Abstract
Natural hybridization is widespread among coral reef fishes. However, the ecological promoters and evolutionary consequences of reef fish hybridization have not been thoroughly evaluated. Butterflyfishes form a high number of hybrids and represent an appropriate group to investigate hybridization in reef fishes. This study provides a rare test of terrestrially derived hybridization theory in the marine environment by examining hybridization between Chaetodon trifasciatus and C. lunulatus at Christmas Island. Overlapping spatial and dietary ecologies enable heterospecific encounters. Nonassortative mating and local rarity of both parent species appear to permit heterospecific breeding pair formation. Microsatellite loci and mtDNA confirmed the status of hybrids, which displayed the lowest genetic diversity in the sample and used a reduced suite of resources, suggesting decreased adaptability. Maternal contribution to hybridization was unidirectional, and no introgression was detected, suggesting limited, localized evolutionary consequences of hybridization.
Comparisons to other reef fish hybridization studies revealed that different evolutionary consequences emerge, despite being promoted by similar factors, possibly due to the magnitude of genetic distance between hybridizing species. This study highlights the need for further enquiry aimed at evaluating the importance and long-term consequences of reef fish hybridization.
Keywords: Christmas island, discriminant analysis of principal components, ecological factors, evolutionary consequences, hybridization, hybrid zone, unidirectional hybridization
Introduction
Natural hybridization occurs when individuals from different species or populations (distinguishable through one or more heritable characters) successfully interbreed, producing viable hybrids (Arnold 1997). To date, more than 25% of plant and 10% of animal species have been reported to hybridize in the wild (Mallet 2005). Certain taxonomic groups tend to hybridize much more than expected (Mallet 2005) and specific geographic regions concentrate hybridization in narrow “hybrid zones” (Barton and Hewitt 1985).
Of all the vertebrate groups, natural hybridization has most commonly been reported in fishes (Hubbs 1955; Allendorf and Waples 1996). More than 160 natural hybrids, involving 93 species across 12 families, have been reported among freshwater fishes (Scribner et al. 2000). In the marine environment 83 natural fish hybrids have been reported, involving 132 species across 17 families (S. Montanari, unpublished data). Where the freshwater literature is constellated with well-studied cases of natural fish hybridization across different climatic regions and levels of commercial interest, marine fish hybrids in the wild have received little attention and this has mainly been focused on commercially important, temperate water species (e.g., Norman 1934; Fujio 1977; Garcia de Leaniz and Verspoor 1989; Ruzzante et al. 2000; Nielsen et al. 2003; Ayllon et al. 2004; Burford et al. 2011). Only naturally occurring hybrids will be referred to as “hybrids” in this study.
Several processes have been proposed to explain the abundance of hybrid fishes, including: external fertilization (Hubbs 1955), competition for limited spawning grounds (Campton 1987), secondary contact of recently diverged sister taxa (McMillan and Palumbi 1995), spatial or dietary overlap in parental species (van Herwerden et al. 2006; Yaakub et al. 2006, 2007; Marie et al. 2007), rarity of one or both parental species (Gosline 1948; Randall et al. 1977; Frisch and van Herwerden 2006; Marie et al. 2007), sneak mating (van Herwerden et al. 2006), and absence of assortative mating (McMillan et al. 1999). Ecological observations of hybridizing reef fishes and comparison of these data to those collected from outside the hybrid zone—where available—may allow identification of ecological conditions that favor hybridization in the hybrid zone.
Genetic investigation of freshwater fish hybridization has revealed evolutionarily significant, though contrasting scenarios. For example, the explosive radiation (speciation) of cichlids in the African lakes has been attributed to protracted hybridization (Shaw et al. 2000; Turner et al. 2001; Verheyen et al. 2003; Seehausen 2004), and so has the collapse (reverse speciation) of the benthic and limnetic species of three-spined stickleback in the lakes of British Columbia (Taylor et al. 2006). The use of molecular genetics, clearly beneficial to understanding the evolutionary consequences of hybridization in freshwater fishes, has only recently been applied to reef fish hybridization (McMillan et al. 1999; van Herwerden et al. 2002, 2006; van Herwerden and Doherty 2006; Yaakub et al. 2006, 2007; Marie et al. 2007; Crow et al. 2010).
At least 75 species of coral reef fish species hybridize (Yaakub et al. 2006). Like most animal hybrids, reef fish hybrids have been traditionally identified through aberrant color patterns and morphological traits, often deemed intermediate between those of putative parent species (e.g., Randall 1956; Pyle and Randall 1994). This approach is still used today, and numerical predictions of hybrid color patterns (Miyazawa et al. 2010) as well as genetic validation of the hybrid status of intermediately colored individuals (e.g., Yaakub et al. 2006, 2007; Marie et al. 2007) have confirmed the soundness of this approach.
Prior reef fish hybridization studies have focused on members of the Acanthuridae (Marie et al. 2007), Chaetodontidae (McMillan et al. 1999), Labridae (Yaakub et al. 2006, 2007), Pomacentridae (van Herwerden and Doherty 2006), and Serranidae (van Herwerden et al. 2002, 2006; Frisch and van Herwerden 2006). Results from these studies have shown that reef fish hybridization can be characterized by unidirectional (e.g., Yaakub et al. 2006) or bidirectional (e.g., Marie et al. 2007) parental contributions, and the presence (e.g., van Herwerden et al. 2006) or absence (e.g., Yaakub et al. 2007) of introgression. Moreover, disparate ecological promoters of reef fish hybridization were identified, and varying levels of evolutionary significance ascribed to the process (McMillan et al. 1999; van Herwerden et al. 2002, 2006; Frisch and van Herwerden 2006; van Herwerden and Doherty 2006; Yaakub et al. 2006, 2007; Marie et al. 2007). Clearly, the combined use of ecological and genetic approaches in the study of reef fish hybridization can help elucidate the contribution of hybridization to the diversity and evolution of this group.
Butterflyfishes have the highest reported incidence of hybridization of all reef fish families (Allen et al. 1998; Kuiter 2002; Yaakub et al. 2006; Hobbs et al. In press): 44 of 130 (34%) species hybridize (Hobbs et al. In press), a proportion higher than most plant or animal taxa (Mallet 2005). The Chaetodontidae are a relatively young reef fish family (Bellwood et al. 2010), in which recently diverged allopatric sister species are common (Blum 1989). Many of these sister species have made secondary contact, setting the scene for hybridization (e.g., McMillan et al. 1999). Moreover, the dietary overlap shown by some species in this family (Pratchett 2005), together with habitat overlap, can increase the frequency of heterospecific encounters (favoring hybridization). In synergy, these characteristics of the Chaetodontidae render butterflyfish a suitable group for reef fish hybridization studies. Further, butterflyfishes are significantly affected by reef degradation (Pratchett et al. 2004, 2006b; Graham 2007), possibly due to the high incidence of corallivory in this group (Cole et al. 2008). Hybridization can result in increased adaptability to altered environments following disturbance (Grant and Grant 2002; Taylor et al. 2006; Riginos and Cunningham 2007) and may potentially be beneficial to butterflyfishes in a time when coral reefs are undergoing significant habitat changes.
Approximately 90% of hybridizing butterflyfishes occur at four specific geographical locations: southern Japan, Hawaii, Papua New Guinea-Micronesia, and the Eastern Indian Ocean (Hobbs et al. In press). Hybridizing reef fishes belonging to other families have been reported from these same locations (Pyle and Randall 1994; Gardner 1997; Kuriiwa et al. 2007; Hobbs et al. 2009). Christmas Island in the Eastern Indian Ocean is a known reef fish hybrid hotspot (Hobbs et al. 2009), where at least eight butterflyfish species hybridize (Hobbs et al. In press), making it an ideal location to study butterflyfish hybridization.
Prior studies of butterflyfish hybridization (McMillan et al. 1999) demonstrated that hybrid phenotypes largely outnumbered parental phenotypes within the hybrid zone, suggesting greater fitness of hybrids in the hybrid zone. Despite the wealth of reported butterflyfish hybrids, studies addressing the ecologies of these intermediate individuals have never been conducted. The results from McMillan et al. (1999) require that other butterflyfish hybrids be studied in the field, to determine whether adaptive disparities consistently result from hybridization among butterflyfishes. This study represents the most comprehensive examination of reef fish hybridization to date combining ecological, behavioral, and genetic approaches to investigate: (1) the ecology of hybridization between butterflyfishes Chaetodon trifasciatus Park 1797 and C. lunulatus Quoy and Gaimard 1824 at Christmas Island by assessing spatial and dietary overlap; (2) abundance of parental and hybrid individuals; (3) presence/absence of assortative pairing in parental species and their hybrids; (4) the directionality and evolutionary consequences of hybridization through molecular genetic analyses of mitochondrial and nuclear (microsatellite) DNA.
Methods
Ecology of hybridization
Study species
The Indian Ocean redfin butterflyfish, C. trifasciatus (Fig. 1A), ranges from East Africa to Bali, Indonesia, Cocos (Keeling), and Christmas Islands (Allen et al. 1998) at the easternmost periphery. The redfin butterflyfish, C. lunulatus (Fig. 1A), is widespread throughout the Western Pacific Ocean, from Eastern Australia north to Japan and east to Hawaii and the Tuamotu Islands (Allen et al. 1998). Christmas Island is at the westernmost edge of its distribution range (Allen et al. 1998; Hobbs and Salmond 2008), where heterospecific pairs are formed with C. trifasciatus (Hobbs et al. 2009). Both species generally form homospecific pairs throughout their range (Allen et al. 1998) and the time of pairing in C. lunulatus—an obligate monogamous species (Yabuta 1997)—corresponds with the onset of sexual maturity (Pratchett et al. 2006a), indicating a reproductive basis for pairing in this complex. Hybrid C. trifasciatus×lunulatus are identified based on coloration of their headband (Fig. 1B) and caudal peduncle (Fig. 1C), both intermediate between those of their parents.
Figure 1.
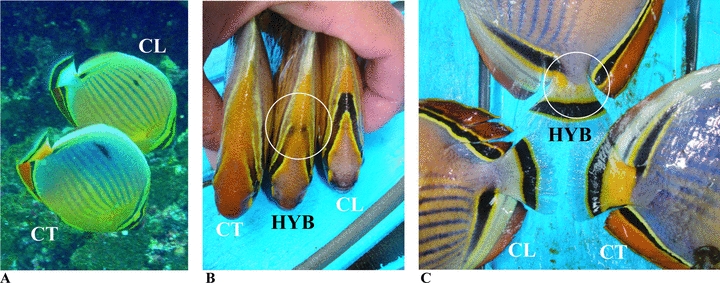
(A) Chaetodon trifasciatus (CT) and C. lunulatus (CL) swimming together in a heterospecific pair at Christmas Island; hybrids (HYB) of this species complex are characterized by their: (B) headband and (C) caudal peduncle, which are intermediate between those of their parents.
Study location
This study was conducted in October 2010–November 2010 at Christmas Island, in the Indian Ocean (10°25′S–10°34′S, 105°32E–105°42E) (Fig. 2A, inset). Christmas Island is located approximately 360 km south of Java and is a recognized suture zone between Pacific and Indian Ocean taxa (Hobbs and Salmond 2008; Hobbs et al. 2009).
Figure 2.
