Abstract
Study of cell signaling often requires examination of the cellular dynamics under variation in the stimulant concentration. Such variation has typically been conducted by dispensing cell populations in a number of chambers or wells containing discrete concentrations. Such practice adds to the complexity associated with experimental or device design and requires substantial labor for implementation. Furthermore, there is also potential risk of missing important results due to the often arbitrary selection of discrete concentration values for testing. In this letter, we study NF-κB activation and translocation at the single cell level using a microfluidic device that generates continuously varying concentration gradient. We use only three device settings to cover stimulant (interleukin-1β) concentrations of four orders of magnitude (0.001-10 ng/ml). Such device allows us to study temporal dynamics of NF-κB in single cells under different stimulant concentrations by real-time imaging. Interestingly, our results reveal that while the percent of cells with NF-κB translocation decreases with lower stimulant concentration in the range of 0.1-0.001 ng/ml, the response time of such translocation remains constant, reflected by the single cell data.
There has been increasing evidence that cell signaling processes are stochastic in nature and cellular responses to extracellular stimuli are best studied at the single cell level. For example, when cells show an all-or-none response to a particular stimulus (bistability), only a subpopulation of cells respond to the signal 1-5. Conventional bulk measurements only yield the lowered population average and do not reveal the subpopulations. In addition, observation of cellular dynamics with distinct temporal patterns is impossible with population-level approaches, due to the lack of synchronization among cells 6.
NF-κB is a prime example for stochastic signaling dynamics 7,8. NF-κB is a family of dimeric transcription factors that control the expression of hundreds of genes involved in cellular stress responses, cell division, apoptosis, and inflammation 9-11. In the canonical activation pathway, NF-κB resides in the cytoplasm of the cell (due to the negative regulation of IκB kinase that is bound to NF-κB heterodimers) until activation in response to a wide range of extracellular stimuli (e.g. tumour-necrosis factor-α (TNFα), interleukin-1 (IL-1), lipopolysaccharide (LPS) and CD40 ligand (CD40L)). Signals are transduced to the IκB kinase (IKKβ) complex. IKKβ then phosphorylates IκB at two conserved serines, leading to its degradation via ubiquitination and proteolysis. This allows librated NF-κB (RELA-p50 or c-REL-p50) to translocate to the nucleus, bind to DNA and regulate gene transcription within. NF-κB signaling presents rich dynamics at the single cell level. For example, NF-κB responds to endotoxin LPS stimulation with heterogeneity in the population (“transient and persistent responders”)5. Furthermore, NF-κB activation triggered by TNF-α and TCR appear to be digital (switch-like, as opposed to be analog or graded) with stochastic element involved in the process 3,12. Not surprisingly, it was found that TNF-α (the stimulant) concentration was a critical parameter that affected NF-κB dynamics 3,13,14.
In this study, we use a microfluidic device that allows exposure of cell culture to a continuous stimulant (IL-1β) concentration gradient. IL-1β is a multifunctional cytokine produced primarily by activated monocytes/macrophages and by a variety of other cell types. IL-1β plays an integral role in the immuno-inflammatory response of the body to a variety of stimuli including infection, trauma and other bodily injuries. IL-1β activates NF-κB via a pathway similar to that of TNF-α, due to the largely overlapping biological activities of the two cytokines 15. The concentration gradient device allows us to investigate concentrations spanning several orders of magnitude with very few experiments. The simple fluid mechanics associated with laminar flows permits accurate computation of the stimulant concentration distribution based on the physical properties of the stimulant molecule and the operational conditions. By combining the stimulant concentration information with the single cell NF-κB dynamics (i.e. nuclear translocation) observed by real-time imaging, we establish the patterns of NF-κB response to IL-1β of various concentrations. Other than confirming that the percentage of responding cells decreases with IL-1β concentration, more interestingly, we discover that the correlation between the response time (the duration from the start of the stimulation to the nuclear translocation) and IL-1β concentration disappears when IL-1β concentration is low (0.001-0.1 ng/ml), revealing an upper bound for the response time.
