Abstract
The efficacy of anti-cancer drugs is often limited by their systemic toxicities and adverse side effects. We report that the EphA2 receptor is over-expressed preferentially in several human cancer cell lines compared to normal tissues and that an EphA2 targeting peptide (YSAYPDSVPMMS) can be effective in delivering anti-cancer agents to such tumors. Hence, we report on the synthesis and characterizations of a novel EphA2-targeting agent conjugated with the chemotherapeutic drug paclitaxel. We found that the peptide-drug conjugate is dramatically more effective than paclitaxel alone at inhibiting tumor growth in a prostate cancer xenograft model, delivering significantly higher levels of drug to the tumor site. We believe these studies open the way to the development of a new class of therapeutic compounds that exploit the EphA2 receptor for drug delivery to cancer cells.
Introduction
Current cancer therapy relies heavily on indiscriminate, highly toxic, chemotherapeutic agents resulting in systemic toxicity and adverse side effects. For instance, the mitotic inhibitor, paclitaxel, is widely utilized in cancer treatment even though it is highly toxic and only a small portion of the delivered dose reaches the tumor1. An ideal solution to such chemotherapeutic limitations would be the selective delivery of anti-cancer drugs to tumor tissues. To this end, recent advances in our understanding of the cell surface proteome of cancer cells as well as cells of the tumor microenvironment have led to the identification of a number of tumor specific cell surface biomarkers 2. Attempts to exploit these targets have thus far focused on developing a variety of agents including antibodies, polymers, polyunsaturated fatty acids, vitamins, hormones, and peptides as selective tumor-homing reagents coupled to a variety of anti-cancer or imaging agents2–3.
The most advanced tumor-homing molecules among these make use of humanized monoclonal antibodies. Such compounds rely on the selective nature of antibodies to specifically bind to targets that have been identified on the surface of cancer cells. These antibodies function as drug delivery agents, serving to increase the local concentration of payload drugs at or near the tumor site. Monoclonal antibody-based cancer therapeutics are currently being evaluated in a number of clinical trials (www.cancer.gov). However, while antibodies can display high affinity and tumor specificity, they suffer from clinical limitations. For example, the formulation and preparation of homogenous antibody-drug conjugates presents challenges due to the many factors that can affect protein stability4. Moreover, humanization of antibodies may reduce the risk of induced immune responses, but it cannot eliminate immunogenicity completely.
In this regard, short peptides that bind to tumor-specific targets show a great deal of promise for selective tumor targeting. Phage display technology and combinatorial chemistry methods have identified highly tumor specific peptide sequences capable of selectively binding cancer cell-specific targets5. Conjugation of known chemotherapeutic agents to these peptides at specific sites results in a homogeneous drug/peptide ratio. Furthermore, some tumor targeting peptides have the ability to not only selectively bind to cancer cells, but also mediate cell-permeabilization of both the peptide and conjugate molecule5a. By possessing the ability to identify tumor cells and mediate drug internalization, such peptides increase drug activity and reduce drug toxicity by overcoming the inherent poor selectivity and limited cellular penetration of many anti-cancer drugs. For example, the synthetic peptides RC-160 and iRGD have been used to target the somatostatin receptor3a and neuropilin-1 receptor2a, respectively. However, many tumor specific peptides that have been characterized are unable to facilitate cell penetration6. In this regard, peptides that are capable of both directly targeting tumor cells and mediating cell permeabilization represent the most attractive molecular entities for use as drug delivery agents.
The Eph family of receptor tyrosine kinases represents a possible target for tumor-specific peptide development7. The Eph receptors play a central role in cellular proliferation and survival processes and act on the actin cytoskeleton influencing cell shape and migration. Several studies have demonstrated that the disruption of binding of one family member, the EphA2 receptor, to ephrin ligands in preclinical mouse tumor models results in decreased tumor growth, likely due to inhibition of tumor angiogenesis 7a, 8. Furthermore, EphA2 is highly expressed in a high proportion of cancer types, and in some cancers the level of EphA2 expression has been correlated with the degree of malignancy7a, 8b, 9. Therefore, EphA2 is being actively studied as a target for tumor diagnosis and treatment9b, 10.
Recently, a chimeric protein consisting of a protein toxin (PE38QQR exotoxin) fused to the natural EphA2 ligand, ephrin-A1, has been shown to cause potent and dose-dependent killing of glioblastoma, breast and prostate cancer cells that express high levels of the EphA2 receptor11. Alternatively, a human EphA2 monoclonal antibody has been developed and conjugated with the tubulin binding agent monomethylauristatin12. This antibody-drug conjugate targets tumors expressing high levels of EphA2 and shows impressive efficacy in mouse xenograft models. However, both the natural ligand and the EphA2 antibody conjugate strategies still have the traditional drawbacks of protein-based therapeutics, such as immunogenic or allergic responses as well as difficulties in insuring homogenous conjugation between drug and protein. It is therefore desirable to develop peptide or small molecule ligands that selectively bind to EphA2 and take advantage of its ability to mediate internalization 7b.
By using phage display, we have previously identified two short peptides, YSA (amino acid sequence: YSAYPDSVPMMS) and SWL (amino acid sequence: SWLAYPGAVSYR), which selectively target EphA2 in preference to other Eph receptors5b. Of the two biotinylated peptides, YSA showed higher affinity for EphA2 (KD of ~200 nM) and was able to inhibit binding of ephrin-A ligands to immobilized EphA2 receptor with IC50 values in the low micromolar range. Consistent with these previous findings, Scarberry et al. recently reported that magnetic cobalt ferrite nanoparticles coated with the YSA peptide can selectively target EphA2 expressing ovarian carcinoma cells 13. Similar to the ephrin ligands, the YSA peptide can also induce EphA2 activation and downstream signaling as well as decrease receptor levels on the cell surface5b, 14. In order to exploit the diagnostic and therapeutic cancer-targeting potential of the YSA peptide toward EphA2 over-expressing cells, we have developed an effective YSA-based targeting agent using a synthetic strategy that allows the conjugation of a variety of anti-cancer and/or imaging reagents to the peptide. In particular, here we report the characterization of the YSA peptide conjugated to paclitaxel and demonstrate its successful delivery to tumors in vivo.
Results
EphA2 receptor is overexpressed in most solid tumors
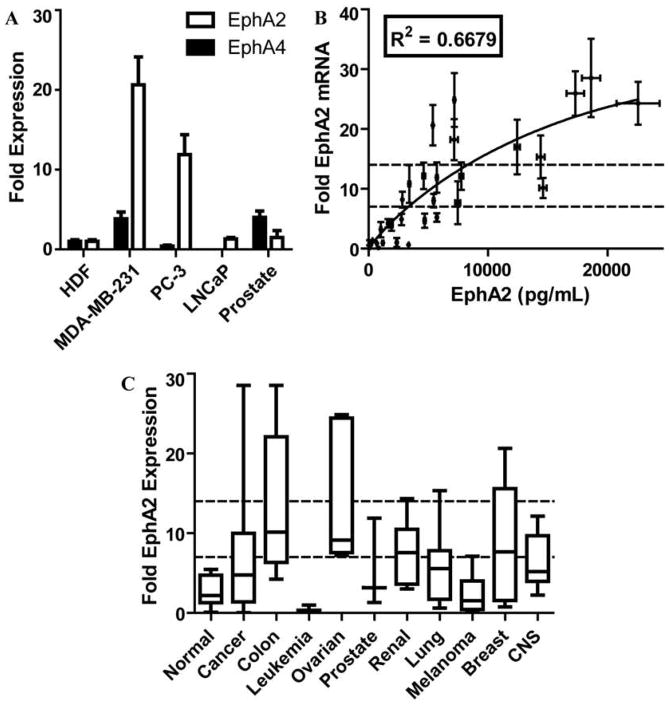
We employed a qPCR (quantitative real-time PCR) screen of total cDNA libraries collected from 68 human cancer cell lines across nine separate cancer types as well as samples extracted from 17 normal tissue types to quantify the expression of the EphA2 receptor (NM_004431.2). The data were compared to the expression of the EphA4 receptor (NM_004438.3), and normalized to the expression levels observed in a cultured human dermal fibroblast (HDF) cell line (supplementary Figure 1). In the 17 normal human tissues total RNA samples we observed an average of 2.83 ± 0.45 (mean ± SEM) folds expression of EphA2 compared to the levels observed in the HDF cell line. As a comparison, the EphA4 expression levels were uniformly higher in 16 of the normal tissue samples while the normal brain total RNA sample showed 45.74 ± 11.87 folds the expression seen in the reference HDF cell line.
