Abstract
Pseudomonas aeruginosa is well adapted to grow in anaerobic environments in the presence of nitrogen oxides by generating energy through denitrification. Environmental cues, such as oxygen and nitrogen oxide concentrations, are important in regulating the gene expression involved in this process. Recent data indicate that P. aeruginosa also employs cell-to-cell communication signals to control the denitrifying activity. The regulation of denitrification by these signalling molecules may control nitric oxide production. Nitric oxide, in turn, functions as a signalling molecule by activating certain regulatory proteins. Moreover, under denitrifying conditions, drastic changes in cell physiology and cell morphology are induced that significantly impact group behaviours, such as biofilm formation.
1. Introduction
It is well acknowledged that bacteria exhibit social behaviours by communicating with each other through signalling molecules or by developing a community known as biofilm. The social behaviour of bacteria is of great interest to researchers, and Pseudomonas aeruginosa is one of the most studied bacterial model organisms.
P. aeruginosa has a flexible metabolism that can utilise nitric oxides as alternative electron acceptors to produce energy when oxygen is depleted [1]. This process is called denitrification and is also performed by many other bacteria. The stepwise process of denitrification in P. aeruginosa is as follows: NO3 − → NO2 − → NO → N2O → N2. The sequential steps are catalysed by the enzymes NO3 − reductase (NAR), NO2 − reductase (NIR), NO reductase (NOR), and N2O reductase (N2OR), respectively [2]. This process is important in the nitrogen cycle to produce nitrogen gases from NO3 − and NO2 −. Moreover, recent studies indicate that the denitrification process is related to the virulence of this bacterial species. P. aeruginosa is notorious as an opportunistic pathogen that infects immunocompromised patients, such as cystic fibrosis (CF) patients. How the bacteria adapt to the host environment is important in terms of its pathogenesis. The CF airway has been described as a microaerobic to anaerobic environment [3, 4]. Independent studies indicate the expression of denitrifying genes in the CF lung, suggesting that denitrification is important for the pathogenicity of P. aeruginosa [5, 6]. Thus, an understanding of the physiology under anaerobic conditions is important for the understanding of bacterial virulence under such conditions.
While there are many excellent reviews available about the social behaviours of P. aeruginosa under aerobic conditions, few have focused on anaerobic conditions. In this paper, we examine the social behaviour of P. aeruginosa under anaerobic conditions.
2. Denitrification Regulation by Physicochemical Conditions
The expression of denitrifying enzymes is controlled by a sophisticated regulatory network that responds to low oxygen conditions and the availability of nitrate or nitrite. The master regulator that monitors oxygen concentration is the ANR (anaerobic regulation of arginine deiminase and nitrate reduction) regulatory protein [7]. The active form of ANR contains an [4Fe-4S]2+ cluster that is destroyed in the presence of oxygen [8]. ANR induces genes that are involved in producing energy under low-oxygen conditions or anaerobic conditions. One of these genes, the cbb3-2 terminal oxidase, has a high affinity for oxygen, indicating that it plays a role under low-oxygen conditions [9, 10]. Other genes induced by ANR include the genes involved in fermentation [11]. In addition to these genes, ANR induces other transcriptional regulators involved in denitrification, NarXL, and DNR (dissimilative nitrate respiration regulator) [12, 13]. NarX and NarL comprise a two-component regulatory system that responds to nitrate. The sensor kinase NarX detects nitrate and activates the response regulator NarL, which regulates the transcription of narK1, nirQ, and dnr [12]. In addition, NarL partially represses the expression of arginine fermentation genes, enabling the bacteria to benefit from the more energetically efficient denitrification instead of low energy-yielding fermentation in the presence of nitrate under anaerobic conditions [14]. DNR is activated by binding to NO, and the active DNR regulator activates transcription of all four denitrifying reductases [12, 13, 15].
