SAS Institute. SAS/STAT User's Guide. Cary, NC: SAS Institute; 1996. [Google Scholar]
- Schnurbusch T, Paillard S, Fossati D, Messmer M, Schachermayr G, Winzeler M, Keller B. Detection of QTLs for Stagonospora glume blotch resistance in Swiss winter wheat. Theoretical and Applied Genetics. 2003;107:1226–1234. doi: 10.1007/s00122-003-1372-3. [DOI] [PubMed] [Google Scholar]
- Sourdille P, Cadalen T, Guyomarc H, Snape JW, Perretant MR, Charmet G, Boeuf C, Bernard S, Bernard M. An update of the Courtot × Chinese Spring intervarietal molecular marker linkage map for the QTL detection of agronomic traits in wheat. Theoretical and Applied Genetics. 2003;106:530–538. doi: 10.1007/s00122-002-1044-8. [DOI] [PubMed] [Google Scholar]
- Sun D, Li W, Zhang Z, Chen Q, Ning H, Qiu L, Sun G. Quantitative trait loci analysis for the developmental behavior of soybean (Glycine max L. Merr.) Theoretical and Applied Genetics. 2006;112:665–673. doi: 10.1007/s00122-005-0169-y. [DOI] [PubMed] [Google Scholar]
- Ungerer MC, Halldorsdottir SS, Purugganan MD, Mackay TFC. Genotype environment interactions at quantitative trait loci affecting inflorescence development in Arabidopsis thaliana. Genetics. 2003;165:353–365. doi: 10.1093/genetics/165.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade MJ. A gene's eye view of epistasis, selection and speciation. Journal of Evolutionary Biology. 2002;15:337–346. [Google Scholar]
- Wang Z, Wu X, Ren Q, Chang X, Li R, Jing R. QTL mapping for developmental behavior of plant height in wheat (Triticum aestivum L.) Euphytica. 2010;21 (on line. DOI: 10.1007/s10681-010-0166-3) [Google Scholar]
- Wu R, Ma C, Zhu J, Casella G. Mapping epigenetic quantitative trait loci (QTL) altering a developmental trajectory. Genome. 2002;45:28–33. doi: 10.1139/g01-118. [DOI] [PubMed] [Google Scholar]
- Wu W, Li W, Tang D, Lu H, Worland AJ. Time related mapping of quantitative trait loci underlying tiller number in rice. Genetics. 1999;151:297–303. doi: 10.1093/genetics/151.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Tan Y, Hua J, Sun X, Xu C, Zhang Q. Characterization of the main effects, epistatic effects and their environmental interactions of QTLs on the genetic basis of yield traits in rice. Theoretical and Applied Genetics. 2002;105:248–257. doi: 10.1007/s00122-002-0952-y. [DOI] [PubMed] [Google Scholar]
- Yan J, Zhu J, He C, Benmoussa M, Wu P. Quantitative trait loci analysis for the developmental behavior of tiller number in rice (Oryza sativa L.) Theoretical and Applied Genetics. 1998a;97:267–274. [Google Scholar]
- Yan J, Zhu J, He C, Benmoussa M, Wu P. Molecular dissection of developmental behavior of plant height in rice (Oryza sativa L.) Genetics. 1998b;150:1257–1265. doi: 10.1093/genetics/150.3.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Jing R, Chang X, Li W. Identification of quantitative trait loci and environmental interactions for accumulation and remobilization of water-soluble carbohydrates in wheat (Triticum aestivum L.) stems. Genetics. 2007;176:571–584. doi: 10.1534/genetics.106.068361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Xing Y, Li S, Ding J, Yue B, Deng K, Li Y, Zhu Y. Molecular dissection of developmental behavior of tiller number and plant height and their relationship in rice (Oryza sativa L.) Hereditas. 2006;143:236–245. doi: 10.1111/j.2006.0018-0661.01959.x. [DOI] [PubMed] [Google Scholar]
- Yang J, Zhu J. Methods for predicting superior genotypes under multiple environments based on QTL effects. Theoretical and Applied Genetics. 2005;110:1268–1274. doi: 10.1007/s00122-005-1963-2. [DOI] [PubMed] [Google Scholar]
- Yang J, Zhu J, Williams RW. Mapping the genetic architecture of complex traits in experimental populations. Bioinformatics. 2007;23:1527–1536. doi: 10.1093/bioinformatics/btm143. [DOI] [PubMed] [Google Scholar]
- Yu S, Li J, Xu C, Tan Y, Li X, Zhang Q. Identification of quantitative trait loci and epistatic interactions for plant height and heading date in rice. Theoretical and Applied Genetics. 2002;104:619–625. doi: 10.1007/s00122-001-0772-5. [DOI] [PubMed] [Google Scholar]
- Zhang K, Tian J, Zhao L, Wang S. Mapping QTLs with epistatic effects and QTL × environment interactions for plant height using a doubled haploid population in cultivated wheat. Journal of Genetics and Genomics. 2008;35:119–127. doi: 10.1016/S1673-8527(08)60017-X. [DOI] [PubMed] [Google Scholar]
- Zhou X, Jing R, Hao Z, Chang X, Zhang Z. Mapping QTL for seedling root traits in common wheat. Agricultural Science in China. 2005;38:1951–1957. [Google Scholar]
- Zhu J. Analysis of conditional genetic effects and variance components in developmental genetics. Genetics. 1995;141:1633–1639. doi: 10.1093/genetics/141.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.