The trend is reversed when observing cancer cell lines, with the expression of the EphA4 receptor substantially unchanged with respect to the HDF reference cell line (0.96 ± 0.16 folds across the 68 cell lines studied; p < 0.0001, Mann-Whitney Test) (Figure 1A and Supplementary Figure 1), while the expression of the EphA2 was markedly higher (>7x compared to HDF) in more than half of the cell lines studied. When grouping the cell lines according to their tissue of origin, over-expression of EphA2 was broadly observed in the colon (13.50 ± 2.95 folds the HDF expression levels; P = 0.0003), ovarian (13.72 ± 3.46 folds the HDF expression levels; P=0.0004) and renal (7.59 ± 1.39 folds the HDF expression levels; P=0.0043) derived cancer cell lines (Figure 1C). For each of these three groups more than 50% of the cell lines showed significant over-expression. The leukemia cell lines showed significant under-expression of EphA2 (0.26 ± 0.13; P = 0.0012) compared to the normal tissue samples (Supplementary Figure 1).
Figure 1. EphA2 and EphA4 expression in 68 human cancer cell lines and 17 normal tissue RNA samples.
(A) EphA2 and EphA4 receptor expression in normal and cancer cell lines. Bar graph shows the fold expression of both EphA2 and EphA4 receptors in MDA-MD-231, PC-3, and LNCaP cancer cell lines compared to cultured human dermal fibroblast (HDF) and to a total human prostate RNA sample (Prostate). (B) Correlation between EphA2 mRNA expression and EphA2 protein levels in 30 selected human cancer cell lines. (C) Box and Whisker plots of the 10–90th percentile of EphA2 expression for the RNA samples grouped according to their tissue of origin. Dotted lines depict cutoffs for 7 fold, and 14 fold the expression levels observed in the reference HDF cell line, respectively.
To further verify that the mRNA expression data collected reflect protein levels in cell, we performed ELISA-based assays for 30 of the cell lines that are designed to quantify the amount of total EphA2 protein present in the cells. The ELISA analysis identified a significant correlation between the mRNA and levels for EphA2 in these 30 cell lines (R2 = 0.6679, P < 0.0001) (Figure 1B; Supplementary data), corroborating our analysis based on mRNA levels.
The YSA peptide targets the EphA2 receptor and induces its internalization
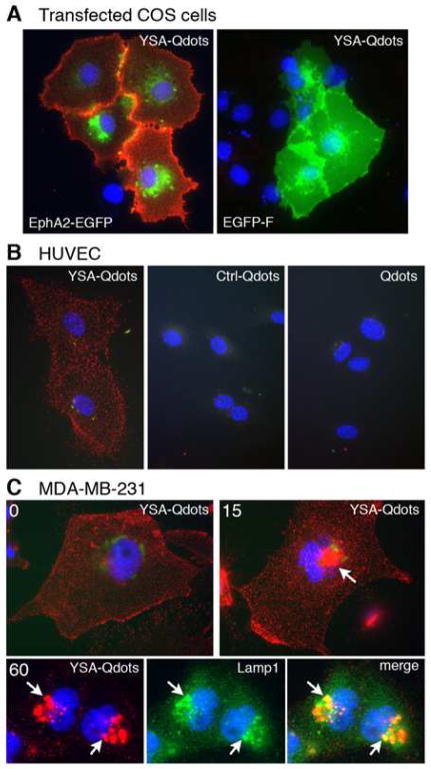
In order to investigate the EphA2 targeting ability of our previously described YSA peptide, we transfected COS cells with a plasmid encoding either the extracellular and transmembrane domains of the EphA2 receptor fused to EGFP (EphA2-EGFP) or membrane-targeted EGFP (EGFP-F). Incubation of COS cells with biotinylated YSA peptide (a YSA peptide conjugated at the C-terminal with Biotin) bound to streptavidin conjugated red fluorescent quantum dots (YSA-Qdots), revealed the presence of quantum dots bound to cells transfected with EphA2-EGFP but not control cells transfected with EGFP-F or untransfected cells (Figure 2A). Although COS cells express endogenous EphA2, overexpressing the receptor provides increased sensitivity. We also observed that the YSA peptide targets quantum dots to endogenous EphA2 located on the surface of human umbilical vein endothelial (HUVE) cells while a control peptide that does not bind to EphA2 or quantum dots without any bound peptide do not bind to the HUVE cell surface (Figure 2B). In addition, we used the lysosomal marker Lamp1 to show that the YSA peptide successfully mediates cellular uptake of quantum dots into lysosomes in the MDA-MB-231 breast cancer cell line, which expresses high levels of EphA215 (Figure 2C). Thus, the YSA peptide not only binds to EphA2, but also is able to initiate the active internalization of the receptor into cancer cells, resulting in the concomitant internalization of its cargo.
Figure 2. The YSA peptide targets cells expressing EphA2.
(A) The YSA peptide targets quantum dots to EphA2 on the cell surface. COS cells transfected with either the extracellular and transmembrane portions of EphA2 fused to enhanced green fluorescent protein (EphA2-EGFP) or membrane targeted EGFP-F were incubated with YSA bound to red fluorescent quantum dots (YSA-Qdots). YSA-Qdots only bind to cells transfected with EphA2-EGFP. EGFP fluorescence is green; nuclei are stained in blue with DAPI. (B) YSA targets quantum dots to endogenous EphA2 on the cell surface. HUVE cells were incubated at 4ºC with YSA conjugated Qdots (YSA-Qdots), an unrelated 12-mer control peptide that does not bind to EphA2 also conjugated to Qdots (Ctrl-Qdots), or unconjugated Qdots (Qdots). Only quantum dots conjugated to the YSA peptide bind to the HUVE cells. (C) YSA targets Qdots to lysosomes. MDA-MB-231 cells, which express high levels of endogenous EphA2, were incubated at 4ºC with YSA-Qdots followed by incubation at 37 ºC for 0, 15, and 60 min. YSA-Qdots are seen on the cell surface at 0 min but become concentrated in structures near the nucleus at 15 and 60 min (arrow). Double labeling with the lysosomal marker Lamp1 (green) shows colocalization of the quantum dots in lysosomes at 60 min (arrows).
Design, synthesis and characterization of a YSA-paclitaxel conjugate (YSA-PTX)
We developed a novel anti-triazole linker for the synthesis of YSA peptide-drug conjugates to avoid the compatibility problems of disulfide and hydrazone linkers. Conjugation at the C-terminus of the peptide was preferred because modifications at the N-terminus of the YSA peptide decrease EphA2 binding activity (data not shown). Hence, the C-terminus of YSA was derivatized with Nε-(5-hexynoyl)-lysine resulting in the amino acid sequence YSAYPDSVPMMS[Nε-(5-hexynoyl)-K], hereafter referred to as “YSA-motif”. This motif provides the alkyne moiety for subsequent coupling of modified paclitaxel using click chemistry 16. To provide the azide counterpart of click chemistry, we attached a diglycolic acid 6-azido-hexanyl monoester to the 23-hydroxyl of paclitaxel by employing a selective protection/deprotection strategy (supplementary material). Coupling of the YSA-motif with the azide linker-derived paclitaxel (PTX) was accomplished under mild click-chemistry reaction conditions, generating the desired YSA-PTX conjugate (Figure 3A). A similar synthetic approach was used to conjugate a YSA scrambled peptide control (DYP-PTX; amino acid sequence: DYPSMAMYSPSV[Nε-(5-hexynoyl)-K])17 to PTX (Figure 3A) as well as for biotinylated peptides, YSA-PTX-Biotin and DYP-PTX-Biotin (supplementary material).