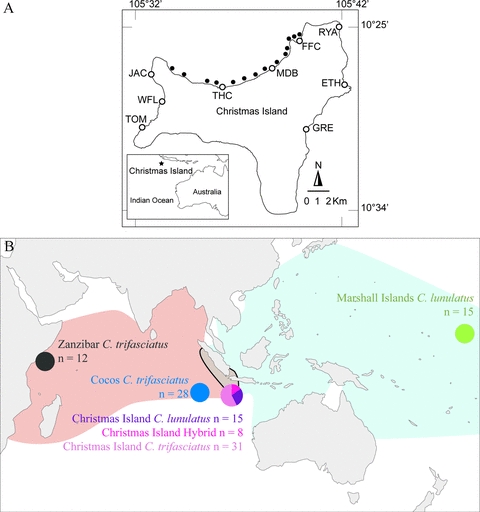
(A) Map of Christmas Island indicating the position of study sites used for the initial surveys (white dots) and the North coast surveys (black dots); Flying Fish Cove (FFC), Million Dollar Bombie (MDB), Thundercliff (THC), Jackson Point (JAC), Stef's Waterfall (WFL), Toms Point (TOM), Greta Beach (GRE), Ethel Beach (ETH), and Ryan's Ravine (RYA); inset shows the location of Christmas Island in the Indian Ocean. (B) Geographical origin and sample size (n) of populations of Chaetodon trifasciatus, C. lunulatus, and their hybrid used to assess phylogenetic relationships and population structure; the parental species ranges (redrawn from Allen et al. 1998) are also shown: C. trifasciatus (light red shaded area), C. lunulatus (light blue shaded area), and their contact zone (gray area with black outline).
Island-wide survey
Underwater visual surveys were conducted at nine sites along the north, east, and west sides of the island (Fig. 2A). The South coast was inaccessible due to prevailing southeasterly winds. At each site, surveys were conducted at three depths: 5, 12, and 20 m, where six belt transects (50 × 5 m) were laid parallel to the depth contour, giving a total of 162 transects. On each transect, the number of each species and their hybrids was recorded. All individuals recorded during these preliminary surveys were encountered along the North coast and, accordingly, all subsequent sampling effort was concentrated there.
Ecological overlap
To assess habitat use of the parent species and hybrids, surveys were conducted along the North coast (Fig. 2A) to record the depth distributions of individuals in the complex. Approximately equal sampling was conducted at all depths from 3 to 25 m, reflecting the maximum depth range recorded for these species during prior reef-wide surveys. The specific depth at which individual fishes were first seen was recorded for 34 individuals of C. trifasciatus, 30 C. lunulatus, and eight putative hybrids. Depth distribution data were analyzed using an analysis of variance (ANOVA) comparing the mean depth at which each of the pure species and the putative hybrids were recorded.
To assess dietary overlap between parental species, as well as compare dietary composition of putative hybrids to that of parental species, in situ feeding observations were conducted for all individuals recorded during depth-based surveys. Three-minute observations were conducted for each individual following Pratchett (2005), recording the number of bites taken from different benthic prey or substrates. Prey items included scleractinian corals categorized based on genus and growth form as follows: Montipora (encrusting), Porites (encrusting or columnar), Porites (massive), Acropora (branching), Acropora (plate), Acropora (corymbose), Galaxea (encrusting), Favites (massive), Pocillopora (corymbose), Echinopora (foliose), Lobophyllia (massive), Astreopora (massive), Diploastrea (massive), Fungia (free living), other (encrusting), other (coral), and epilithic algal matrix (EAM).
Variation in dietary composition was analyzed using a multivariate analysis of variance (MANOVA), comparing the relative number of bites taken from each of the 17 prey categories by each species and the hybrid. To further test for similarities in feeding behavior, feeding rates (bites per 3 min) were compared between species and hybrids, using a one-way ANOVA.
Abundance
To obtain more precise estimates of abundance for rarer butterflyfish species, transect size was increased for additional North coast surveys (Thompson 2004). Abundances of all focal species were recorded at 14 sites along the North coast (Fig. 2A) while swimming along depth contours. The total area sampled for each replicate transect was calculated based on Global Positioning System (GPS) tracks. Recorded tracks were divided into deep and shallow transects which were independently measured, excluding the distances swum to reach the initial depth and the distance swum while moving between the deep and shallow waters. For the deep part of the dive (10–25 m), transect width was 10 m and average length was 323.56 m (average transect area 3235.64 m2). For the shallow part (3–9 m), the transect width was increased to 20 m (due to low abundances, and the increased width was appropriate given the high visibility and flat topography) and average length was 314.76 m (average transect area 6295.14 m2). Abundance data were analyzed using t-tests.
Assortative pairing
During large-scale abundance surveys, the composition of all pairs was recorded to assess whether pairing within this species complex is assortative or nonassortative. Partners were recorded for all individuals surveyed, regardless of whether the partner was encountered, and therefore counted, within the transect area. Fish not yet paired up were recorded as single individuals and most of these were of small size (< 100 mm total length) and probably juveniles. Expected pairing frequencies were calculated for each taxon by multiplying the total number of paired individuals by the proportional observed abundances of each of the three members of the complex. Pairing data were analyzed separately for each taxon in the complex using χ2-test between the expected and observed pairing frequencies.
Genetics of hybridization
Sampling
Samples of C. trifasciatus, C. lunulatus, and hybrids were collected from within the hybrid zone at Christmas Island (but outside the survey locations), between 2005 and 2008 (Fig. 2B). Samples of the Indian Ocean species C. trifasciatus from outside the hybrid zone were collected at Cocos (Keeling) Islands (2005–2008) and Zanzibar (Fig. 2B). Similarly, C. lunulatus from outside the hybrid zone were collected from the Marshall Islands in the Pacific Ocean (2008–2009) (Fig. 2B). To obtain genetic samples, individual fishes were speared by scuba divers and, following capture, fin clips were immediately placed in 80% ethanol for subsequent analyses. The inclusion of samples of each “purebred” parental species from outside the hybrid zone allowed identification of species-specific genetic signals for comparison with the signal obtained at Christmas Island, where hybridization is apparent. Fin clips of C. citrinellus from Lizard Island, stored in 80% ethanol, were obtained from M. Pratchett and utilized as an outgroup taxon in all subsequent phylogenetic analyses (Fessler and Westneat 2007).
Laboratory procedures
For all subsequently described laboratory procedures and analyses, DNA was extracted from fin clips using a 5% Chelex extraction protocol (Walsh et al. 1991).
Approximately 600 bp of the mitochondrial gene cytochrome (cyt) b were amplified in both species and hybrids using cyt b specific primers (CBMP95–1; 5′-ATTCTAACTGGACTATTCCTTGCC-3′ and CBMP95–2; 5′-ATTATCTGGGTCTCCGAA(C/T)AGGTT-3′) previously utilized to study color pattern evolution in butterflyfishes of genus Chaetodon (McMillan and Palumbi 1995).
Polymerase chain reactions (PCR) were conducted as follows: twenty microliters reactions containing 2.5 mM Tris-Cl (pH 8.7), 5 mM KCl, 5 mM (NH4)2SO4, 200 µM each dNTP, 1.5–2 mM MgCl2, 0.25 µM each primer, 1 unit of Biotaq DNA polymerase (BIOLINE™), and 2 µl of Chelex extracted DNA template. Thermocycling was carried out with an initial denaturation step of 2 min at 94°C, followed by 35 cycles of denaturation, annealing, and extension (94°C for 30 sec, 50°C for 30 sec, 72°C for 90 sec) with a final extension of 10 min at 72°C. PCR products were visually confirmed using 1.5% agarose gel electrophoresis and amplicon sizes estimated with a 100-bp standard marker (BIOLINE™ Hyperladder IV).
PCR products were purified using either a standard isopropanol precipitation protocol (Sambrook et al. 1989) or a Sephadex G-25 resin 350-µl column spin protocol. Purified PCR products were sequenced with both primers using ABI (Applied Biosystems Incorporated) technologies either at Macrogen Seoul, South Korea or at the Australian Genome Research Facility (AGRF) Brisbane, Australia. GenBank accession numbers for all sequences are JQ012110 - JQ012216.
Sixteen C. lunulatus microsatellite markers (Lawton et al. 2010) were tested on two individuals each of C. lunulatus and C. trifasciatus and on three individuals of the putative hybrid. Detailed information about the primers used is presented in Table 1. PCR was conducted as described above, except primer concentration was increased to 0.5 µM. Thermocycling was carried out with an initial denaturation step of 3 min at 94°C, followed by a touchdown protocol of denaturation, annealing, and extension constituted of five cycles (94°C for 30 sec, primer-specific temperature (Ta) °C(Table 1) for 30 sec, 72°C for 90 sec) plus 30 cycles (94°C for 30 sec, Ta- 2°C for 30 sec, 72°C for 90 sec) and a final extension of 10 min at 72°C.
Table 1.
Details of 16 microsatellite loci developed for Chaetodon lunulatus by Lawton et al. (2010). Primer sequences and repeat motifs are provided as given by the authors of the original study (Lawton et al. 2010). Annealing temperatures (Ta) and size ranges are the ones utilized and found in the present investigation
| Locus (label) | Primer sequence (5′-3′) | Repeat motif | Ta (°C) | Size range (bp) |
|---|---|---|---|---|
| Lun01 (FAM) | TGAACTGCAAAGCAACAACC | (CAT)17 | 55 | 2 |
| CTGCTTCTCTTTGGTGAGGAG | ||||
| Lun03 (TET) | TGTGTGTCACCACCTGGTCT | (AG)29 | 58 | 175–239 |
| ACTCAGTTTTGAGCCGCTTC | ||||
| Lun05 (FAM) | GCAACCCAGTCTCACATCAA | (CAA)30 | 55 | 155–191 |
| TCTGCTATTTCACAATTTTAGAGCA | ||||
| Lun07 (HEX) | AAGTGCCCTTTAGCAAAGCA | (TG)17 | 58 | 153–212 |
| CTCCAGTCGCTTTCTGTGTG | ||||
| Lun08 (HEX) | GGCCTTTGTTTGTGGTCATT | (CA)26 | 55 | 174–226 |
| CCTGAAGAGAGAGCTGCTCAA | ||||
| Lun09 (TET) | CCTGTGTTTGTCATCCAACG | (TG)15 | 58 | 143–167 |
| CTTTGGGACACACACTTCCA | ||||
| Lun10 (TET) | TTGTGTTGTTTTAGTGTTCCCTTT | (AC)24 | 58 | 223–285 |
| TGAGTGGTTATGATACATTAGATTTTG | ||||
| Lun14 (HEX) | TACGTTGGACAGTGGCTGTG | (TCA)11 | 58 | 207–240 |
| TGGCTCTGTGGCATGTATGT | ||||
| Lun17 (*) | TCAGAGGTCGCTAACGTGTG | (GAT)12 | 55 | 1 |
| CTCTAACGCGTCCTCTGTCC | ||||
| Lun19 (FAM) | TCCAGTTCCATTCTGCCTTT | (GAT)16 | 55 | 125–188 |
| CCGTCATTAACCTCCAGCAG | ||||
| Lun20 (TET) | CAGTGTCGGAGAACAACGAA | (CTT)12 | 58 | 2 |
| TCACTGTGTCACCAATGCAC | ||||
| Lun21 (FAM) | CAGGGAAAATCACACTTTCACA | (TGCC)14 | 55 | 223–283 |
| TGTCAAGCTGTGTGGGACAT | ||||
| Lun22 (HEX) | GGATGATGCAACTGATGGAA | (ATC)15 | 58 | 2 |
| TGTAGCATTTCATCTTTGACACTG | ||||
| Lun29 (HEX) | CACCCACAGGCAGTGTATTG | (AC)33 | 55 | 233–277 |
| GCCAGCCTGTCAAAACTTTA | ||||
| Lun34 (TET) | CATGCTTGGGTGAGCATGTA | (CA)36 | 58 | 165–185 |
| TGTGCGTTTGTGCAAGTGTA | ||||
| Lun36 (FAM) | GCGTTTGACTTCACGTTTCA | (GT)30 | 58 | 188–238 |
| TGCAAAACAACAACCTACGG |
Discarded prior to genotyping.