3. Cell-Cell Communication Signals in P. aeruginosa
In P. aeruginosa, two chemically distinct types of signalling molecules have been characterised in detail. One type consists of the N-acyl-L-homoserine lactone (AHL) signals. AHLs are produced widely in gram-negative bacteria, and P. aeruginosa is known to possess two AHL signaling systems, the LasR-LasI (las) and the RhlR-RhlI (rhl) systems [16]. LasI directs the synthesis of the AHL signal N-(3-oxododecanoyl)-L-homoserine lactone (3-oxo-C12-HSL) [17, 18], and RhlI directs the synthesis of another AHL signal, N-butyryl-L-homoserine lactone (C4-HSL) [19, 20]. These AHL signals have cognate receptors (LuxR proteins), LasR [21] and RhlR [22], that are activated by 3-oxo-C12-HSL and C4-HSL, respectively. In addition to the LasR and RhlR receptors, two additional LuxR homologues, QscR and VqsR, which are orphan LuxR proteins and not associated with a cognate AHL synthase, have been found. QscR binds to a variety of AHL molecules with different side chains, while the AHL molecule that binds to VqsR is unknown [23, 24]. The other type of signalling molecules in P. aeruginosa include 2-alkyl-4-quinolone (AQ). The AQ signalling molecules are produced by the product of the pqsABCDE operon together with the product of the pqsH gene and converts 2-heptyl-4-quinolone (HHQ) to the Pseudomonas quinolone molecule (PQS; 2-heptyl-3-hydroxy-4-quinolone) [25]. HHQ is also produced in bacteria other than P. aeruginosa [26]. In P. aeruginosa, both PQS and HHQ are able to regulate the pqsABSDE operon via a transcriptional regulator, PqsR (MvfR) [27]. Interestingly, PQS is carried by outer membrane vesicles (OMVs), which are thought to target neighbouring cells [28, 29].
4. Denitrification Regulation by Signalling Molecules
As mentioned above, denitrification is well regulated by the physiochemical environment. In addition, denitrification is regulated by cell-cell communication molecules. Interestingly, all three cell-cell signalling molecules, C4-HSL, 3-oxo-C12-HSL, and PQS, repress denitrification. AHLs and PQS affect denitrification in different manners. In the study conducted by Yoon et al. [30], it was demonstrated that the activity of the denitrifying enzymes is higher in rhlR mutants compared to its parent strain. A microarray study suggested that the denitrifying genes are regulated by AHLs [31]. Following these observations, it was demonstrated in detail that, indeed, AHLs regulate denitrifying activity. Both C4-HSL and 3-oxo-C12-HSL repressed denitrifying activity via their cognate regulator, RhlR or LasR, by regulating the expression of the denitrifying genes [32]. Regulation by the las quorum-sensing system was dependent on the rhl quorum-sensing system, suggesting hierarchical regulation by the las system over the rhl system in denitrification regulation, although the precise mechanism of denitrification regulation by AHLs has yet to be identified. In P. aeruginosa isolates from CF patients, mutations in the lasR gene are often found [33, 34]. Considering the fact that there are microenvironments with low oxygen tension inside the CF lung, it is possible that the lasR mutation confers a growth advantage by the activation of denitrification Figure 1 [35].
Figure 1.
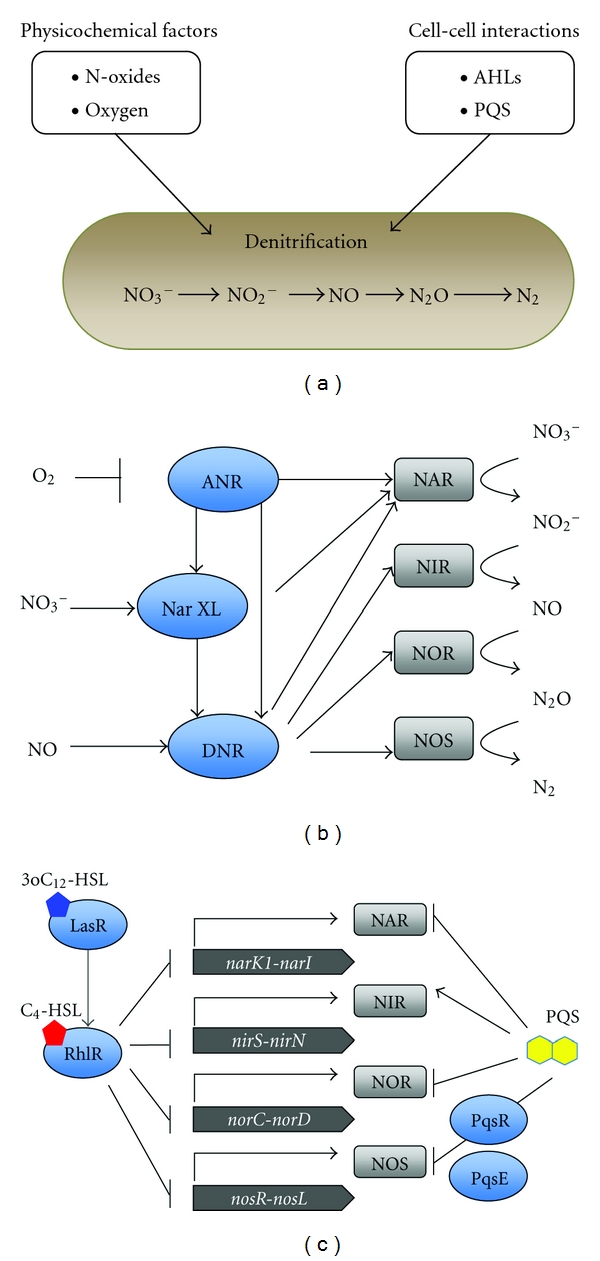