Previous studies on stimulant concentration effect applied chambers or wells with discrete concentrations 3,13,14,16. These studies required using a large number of units for various concentrations and there was potential risk for not covering the important concentration values. In comparison, we use a microfluidic device with a network of channels for generating concentration gradient by controlled diffusive mixing 17-19. We use three settings (i.e. inlet concentrations) to cover IL-1β concentrations that continuously span 4 orders of magnitude (0.001-10 ng/ml). As shown in Fig. 1, with the stimulant (IL-1β) solution coming in from one inlet and buffer in from the other inlet, the branched array of channels repeatedly splits and combines microscale streams to result in multiple streams with different stimulant concentrations that flow into a downstream microfluidic chamber. This generates a stable concentration gradient that is perpendicular to the flow direction and maintained over time at steady state. We culture a monolayer of Chinese hamster ovary (CHO)/GFP-NFκBp65 cells (Panomics), created by co-transfection of an expression vector for a fusion protein of turboGFP (Evrogen) and human NF-κBp65, as well as pHyg into CHO cells, in the microfluidic chamber 20. Cells are flowed into the chamber for seeding from the outlet and there is a microfluidic valve that is half-closed during cell seeding for preventing cell entry into the channel network. The inset plots show that IL-1β concentration profile along the width of the culture chamber at any specific location along the stream can be modeled using COMSOL (ignoring the effects of the seeded cells on the flow). The modeled values match experimental results well as shown by the example of labeled β-casein (Fig. S1 in SI). At Z=1 mm (i.e. 1 mm from where the channel network interfaces with the culture chamber), the maximal IL-1β concentration along the chamber width (at the very left) is the same as its inlet concentration and the minimal concentration (at the very right) is 0. In comparison, the maximal concentration becomes 93% of the inlet concentration and the minimum is 8% in the downstream at Z=11 mm. As shown in the inset image series and Movie S1 in SI, with inlet IL-1β concentration being 1 ng/ml, the cells at the left side of the culture chamber present higher percentage of activation (NF-κB nuclear translocation) and faster response than the ones at the right, due to the concentration gradient.
Figure 1.
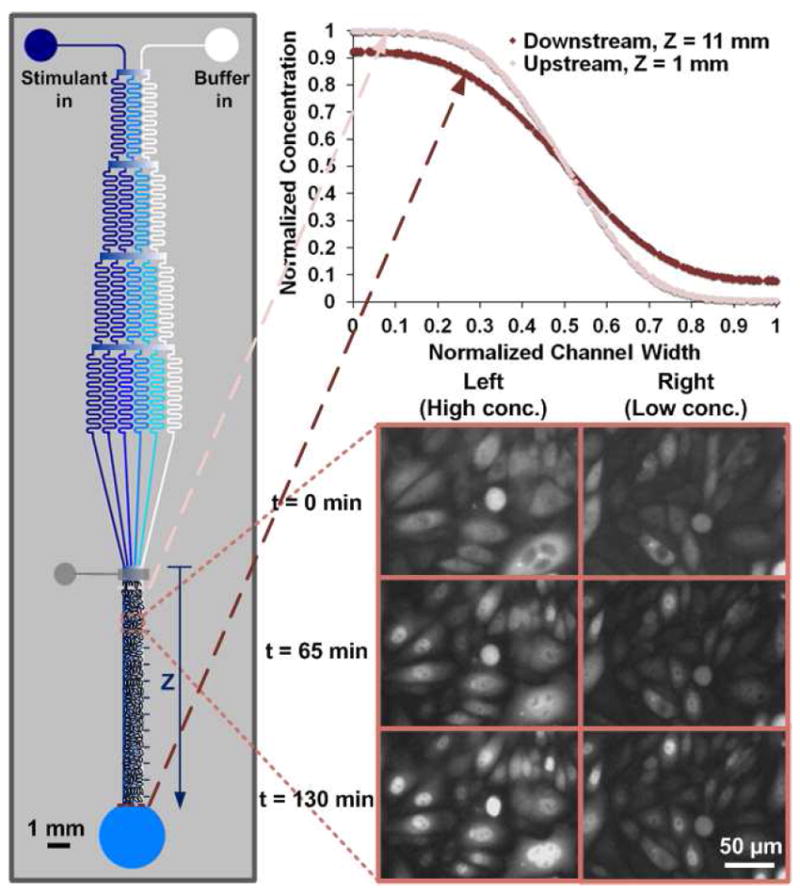
The design of the microfluidic concentration gradient device for studying single cell NF-κB dynamics. CHO/GFP-NFκBp65 cells are seeded in one monolayer in the microfluidic culture chamber (having a depth of 60 μm, a width of 0.9 mm, and a length of 11 mm). For stimulation of cells, IL-1β of a specific concentration (0.1, 1, or 10 ng/ml) flows into the device at 0.05 μl/min from the left inlet while neat PBS buffer flows in from the right inlet at the same flow rate. The concentration profiles along the width of the culture chamber can be modeled by COMSOL and the concentration profiles at Z=1 and 11 mm are shown in the upper inset graph. The lower inset images show the fluorescence images of the cells after stimulation by IL-1β (inlet concentration of 1 ng/ml) for 0, 65, 130 min. The images show that at the left side of the culture chamber with high IL-1β concentration, the majority of the cells experience NF-κB translocation. In comparison, at the right side, few cells show translocation.