Could not be scored reliably.
PCR products were visually confirmed by 1.5% agarose gel electrophoresis and amplicon sizes estimated with a 100-bp standard marker (BIOLINE™ Hyperladder IV). All markers reliably amplified in all individuals and product sizes were consistent with expectations (Lawton et al. 2010).
In light of the successful testing, the best 15 markers were labeled with fluorescent tags (HEX, FAM, or TET) (Table 1). One marker (Lun17) was discarded during this step because it was more economical in both time and funds to process five sets of three loci. PCR cocktails were made as described above except the forward primers were substituted with their labeled equivalents. Thermocycling was performed as described above.
PCR products were purified by centrifugation through 350-µl Sephadex G-25 resin columns and subsequently inspected for quality via 1.5% agarose gel electrophoresis prior to genotyping. Genotypes were run on an Amersham Biosciences Megabase Capillary Sequencer with a 400 bp standard at the Advanced Analytical Centre (AAC) at James Cook University, Townsville, Australia. All genotypic data were deposited in the Dryad Repository: doi:10.5061/dryad.20fc5v4j
Data compilation
Nucleotide sequences were aligned using the ClustalW (Thompson et al. 1994) algorithm and manually edited in Geneious Pro v5.3.3 (Biomatters Ltd.). Microsatellite genotypes were scored using Fragment Profiler v1.2 (Amersham Biosciences) and subsequently edited using Genalex 6.1 (Peakall and Smouse 2006).
Phylogenetic analyses
Phylogenetic relationships were inferred in order to establish whether the two parental species had fixed sequence differences when considering samples from outside the hybrid zone, allowing the identification of clades representative of the genetically unique parental species. This was done using four approaches: neighbor-joining (NJ) (Saitou and Nei 1987) and maximum parsimony (MP) (Eck and Dayhoff 1966) algorithms, implemented in Mega 4 (Tamura et al. 2007), Bayesian inference (BI), run through the Mr. Bayes (Huelsenbeck and Ronquist 2001) plug-in (compiled by Marc Suchard and the Geneious Team) from within Geneious Pro v5.3.3 (Biomatters Ltd.), and maximum likelihood (ML) analysis, performed using Garli v0.95 (Zwickl 2006) and Bootscore v3.11 (Sukumaran 2007). In all analyses described here, trees were outgroup rooted with two individuals of C. citrinellus.
In the NJ algorithm, evolutionary distances were computed using the Maximum Composite Likelihood method (Tamura et al. 2004) with 1000 Bootstrap replicates. All gaps and missing data were pairwise deleted.
Ten independent MP analyses were run and the overall shortest tree selected. The tree was obtained using a Close-Neighbour-Interchange algorithm (Nei and Kumar 2000) with search level 2 (Eck and Dayhoff 1966; Nei and Kumar 2000) with initial trees inferred by random addition (10 replicates). All gaps and missing data were discarded.
BI analysis used the JC69 substitution model (Jukes and Cantor 1969)—selected using jModelTest v0.1.1 (Guindon and Gascuel 2003; Posada 2008)—and was run using Markov chain Monte Carlo (MCMC) simulations with four chains of 100,000 generations each, sampling trees every 100 generations. A 50% majority rule consensus tree was computed using the 1000 best post-burn-in trees.
ML analysis was repeated independently 10 times and the resulting best trees compared to ensure consistency of topology. A bootstrap ML analysis with 100 replicates was also run to compute a consensus tree based on the best topology previously obtained.
Population genetic analyses
A minimum spanning network (MSN) of cyt b haplotypes was constructed in Hapstar v0.6 (Prim 1957; Excoffier et al. 2005) and subsequently edited in Illustrator CS (Adobe Systems Inc.). Genetic diversity indices of cyt b haplotypes, including haplotype diversity (h) and nucleotide diversity (π), were calculated for all populations sampled using Arlequin v3.1 (Excoffier et al. 2005). Spatial heterogeneity for cyt b was assessed through analysis of molecular variance (AMOVA) and pairwise Fst, performed in Arlequin with 1000 permutations.
Twelve of 15 markers genotyped were successfully scored, while three (Lun01, Lun20, and Lun22) could not be scored confidently and were therefore excluded from further analyses. Microsatellite metrics including number of alleles (Na), private alleles (Pa), observed (HO), and expected (HE) heterozygosities and average inbreeding coefficient (FIS) were calculated in Genalex (Peakall and Smouse 2006) and Fstat v2.9 (Goudet 1995). Probabilities of departure from Hardy–Weinberg equilibrium (HWE) and linkage disequilibrium (LD) were estimated in Genepop v4.0 (Rousset 2008) using Markov chains with dememorization 10,000, 20 batches, and 5000 iterations per batch. The presence of null alleles, large allele dropout, and scoring bias were assessed using Microchecker v2.2.3 (van Oosterhout et al. 2004).
Raw estimates of population structure were calculated locus-by-locus and as an average over 11 loci (Lun34 was excluded due to >5% missing data) using AMOVA in Arlequin with 1000 permutations (Excoffier et al. 1992). Four individuals missing data at more than three loci were excluded from raw estimates of population differentiation, leaving a sample (n = 105) in which all individuals had at least eight loci and each locus <5% missing data.
Null allele frequencies and associated Excluding Null Alleles (ENA) corrected estimates of population structure were calculated in Freena (Chapuis and Estoup 2007). In this analysis, missing data were regarded as null homozygotes. Estimators of actual differentiation (Dest), which have been indicated as particularly suitable in estimating population structure in the presence of high heterozygosities and small sample size (Jost 2008), were also calculated, using the web-based algorithm Smogd v1.2.5 (Crawford 2010).
To further investigate population structures inferred through conventional analytical approaches, a discriminant analysis of principal components (DAPC) (Jombart et al. 2010) was run on all 12 microsatellite loci. This multivariate method is designed to extract information from genetic datasets and assign genotypes to predefined clusters (Jombart et al. 2010). In DAPC, a linear discriminant analysis (DA) is conducted on genotypic information, previously transformed into uncorrelated components through a principal component analysis (PCA) (Jombart et al. 2010). In doing so, the shortcomings of both PCA and DA in their applicability to genotypic datasets are overcome (Jombart et al. 2010). This method allows the retention of a significant proportion of the genetic variability while it minimizes within- and maximizes between-population variance (Jombart et al. 2010). Furthermore, DAPC is robust to deviations from HWE and LD and is as sensitive—but not as computationally intensive—as Bayesian clustering approaches (Jombart et al. 2010). In DAPC, the tradeoff between obtaining stable results and explaining most genetic variability is still under debate (T. Jombart, pers. comm.). In the present analysis, 73 PCs were retained in the DA, accounting for 90% of the genotypic variability. DAPC was implemented in R v2.12 (http://www.Rproject.org) using functions dudi.pca and dapc from the R packages ade4, adegenet, and MASS. The results were visualized in a scatterplot generated by adegenet (Jombart 2008; Jombart et al. 2010).
Results
Ecology of hybridization
Chaetodon trifasciatus and C. lunulatus have very similar ecologies, reflected in strongly overlapping patterns of abundance and dietary composition. Chaetodon trifasciatus (average depth 7 m ± 0.62 SE) and C. lunulatus (average depth 7.1 m ± 0.66 SE) occupied relatively narrow, largely overlapping depth ranges (4–15 m and 3–18 m, respectively) and were most frequently (70% and 67% of individuals, respectively) encountered at 5–8 m depth (Fig. 3A). There was no significant difference in the depth distributions of C. trifasciatus (n = 34) and C. lunulatus (n = 30) (ANOVA, df = 1, F = 0.006, P = 0.94) (Fig. 3A). While hybrids tended to be found slightly deeper than the parent species (8.4 m ± 1.3 SE), the depth distribution of hybrids (n = 8) did not significantly differ from those of their parents (ANOVA, df = 2, F = 0.489, P = 0.615) (Fig. 3A).
Figure 3.
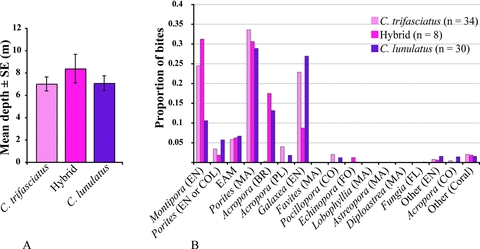
(A) Mean depth distribution (± SE) of C. trifasciatus (n = 34), C. lunulatus (n = 30), and hybrid (n = 8) on the North coast of Christmas Island. (B) Proportional dietary composition of the parental species and hybrid at Christmas Island. Data are based on direct 3-min feeding observations of each species and the hybrid. Coral food categories are grouped by genus and growth form: encrusting (EN), columnar (COL), massive (MA), branching (BR), plate (PL), corymbose (CO), foliose (FO), and free living (FL). The category other (EN) includes encrusting corals not belonging to the specified encrusting genera. The category other (Coral) includes all other, nonencrusting coral.
Chaetodon trifasciatus (n = 34) and C. lunulatus (n = 30) each fed on a broad range (11 and 13, out of 17, respectively) of prey items, and there was no significant difference in the relative use of these preys (MANOVA, Pillai's Trace = 0.715, hypothesis df = 32, P = 0.062) (Fig. 3B). Over 50% of the diet of C. trifasciatus and C. lunulatus was constituted of massive Porites and encrusting Galaxea corals (Fig. 3B). Chaetodon trifasciatus and C. lunulatus also frequently consumed encrusting Montipora corals (25% and 10%, respectively). The hybrids (n = 8) were observed feeding mostly on encrusting Montipora (> 30%) and massive Porites (> 30%), and utilized nine of 17 food categories (eight corals and EAM) (Fig. 3B). Other major coral genera consumed by the hybrids included branching Acropora (> 15%), Galaxea (< 10%), and Echinopora (< 5%).
The feeding rates of both species and their hybrid were also comparable. Chaetodon trifasciatus took an average of 29 (± 2.8 SE) bites over 3 min, C. lunulatus 24 (± 2.1 SE), and the hybrids 36 (± 5 SE) bites (ANOVA, df = 2, F = 2.436, P = 0.09). Furthermore, both species and the hybrid consumed an average of four (± 0.29 SE, ± 0.26 SE, ± 0.18 SE, respectively) prey types over 3 min. Moreover, when observed in mixed pairs, individuals of different species were often feeding on the same prey in largely overlapping areas.
Abundance
Underwater visual surveys indicated both parent species were relatively uncommon, and there was no significant difference in abundance of the parental species (t-test, t = 1.285, df = 38, P = 0.207) (Fig. 4A). The average abundance (individuals per 3000 m2) of C. trifasciatus was two (± 0.60 SE) and C. lunulatus was one (± 0.48 SE) (Fig. 4A). Hybrids were even less abundant than their parents (t-test, t = 3.552, df = 42, P = 0.001) (Fig. 4A). The average density of hybrids was approximately one individual per 12,000 m2 (Fig. 4A). Overall, only eight hybrids were encountered in all surveys—which covered >170,000 m2 of reef habitat at Christmas Island.
Figure 4.
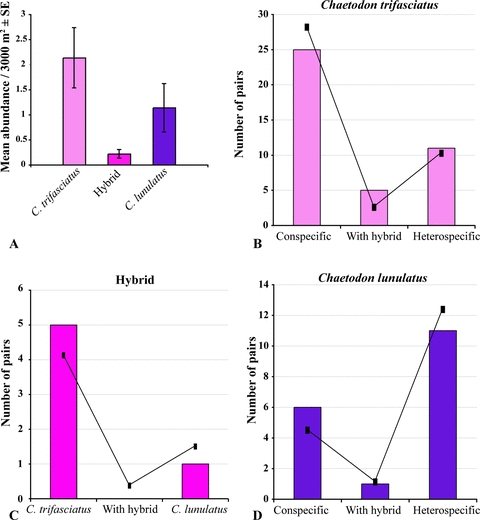
(A) Mean abundance (per 3000 m2± SE) of the parental species and hybrid along the North coast of Christmas Island. (B), (C), and (D) Expected (lines) and observed (bars) pairing frequencies of (B) C. trifasciatus, (C) hybrids, and (D) C. lunulatus at Christmas Island. Expected frequencies were calculated based on observed abundances of paired individuals.