(a) Denitrification regulation in P. aeruginosa. Denitrification is regulated by physiochemical conditions, such as oxygen concentration and the availability of nitric oxides as well as cell-cell communication molecules. (b) Denitrification regulation by physiochemical factors in P. aeruginosa. ANR is activated under low-oxygen tension conditions, which increases the transcription of the NarXL two-component system and DNR. NarXL responds to nitrate and activates the expression of DNR together with ANR. DNR is activated in the presence of NO and induces the transcription of the four reductases. The narK1K2GHJI operon, which encodes nitrate/nitrite transporters and the structural gene for NAR, is also induced by ANR and NarL. (c) Denitrification regulation by cell-cell communication. C4-HSL and 3-o-C12-HSL repress the transcription of denitrifying reductases via their cognate receptors RhlR and LasR. Regulation by LasR is dependent on RhlR. PQS represses the activity of the NAR, NOR, and NOS enzymes while activating the NIR enzyme. PqsR and PqsE are involved in the PQS effect on NOR, while the effect on the other three enzymes does not require PqsR or PqsE.
While the AHLs regulate the transcription of denitrifying genes, PQS affects the activity of denitrifying enzymes posttranscriptionally. It has been shown that NAR and NOR activity is repressed and NIR activity is increased in the presence of PQS [36]. Furthermore, when the PQS molecule was added to a crude extract containing denitrifying enzymes, NO3 −-respiration activity and NOR activity were repressed [36]. This was the first study to demonstrate that a signalling molecule affects enzyme activity in a direct manner. The transcription of denitrifying genes was unaffected by PQS under anaerobic conditions [36], while microarray studies suggested that PQS represses nar gene transcription under aerobic conditions [37], indicating that there may be several pathways in denitrification regulation by PQS. In addition to working as a signalling molecule that activates its cognate receptor, the PQS molecule is shown to have multiple functions, such as chelating iron, balancing the production of reacting oxygen species, and inducing outer membrane production [28, 38–40]. Iron concentration is a key environmental condition for the PQS effect on denitrification because excess amounts of iron inhibit this effect [36]. Interestingly, PQS affects the aerobic and anaerobic growth of bacterial species other than P. aeruginosa, indicating its broad impact on the bacterial community [41].
5. Cell-Cell Signalling under Denitrifying Conditions
While cell-cell communication has a wide impact on cell physiology, most of the studies in P. aeruginosa have been performed under aerobic conditions, and there have been only a limited number of studies concerning the impact of the cell-cell communication systems under anaerobic conditions. Expression of lasR, lasI, rhlR, and rhlI was shown to be altered under denitrifying conditions [31, 42]. Furthermore, a recent study measuring the AHLs under denitrifying conditions revealed that the production of AHLs is significantly lower compared to aerobic cultures [43]. The exact mechanism of this attenuation in signal production has not been revealed, but it is proposed that the limitation of the acyl carrier proteins leads to the low level of signalling molecules [43]. Interestingly, the AHL signalling systems under denitrifying conditions still actively regulate genes to some extent, as has been demonstrated by the regulation of denitrifying activity using AHL production-defective mutant strains [30, 32]
PQS production is also suppressed under anaerobic conditions [36, 44]. In this case, the enzymes that convert HHQ to PQS require oxygen. Therefore, under anaerobic conditions, hydroxylation of HHQ does not occur, and thus, PQS synthesis is prevented [44]. However, under these conditions, a sufficient amount of HHQ that can induce pqsABCDE transcription is present [44]. It was shown that both PQS and HHQ bind to the PqsR transcriptional regulator, although PQS is approximately 100-fold more potent in stimulating the activation of PqsR [27]. These results imply that HHQ may play an important role in cell-cell communication under denitrifying conditions. In fact, HHQ is used as a signalling molecule in several other bacteria [26], but the impact of HHQ as a signalling molecule has yet to be fully understood in P. aeruginosa.