There are two types of measurements we conduct using the gradient generation device. First, we examine the per cent activation of CHO cells at various IL-1β concentrations. In this case, we divide the observation frame (900 μm × 685 μm in the size) into 9 or 10 vertical slabs with each slab roughly covering a concentration range of equal size. The IL-1β concentration at the center of the slab is used to represent the concentration of the slab. The population of cells within each slab (N~ 30-50 cells) was observed for NF-κB translocation during a period of 3 h under constant stimulation by IL-1β. The cells undergoing NF-κB translocation (indicated by movement of the fluorescence intensity center from the cytosol to the nucleus) are enumerated and the per cent activation is calculated by dividing the number of activated cells by the total cell number. Second, we also examine the response time for every single cell in the observation frame. The response time is determined by how long it takes for NF-κB translocation to occur since the start of the IL-1β stimulation. We are able to determine the IL-1β concentration that each single cell is exposed to, based on the cell location using COMSOL modeling, and correlate it to the response time for the particular cell.
Fig. 2 (a-c) show how the per cent cell activation varies with the IL-1β concentration, examined by having the inlet IL-1β concentrations at 10, 1 and 0.1 ng/ml, respectively. We find that the percentage of activated cells (exhibiting NF-κB translocation) does not vary in the range of 1-10 ng/ml, due to the saturation of cell surface receptors by the stimulant. The percentage of activated cells increases with higher IL-1β concentration in the range of 0.001-1 ng/ml. Such trend can also be seen clearly in the Fig. 2d with all the data compiled and IL-1β concentration shown in a logarithmic scale. Fig. 2 (e-g) show single cell dynamics in the NF-κB signaling dependent on the stimulant concentration. Each marker in the figures represents correlation between the response time and the IL-1β concentration, generated by analyzing a single cell. Fig. 2e shows that there is no strong correlation between the IL-1β concentration and the response time when the IL-1β concentration is in the range of 5-10 ng/ml. Then the response time decreased with higher IL-1β concentration in the range of 0.1-5 ng/ml (Fig. 2e and 2f). However, the response time becomes independent of the IL-1β concentration again in the range of 0.001-0.1 ng/ml again (Fig. 2g). We did not observe NF-κB nuclear oscillations3,21 (i.e. in and out of the nucleus) within the observation time (~3 h). Since the expression level of NF-κB fused with the fluorescent protein marker is known to strongly affect the dynamics and oscillations 22,23, such lack of nuclear oscillation is possibly due to the characteristics of the CHO cell line used.
Figure 2.