Assortative pairing
The pairing frequencies of both species and the hybrids did not differ significantly from expectations based on relative abundances (C. trifasciatus: χ2 = 2.73, df = 2, P = 0.26; C. lunulatus: χ2 = 0.67, df = 2, P = 0.71; Hybrids: χ2 = 0.73, df = 2, P = 0.69) (Figs. 4B–D), indicating that members of this complex pair nonassortatively. Chaetodon trifasciatus (paired individuals, n = 41) paired conspecifically in more than 60% of cases, heterospecifically with C. lunulatus in almost 27% and with hybrids in 12% of pairs (Fig. 4B).
Chaetodon lunulatus (paired individuals, n = 18) paired conspecifically in 33% of cases, more frequently with C. trifasciatus (61%) and with hybrids in 5% of pairs (Fig. 4D). The hybrids (paired individuals, n = 6) were never observed together in a pair, but formed pairs with both parents, most frequently with C. trifasciatus (83%) (Fig. 4C).
Genetics of hybridization
Five hundred and fifty-two basepair of the mitochondrial cyt b region were resolved for a total of 105 individuals in the complex (details provided in Table 2). Of the 522 bp sequenced, 114 parsimony informative sites and 29 individual haplotypes were identified (Fig. 5A).
Table 2.
Sample sizes (total n = 105) for cyt b analyses, number of haplotypes (nh), haplotype diversities (h), nucleotide diversities (π) of cyt b for all populations in the species complex
| Population | n | nh | h | π |
|---|---|---|---|---|
| Zanzibar C. trifasciatus | 10 | 6 | 0.89 | 0.007 |
| Cocos Is. C. trifasciatus | 28 | 10 | 0.82 | 0.002 |
| Christmas Is. C. trifasciatus | 28 | 8 | 0.71 | 0.001 |
| Christmas Is. Hybrid | 9 | 4 | 0.58 | 0.001 |
| Christmas Is. C. lunulatus | 15 | 8 | 0.83 | 0.005 |
| Marshall Is. C. lunulatus | 15 | 8 | 0.83 | 0.006 |
Figure 5.
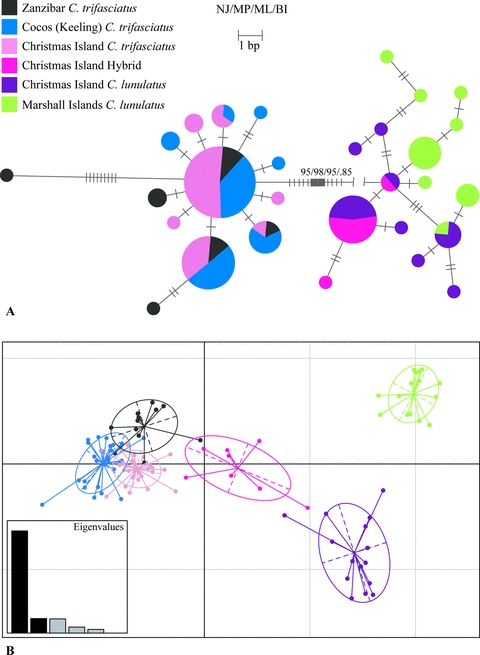
(A) Scaled branch length minimum spanning network (MSN) depicting the genealogical relationships between haplotypes in the Chaetodon trifasciatus complex. Haplotypes are represented by scaled circles showing the origin and number of individuals sharing a haplotype. Branches are to scale with number of substitutions (bp) and perpendicular bars on branches are substitution counts (thin line = 1 substitution; thick line = 10 substitutions). The branch separating the two clades has been truncated for ease of representation. Bootstrap support values for phylogenetic relationships inferred by neighbor-joining (NJ), maximum parsimony (MP) and maximum likelihood (ML), and posterior probabilities from Bayesian Inference of phylogeny (BI) are shown along the main branch. (B) Scatterplot of the discriminant analysis of principal components (DAPC) (Jombart et al. 2010) performed on 12 microsatellite loci for six populations of the C. trifasciatus complex. Taxon and geographical origin of each population are indicated in the legend and depicted in the plot by colors and 95% inertia ellipses. Individual genotypes are represented by dots. The x and y axes represent the first two discriminant functions, respectively. The plot of eigenvalues shows the amount of genetic information retained by each successive discriminant function.
Summary statistics for 12 microsatellite loci are presented in Table 3. Hybrids showed the lowest allelic diversity and Christmas Island C. trifasciatus the highest. Significant single-locus departures from HWE were detected in 12 of 72 tests at population level before sequential Bonferroni correction and nine afterwards (α = 0.0083) (Table 3). Null alleles might contribute to departures from HWE in loci Lun10, Lun29, and Lun36. Chaetodon trifasciatus populations of Christmas and Cocos (Keeling) Islands had the highest number of private alleles (13 and 10, respectively), while hybrids had none (Table 3).
Table 3.
Summary statistics for 12 microsatellite loci used in population genetic analyses. Sample sizes (n), observed number of alleles (Na), the average inbreeding coefficient (FIS), observed number of private alleles (Pa), observed heterozygosity (HO), expected heterzygosity (HE), and probability of departure from HWE for each locus at each population (p)
| Locus | Christmas Island C. lunulatus (n = 15) | Christmas Island Hybrids (n = 8) | Christmas Island C. trifasciatus (n = 31) | Marshall Islands C. lunulatus (n = 15) | Cocos (Keeling) Islands C. trifasciatus (n = 28) | Zanzibar C. trifasciatus (n = 12) |
|---|---|---|---|---|---|---|
| Lun03 | Na = 13 | Na = 13 | Na = 21 | Na = 11 | Na = 19 | Na = 15 |
| Pa = 1 | Pa = 0 | Pa = 3 | Pa = 0 | Pa = 4 | Pa = 1 | |
| HO = 0.867 | HO = 0.875 | HO = 0.964 | HO = 0.733 | HO = 0.821 | HO = 0.750 | |
| HE = 0.818 | HE = 0.914 | HE = 0.940 | HE = 0.891 | HE = 0.907 | HE = 0.913 | |
| FIS = 0.057 | FIS = 0.109 | FIS = –0.008 | FIS = 0.210 | FIS = 0.112 | FIS = 0.220 | |
| P = 0.132 | P = 0.178 | P = 0.426 | P = 0.169 | P = 0.112 | P = 0.01* | |
| Lun05 | Na = 9 | Na = 7 | Na = 8 | Na = 6 | Na = 9 | Na = 8 |
| Pa = 0 | Pa = 0 | Pa = 1 | Pa = 0 | Pa = 0 | Pa = 0 | |
| HO = 0.857 | HO = 0.875 | HO = 0.815 | HO = 0.867 | HO = 0.786 | HO = 0.833 | |
| HE = 0.839 | HE = 0.836 | HE = 0.774 | HE = 0.807 | HE = 0.810 | HE = 0.802 | |
| FIS = 0.016 | FIS = 0.020 | FIS = –0.033 | FIS = –0.040 | FIS = 0.048 | FIS = 0.005 | |
| P = 0.670 | P = 0.836 | P = 0.003** | P = 0.698 | P = 0.097 | P = 0.705 | |
| Lun07 | Na = 11 | Na = 12 | Na = 22 | Na = 12 | Na = 18 | Na = 14 |
| Pa = 0 | Pa = 0 | Pa = 1 | Pa = 1 | Pa = 0 | Pa = 0 | |
| HO = 0.667 | HO = 1.000 | HO = 0.806 | HO = 1.000 | HO = 0.929 | HO = 1.000 | |
| HE = 0.818 | HE = 0.898 | HE = 0.932 | HE = 0.876 | HE = 0.927 | HE = 0.910 | |
| FIS = 0.218 | FIS = –0.047 | FIS = 0.151 | FIS = –0.108 | FIS = 0.017 | FIS = –0.056 | |
| P = 0.000** | P = 1.000 | P = 0.17 | P = 0.855 | P = 0.045* | P = 0.627 | |
| Lun08 | Na = 14 | Na = 8 | Na = 15 | Na = 14 | Na = 16 | Na = 15 |
| Pa = 0 | Pa = 0 | Pa = 0 | Pa = 1 | Pa = 1 | Pa = 1 | |
| HO = 0.867 | HO = 0.750 | HO = 0.929 | HO = 1.000 | HO = 1.000 | HO = 0.917 | |
| HE = 0.902 | HE = 0.844 | HE = 0.913 | HE = 0.900 | HE = 0.917 | HE = 0.899 | |
| FIS = 0.074 | FIS = 0.176 | FIS = 0.001 | FIS = –0.077 | FIS = –0.071 | FIS = 0.024 | |
| P = 0.259 | P = 0.346 | P = 0.235 | P = 0.267 | P = 0.961 | P = 0.733 | |
| Lun09 | Na = 2 | Na = 3 | Na = 8 | Na = 3 | Na = 7 | Na = 3 |
| Pa = 0 | Pa = 0 | Pa = 2 | Pa = 1 | Pa = 1 | Pa = 0 | |
| HO = 0.286 | HO = 0.875 | HO = 0.600 | HO = 0.533 | HO = 0.536 | HO = 0.333 | |
| HE = 0.245 | HE = 0.633 | HE = 0.571 | HE = 0.504 | HE = 0.605 | HE = 0.288 | |
| FIS = –0.130 | FIS = –0.324 | FIS = –0.034 | FIS = –0.023 | FIS = 0.132 | FIS = –0.114 | |
| P = 1.000 | P = 0.608 | P = 0.742 | P = 1.000 | P = 0.037* | P = 1.000 | |
| Lun10 | Na = 12 | Na = 6 | Na = 13 | Na = 15 | Na = 15 | Na = 12 |
| Pa = 0 | Pa = 0 | Pa = 0 | Pa = 3 | Pa = 4 | Pa = 2 | |
| HO = 0.786 | HO = 0.571 | HO = 0.393 | HO = 1.000 | HO = 0.480 | HO = 0.500 | |
| HE = 0.890 | HE = 0.776 | HE = 0.873 | HE = 0.907 | HE = 0.840 | HE = 0.861 | |
| FIS = 0.154 | FIS = 0.333 | FIS = 0.563 | FIS = –0.069 | FIS = 0.445 | FIS = 0.455 | |
| P = 0.297 | P = 0.242 | P = 0.000** | P = 0.870 | P = 0.000** | P = 0.000** | |
| Lun14 | Na = 5 | Na = 3 | Na = 5 | Na = 2 | Na = 5 | Na = 4 |
| Pa = 2 | Pa = 0 | Pa = 0 | Pa = 0 | Pa = 0 | Pa = 0 | |
| HO = 0.533 | HO = 0.750 | HO = 0.571 | HO = 0.400 | HO = 0.571 | HO = 0.455 | |
| HE = 0.538 | HE = 0.508 | HE = 0.551 | HE = 0.391 | HE = 0.591 | HE = 0.442 | |
| FIS = 0.043 | FIS = –0.424 | FIS = –0.019 | FIS = 0.012 | FIS = 0.052 | FIS = 0.020 | |
| P = 0.681 | P = 0.627 | P = 0.520 | P = 1.000 | P = 0.336 | P = 0.405 | |
| Lun19 | Na = 11 | Na = 11 | Na = 15 | Na = 10 | Na = 13 | Na = 10 |
| Pa = 1 | Pa = 0 | Pa = 0 | Pa = 0 | Pa = 0 | Pa = 0 | |
| HO = 0.800 | HO = 0.875 | HO = 0.871 | HO = 0.867 | HO = 0.893 | HO = 0.818 | |
| HE = 0.887 | HE = 0.875 | HE = 0.887 | HE = 0.864 | HE = 0.902 | HE = 0.851 | |
| FIS = 0.132 | FIS = 0.067 | FIS = 0.035 | FIS = 0.032 | FIS = 0.028 | FIS = 0.086 | |
| P = 0.192 | P = 0.576 | P = 0.455 | P = 0.497 | P = 0.014* | P = 0.321 | |
| Lun21 | Na = 14 | Na = 10 | Na = 17 | Na = 11 | Na = 13 | Na = 11 |
| Pa = 0 | Pa = 0 | Pa = 2 | Pa = 1 | Pa = 0 | Pa = 1 | |
| HO = 0.929 | HO = 1.000 | HO = 0.929 | HO = 0.800 | HO = 0.889 | HO = 0.917 | |
| HE = 0.908 | HE = 0.844 | HE = 0.899 | HE = 0.860 | HE = 0.905 | HE = 0.878 | |
| FIS = 0.015 | FIS = –0.120 | FIS = –0.015 | FIS = 0.104 | FIS = 0.037 | FIS = 0.000 | |
| P = 0.413 | P = 0.800 | P = 0.777 | P = 0.278 | P = 0.372 | P = 0.622 | |
| Lun29 | Na = 8 | Na = 10 | Na = 20 | Na = 13 | Na = 18 | Na = 13 |
| Pa = 0 | Pa = 0 | Pa = 0 | Pa = 1 | Pa = 0 | Pa = 0 | |
| HO = 0.857 | HO = 0.875 | HO = 0.714 | HO = 1.000 | HO = 0.704 | HO = 0.833 | |
| HE = 0.821 | HE = 0.859 | HE = 0.932 | HE = 0.884 | HE = 0.931 | HE = 0.906 | |
| FIS = –0.006 | FIS = 0.049 | FIS = 0.251 | FIS = –0.097 | FIS = 0.262 | FIS = 0.124 | |