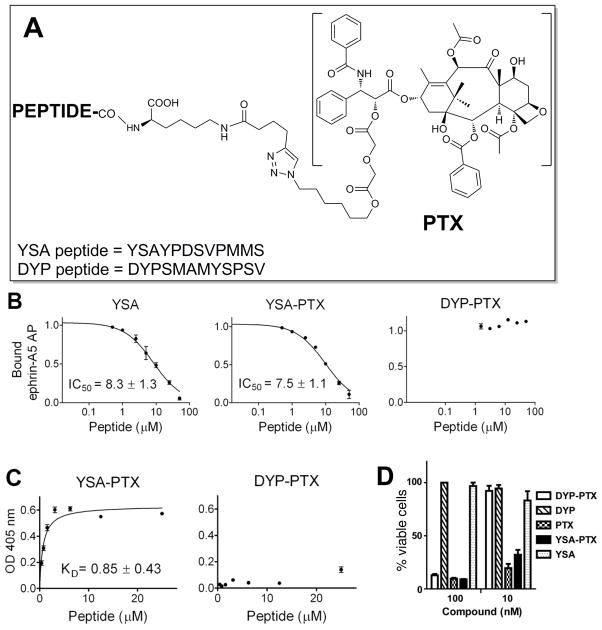
Figure 3. YSA-PTX retains high potency for inhibition of EphA2-ephrin-A5 binding and high EphA2 binding affinity.
(A) General chemical structures of YSA and DYP peptide-drug conjugates. (B) YSA, YSA-PTX and control DYP-PTX were incubated at the indicated concentrations together with a constant concentration of ephrin-A5 AP in ELISA wells pre-coated with EphA2 Fc. The ratio of ephrin-A5 AP bound in the presence and in the absence of peptide is shown. (C) Biotinylated YSA-PTX and control DYP-PTX were incubated at the indicated concentrations in EphA2 Fc-coated ELISA wells and were then detected with streptavidin-HRP. The graphs show averages ± SE from quadruplicate measurements in representative experiments, while the KD and IC50 values are calculated from 3 to 5 experiments. (D) ATP-Lite analysis of viability of PC-3 cells 72 hours after the addition of DYP, DYP-PTX, YSA, YSA-PTX or PTX at 10 nM (n = 3).
After preparation of the YSA-PTX conjugate, we confirmed that the conjugation does not interfere with the ability of YSA to bind EphA2 and cause its internalization. The ability of YSA-PTX to efficiently compete, in a dose dependent manner, with ephrin-A5 fused to alkaline phosphatase (ephrin-A5 AP) for binding to the EphA2 extracellular domain fused to Fc (EphA2 Fc) in ELISA assays demonstrates that the conjugate retains its EphA2 binding ability (Table 1 and Figure 3B). In contrast, neither of the control compounds, DYP motif nor DYP-PTX conjugate, show any detectable displacement of ephrin-A5. We measured a KD of ~0.8 μM for the binding of biotinylated YSA-PTX to EphA2 Fc immobilized in ELISA wells, whereas binding of the biotinylated control DYP-PTX was undetectable (Figure 3C).
Table 1.
EphA2 inhibition
| Compounds | Sequence | IC50 (μM) | n |
|---|---|---|---|
| YSA | YSAYPDSVPMMS | 12.6 ± 1.03 | 15 |
| YSA-motif | YSAYPDSVPMMS-K* | 1.68 ± 0.26 | 6 |
| YSA-PTX | YSAYPDSVPMMS-K*-PTX | 6.82 ± 0.81 | 6 |
| DYP-motif | DYPSMAMYSPSV- K* | >200 | 2 |
| DYP-PTX | DYPSMAMYSPSV- K*-PTX | >200 | 2 |
K* a modified lysine with an alkyne linker
In vitro cell viability assays confirmed that the YSA-PTX conjugate retained the ability of paclitaxel to induce cell death at low nanomolar concentrations in the EphA2 overexpressing prostate cancer cell line PC3 (Figure 3D), while paclitaxel conjugated to the scrambled peptide, DYP-PTX, showed no significant cell killing in the same assay (Figure 3D). These data suggest that the peptide has sufficient affinity for the receptor to deliver an effective dose of drug.
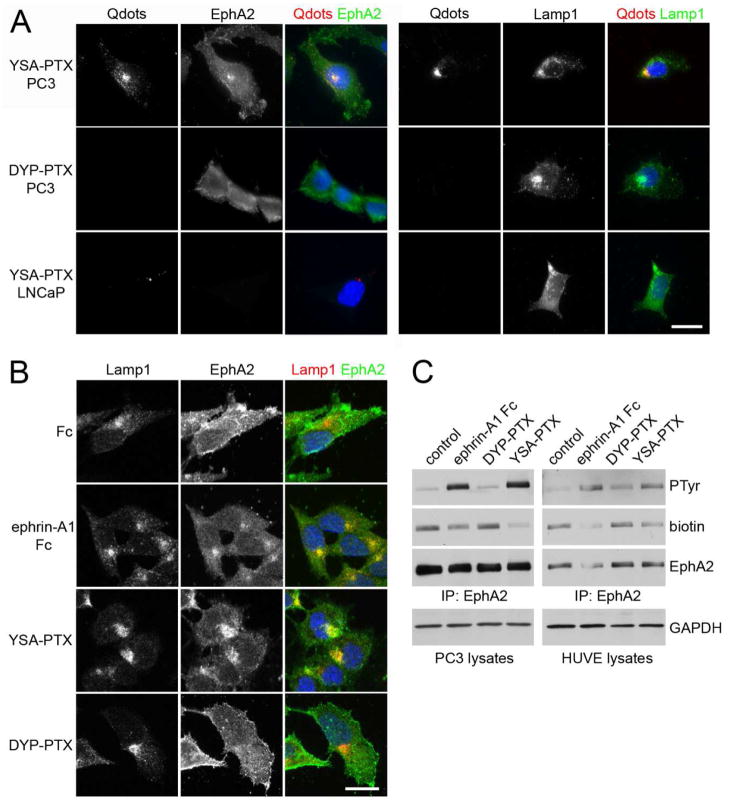
Further analysis of YSA-PTX-Qdots revealed that the peptide conjugate facilitates Qdot uptake into the lysosomes of the highly EphA2 expressing prostate cancer cell line, PC318. In contrast, YSA-PTX-Qdots does not target the LNCaP prostate cancer cell line (Figure 4A), which, according to both our qPCR data (Figure 1A) and previous literature data,19 does not express detectable levels of EphA2. As a further control, the DYP-PTX control scrambled peptide does not target PC3 cells and does not trigger EphA2 internalization (Figure 4A). Moreover, we confirmed that even when not coupled to quantum dots, YSA-PTX causes EphA2 internalization into the lysosomes of PC3 cells, similar to the ephrin-A1 Fc ligand (Figure 4B). YSA-PTX, but not DYP-PTX, also causes EphA2 tyrosine phosphorylation (which is indicative of activation) and loss of the receptor from the surface of not only PC3 cancer cells but also HUVE endothelial cells (Figure 4C). These data indicate that the YSA-PTX conjugate retains the ability of YSA to bind EphA2 and allows effective internalization of PTX into cells. Furthermore, they suggest that YSA-PTX may be useful to target not only cancer cells but also the tumor vasculature.
Figure 4. YSA-PTX is internalized in cancer and endothelial cells expressing EphA2.
(A) YSA-PTX coupled to fluorescent quantum dots, but not DYP-PTX, is internalized with EphA2 into lysosomes of prostate cancer cells expressing EphA2. PC3 prostate cancer cells, which express high levels of EphA2, or LNCaP prostate cancer cells, which do not detectably express EphA2 protein, were treated for 20 min with 100 μM YSA-PTX or control DYP-PTX, followed by a 20 min incubation with streptavidin-conjugated quantum dots (Qdots). After removing the solution containing the peptides and the quantum dots, the cells were incubated for 2 h at 37°C to allow EphA2 internalization induced by YSA-PTX binding. The cells were stained for EphA2 (left) or the lysosomal marker Lamp1 (right). Qdots were also imaged and nuclei were labelled with DAPI in blue. Representative fluorescence micrographs are shown. Scale bar = 25 μM. (B) Ephrin-A1 Fc and YSA-PTX, but not DYP-PTX, causes EphA2 internalization into lysosomes. PC3 cells were treated for 2 h with 0.2 μg/mL ephrin-A1 Fc, 100 μM YSA-PTX or 100 μM control DYP-PTX. The cells were stained for Lamp1 (red) and EphA2 (green). Nuclei were labelled with DAPI (blue). Representative confocal micrographs are shown. Scale bar = 25 μm. (C) Ephrin-A1 Fc and YSA-PTX, but not DYP-PTX, cause EphA2 internalization into cells. PC3 prostate cancer and HUVE endothelial cells were treated for 1 h with 0.2 μg/mL ephrin-A1 Fc, 100 μM YSA-PTX or 100 μM control DYP-PTX. Proteins present on the cell surface were then labeled with biotin. EphA2 immunoprecipitates were probed with anti-phosphotyrosine antibody (PTyr), streptavidin-HRP (biotin) and EphA2 antibody. The amount of GAPDH in the cell lysates used for the immune-precipitations is also shown as a control for equal amounts of protein in the lysates used for immunoprecipitation.