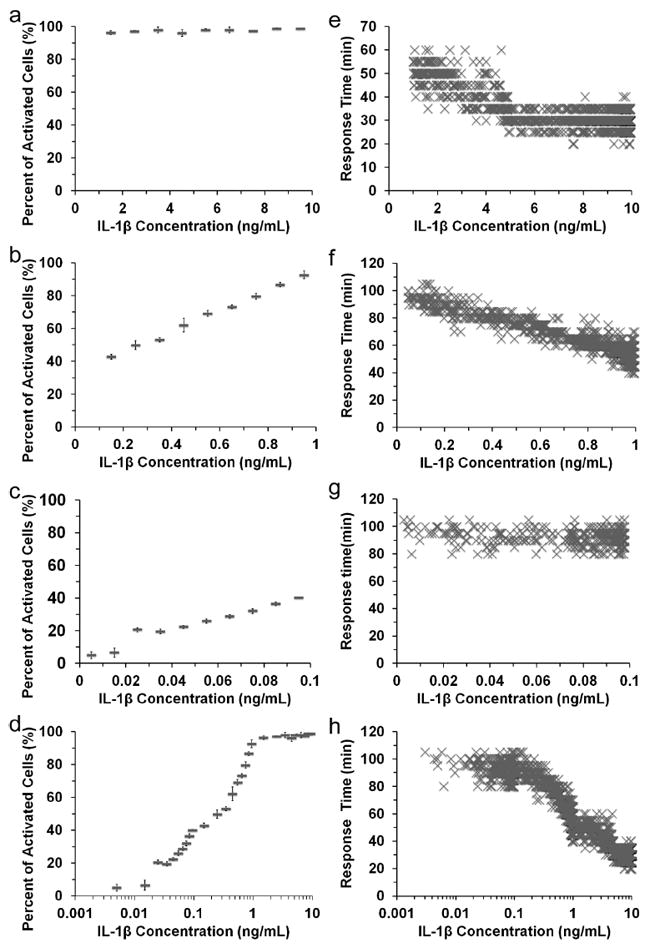
The variations of the per cent cell activation and the response time of single cells with IL-1β concentration. The percentage of activated cells (with NF-κB translocation) varies with different IL-1β concentrations when the device has IL-1β inlet concentrations at (a) 10, (b) 1, and (c) 0.1 ng/ml. The data from (a-c) are also compiled in (d) with the concentrations in a logarithmic scale. Each data point in (a-d) is generated by three separate trials. The response time (the time from application of IL-1β stimulation to NF-κB translocation) of single cells is plotted against the concentration that the particular cell is exposed to, when the device has IL-1β inlet concentrations at (e) 10, (f) 1, and (g) 0.1 ng/ml. The data from (e-g) are also compiled in (h) with the concentrations in a logarithmic scale.
Our observation of NF-κB activation and translocation yields interesting insights. Although it is known that the lower stimulant concentration induces a lower percentage of cells that respond with NF-κB activation, it is surprising to reveal that the response time of these responding cells does not depend on IL-1β concentration in the very low concentration regime. Our results show that while the percentage of activated cells experiences a significant and continuous decline from 40% to 5% as IL-1β concentration varies from 0.1 to 0.005 ng/ml (Fig. 2c), there is no significant change in the response time at the single cell level in the same concentration range (R2 = 0.0372 in Fig. 2g). This finding is consistent with the previous bulk measurements revealing that lower TNFα dose reduces the amount of nuclear NF-κB while maintaining the same temporal profile regardless of the dose 13.
Microfluidic concentration gradient device provides a simple platform for studying NF-κB activity and cell signaling in general at the single cell level. Stimulant concentrations spanning multiple orders of magnitude can be easily covered by a small set of experiments. Single cells in a monolayer that are treated by continuously varying stimulant concentration can be observed by real-time imaging for their dynamics. Because we are able to treat each single cell as an individual experimental subject, the results yield high statistical significance. We expect increased application of this approach for single cell signaling studies in the future.
Supplementary Material
Acknowledgments
This work was supported by NSF grants CBET 1016547, 0967069 (to C.L.) and NIH AI35098 (to A.S.B.).