| P = 0.438 | P = 0.734 | P = 0.001** | P = 0.480 | P = 0.001** | P = 0.282 | |
| Lun34 | Na = 3 | Na = 6 | Na = 9 | Na = 3 | Na = 7 | Na = 8 |
| Pa = 0 | Pa = 0 | Pa = 2 | Pa = 0 | Pa = 0 | Pa = 1 | |
| HO = 0.600 | HO = 0.875 | HO = 0.840 | HO = 0.467 | HO = 0.833 | HO = 0.909 | |
| HE = 0.558 | HE = 0.750 | HE = 0.821 | HE = 0.451 | HE = 0.809 | HE = 0.814 | |
| FIS = –0.041 | FIS = –0.101 | FIS = –0.003 | FIS = 0.000 | FIS = –0.009 | FIS = –0.070 | |
| P = 0.632 | P = 0.707 | P = 0.09 | P = 1.000 | P = 0.386 | P = 0.727 | |
| Lun36 | Na = 17 | Na = 7 | Na = 15 | Na = 15 | Na = 9 | Na = 8 |
| Pa = 1 | Pa = 0 | Pa = 2 | Pa = 0 | Pa = 0 | Pa = 0 | |
| HO = 0.933 | HO = 0.625 | HO = 0.621 | HO = 0.600 | HO = 0.667 | HO = 0.750 | |
| HE = 0.929 | HE = 0.648 | HE = 0.705 | HE = 0.916 | HE = 0.580 | HE = 0.663 | |
| FIS = 0.030 | FIS = 0.103 | FIS = 0.136 | FIS = 0.375 | FIS = –0.130 | FIS = –0.088 | |
| P = 0.693 | P = 0.454 | P = 0.008** | P = 0.000** | P = 0.837 | P = 0.971 |
P < 0.05
P < 0.0083 after sequential Bonferroni correction (highlighted in bold).
Phylogenetic analyses
Phylogenetic relationships were inferred using four methods, which all produced highly congruent tree topologies. The bootstrap support values from NJ, MP, ML, and the posterior probabilities from BI are reported on the scaled branch MSN depicting genealogical relationships between haplotypes (Fig. 5A). The main partition was well supported across all analyses, identifying a clear separation between the two parental clades (C. trifasciatus and C. lunulatus) with 29 fixed nucleotide changes separating the species (5% divergence at cyt b). Hybrids shared haplotypes with the Pacific Ocean C. lunulatus parental clade only, indicating a unidirectional maternal contribution to hybridization (Fig. 5A). No evidence of introgression was found between parental species in this complex (Fig. 5A).
Population genetic analyses
Molecular diversity indices for mitochondrial cyt b are presented in Table 2. The Zanzibar population of C. trifasciatus had the highest haplotype (h) and nucleotide (π) diversities, C. lunulatus from within the Christmas Island hybrid zone had the second highest h and third highest π (Table 2). For the Indian Ocean C. trifasciatus, h and π were lower as geographic distance from the hybrid zone decreased, and lowest at Christmas Island (Table 2). Conversely, the Pacific Ocean sister species, C. lunulatus, had the lowest π in the population furthest away from the hybrid zone—but the two populations had same h (Table 2). Hybrids had the lowest π and the second lowest h of all populations in this complex (Table 2).
The level of genetic population differentiation was high for all comparisons (Tables 4, 5). The AMOVA fixation index for the mitochondrial cyt b marker was Φst = 0.905, P < 0.0001. The microsatellite data also revealed a high level of population structure (raw Φst = 0.055, P < 0.0001; Dest = 0.194 P < 0.0001) and raw values were comparable to those corrected for null alleles (Table 5). For cyt b and the 12 microsatellite loci, nearly all pairwise comparisons were significant (Fst and Dest, respectively) (Table 4). Not surprisingly, there was clear genetic structuring between the parental species (Table 4). Conversely, cyt b analyses failed to detect significant population structure between C. trifasciatus samples from Christmas and Cocos (Keeling) Islands (Table 4A). The microsatellite data, in this respect, confirm the lack of structure between these two populations, and indicate that such is the case for C. trifasciatus populations across the whole Indian Ocean, including Zanzibar (Table 4B). Populations of C. lunulatus from separate ocean basins showed significant population structure, according to cyt b and microsatellite data (Table 4). The hybrid population significantly differed from all other populations (Table 4). Also to note is the clear structure in both mitochondrial and nuclear data between the parental populations from within the hybrid zone (Table 4), despite the apparent hybridization—indicating lack of introgression.
Table 4.
Pairwise population comparisons: (A) Fst generated from 552 bp of mitochondrial cyt b gene (hybrid population N = 9 was omitted due to sample size); (B) estimator of actual differentiation (Dest) (Jost 2008) based on 12 microsatellite loci and corresponding P values (upper diagonal)
| CL (CI) | CL (RMI) | HYB (CI) | CT (CK) | CT (CI) | CT (Z) | |
|---|---|---|---|---|---|---|
| A | ||||||
| CL (CI) | 0.009 | 0.017 | <0.001 | <0.001 | <0.001 | |
| CL (RMI) | 0.168 | 0.006 | <0.001 | <0.001 | <0.001 | |
| HYB (CI) | 0.190 | 0.284 | <0.001 | <0.001 | <0.001 | |
| CT (CK) | 0.945 | 0.958 | 0.968 | 0.428 | 0.037 | |
| CT (CI) | 0.950 | 0.963 | 0.975 | 0.000 | 0.015 | |
| CT (Z) | 0.901 | 0.922 | 0.931 | 0.076 | 0.099 | |
| B | ||||||
| CL (CI) | 0.018 | <0.001 | <0.001 | <0.001 | <0.001 | |
| CL (RMI) | 0.005 | <0.001 | <0.001 | <0.001 | <0.001 | |
| HYB (CI) | 0.096 | 0.121 | <0.001 | 0.009 | <0.001 | |
| CT (CK) | 0.328 | 0.375 | 0.050 | 0.135 | 0.117 | |
| CT (CI) | 0.301 | 0.363 | 0.047 | 0.016 | 0.694 | |
| CT (Z) | 0.289 | 0.241 | 0.011 | 0.003 | 0.000 | |
Significant comparisons are highlighted in bold.
CL = C. lunulatus; CT = C. trifasciatus and HYB = hybrids; CI = Christmas Island; CK = Cocos (Keeling) Islands; RMI = Marshall Islands and Z = Zanzibar.
Table 5.
Raw population differentiation from microsatellite allele frequencies , population differentiation corrected for null allele frequencies using the ENA correction of Chapuis and Estoup (2007), and estimator of actual differentiation (Jost 2008) (Dest); results are presented locus-by-locus and as an average over 12 loci. All values are significant to the 95% confidence interval
| Locus | Raw | ENA corrected | Dest |
|---|---|---|---|
| Lun05 | 0.033 | 0.037 | 0.161 |
| Lun08 | 0.009 | 0.008 | 0.112 |
| Lun09 | 0.298 | 0.296 | 0.472 |
| Lun21 | 0.010 | 0.009 | 0.115 |
| Lun14 | 0.048 | 0.049 | 0.052 |
| Lun19 | 0.017 | 0.016 | 0.164 |
| Lun34 | 0.096 | 0.100 | 0.291 |
| Lun10 | 0.053 | 0.033 | 0.475 |
| Lun29 | 0.021 | 0.023 | 0.294 |
| Lun03 | 0.017 | 0.016 | 0.258 |
| Lun07 | 0.035 | 0.032 | 0.374 |
| Lun36 | 0.080 | 0.081 | 0.260 |
| Average over 12 loci | 0.0551 | 0.054 | 0.194 |
11 loci only: locus Lun34 was excluded due to >5% missing data.
The clear genetic population structuring shown by traditional analyses of both mtDNA and nuclear DNA was also visible in the DAPC of 12 microsatellite loci (Fig. 5B). The two parental species were clearly clustered within species and separated from each other regardless of sampling location (Fig. 5B). Populations of C. lunulatus—Christmas and Marshall Islands—sampled from separate ocean basins showed clear separation, while populations of C. trifasciatus from the Indian Ocean—as far apart as Christmas Island and Zanzibar—were much less clearly separated irrespective of geographical distance (Fig. 5B). The hybrid population was distinct from all others, and genotypes of this population were intermediate between those of the parental species (Fig. 5B).
Discussion
This study genetically confirmed hybridization between the butterflyfishes C. trifasciatus and C. lunulatus at Christmas Island, determined the evolutionary consequences of this process and identified ecological conditions that favor hybridization. Direct field observations suggest that low abundances of both species and nonassortative mating may have promoted interbreeding. Furthermore, genetic analyses of mtDNA and microsatellite loci have confirmed that distinct and intermediate color morphs are hybrids, and shown that there is a unidirectional maternal contribution to hybridization and no introgression between the parental species. These findings suggest that the production of viable hybrids between C. trifasciatus and C. lunulatus is relatively rare. Apparently hybridization may facilitate persistence of the Pacific Ocean species at Christmas Island, but this may be at the expense of the Indian Ocean species at this isolated location.
Ecology of hybridization
The recently diverged, sister species C. trifasciatus and C. lunulatus (Bellwood et al. 2010) have largely allopatric distributions (Allen et al. 1998) and occur in sympatry at Christmas Island, where they form heterospecific pairs. In butterflyfishes, cases of hybridization between allopatric sister species that come into secondary contact, such as this one, represent the minority—13 of 35 (37%) (Hobbs et al. In press). Sympatry of Indian and Western Pacific Ocean species at Christmas Island has been considered a potential indication of westward dispersal of Pacific Ocean taxa (Allen and Steene 1979; Blum 1989), has set the scene for previously reported cases of reef fish hybridization (Marie et al. 2007; Hobbs et al. 2009) and is a precursor for hybridization between C. trifasciatus and C. lunulatus.