YSA-PTX targets tumors in vivo
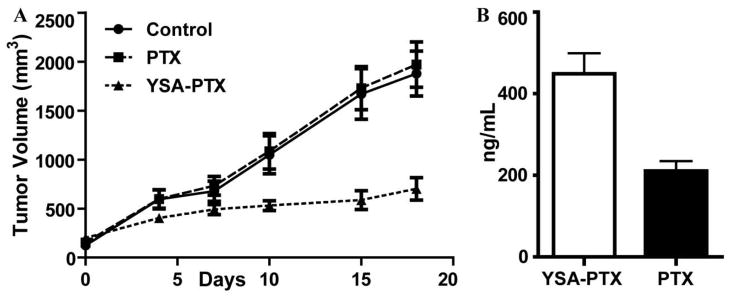
In preparation for in vivo studies of the YSA-PTX conjugate, we found that YSA-PTX shows favorable plasma and microsomal half-lives (43 and 23 minutes, respectively), and is able to release native paclitaxel in the presence of PC3 cells with nearly complete release in about 60 minutes (data not shown). Therefore, we examined the effects of YSA-PTX in preliminary in vivo studies using PC3 tumor xenografts. Tumor-bearing mice were treated twice weekly with an intravenous injection of PTX or YSA-PTX for a total of 3 weeks. Remarkable tumor growth inhibition (p<0.05) was observed with YSA-PTX (Figure 5A) while un-conjugated PTX administered at an equimolar dose did not significantly affect tumor growth as compared to vehicle control. Moreover, mouse body weights were not affected by YSA-PTX administration in comparison to administration of vehicle control, and no adverse signs of toxicity in the YSA-PTX-treated mice were observed. To confirm preferential delivery of paclitaxel to tumor xenografts, we analyzed tumors dissected from mice treated with PTX or YSA-PTX to determine PTX tumor delivery using mass spectrometry. Tumor-bearing mice were treated with a single dose of PTX or YSA-PTX at equimolar doses, and PTX levels were measured in tumor tissue and plasma 30 minutes after administration. This revealed that the tumor tissue from the YSA-PTX group shows significantly higher PTX levels compared to the group where unconjugated PTX was administered, while plasma PTX concentrations were equivalent (Figure 5B).
Figure 5. Expression of EphA2 and cellular and in vivo activity of drug conjugates.
(A) Groups of 5–7 SCID-beige mice bearing pre-established subcutaneous PC3 tumors were treated with twice weekly with intravenous doses of vehicle (CT), paclitaxel (5 mg/Kg), YSA-taxol (equimolar doses of the taxol dose), starting at day 0. Tumor sizes were measured and averages ± SEM are shown. p<0.05 for the comparison of YSA-taxol with control, by repeated-measures two-way ANOVA using all the measurements as well as by one-way ANOVA and Dunnett's posthoc test using the measurements at day 18. (B) Bio-distribution analysis. PC3 tumors (n = 3; average volume 200 mm ) were excised 30 min after equimolar amounts of YSA-PTX or PTX (25 mg/kg) were injected intravenously. LC/MS analysis was performed to quantify PTX levels from tumor extracts. The histogram shows averages ± SEM. p<0.05 for the comparison of PTX levels, by t-test analysis. Plasma levels of PTX were not significantly different (YSA-PTX = 998 ± 742 ng/mL; PTX = 771 ± 585 ng/mL).
Discussion
The development of tumor specific, cell penetrating molecules remains a primary focus in cancer research. Our data demonstrate that the YSA peptide is capable of specifically targeting the EphA2 receptor and of causing receptor activation and internalization, which also results in the internalization of the YSA peptide and any conjugated biomolecule. As expected, YSA-PTX can induce activation of EphA2, thus mediating its internalization into cancer and endothelial cells. In addition, conjugation of PTX to the YSA peptide does not interfere with the chemotherapeutic function of PTX.
Our qPCR studies show that overexpression of EphA2 is a common (occurring in one third of the solid tumor-derived cell lines studies), but not universal characteristic of solid tumor types. Recent studies have reported that EphA2 expression levels may be upregulated in response to hypoxic stress 20. As a result, the over expression of EphA2 in tumor tissues in vivo may be much higher than the levels observed in tissue culture environment, where oxygen levels are abnormally high compared to not only the hypoxic regions of tumors but also the levels in normal tissues. In addition, EphA2 is widely up-regulated in the tumor vasculature but is not expressed in normal quiescent vasculature7a, 8a. Our results suggest that the YSA peptide could be used as a vehicle for the delivery of diagnostic and chemotherapeutic agents to EphA2 expressing tumors.
Our preliminary paclitaxel bio-distribution studies demonstrate that the YSA-PTX conjugate affords a significant increase in the amount of drug delivered to the tumor compared with administration of free paclitaxel. Consistent with this, our results show that YSA-PTX is much more effective in inhibiting PC3 tumor xenograft growth than free paclitaxel, with no overt signs of toxicity. We speculate that the enhanced anti-tumor activity of YSA-PTX can be attributed to its two sought-after traits: tumor selectivity and tumor tissue penetration. Conjugation of the YSA peptide to paclitaxel endows YSA-PTX with the ability to selectively bind to and activate EphA2, leading to internalization of the EphA2-YSA-PTX complex. As a result, paclitaxel is actively transported into cells, rather than relying on passive transport as is the case when a tumor selective molecule lacks the ability to mediate internalization. Our data demonstrate that for equivalent molar dosing, YSA-PTX delivers a higher proportion of active drug molecules into the tumors, leading to a dramatic decrease in tumor growth.
Along with increasing tumor selectivity and tumor tissue penetration, conjugation to YSA may increase the water solubility of paclitaxel, thus making it more bioavailable 21, as well as allow an increase in the therapeutic window of the drug. In fact, EphA2 expression is limited in normal tissues7a, 8b, 9, and in normal epithelial cells EphA2 is likely found in complex with ephrin ligands and therefore cannot be efficiently targeted by YSA, an ephrin antagonist, in contrast to tumor tissue where EphA2 is not frequently co-expressed with ephrin ligands 15, 22. The low EphA2 expression observed in normal tissues and its engagement with ephrin ligands, as well as our bio-distribution data, highlights the fact that the use of the YSA peptide for drug delivery may reduce exposure of normal tissues to cytotoxic drugs, thus presumably reducing toxic side-effects. As a result, YSA-PTX may broaden the therapeutic window of paclitaxel to an extent that elevated doses could still be safely used. Administering larger doses of YSA-PTX to patients may increase the chances of eradicating tumors while decreasing the cancer cells’ ability to evade either the YSA targeting molecule or the cell stress caused by the chemotherapeutic.
The use of a targeting peptide, as opposed to a protein or antibody drug conjugate, for delivery of anti-cancer drugs has also a number of significant advantages over other the use of full length ephrins or antibodies. The short peptide sequence is a single, relatively small molecule that lacks the size necessary to mount an immune response during therapy. Also not to be overlooked is the significant lower cost for the synthesis of a homogenous short peptide when compared to the preparation of proteins and humanized antibodies.
In summary, YSA-PTX has been demonstrated to have a powerful anti-tumor activity in vitro and in vivo and could serve as a valuable candidate for further pre-clinical and eventually clinical evaluation in humans. The use of small peptide ligands targeting EphA2-expressing tumors for drug delivery could provide a new arsenal of diagnostic and therapeutic options for cancer patients. With this report we intend to share our compelling preliminary findings, which we believe open the way to future studies to optimize the peptide sequence for binding affinity and in vivo stability, investigate additional YSA-drug conjugates, conjugate YSA and optimized derivatives with MRI or PET imaging agents such as DTPA or DOTA, and perform detailed studies on the pharmacokinetics of the conjugates.