Footnotes
Supporting Information. Additional information including experimental section, supplemental data, and video. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Ferrell JE, Jr, Machleder EM. Science. 1998;280:895. doi: 10.1126/science.280.5365.895. [DOI] [PubMed] [Google Scholar]
- 2.Ferrell JE, Xiong W. Chaos. 2001;11:227. doi: 10.1063/1.1349894. [DOI] [PubMed] [Google Scholar]
- 3.Tay S, Hughey JJ, Lee TK, Lipniacki T, Quake SR, Covert MW. Nature. 2010;466:267. doi: 10.1038/nature09145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Covert MW, Leung TH, Gaston JE, Baltimore D. Science. 2005;309:1854. doi: 10.1126/science.1112304. [DOI] [PubMed] [Google Scholar]
- 5.Lee TK, Denny EM, Sanghvi JC, Gaston JE, Maynard ND, Hughey JJ, Covert MW. Sci Signal. 2009;2:ra65. doi: 10.1126/scisignal.2000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lahav G, Rosenfeld N, Sigal A, Geva-Zatorsky N, Levine AJ, Elowitz MB, Alon U. Nat Genet. 2004;36:147. doi: 10.1038/ng1293. [DOI] [PubMed] [Google Scholar]
- 7.Lee TK, Covert MW. Curr Opin Genet Dev. 2010;20:677. doi: 10.1016/j.gde.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paszek P, Ryan S, Ashall L, Sillitoe K, Harper CV, Spiller DG, Rand DA, White MR. Proc Natl Acad Sci U S A. 2010;107:11644. doi: 10.1073/pnas.0913798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann A, Baltimore D. Immunol Rev. 2006;210:171. doi: 10.1111/j.0105-2896.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- 10.Hoffmann A, Natoli G, Ghosh G. Oncogene. 2006;25:6706. doi: 10.1038/sj.onc.1209933. [DOI] [PubMed] [Google Scholar]
- 11.Kim HJ, Hawke N, Baldwin AS. Cell Death Differ. 2006;13:738. doi: 10.1038/sj.cdd.4401877. [DOI] [PubMed] [Google Scholar]
- 12.Kingeter LM, Paul S, Maynard SK, Cartwright NG, Schaefer BC. J Immunol. 2010;185:4520. doi: 10.4049/jimmunol.1001051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheong R, Bergmann A, Werner SL, Regal J, Hoffmann A, Levchenko A. J Biol Chem. 2006;281:2945. doi: 10.1074/jbc.M510085200. [DOI] [PubMed] [Google Scholar]
- 14.Turner DA, Paszek P, Woodcock DJ, Nelson DE, Horton CA, Wang Y, Spiller DG, Rand DA, White MR, Harper CV. J Cell Sci. 2010;123:2834. doi: 10.1242/jcs.069641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathias S, Younes A, Kan CC, Orlow I, Joseph C, Kolesnick RN. Science. 1993;259:519. doi: 10.1126/science.8424175. [DOI] [PubMed] [Google Scholar]
- 16.Cheong R, Wang CJ, Levchenko A. Mol Cell Proteomics. 2009;8:433. doi: 10.1074/mcp.M800291-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeon NL, Dertinger SKW, Chiu DT, Choi IS, Stroock AD, Whitesides GM. Langmuir. 2000;16:8311. [Google Scholar]
- 18.Li Jeon N, Baskaran H, Dertinger SK, Whitesides GM, Van de Water L, Toner M. Nat Biotechnol. 2002;20:826. doi: 10.1038/nbt712. [DOI] [PubMed] [Google Scholar]
- 19.Gunawan RC, Silvestre J, Gaskins HR, Kenis PJA, Leckband DE. Langmuir. 2006;22:4250. doi: 10.1021/la0531493. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, Fei B, Zhan YH, Geahlen RL, Lu C. Lab Chip. 2010;10:2911. doi: 10.1039/c0lc00094a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nelson DE, Ihekwaba AEC, Elliott M, Johnson JR, Gibney CA, Foreman BE, Nelson G, See V, Horton CA, Spiller DG, Edwards SW, McDowell HP, Unitt JF, Sullivan E, Grimley R, Benson N, Broomhead D, Kell DB, White MRH. Science. 2004;306:704. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- 22.Nelson G, Paraoan L, Spiller DG, Wilde GJC, Browne MA, Djali PK, Unitt JF, Sullivan E, Floettmann E, White MRH. J Cell Sci. 2002;115:1137. doi: 10.1242/jcs.115.6.1137. [DOI] [PubMed] [Google Scholar]
- 23.Barken D, Wang CJ, Kearns J, Cheong R, Hoffmann A, Levchenko A. Science. 2005:308. doi: 10.1126/science.1107904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