At Christmas Island, C. trifasciatus and C. lunulatus have very similar ecologies (dietary composition and habitat use), which increases the chance of heterospecific encounters. Ecological similarities and habitat overlap among recently diverged species are not uncommon, and act to increase (or at least do not limit) social interactions between hybridizing reef fishes (Randall 1956; Fischer 1980; Frisch and van Herwerden 2006; Yaakub et al. 2006, 2007; Marie et al. 2007). Both C. trifasciatus and C. lunulatus are known obligate corallivores (Harmelin–Vivien 1989; Pratchett et al. 2004; Berumen et al. 2005; Pratchett 2005; Cole et al. 2008) and accordingly, at Christmas Island, both species feed almost exclusively on scleractinian corals, mostly Porites, Galaxea, and Montipora. The dietary composition and feeding rates were not significantly different between these two species, indicating a high degree of dietary overlap (see also Randall 1956; Feddern 1968; Fischer 1980). If there were inconsistencies in the dietary composition, combined with broadly nonoverlapping distributions of respective prey, this might prevent pairing and subsequent reproduction between these species.
Chaetodon trifasciatus and C. lunulatus were rare at Christmas Island and in both cases their abundances were one to three orders of magnitude lower compared to any other location for which abundance data are available (Adrim and Hutomo 1989; Findley and Findley 2001; Pratchett et al. 2004, 2006b; Pereira and Videira 2005). Rarity of one or both parental species plays a significant role in hybrid formation in reef fishes (Randall et al. 1977; Fisher 1980; Pyle and Randall 1994; Frisch and van Herwerden 2006; Yaakub et al. 2006; Marie et al. 2007; Maruska and Peyton 2007; Hobbs et al. 2009). Furthermore, one or both hybridizing species have been reported as rare in 11 (58%) of 19 locations where other butterflyfishes hybridize (Hobbs et al. In press).
Chaetodon trifasciatus and C. lunulatus form heterospecific pairs at Christmas Island indicating that assortative mating has broken down between these two sister species, probably as a consequence of their local rarity. Pairing in this species complex most likely occurs for reproduction, because C. lunulatus has been deemed a monogamous breeder (Yabuta 1997) and pair formation corresponds with the onset of sexual maturity (Pratchett et al. 2006a).
The nominal hybrids in this complex—initially identified through aberrant markings, intermediate to those of their parents (Hobbs et al. 2009)—had spatial and dietary ecologies comparable to those of their putative parents. Moreover, hybrids were observed in breeding pairs with both parent species, potentially facilitating gene transfer between these species—if hybrids are fertile. Importantly, hybrids in this complex were relatively rare and the statistical power associated with their data is therefore limited. Nevertheless, rarity of hybrids is, per se, interesting and might simply reflect the relative rarity of their parents, or be indicative of some selective pressure on hybrids.
Genetics of hybridization
The use of both mtDNA and microsatellite loci allowed confirmation of the hybrid status of intermediately colored individuals, and shed light on the potential evolutionary consequences of hybridization in this complex. The mitochondrial cyt b identified two distinct parental clades, separated by 5% (29 fixed substitutions out of 522 nucleotides). The genetic distance between the species clades is consistent with divergence of these species, some 2.5–4 Ma (Bellwood et al. 2010), given the cyt b mutation rate of 1–2.5% Ma−1 (McMillan and Palumbi 1997) and assuming a molecular clock. Hybridization is most common between species that diverge by less than 10% because the genetic similarity increases the chance of viable hybrid formation (Mallet 2005). Each parental clade contained all individuals of the respective species, from all geographical locations. Hybrids shared cyt b haplotypes with the Pacific Ocean C. lunulatus only, indicating a unidirectional contribution to hybridization, in which C. lunulatus appears to always be the mother.
Despite informing about the matrilineal contribution, the results from mtDNA analyses were not sufficient to confirm the hybrid status of the intermediately colored individuals, which could simply be an aberrant color pattern of C. lunulatus. However, nuclear microsatellite markers ruled out this scenario and showed that hybrid genotypes were distinct from (and intermediate to) those of C. trifasciatus and C. lunulatus. This confirms the hybrid status and suggests that intermediately colored individuals were most likely F1 hybrids. The unidirectional maternal contribution contrasts with findings of other reef fish hybridization studies (McMillan et al. 1999; van Herwerden et al. 2006; Yaakub et al. 2006; Marie et al. 2007), which found that maternal contribution was either bidirectional or biased toward the more abundant species. This may suggest that hybrids resulting from the opposite cross (C. trifasciatus females) are subject to some negative selection or that female C. lunulatus are actively choosing (i.e., female choice, Wirtz 1999) to mate with C. trifasciatus males, because of lack of conspecifics. This latter explanation seems more likely because, even though abundances of the parental species in this complex were not statistically different, C. lunulatus was observed less frequently than C. trifasciatus, and in previous censuses of the ichthyofauna of Christmas Island it was not recorded (Allen et al. 2007; but see Hobbs et al. 2009; Hobbs et al. 2010).
The apparent lack of introgression between C. trifasciatus and C. lunulatus could be explained by the extreme rarity of the hybrids or their infertility. F1 hybrids of this complex sampled at Christmas Island previously to this study were sexually mature (J-P. Hobbs, unpublished data), henceforth the rarity of these individuals seems to be the most likely explanation. Three conditions are required for introgression to take place: (1) a fertile hybrid with the mtDNA of one species must (2) pair with an individual of the other species and (3) produce viable offspring (see Fig. 8 in Yaakub et al. 2006). These three conditions have to be sequentially satisfied frequently enough for the genetic signal of introgression to perpetuate and be detected. The hybrids in this complex may not be abundant enough to meet the conditions for introgression. In addition, C. lunulatus appears to be a recent colonist to Christmas Island (Hobbs et al. 2010) and there may not have been sufficient time for introgression to be evident in the genetic composition of these species. Lack of introgression contrasts with the results of most reef fish hybridization studies (van Herwerden and Doherty 2006—northern hybrid zone; van Herwerden et al. 2006; Yaakub et al. 2006; Marie et al. 2007; but see van Herwerden and Doherty 2006—southern hybrid zone; Yaakub et al. 2007) and, more importantly, with the findings of other studies of butterflyfish hybridization (McMillan et al. 1999).
The different divergence ages of parental species might account for the dissimilarities found in the outcome of hybridization in reef fishes. At the Solomon Islands, C. punctatofasciatus hybridizes with C. pelewensis (McMillan et al. 1999) from which it differs by only 0.7% of the mitochondrial cyt b (McMillan and Palumbi 1995). Bidirectional introgression between these species is so extensive that, within the hybrid zone, hybrid phenotypes account for over 70% of the individuals (McMillan et al. 1999). Moreover, despite phenotypic differentiation of the two species evident outside the hybrid zone, their genetic homogeneity extends for thousands of kilometers beyond the hybrid zone (McMillan et al. 1999) as a result of extensive bidirectional introgression. Conversely, taxa in the C. trifasciatus complex show a clear mtDNA break and hybridization between species is unidirectional and lacks introgression, with apparently more localized, limited evolutionary consequences. From the data available, it appears that increased genetic divergence between hybridizing butterflyfishes signifies a higher fitness cost to hybridization and limits introgression.
This is consistent with other reef fish hybridization studies in which the authors found that introgression was characteristic of more recently diverged taxa (Doherty et al. 1994; Planes and Doherty 1997a, b; van Herwerden and Doherty 2006; van Herwerden et al. 2006; Yaakub et al. 2006, 2007; Marie et al. 2007). For example in the Labridae, hybridization between Thalassoma jansenii and T. quinquevittatum-–for which divergence is dated at 5–10 Ma and genetic distance less than 2% at cyt b (Bernardi et al. 2004)—was bidirectional and introgressive (Yaakub et al. 2006), whereas in the relatively older species Halichoeres garnoti and H. bivittatus (11–18 Ma, genetic break >5.5% based on three mtDNA markers) (Barber and Bellwood 2005) hybridization was deemed to have limited evolutionary significance, due to the apparent lack of introgression and rarity of hybrids (Yaakub et al. 2007).
Similarly, the hybridizing color morphs of Acanthochromis polyacanthus in the northern hybrid zone on the Great Barrier Reef (GBR), not differentiated by allozyme data (Planes and Doherty 1997b), showed high levels of mtDNA Hypervariable Region 1 (HVR1) introgression (van Herwerden and Doherty 2006). In contrast, the hybridizing southern GBR A. polyacanthus color morph, divergent from all other populations by 2.8% mtDNA cyt b (Planes et al. 2001), showed a distinct allozyme structure (Planes and Doherty 1997a) and negligible introgression in the HVR1 (van Herwerden and Doherty 2006).
Further, introgression and bidirectional maternal contribution were found in hybridizing surgeonfishes Acanthurus leucosternon and A. nigricans, for which 1% genetic break was reported in the mtDNA COI marker used (Marie et al. 2007). In hybridizing groupers, Plectropomus leopardus and P. maculatus, separated by 1% genetic break based on two nuclear and two mtDNA markers (Craig and Hastings 2007), hybridization was highly introgressive, but the maternal contribution was unidirectional (van Herwerden et al. 2006). Based on the available data, it seems that for reef fishes a genetic break smaller than 2%—at a range of mtDNA loci other than HVR1—results in introgressive hybridization, which can have evolutionary consequences extending outside the hybrid zone and may result in increased genetic diversity and adaptability (e.g., McMillan et al. 1999). Conversely, a genetic break larger than 5%, albeit still lower than the proposed 10% cutoff for successful hybrid formation in the terrestrial environment (Mallet 2005), seems to result in a higher cost to hybridization and a reduction in the number of viable, fertile hybrids.
In addition to evolutionary relatedness, ecological factors may influence the occurrence of hybridization. Sustained pressure on coral reefs worldwide, which decreases available habitat and negatively affects the abundance of some reef fishes (e.g., Pratchett et al. 2006b; Graham 2007), might result in increased habitat overlap and local rarity of species—the ecological conditions most frequently ascribed a role conducive to hybridization in reef fishes. Further, ocean acidification negatively impacts mate recognition in reef fishes (Munday et al. 2009), increasing the chances of heterospecific breeding. While increased cases of hybridization between recently diverged reef fishes may prove beneficial to the adaptability of the species involved, a greater number of cases of hybridization among more distantly related species might have a detrimental effect and it may also result in reverse speciation (two species become one).
Evolutionary consequences of hybridization
The genetic diversity of C. trifasciatus is relatively low within the hybrid zone. Low genetic diversity can have a detrimental effect on the adaptability of species to novel environments (Rhymer and Simberloff 1996), and in this case might result in the local extinction of C. trifasciatus. For this species, hybridization might represent a significant cost. Indicative of this is the apparent lack of maternal contribution to hybridization from C. trifasciatus. Female C. trifasciatus form pairs with C. lunulatus males (J-P. Hobbs, unpublished data) and it seems therefore possible that the resulting hybrids may not be viable, and heterospecific mating may represent a significant reproductive cost. Moreover, the hybrids of this complex that do survive (those with C. lunulatus mothers) have the lowest genetic diversity and, compared to their parents, may be less well adapted to the environmental conditions of Christmas Island (as hinted at by their more limited resource use and significantly lower abundance). These findings are in sharp contrast with those of the only previous genetic studies of hybridization in butterflyfishes (McMillan et al. 1999). McMillan et al. (1999) found that, within the hybrid zone at the Solomon Islands and Papua New Guinea, hybrid phenotypes largely outnumbered the pure C. pelewensis and C. punctatofasciatus suggesting better survival (and hence fitness) of hybrids than parents within the hybrid zone environment.