Experimental Section
Chemical synthesis and purification
Unless otherwise noted, all reagents and anhydrous solvents were obtained from commercial sources and used without purification, and the fully protected peptides on resin were purchased from Biopeptide Co. The YSA and DYP peptides were purchased from either Medical College of Wisconsin or Abgent, Inc. (San Diego). All reactions were performed in oven-dried glassware. All reactions involving air or moisture sensitive reagents were performed under a nitrogen atmosphere. Silica gel chromatography was performed using pre-packed silica gel or C-18 cartridges (RediSep). All final compounds were purified to >95% purity, as determined by a HPLC Breeze from Waters Co. using an Atlantis T3 5.0 μM 4.6 mm ×150 mm reverse phase column. 1D and 2D NMR spectra were recorded on a Bruker 600 MHz instruments. Chemical shifts are reported in ppm (δ) relative to 1H (Me4Si at 0.00 ppm), coupling constant (J) are reported in Hz throughout, and NMR signal assignments were based on DEPT, 2D [13C, 1H]-HSQC, 2D [1H, 1H]-COSY, 2D [1H, 1H]-TOCSY, and 2D [1H, 1H]-HMBC experiments. Low resolution and high resolution mass spectral data were acquired on an Esquire LC00066 Mass Spectrometer, an Agilent ESI-TOF Mass Spectrometer, or a Bruker Daltonic Autoflex Maldi-Tof/Tof Mass Spectrometer. Detailed synthetic procedures and analytical data for compound intermediates and final compounds are reported in the Supplementary Information section.
ELISA-based displacement and binding assays
EphA2 Fc (R&D Systems, Minneapolis, MN) diluted to 1 μg/mL in TBST buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.5 with 0.01% Tween 20) was immobilized in protein A-coated wells. The wells were then incubated for 3 hours with 0.01 nM ephrin-A5 fused to alkaline phosphatase (ephrin-A5 AP) 5b in culture medium diluted in TBST in the presence of different concentrations of YSA, YSA-PTX or DYP-PTX. The concentration of ephrin-A5 AP was calculated based on AP activity measurements 23. Bound ephrin-A5 AP was quantified by adding pNPP as the substrate and measuring the absorbance at 405 nm. Absorbance from wells where human Fc (R&D Systems, Minneapolis, MN) was added instead of EphA2 Fc was subtracted as the background. Curves for the binding of biotinylated YSA-PTX and DYP-PTX to EphA2 Fc were measured by adding different concentrations of the peptides to ELISA wells pre-coated with protein A and EphA2 Fc. Bound biotinylated peptides were detected with horseradish peroxidase (HRP)-conjugated streptavidin (Thermo Scientific/Pierce, Rockford, IL; 1:2,000 dilution in TBST). Absorbance at 405 nm was measured following incubation with 0.2 mg/ml 2,2’-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS) (Sigma-Aldrich, Steinheim, Germany) in citric acid as a substrate, and the absorbance in wells without peptide was subtracted as background. The inhibition and binding curves were analyzed using non-linear regression and the program Prism (GraphPad Software Inc.).
Fluorescence cell imaging
COS cells and MDA-MB-231 breast cancer cells (ATCC) were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) (Mediatech, Inc, Herndon, VA) with 10% fetal bovine serum (FBS) (Hyclone, Logan, UT) and Pen/Strep (Omega Scientific, Tarzana, CA). PC3-3M-luc-C6 Bioware® (Caliper) and LNCaP prostate cancer cells (ATCC) were grown in RPMI 1640 medium (Mediatech, Inc, Herndon, VA) with 10% FBS and Pen/Strep. Human umbilical vein endothelial cells (HUVECs, Cascade Biologics, Portland, OG) were grown in Medium 200 supplemented with low serum growth supplements (Cascade Biologics), 10% FBS, Pen/Strep, and fungizone (Omega Scientific, Tarzana, CA).
To label cells expressing transfected EphA2 with fluorescent quantum dots, COS cells grown in 6 cm plates were transfected with 3 μg of a plasmid encoding EphA2 extracellular and transmembrane domains fused to enhanced green fluorescent protein (EGFP) or 3 μg of a vector encoding farnesylated EGFP (F-EGFP) (BD Biosciences Clontech) as control, using SuperFect Transfection Reagent (Qiagen, Inc., Valencia, CA). The cells were plated on glass coverslips and labeled 2 days after transfection. For the labeling experiments, 20 nM streptavidin-conjugated Qdot 655 quantum dots (Invitrogen/ Molecular Probes, Eugene, OR) were preincubated for 20 min at 4°C with 500 nM biotinylated YSA peptide (YSAYPDSVPMMS-GSGSK-biotin), or a biotinylated control 12-mer peptide that does not bind to EphA2, in quantum dots-binding-buffer (1 mM CaCl2, 2% BSA in PBS). The cells were then incubated for 20 min at 4°C with quantum dots containing the YSA peptide or the control peptide, or quantum dots without peptide as a further control, and then washed with ice cold 1 mM CaCl2 in PBS. To label human umbilical endothelial (HUVE) cells expressing endogenous EphA2, the cells were plated on glass coverslips coated with fibronectin (10 μg/ml) and incubated with 100 μM biotinylated YSA peptide in quantum dots-binding-buffer for 20 min at 4°C. The cells were then washed with ice cold 1 mM CaCl2 in PBS, followed by incubation with 20 nM streptavidin quantum dots for 20 min at 4°C in quantum dots-binding-buffer.
For quantum dot internalization experiments, MDA-MB-231, PC3 and LNCaP cells were grown on glass coverslips. For MDA-MB-231 cells, 20 nM streptavidin-conjugated QDot 655 quantum dots were pre-incubated for 20 min on ice with 500 nM biotinylated YSA peptide in conditioned medium supplemented with 10 mM HEPES. The cells were then incubated for 20 min on ice with the quantum dot/YSA peptide mixture, or quantum dots without peptide (negative control), and then in a 37°C CO2 incubator for various periods of time. For PC3 and LNCaP cells, the conditioned medium was removed from the cells, supplemented with 10 mM HEPES and stored at 4 °C, and the cells were serum starved for 3 hours in serum-free medium. The cells were then treated with 100 μM biotinylated YSA-PTX or DYP-PTX in quantum dots-binding buffer for 20 min on ice, followed by 20 nM streptavidin-conjugated Qdot 655 in binding buffer for 20 min on ice. After removing the quantum dot solution, the cells were washed with the binding buffer and incubated with the stored conditioned medium in a 37°C CO2 incubator for 2 hours to allow internalization of the EphA2-YSA-PTX-quantum dots complexes. The cells were then washed with ice cold PBS, fixed in 4% formaldehyde with 4% sucrose for 10 min, permeabilized for 5 min with 0.05% Triton-X100 in PBS, and incubated for 1 hour with PBS containing 10% goat serum and 2% BSA. For EphA2 staining, the coverslips were incubated with a rabbit anti-EphA2 antibody (Life Technologies/Invitrogen) followed by a secondary anti-rabbit antibody conjugated with Alexa Fluor 488 (Life Technologies/Molecular Probes). For staining of lysosomes, the coverslips were incubated with polyclonal rabbit anti-human Lamp1 antibody 24 followed by a secondary anti-rabbit antibody conjugated with Alexa Fluor 568. The nuclei were counterstained with DAPI. Images were obtained with an Inverted TE300 Nikon fluorescence microscope and processed using Adobe Photoshop.
To image EphA2 internalization and co-localization with lysosomes after stimulation PC3 cells plated on glass coverslips were serum starved for 1 hour in serum-free medium and then stimulated with 0.2 μg/mL Fc (as a negative control), 0.2 μg/mL ephrin-A1 Fc (as a positive control), 100 μM YSA-PTX-Biotin or 100 μM DYP-PTX-Biotin for 2 hours. The cells were then fixed, permeabilized and labeled as described above. Images were acquired using a Radiance 2100 MP confocal microscope.
Immunoprecipitation and immunoblotting
PC3 cells were serum-starved for 1 hour in serum-free medium and treated for 1 hour with 0.2 μg/mL human Fc (as a negative control), 0.2 μg/mL ephrin-A1 Fc (as a positive control), 100 μM YSA-PTX or 100 μM DYP-PTX. The cells were then placed on ice, rinsed once with cold PBS and incubated for 20 min at 4ΰC with a 0.5 mg/mL EZ-link sulfo-NHS-biotin (Thermo Scientific/Pierce, Rockford, IL) in PBS. The cells were then washed 3 times with a 100 mM glycine in PBS to quench the biotinylation reaction, followed by PBS. The cells were lysed in modified RIPA lysis buffer (150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 20 mM Tris, pH 8.0) containing protease inhibitors and 1 mM sodium orthovanadate. For immunoprecipitations, cell lysates were incubated with 2 μg anti-EphA2 antibody (Millipore-Upstate, Inc. Temecula, CA) immobilized on GammaBind Sepharose beads (GE Healthcare Life Sciences). Immunoprecipitates and lysates were probed by immunoblotting with anti-phosphotyrosine antibody (Millipore, Inc, Temecula, CA), streptavidin coupled to HRP (Thermo Scientific/Pierce, Rockford, IL), anti-EphA2 antibody (Life Technologies/Invitrogen) and anti-GAPDH antibody (Sigma-Aldrich, Steinheim, Germany).
RNA expression of tumor cells
Cancer cell lines and human dermal fibroblast cells were cultured and total RNA was isolated from each as previously described 25. The Firstchoice Human Total RNA Survey Panel (Agilent Technologies) was purchased to serve as control for normal tissue RNA expression levels. Total RNA was extracted from 3 × 106 cells from each line harvested at 70–80% confluence and prepared using the Illustra RNAspin mini total RNA extraction kit (GE Healthcare) according to the manufacturer’s recommendations for cultured cells. DNA contamination and sample concentration for each sample was determined using a NanoDrop ND-1000 Spectrophotometer. The RNA integrity of each sample was further confirmed using a BioRad Experion Bioanalyzer to assess the ratio of 28s and 18s RNA. All samples exhibited 28s/18s ratios of 1.8 – 2.2 and RIN values of 9 or greater, which indicate the presence of high quality total RNA. cDNA was produced using qScript cDNA Supermix (Quanta biosciences) according to manufacturer’s recommendations in 20 μL reaction mixtures using 400 ng of total RNA as the template. The resultant cDNA was stored at −40ΰC and diluted 1:7 prior to use in qPCR.
Primers for use in qPCR were identified using the Universal Probe Library design tools (Roche Applied Sciences) for both EphA2 receptor (NM_004431.2; sense: ggcaggagttggcttctttat; antisense: tgttctgacttggagaagtaaacg) and EphA4 receptor (NM_004438.3; sense: gatagcaagccctctggaag; antisense: ggtaggttcggattggtgtatt) in combination with the universal probe #69 (Roche Applied Sciences). Each qPCR reaction also contained the primers and probe necessary to run a simultaneous internal G6PH multiplexed control (Roche Applied Sciences). Primers were purchased from Invitrogen. All qPCR reactions were performed in 17 μL reaction mixtures containing 2 μL of the diluted cDNA, 0.5 μL of G6PH primer mix, 0.5 μL G6PH Probe, 1.0 μL EphA2 or EphA4 primer mix, 0.5 μL Probe #69, 10 μL mastermix (Roche Applied Sciences), and 2.5 μL molecular grade water. Each sample was run in triplicate in a 96 well polypropylene plate (Stratagene) with the Mx3000P qPCR system (Stratagene). Thermocycling conditions consisted of an initial polymerase inactivation step at 95°C for 10 minutes, followed by 40 cycles at 95°C for 30 s, 55°C for 1 minute, and 72°C for 1 minute. Baselines and thresholds were automatically set by the software. Serial dilution showed that the Cq value for all data was linear over 5 orders of magnitude (R2 of 0.996). Control experiments without primers, template or probe exhibited no detectable signal.
Calculated Cq values were exported to Excel 2007 (Microsoft) and Prism 5.04 (Graphpad Softward Inc.) for further analysis. Each cell type’s data was normalized to the average level of mRNA expression (Cq value) of G6PH across all cell lines. SEM values for the three replicates for each gene in all 68 cell lines and 17 normal tissue cDNA pools were then calculated from these normalized Cq values. The Cq value for each cell line was compared to that observed in the cultured human dermal fibroblast cell line. The average +/− the SEM values were calculated and transformed into fold expression values for the purpose of displaying the error bars in graphical representations of the data.
EphA2 Protein Quantification
The amount of EphA2 protein in each cell line was assayed using the Human Total EphA2 DuoSet Elisa kit (R&D Systems, Minneapolis, MN). Lysates we were collected from a selection of the cell lines studied using a concentration of 1 million cells/ mL of Lysis Buffer #9. Total protein samples were flash frozen and stored at −80°C until tested. The protein concentration from each sample was determined by BCA protein analysis (Pierce Biotechnology Inc., Rockford, IL) and 100 and/or 30 μg of total protein was analyzed as outlined by the manufacturer’s directions. Each protein sample dilution was tested in duplicate. A standard curve of recombinant EphA2 protein (R2 = 0.996) was used to quantify the resulting absorbance values.
ATP-Lite based analysis of cell viability
PC3 cells were cultured and tested using the ATP-Lite Onestep kit (Perkin-Elmer) as outlined previously 25. A fixed concentration of 0.5% DMSO was present in each 100 μL culture volume upon addition of the compounds and ATP concentration was recorded 72 hours after addition of the compounds.
Microsomal and plasma stability assays
To assess microsomal stability of YSA-PTX, the conjugate was incubated with rat liver microsomes (RLM) for 60 min at 37ºC. The final incubation solutions contained 4 μM YSA-PTX, 2 mM NADPH, 1 mg/mL (total protein) microsomes, and 50 mM phosphate (pH 7.2). Compound solutions, protein, and phosphate were pre-incubated at 37 ºC for 5 min and the reactions were initiated by the addition of NADPH and incubated for 1 hour at 37 ºC. Aliquots were taken at 30 minute time-points and quenched with the addition of methanol containing an internal standard. Following protein precipitation and centrifugation, the samples were analyzed by LC-MS. YSA-PTX was run in duplicate with a control compound of known half-life. HLM t1/2 ≥ 30 min is generally classified as good microsomal stability.
For measurements of plasma stability, 1 μM YSA-PTX was incubated at 37 ºC with fresh rat plasma, 2.5% final DMSO concentration. The reactions were terminated at 0, 30, and 60 min by the addition of two volumes of methanol containing an internal standard. Following protein precipitation and centrifugation, the samples were analyzed by LC-MS. The percentage of YSA-PTX remaining at each time point relative to the 0 min sample is calculated from peak area ratios in relation to the internal standard. YSA-PTX was run in duplicate with a positive control known to be degraded in plasma.
In vivo xenograft studies
PC3-3M-luc-C6 cells (1 × 106) were injected subcutaneously in SCID-beige mice and tumor sizes were monitored twice per week using calipers. Once the tumors reached sizes of ~100 mm3, the mice were treated twice a week with intravenous doses of vehicle, PTX (5 mg/Kg), and YSA-PTX (at a dose that is equimolar to the taxol dose, i.e. 15 mg/Kg). Both YSA-PTX and PTX were dissolved in a mixture of 84% PBS, 8% DMSO, and 8% water and injected in a 100 μL final volume.
Tissue drug level determination
Tumor samples were weighted, homogenized and extracted with 4 equal volumes (5 fold dilution) of quenching solution: water: acetonitrile (20:80) with 0.05% formic acid. The plasma samples were extracted with 1 equal volume (2 fold dilution) of quenching solution. The samples were centrifuged at approximately 14,000 x g for 15 min and the supernatants were mixed 1:1 with quenching solution containing an internal standard (2.5 μM indomethacin). After a second centrifugation, the supernatants were transferred to an autosampler vial and the relative concentrations were assessed by LC-MS/MS in multiple reaction monitoring (MRM) mode. LC-MS/MS was performed using a Prominence liquid chromatography system (Shimadzu) directly coupled to an API3000 triple quadrupole mass spectrometer. (Applied Biosystems) using MRM. Chromatography was carried out using gradient elution (water:acetonitrile) on a C18 reversed-phase column (Kromasil 100-5 μm-C18 50 mm x 2.1 mm ID, Peeke Scientific) at a flow rate of 0.4 mL/min.
PTX concentrations were determined using an 8 point calibration curve derived from peak areas obtained from serially-diluted solutions of the compound. The standard solutions were prepared using the same procedure outlined above. PTX concentrations in tissue extracts were determined using Analyst 1.4.2 software (Applied Biosystems). This gives a very accurate quantification of drug levels in tissues.
Supplementary Material
Acknowledgments
Financial support was obtained in part by NIH grant CA138390 to MP and EBP.