Hybridization between C. trifasciatus and C. lunulatus at Christmas Island could be rare and relatively recent (and this could explain the lack of introgression), but, protracted through time, this may facilitate the local persistence of at least one of these rare species (C. lunulatus). Moreover, this study detected high levels of intrabasin connectivity in the Indian Ocean, with populations of C. trifasciatus from locations as far apart as Zanzibar and Christmas Island showing no significant genetic differentiation. According to previous studies of reef fish hybridization (Yaakub et al. 2006; Marie et al. 2007; Hobbs et al. 2009), hybrid individuals could stray further west to Cocos (Keeling) Islands. However, C. trifasciatus×C. lunulatus hybrids seem to be restricted to the hybrid zone, with limited, more localized consequences, a scenario also found in Caribbean wrasses of genus Halichoeres (Yaakub et al. 2007).
Conclusions
Hybridization between C. trifasciatus and C. lunulatus at Christmas Island is facilitated by a combination of similarities in habitat and resource use, low abundance, apparently random mating behavior, and limited genetic divergence. Hybrids resulting from at least one of the alternative possible crosses are viable. However, the apparent lack of introgression between C. trifasciatus and C. lunulatus and the rarity of the hybrids suggest that hybridization in this complex is rare and relatively recent, and so far has had limited evolutionary consequences. Hybridization between C. trifasciatus and C. lunulatus might facilitate persistence of at least one of these locally rare species at Christmas Island (C. lunulatus), while it may result in the local extinction of the Indian Ocean species C. trifasciatus, for which hybridization appears to represent a significant cost. Future research should be directed toward hybrid fitness in this complex to ascertain why hybrids are so rare and why crosses in which C. trifasciatus is the mother were not detected in the genetic analyses. Also, the hybridization scenario presented here warrants careful temporal monitoring of the Christmas Island hybrid zone, to determine whether hybridization between C. trifasciatus and C. lunulatus is indeed an isolated process or whether it is the start of the replacement of an Indian Ocean species by a Pacific Ocean species at this location.
In the face of potential future increased instances of hybridization in reef fishes, trends identified here should be further evaluated. The inclusion and similar characterization of other butterflyfish species pairs that hybridize could further help elucidate the relative importance of different factors in promoting hybridization and identify common trends in the evolutionary (genetic) outcomes. Clearly more studies are required to further elucidate the mechanisms conducive to hybridization in this group and also to determine the evolutionary consequences. Further, the inclusion of taxa belonging to different families could help explain the disproportionate incidence of hybridization in the Chaetodontidae and in reef fishes generally. Long-term monitoring of known hybrid zones (Arnold and Martin 2010), such as Christmas Island (Hobbs et al. 2009), including changes in coral reef health and abundance of live corals over time, could provide insights into the adaptive consequences of hybridization in a changing world.
Acknowledgments
This research was financially supported by the ARC Centre of Excellence for Coral Reef Studies. Fieldwork was made possible by the logistic support of Parks Australia Christmas Island. The authors wish to thank B. Danastas, D. Jones, P. Momigliano, and H. Veilleux for their assistance during work at MEEL; S. Blowes and two anonymous reviewers for their contribution to the improvement of this manuscript.
References
- Adrim M, Hutomo M. Species composition, distribution and abundance of chaetodontidae along reef transects in the Flores Sea. Neth. J. Sea Res. 1989;23:85–93. [Google Scholar]
- Allen GR, Steene R. The fishes of Christmas Island, Indian Ocean. Australian Parks Service, Special Publication. 1979;2:1–81. [Google Scholar]
- Allen GR, Steene R, Allen M. A guide to angelfishes and butterflyfishes. Perth: Odissey Publishing; 1998. [Google Scholar]
- Allen GR, Steene R, Orchard M. Fishes of Christmas Island. 2nd ed. Indian OceanAustralia: Christmas Island Natural History Association, Christmas Island; 2007. [Google Scholar]
- Allendorf FW, Waples RS. Conservation and genetics of salmonid fishes. In: Avise JC, Hamrick JL, editors. Conservation genetics: case histories from nature. New York: Chapman and Hall; 1996. pp. 238–280. [Google Scholar]
- Arnold ML. Natural hybridization and evolution. New York: Oxford Univ. Press; 1997. [Google Scholar]
- Arnold ML, Martin NH. Hybrid fitness across time and habitats. Trends Ecol. Evol. 2010;25:530–536. doi: 10.1016/j.tree.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Ayllon F, Martinez JL, Davaine P, Beall E, Garcia-Vazquez E. Interspecific hybridization between Atlantic salmon and brown trout introduced in the subantarctic Kerguelen islands. Aquaculture. 2004;230:81–88. [Google Scholar]
- Barber PH, Bellwood DR. Biodiversity hotspots: evolutionary origins of biodiversity in wrasses (Halichoeres: Labridae) in the Indo-Pacific and new world tropics. Mol. Phylogenet. Evol. 2005;35:235–253. doi: 10.1016/j.ympev.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Barton NH, Hewitt GM. Analysis of hybrid zones. Annu. Rev. Ecol. Syst. 1985;16:113–148. [Google Scholar]
- Bellwood DR, Klanten S, Cowman PF, Pratchett MS, Konow N, van Herwerden L. Evolutionary history of the butterflyfishes (f: Chaetodontidae) and the rise of coral feeding fishes. J. Evolution. Biol. 2010;23:335–349. doi: 10.1111/j.1420-9101.2009.01904.x. [DOI] [PubMed] [Google Scholar]
- Bernardi G, Bucciarelli G, Costagliola D, Robertson D, Heiser J. Evolution of coral reef fish Thalassoma spp. (Labridae). 1. Molecular phylogeny and biogeography. Mar. Biol. 2004;144:369–375. [Google Scholar]
- Berumen ML, Pratchett MS, McCormick MI. Within-reef differences in diet and body condition of coral-feeding butterflyfishes (Chaetodontidae) Mar. Ecol. Prog. Ser. 2005;287:217–227. [Google Scholar]
- Blum S. Biogeography of the chaetodontidae: an analysis of allopatry among closely related species. Environ. Biol. Fish. 1989;25:9–31. [Google Scholar]
- Burford MA, Bernardi G, Carr MH. Analysis of individual year-classes of a marine fish reveals little evidence of first-generation hybrids between cryptic species in sympatric regions. Mar. Biol. 2011;158:1815–1827. [Google Scholar]
- Campton DE. Natural hybridization and introgression in fishes: methods of detection and genetic interpretations. In: Ryman N, Utter F, editors. Population genetics and fishery management. Seattle, WA: Univ. of Washington Press; 1987. pp. 161–192. [Google Scholar]
- Chapuis M-P, Estoup A. Microsatellite null alleles and estimation of population differentiation. Mol. Biol. Evol. 2007;24:621–631. doi: 10.1093/molbev/msl191. [DOI] [PubMed] [Google Scholar]
- Cole AJ, Pratchett MS, Jones GP. Diversity and functional importance of coral-feeding fishes on tropical coral reefs. Fish and Fisheries. 2008;9:286–307. [Google Scholar]
- Craig MT, Hastings PA. A molecular phylogeny of the groupers of the subfamily Epinephelinae (Serranidae) with a revised classification of the Epinephelini. Ichthyol. Res. 2007;54:1–17. [Google Scholar]
- Crawford NG. SMOGD: software for the measurement of genetic diversity. Mol. Ecol. Resour. 2010;10:556–557. doi: 10.1111/j.1755-0998.2009.02801.x. [DOI] [PubMed] [Google Scholar]
- Crow K, Munehara H, Bernardi G. Sympatric speciation in a genus of marine reef fishes. Mol. Ecol. 2010;19:2089–2105. doi: 10.1111/j.1365-294X.2010.04611.x. [DOI] [PubMed] [Google Scholar]
- Doherty PJ, Mather P, Planes S. Acanthochromis polyacanthus, a fish lacking larval dispersal, has genetically differentiated populations at local and regional scales on the Great Barrier Reef. Mar. Biol. 1994;121:11–21. [Google Scholar]
- Eck RV, Dayhoff MO. Atlas of protein sequence and structure. Silver Springs, Maryland: National Biomedical Research Foundation; 1966. [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Feddern HA. Hybridization between the Western Atlantic Angelfishes, Holacanthus isabelita and H. ciliaris. Bull. Mar. Sci. 1968;18:351–382. [Google Scholar]
- Fessler JL, Westneat MW. Molecular phylogenetics of the butterflyfishes (Chaetodontidae): taxonomy and biogeography of a global coral reef fish family. Mol. Phylogenet. Evol. 2007;45:50–68. doi: 10.1016/j.ympev.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Findley JS, Findley MT. Global, regional, and local patterns in species richness and abundance of butterflyfishes. Ecol. Monogr. 2001;71:69–91. [Google Scholar]
- Fischer EA. Speciation in the hamlets (Hypoplectrus: Serranidae): a continuing enigma. Copeia. 1980;1980:649–659. [Google Scholar]
- Frisch AJ, van Herwerden L. Field and experimental studies of hybridization between coral trouts, Plectropomus leopardus and Plectropomus maculatus (Serranidae), on the Great Barrier Reef, Australia. J. Fish Biol. 2006;68L:1013–1025. [Google Scholar]
- Fujio Y. Natural hybridization between Platichthys stellatus and Kareius bicoloratus. JPN. J. Genet. 1977;52:117–124. [Google Scholar]
- Garcia de Leaniz C, Verspoor E. Natural hybridization between Atlantic salmon, Salmo salar, and brown trout, Salmo trutta, in northern Spain. J. Fish Biol. 1989;34:41–46. [Google Scholar]
- Gardner JPA. Hybridization in the sea. Adv. Mar. Biol. 1997;31:1–78. [Google Scholar]
- Gosline WA. Speciation in the fishes of the Genus Menidia. Evolution. 1948;2:306–313. doi: 10.1111/j.1558-5646.1948.tb02748.x. [DOI] [PubMed] [Google Scholar]
- Goudet J. Fstat version 1.2: a computer program to calculate Fstatistics. J. Hered. 1995;86:485–486. [Google Scholar]
- Graham N. Ecological versatility and the decline of coral feeding fishes following climate driven coral mortality. Mar. Biol. 2007;153:119–127. [Google Scholar]
- Grant PR, Grant BR. Unpredictable evolution in a 30-year study of Darwin's finches. Science. 2002;296:707–711. doi: 10.1126/science.1070315. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Harmelin-Vivien M. Implications of feeding specialization on the recruitment processes and community structure of butterflyfishes. Environ. Biol. Fish. 1989;25:101–110. [Google Scholar]
- Hobbs J-PA, Salmond J. Cohabitation of Indian and Pacific ocean species at Christmas and Cocos (Keeling) islands. Coral Reefs. 2008;27:933–933. [Google Scholar]
- Hobbs J-PA, Frisch AJ, Allen GR, van Herwerden L. Marine hybrid hotspot at Indo-Pacific biogeographic border. Biol. Lett. 2009;5:258–261. doi: 10.1098/rsbl.2008.0561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs J-PA, Ayling AM, Choat JH, Gilligan JJ, McDonald CA, Neilson J, Newman SJ. New records of marine fishes illustrate the biogeographic importance of Christmas Island, Indian Ocean. Zootaxa. 2010;2422:63–68. [Google Scholar]