Abbreviations used
- YSA
A linear peptide of sequence YSAYPDSVPMMS
- DYP
A linear peptide of sequence DYPSMAMYSPSV
- PTX
paclitaxel
- qPCR
quantitative real-time PCR
- DTPA
Diethylene-triamine-penta-acetic acid
- DOTA
1,4,7,10-Tetraazacyclododecane-N,N′,N″,N′″-tetraacetic acid
- ELISA
Enzyme Linked Immunosorbent Assay
- DMEM
Dulbecco’s Modified Eagle’s Medium
- HDF
Human dermal fibroblast
Footnotes
Authors’ Contributions
MP designed the research strategy for the peptide conjugates; SW synthesized the peptide-PTX conjugates; WJP performed the qPCR experiments, EphA2 protein quantification and characterization of peptide-PTX conjugates in cultured PC3 cells; JLS performed the tumor xenograft experiments; SM and RN performed the ELISA and cell culture experiments; MK performed the YSA cell labeling and internalization experiments; RD performed the plasma and microsomal stability experiments; EPB and MP helped the other authors design and interpret experiments and MP wrote the manuscript with help from the other authors.
Detailed experimental procedures and spectral and purity analysis all compounds. Data relative to EphA2 and EphA4 RNA expression from 68 cancer cell lines is also reported. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Gangloff A, Hsueh WA, Kesner AL, Kiesewetter DO, Pio BS, Pegram MD, Beryt M, Townsend A, Czernin J, Phelps ME, Silverman DH. Estimation of paclitaxel biodistribution and uptake in human-derived xenografts in vivo with (18)F-fluoropaclitaxel. Journal of Nuclear Medicine : official publication, Society of Nuclear Medicine. 2005;46(11):1866–1871. [PubMed] [Google Scholar]
- 2.(a) Sugahara KN, Teesalu T, Karmali PP, Kotamraju VR, Agemy L, Greenwald DR, Ruoslahti E. Coadministration of a tumor-penetrating peptide enhances the efficacy of cancer drugs. Science. 2010;328(5981):1031–1035. doi: 10.1126/science.1183057. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wu AM, Senter PD. Arming antibodies: prospects and challenges for immunoconjugates. Nat Biotechnol. 2005;23(9):1137–1146. doi: 10.1038/nbt1141. [DOI] [PubMed] [Google Scholar]
- 3.(a) Jaracz S, Chen J, Kuznetsova LV, Ojima I. Recent advances in tumor-targeting anticancer drug conjugates. Bioorg Med Chem. 2005;13(17):5043–54. doi: 10.1016/j.bmc.2005.04.084. [DOI] [PubMed] [Google Scholar]; (b) Langer R. New methods of drug delivery. Science. 1990;249(4976):1527–1533. doi: 10.1126/science.2218494. [DOI] [PubMed] [Google Scholar]; (c) Schrama D, Reisfeld RA, Becker JC. Antibody targeted drugs as cancer therapeutics. Nat Rev Drug Discov. 2006;5(2):147–159. doi: 10.1038/nrd1957. [DOI] [PubMed] [Google Scholar]; (d) van Vlerken LE, Amiji MM. Multi-functional polymeric nanoparticles for tumour-targeted drug delivery. Expert Opin Drug Deliv. 2006;3(2):205–216. doi: 10.1517/17425247.3.2.205. [DOI] [PubMed] [Google Scholar]
- 4.(a) Leader B, Baca QJ, Golan DE. Protein therapeutics: a summary and pharmacological classification. Nat Rev Drug Discov. 2008;7(1):21–39. doi: 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]; (b) Marx JL. Monoclonal antibodies in cancer. Science. 1982;216(4543):283–285. doi: 10.1126/science.7063886. [DOI] [PubMed] [Google Scholar]
- 5.(a) Aina OH, Liu R, Sutcliffe JL, Marik J, Pan CX, Lam KS. From combinatorial chemistry to cancer-targeting peptides. Mol Pharm. 2007;4(5):631–651. doi: 10.1021/mp700073y. [DOI] [PubMed] [Google Scholar]; (b) Koolpe M, Dail M, Pasquale EB. An ephrin mimetic peptide that selectively targets the EphA2 receptor. J Biol Chem. 2002;277(49):46974–46979. doi: 10.1074/jbc.M208495200. [DOI] [PubMed] [Google Scholar]; (c) Hatakeyama S, Sugihara K, Shibata TK, Nakayama J, Akama TO, Tamura N, Wong SM, Bobkov AA, Takano Y, Ohyama C, Fukuda M, Fukuda MN. Targeted drug delivery to tumor vasculature by a carbohydrate mimetic peptide. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(49):19587–19592. doi: 10.1073/pnas.1105057108. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Cutrera J, Dibra D, Xia X, Hasan A, Reed S, Li S. Discovery of a linear peptide for improving tumor targeting of gene products and treatment of distal tumors by IL-12 gene therapy. Molecular Therapy : The Journal of the American Society of Gene Therapy. 2011;19(8):1468–1477. doi: 10.1038/mt.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Corti A, Curnis F. Tumor vasculature targeting through NGR peptide-based drug delivery systems. Current Pharmaceutical Biotechnology. 2011;12(8):1128–1134. doi: 10.2174/138920111796117373. [DOI] [PubMed] [Google Scholar]; (f) Wu HC, Chang DK. Peptide-mediated liposomal drug delivery system targeting tumor blood vessels in anticancer therapy. Journal of Oncology. 2010;2010:723798. doi: 10.1155/2010/723798. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Brown KC. Peptidic tumor targeting agents: the road from phage display peptide selections to clinical applications. Current Pharmaceutical Design. 2010;16(9):1040–1054. doi: 10.2174/138161210790963788. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Newton JR, Deutscher SL. In vivo bacteriophage display for the discovery of novel peptide-based tumor-targeting agents. Methods in Molecular Biology. 2009;504:275–290. doi: 10.1007/978-1-60327-569-9_17. [DOI] [PubMed] [Google Scholar]; (i) Yang W, Luo D, Wang S, Wang R, Chen R, Liu Y, Zhu T, Ma X, Liu R, Xu G, Meng L, Lu Y, Zhou J, Ma D. TMTP1, a novel tumor-homing peptide specifically targeting metastasis. Clinical Cancer Research : An Official Journal of the American Association for Cancer Research. 2008;14(17):5494–5502. doi: 10.1158/1078-0432.CCR-08-0233. [DOI] [PubMed] [Google Scholar]; (j) Laakkonen P, Porkka K, Hoffman JA, Ruoslahti E. A tumor-homing peptide with a targeting specificity related to lymphatic vessels. Nature Medicine. 2002;8(7):751–755. doi: 10.1038/nm720. [DOI] [PubMed] [Google Scholar]; (k) Ruoslahti E. Targeting tumor vasculature with homing peptides from phage display. Seminars in Cancer Biology. 2000;10(6):435–442. doi: 10.1006/scbi.2000.0334. [DOI] [PubMed] [Google Scholar]; (l) Koivunen E, Arap W, Valtanen H, Rainisalo A, Medina OP, Heikkila P, Kantor C, Gahmberg CG, Salo T, Konttinen YT, Sorsa T, Ruoslahti E, Pasqualini R. Tumor targeting with a selective gelatinase inhibitor. Nature Biotechnology. 1999;17(8):768–774. doi: 10.1038/11703. [DOI] [PubMed] [Google Scholar]
- 6.Gersuk GM, Corey MJ, Corey E, Stray JE, Kawasaki GH, Vessella RL. High-affinity peptide ligands to prostate-specific antigen identified by polysome selection. Biochem Biophys Res Commun. 1997;232(2):578–582. doi: 10.1006/bbrc.1997.6331. [DOI] [PubMed] [Google Scholar]