- Hobbs J-PA, van Herwerden L, Pratchett MS, Allen GR. Hybridization among coral reef butterflyfishes. In: Pratchett MS, Berumen ML, Kapoor BG, editors. Biology of butterflyfish. Enfield, New Hampshire: Science Publishers Inc; (In press) [Google Scholar]
- Hubbs CL. Hybridization between fish species in nature. Syst. Biol. 1955;4:1–20. [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Jombart T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- Jombart T, Devillard S, Balloux F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genetics. 2010;11:94–108. doi: 10.1186/1471-2156-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost L. GST and its relatives do not measure differentiation. Mol. Ecol. 2008;17:4015–4026. doi: 10.1111/j.1365-294x.2008.03887.x. [DOI] [PubMed] [Google Scholar]
- Jukes TH, Cantor CR. Evolution of protein molecules. New York: Academic Press; 1969. [Google Scholar]
- Kuiter RH. Butterflyfishes. Bannerfishes and their relatives. ChorleywoodU.K: TMC Publishing; 2002. [Google Scholar]
- Kuriiwa K, Hanzawa N, Yoshino T, Kimura S, Nishida M. Phylogenetic relationships and natural hybridization in rabbitfishes (Teleostei: Siganidae) inferred from mitochondrial and nuclear DNA analyses. Mol. Phylogenet. Evol. 2007;45:69–80. doi: 10.1016/j.ympev.2007.04.018. [DOI] [PubMed] [Google Scholar]
- Lawton R, Pratchett MS, Bay L. Isolation and characterization of 29 microsatellite loci for studies of population connectivity in the butterflyfishes Chaetodon trifascialis and Chaetodon lunulatus. Conserv. Genetics Resour. 2010;2:209–213. [Google Scholar]
- Mallet J. Hybridization as an invasion of the genome. Trends Ecol. Evol. 2005;20:229–237. doi: 10.1016/j.tree.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Marie A, van Herwerden L, Choat J, Hobbs J-PA. Hybridization of reef fishes at the Indo-Pacific biogeographic barrier: a case study. Coral Reefs. 2007;26:841–850. [Google Scholar]
- Maruska KP, Peyton KA. Interspecific spawning between a recent immigrant and an endemic damselfish (Pisces: Pomacentridae) in the Hawaiian Islands. Pac. Sci. 2007;61:211–221. [Google Scholar]
- McMillan WO, Palumbi SR. Concordant evolutionary patterns among Indo-West Pacific butterflyfishes. Proc. Roy. Soc. Lond. B. Biol. Sci. 1995;260:229–236. doi: 10.1098/rspb.1995.0085. [DOI] [PubMed] [Google Scholar]
- McMillan WO, Palumbi SR. Rapid rate of control-region evolution in Pacific butterflyfishes (Chaetodontidae) J. Mol. Evol. 1997;45:473–484. doi: 10.1007/pl00006252. [DOI] [PubMed] [Google Scholar]
- McMillan WO, Weigt LA, Palumbi SR. Color pattern evolution, assortative mating, and genetic differentiation in brightly colored butterflyfishes (Chaetodontidae) Evolution. 1999;53:247–260. doi: 10.1111/j.1558-5646.1999.tb05350.x. [DOI] [PubMed] [Google Scholar]
- Miyazawa S, Okamoto M, Kondo S. Blending of animal colour patterns by hybridization. Nature Communications. 2010;1:66. doi: 10.1038/ncomms1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munday PL, Dixson DL, Donelson JM, Jones GP, Pratchett MS, Devitsina G, Døving KB. Ocean acidification impairs olfactory discrimination and homing ability of a marine fish. Proc. Natl. Acad. Sci. 2009;106:1848–1852. doi: 10.1073/pnas.0809996106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Kumar S. Molecular evolution and phylogenetics. New York: Oxford Univ. Press; 2000. [Google Scholar]
- Nielsen EE, Hansen MM, Ruzzante DE, Meldrup D, Grønkjær P. Evidence of a hybrid-zone in Atlantic cod (Gadus morhua) in the Baltic and the Danish Belt Sea revealed by individual admixture analysis. Mol. Ecol. 2003;12:1497–1508. doi: 10.1046/j.1365-294x.2003.01819.x. [DOI] [PubMed] [Google Scholar]
- Norman JR. A systematic monograph of the flatfishes (Heterostomata) LondonU.K: Natural History Section of the British Museum; 1934. [Google Scholar]
- Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira MAM, Videira EJS. Distribution and community structure of butterflyfishes (Pisces: Chaetodontidae) in Southern Mozambique. Western Indian Ocean J. Mar. Sci. 2005;4:39–46. [Google Scholar]
- Planes S, Doherty PJ. Genetic and color interactions at a contact zone of Acanthochromis polyacanthus: a marine fish lacking pelagic larvae. Evolution. 1997a;51:1232–1243. doi: 10.1111/j.1558-5646.1997.tb03970.x. [DOI] [PubMed] [Google Scholar]
- Planes S, Doherty PJ. Genetic relationships of the colour morphs of Acanthochromis polyacanthus (Pomacentridae) on the northern Great Barrier Reef. Mar. Biol. 1997b;130:109–117. [Google Scholar]
- Planes S, Doherty PJ, Bernardi G. Strong genetic divergence among populations of a marine fish with limited dispersal, Acanthochromis polyacanthus, within the Great Barrier Reef and the Coral Sea. Evolution. 2001;55:2263–2273. doi: 10.1111/j.0014-3820.2001.tb00741.x. [DOI] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Pratchett MS. Dietary overlap among coral-feeding butterflyfishes (Chaetodontidae) at Lizard Island, Northern Great Barrier Reef. Mar. Biol. 2005;148:373–382. [Google Scholar]
- Pratchett MS, Wilson SK, Berumen ML, McCormick MI. Sublethal effects of coral bleaching on an obligate coral feeding butterflyfish. Coral Reefs. 2004;23:352–356. [Google Scholar]
- Pratchett MS, Pradjakusuma O, Jones G. Is there a reproductive basis to solitary living versus pair-formation in coral reef fishes? Coral Reefs. 2006a;25:85–92. [Google Scholar]
- Pratchett MS, Wilson SK, Baird AH. Declines in the abundance of Chaetodon butterflyfishes following extensive coral depletion. J. Fish. Biol. 2006b;69:1269–1280. [Google Scholar]
- Prim RC. Shortest connection networks and some generalizations. Bell System Technical Journal. 1957;36:1389–1401. [Google Scholar]
- Pyle RL, Randall JE. A review of hybridization in marine angelfishes (Perciformes: Pomacanthidae) Environ. Biol. Fish. 1994;41:127–145. [Google Scholar]
- Randall JE. Acanthurus rackliffei, a possible hybrid surgeon fish (A. achilles x A. glaucopareius) from the Phoenix Islands. Copeia. 1956;1956:21–25. [Google Scholar]
- Randall JE, Allen GR, Steene R. Five probable hybrid butterfly fishes of the genus Chaetodon from the central and western Pacific. Record. West. Aust. Mus. 1977;6:3–26. [Google Scholar]
- Rhymer JM, Simberloff D. Extinction by hybridization and introgression. Annu. Rev. Ecol. Syst. 1996;27:83–109. [Google Scholar]
- Riginos C, Cunningham CW. Hybridization in postglacial marine habitats. Mol. Ecol. 2007;16:3971–3972. doi: 10.1111/j.1365-294X.2007.03505.x. [DOI] [PubMed] [Google Scholar]
- Rousset F. Genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- Ruzzante DE, Wroblewski JS, Taggart CT, Smedbol RK, Cook D, Goddaard SV. Bay-scale population structure in coastal Atlantic cod in Labrador and Newfoundland, Canada. J. Fish. Biol. 2000;56:431–447. [Google Scholar]
- Saitou N, Nei M. The Neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd edn. Cold Spring Harbour, New York: Cold Spring Harbour Laboratory Press; 1989. [Google Scholar]
- Scribner KT, Page KS, Bartron ML. Hybridization in freshwater fishes: a review of case studies and cytonuclear methods of biological inference. Rev. Fish. Biol. Fisher. 2000;10:293–323. [Google Scholar]
- Seehausen O. Hybridization and adaptive radiation. Trends Ecol. Evol. 2004;19:198–207. doi: 10.1016/j.tree.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Shaw PW, Turner GF, Rizman Idid M, Robinson RL, Carvalho GR. Genetic population structure indicates sympatric speciation of Lake Malawi pelagic cichlids. Proc. Roy. Soc. Lond. B. Biol. Sci. 2000;267:2273–2280. doi: 10.1098/rspb.2000.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumaran J. 2007. Bootscore: a bootstrap tree scoring utility. Version 3.11. Available at: http://sourceforge.net/projects/bootscore.
- Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U. S. A. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Taylor EB, Boughman JW, Groenenboom M, Sniatynski M, Schluter D, Gow JL. Speciation in reverse: morphological and genetic evidence of the collapse of a three-spined stickleback (Gasterosteus aculeatus) species pair. Mol. Ecol. 2006;15:343–355. doi: 10.1111/j.1365-294X.2005.02794.x. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson WL. Sampling rare or elusive species: concepts, designs, and techniques for estimating population parameters. Washington, D. C: Island Press; 2004. [Google Scholar]
- Turner GF, Seehausen O, Knight ME, Allender CJ, Robinson RL. How many species of cichlid fishes are there in African lakes? Mol. Ecol. 2001;10:793–806. doi: 10.1046/j.1365-294x.2001.01200.x. [DOI] [PubMed] [Google Scholar]
- van Herwerden L, Doherty PJ. Contrasting genetic structures across two hybrid zones of a tropical reef fish, Acanthochromis polyacanthus (Bleeker 1855) J. Evolution. Biol. 2006;19:239–252. doi: 10.1111/j.1420-9101.2005.00969.x. [DOI] [PubMed] [Google Scholar]
- van Herwerden L, Davies CR, Choat JH. Phylogenetic and evolutionary perspectives of the Indo-Pacific grouper Plectropomus species on the Great Barrier Reef, Australia. J. Fish. Biol. 2002;60:1591–1596. [Google Scholar]
- van Herwerden L, Choat JH, Dudgeon CL, Carlos G, Newman SJ, Frisch A, van Oppen M. Contrasting patterns of genetic structure in two species of the coral trout Plectropomus (Serranidae) from east and west Australia: Introgressive hybridization or ancestral polymorphisms. Mol. Phylogenet. Evol. 2006;41:420–435. doi: 10.1016/j.ympev.2006.04.024. [DOI] [PubMed] [Google Scholar]
- van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes. 2004;4:535–538. [Google Scholar]
- Verheyen E, Salzburger W, Snoeks J, Meyer A. Origin of the superflock of Cichlid fishes from Lake Victoria, East Africa. Science. 2003;300:325–329. doi: 10.1126/science.1080699. [DOI] [PubMed] [Google Scholar]
- Walsh PS, Metzger DA, Higuchi R. Chelex-100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques. 1991;10:506–513. [PubMed] [Google Scholar]
- Wirtz P. Mother species-father species: unidirectional hybridization in animals with female choice. Anim. Behav. 1999;58:1–12. doi: 10.1006/anbe.1999.1144. [DOI] [PubMed] [Google Scholar]
- Yaakub S, Bellwood D, van Herwerden L. A rare hybridization event in two common Caribbean wrasses (genus Halichoeres; family Labridae) Coral Reefs. 2007;26:597–602. [Google Scholar]
- Yaakub SM, Bellwood DR, van Herwerden L, Walsh FM. Hybridization in coral reef fishes: introgression and bi-directional gene exchange in Thalassoma (family Labridae) Mol. Phylogenet. Evol. 2006;40:84–100. doi: 10.1016/j.ympev.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Yabuta S. Spawning migrations in the monogamous butterflyfish Chaetodon trifasciatus. Ichthyol. Res. 1997;44:177–182. [Google Scholar]
- Zwickl DJ. 2006. Garli – genetic algorithm for rapid likelihood inference. Available at http://www.bio.utexas.edu/faculty/antisense/garli/Garli.html.