- 7.(a) Pasquale EB. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat Rev Cancer. 2010;10(3):165–180. doi: 10.1038/nrc2806. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Noberini R, Lamberto I, Pasquale EB. Targeting Eph receptors with peptides and small molecules: Progress and challenges. Semin Cell Dev Biol. 2011 doi: 10.1016/j.semcdb.2011.10.023. (epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Ireton RC, Chen J. EphA2 receptor tyrosine kinase as a promising target for cancer therapeutics. Curr Cancer Drug Targets. 2005;5(3):149–157. doi: 10.2174/1568009053765780. [DOI] [PubMed] [Google Scholar]; (b) Wykosky J, Debinski W. The EphA2 receptor and ephrinA1 ligand in solid tumors: function and therapeutic targeting. Mol Cancer Res. 2008;6(12):1795–1806. doi: 10.1158/1541-7786.MCR-08-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Landen CN, Kinch MS, Sood AK. EphA2 as a target for ovarian cancer therapy. Expert Opin Ther Targets. 2005;9(6):1179–1187. doi: 10.1517/14728222.9.6.1179. [DOI] [PubMed] [Google Scholar]; (b) Tandon M, Vemula SV, Mittal SK. Emerging strategies for EphA2 receptor targeting for cancer therapeutics. Expert Opin Ther Targets. 2011;15(1):31–51. doi: 10.1517/14728222.2011.538682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biao-Xue R, Xi-Guang C, Shuan-Ying Y, Wei L, Zong-Juan M. Epha2-Dependent Molecular Targeting Therapy for Malignant Tumors. Curr Cancer Drug Targets. 2011;11(9):1082–1097. doi: 10.2174/156800911798073050. [DOI] [PubMed] [Google Scholar]
- 11.(a) Wykosky J, Gibo DM, Debinski W. A novel, potent, and specific ephrinA1-based cytotoxin against EphA2 receptor expressing tumor cells. Mol Cancer Ther. 2007;6(12 Pt 1):3208–3218. doi: 10.1158/1535-7163.MCT-07-0200. [DOI] [PubMed] [Google Scholar]; (b) Sun XL, Xu ZM, Ke YQ, Hu CC, Wang SY, Ling GQ, Yan ZJ, Liu YJ, Song ZH, Jiang XD, Xu RX. Molecular targeting of malignant glioma cells with an EphA2-specific immunotoxin delivered by human bone marrow-derived mesenchymal stem cells. Cancer Lett. 2011;312(2):168–177. doi: 10.1016/j.canlet.2011.07.035. [DOI] [PubMed] [Google Scholar]
- 12.Jackson D, Gooya J, Mao S, Kinneer K, Xu L, Camara M, Fazenbaker C, Fleming R, Swamynathan S, Meyer D, Senter PD, Gao C, Wu H, Kinch M, Coats S, Kiener PA, Tice DA. A human antibody-drug conjugate targeting EphA2 inhibits tumor growth in vivo. Cancer Res. 2008;68(22):9367–9374. doi: 10.1158/0008-5472.CAN-08-1933. [DOI] [PubMed] [Google Scholar]
- 13.Scarberry KE, Dickerson EB, McDonald JF, Zhang ZJ. Magnetic nanoparticle-peptide conjugates for in vitro and in vivo targeting and extraction of cancer cells. J Am Chem Soc. 2008;130(31):10258–10262. doi: 10.1021/ja801969b. [DOI] [PubMed] [Google Scholar]
- 14.Mitra S, Duggineni S, Koolpe M, Zhu X, Huang Z, Pasquale EB. Structure-activity relationship analysis of peptides targeting the EphA2 receptor. Biochemistry. 2010;49(31):6687–6695. doi: 10.1021/bi1006223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macrae M, Neve RM, Rodriguez-Viciana P, Haqq C, Yeh J, Chen C, Gray JW, McCormick F. A conditional feedback loop regulates Ras activity through EphA2. Cancer Cell. 2005;8(2):111–118. doi: 10.1016/j.ccr.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 16.(a) Kolb HC, Finn MG, Sharpless KB. Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angewandte Chemie. 2001;40(11):2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]; (b) Deshayes S, Maurizot V, Clochard MC, Baudin C, Berthelot T, Esnouf S, Lairez D, Moenner M, Deleris G. "Click" conjugation of peptide on the surface of polymeric nanoparticles for targeting tumor angiogenesis. Pharmaceutical Research. 2011;28(7):1631–1642. doi: 10.1007/s11095-011-0398-5. [DOI] [PubMed] [Google Scholar]; (c) Dijkgraaf I, Rijnders AY, Soede A, Dechesne AC, van Esse GW, Brouwer AJ, Corstens FH, Boerman OC, Rijkers DT, Liskamp RM. Synthesis of DOTA-conjugated multivalent cyclic-RGD peptide dendrimers via 1,3-dipolar cycloaddition and their biological evaluation: implications for tumor targeting and tumor imaging purposes. Organic & Biomolecular Chemistry. 2007;5(6):935–944. doi: 10.1039/b615940k. [DOI] [PubMed] [Google Scholar]; (d) von Maltzahn G, Ren Y, Park JH, Min DH, Kotamraju VR, Jayakumar J, Fogal V, Sailor MJ, Ruoslahti E, Bhatia SN. In vivo tumor cell targeting with "click" nanoparticles. Bioconjug Chemistry. 2008;19(8):1570–1578. doi: 10.1021/bc800077y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blackburn WH, Dickerson EB, Smith MH, McDonald JF, Lyon LA. Peptide-Functionalized Nanogels for Targeted siRNA Delivery. Bioconjug Chem. 2009;20(5):960–968. doi: 10.1021/bc800547c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai W, Ebrahimnejad A, Chen K, Cao Q, Li ZB, Tice DA, Chen X. Quantitative radioimmunoPET imaging of EphA2 in tumor-bearing mice. Eur J Nucl Med Mol Imaging. 2007;34(12):2024–2036. doi: 10.1007/s00259-007-0503-5. [DOI] [PubMed] [Google Scholar]
- 19.Fox BP, Tabone CJ, Kandpal RP. Potential clinical relevance of Eph receptors and ephrin ligands expressed in prostate carcinoma cell lines. Biochem Biophys Res Commun. 2006;342(4):1263–1272. doi: 10.1016/j.bbrc.2006.02.099. [DOI] [PubMed] [Google Scholar]
- 20.Vihanto MM, Plock J, Erni D, Frey BM, Frey FJ, Huynh-Do U. Hypoxia up-regulates expression of Eph receptors and ephrins in mouse skin. The FASEB journal : Official Publication of the Federation of American Societies for Experimental Biology. 2005;19(12):1689–1691. doi: 10.1096/fj.04-3647fje. [DOI] [PubMed] [Google Scholar]
- 21.(a) Liu Y, Zhang B, Yan B. Enabling anticancer therapeutics by nanoparticle carriers: the delivery of Paclitaxel. Int J Mol Sci. 2011;12(7):4395–4413. doi: 10.3390/ijms12074395. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Singh S, Dash AK. Paclitaxel in cancer treatment: perspectives and prospects of its delivery challenges. Crit Rev Ther Drug Carrier Syst. 2009;26(4):333–372. doi: 10.1615/critrevtherdrugcarriersyst.v26.i4.10. [DOI] [PubMed] [Google Scholar]
- 22.(a) Coffman KT, Hu M, Carles-Kinch K, Tice D, Donacki N, Munyon K, Kifle G, Woods R, Langermann S, Kiener PA, Kinch MS. Differential EphA2 epitope display on normal versus malignant cells. Cancer Res. 2003;63(22):7907–7912. [PubMed] [Google Scholar]; (b) Wakayama Y, Miura K, Sabe H, Mochizuki N. EphrinA1-EphA2 signal induces compaction and polarization of Madin-Darby canine kidney cells by inactivating ezrin through negative regulation of RhoA. J Biol Chem. 2011 doi: 10.1074/jbc.M111.267047. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Miura K, Nam JM, Kojima C, Mochizuki N, Sabe H. EphA2 engages Git1 to suppress Arf6 activity modulating epithelial cell-cell contacts. Mol Biol Cell. 2009;20(7):1949–1959. doi: 10.1091/mbc.E08-06-0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flanagan JG, Cheng HJ, Feldheim DA, Hattori M, Lu Q, Vanderhaeghen P. Alkaline phosphatase fusions of ligands or receptors as in situ probes for staining of cells, tissues, and embryos. Methods in Enzymology. 2000;327:19–35. doi: 10.1016/s0076-6879(00)27264-9. [DOI] [PubMed] [Google Scholar]
- 24.Carlsson SR, Roth J, Piller F, Fukuda M. Isolation and characterization of human lysosomal membrane glycoproteins, h-lamp-1 and h-lamp-2. Major sialoglycoproteins carrying polylactosaminoglycan. J Biol Chem. 1988;263(35):18911–18919. [PubMed] [Google Scholar]
- 25.Placzek WJ, Wei J, Kitada S, Zhai D, Reed JC, Pellecchia M. A survey of the anti-apoptotic Bcl-2 subfamily expression in cancer types provides a platform to predict the efficacy of Bcl-2 antagonists in cancer therapy. Cell Death & Disease. 2010;1:e40. doi: 10.1038/cddis.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.