Although production of all three signalling molecules is attenuated under anaerobic conditions, the exogenous addition of these signalling molecules restore the transcription of target genes [36, 43]. These results indicate that the cells under denitrifying conditions are altered in producing signalling molecules, but they are still able to respond to them. It is important to consider that the natural habitat of bacteria is not a stable environment and conditions, such as oxygen concentration, are likely to fluctuate. Moreover, even under the same environmental conditions, oxygen-limited patches are produced by the cells itself as observed in biofilms [45]. In these cases, it is possible that the signalling molecules produced in one growth condition affect the cells in other growth conditions. One example is the PQS effect on denitrification. As mentioned above, PQS is not produced at a sufficient amount under anaerobic conditions [36, 44]; however, when bacteria were grown under oxygen-limiting conditions in which oxygen was depleted depending on cell growth, denitrifying activity was higher in the non-PQS-producing mutants [36]. This result demonstrates that a signalling molecule produced in one environment can affect the cells in another environment. Because it is known that cell-cell signalling is modulated by environmental factors [46], it would also be interesting to further investigate whether there are any differences in the regulon regulated by the same molecules under aerobic and anaerobic conditions.
6. NO Signalling in P. aeruginosa
NO has been studied extensively as a signalling molecule in eukaryotic cells that plays important roles in many biological processes. However, the role of NO in bacteria has not been fully understood. Some recent studies have demonstrated that NO is produced through the oxidation of L-arginine in certain gram-positive bacteria [47, 48], as it is in mammals, and it can protect the bacteria from reactive oxygen species. During denitrification, NO is produced as an intermediate by the reduction of nitrite, a process catalysed by the NIR enzyme. In P. aeruginosa, the denitrifying pathway is the only biological pathway known to produce NO. This implies that under denitrifying conditions, NO may become a signal that, when produced by one cell, can affect the other cell. A number of regulators that respond to NO have been revealed. As explained above, NO is an important signal for inducing the denitrifying genes. In this case, NO is recognised by the regulatory protein DNR, which regulates the denitrifying genes [15, 49]. The activated DNR recognises the conserved DNA binding site (ANR box) in promoters to regulate transcription. A recent comprehensive study to determine ANR and DNR regulons suggested that, in addition to the denitrifying genes, the transcription of three genes, C4-dicarboxylate transport (PA1183), a hypothetical protein (PA3519), and the RND-type efflux pump mexG (PA4205), is influenced by DNR, although no ANR box has been found in the promoter regions of these genes [50]. Still, the roles of these genes under denitrifying conditions or in NO response are not known. Another type of RND efflux pump (mexEF-oprN) was suggested to be induced by NO [51]. In this study, the MexT regulator was required for mexEF-oprN induction by NO, while the mechanism is still obscure. Nevertheless, it is becoming evident that the expression of efflux pumps is involved in diverse cellular activities that affect the expression of several genes. A recent study has demonstrated that the expression of the MexEF-oprN efflux pump downregulates several genes [52]; thus, NO may impact the expression of these genes through the expression of this efflux pump.
FhpR is another NO-responsive regulator that, under aerobic conditions, regulates the flavohaemoglobin (fhp) gene, the product of which oxidises NO. FhpR is an orthologue of NorR found in E.coli and belongs to the σ 54-dependent family of transcriptional activators [53].
While high amounts of NO induce genes for NO detoxification [54] and the DNR and FhpR regulatory proteins are likely to regulate these genes, a series of studies have revealed that NO in nontoxic levels regulates the social behaviour of bacteria, such as the dispersal of P. aeruginosa in biofilms [55]. The underlying mechanism is not yet fully understood, but a secondary messenger (c-di-GMP) is involved in this process. Low levels of c-di-GMP induce bacterial motility in P. aeruginosa, which in turn induce dispersal in biofilms. NO has been shown to increase the activity of enzymes that degrade c-di-GMP. This process requires the chemotaxis transducer BdlA [56]. These data indicate that there is a biological pathway independent of the toxic response that responds to NO.
Although it is not fully characterised, another NO-responsive gene that regulates biofilm formation has been suggested. This gene product (PA2663) increases biofilm formation by inducing the production of psl exopolysaccharides while reducing swarming and swimming motility [57]. It also increases the production of virulence-related factors, such as pyoverdine, PQS, and elastase. The gene is located within the same operon as the fhp gene, indicating that NO will induce its expression. Interestingly, PA2663 has two transmembrane regions and may sense NO, although further study is needed.
NO has also been suggested to induce virulence factors. A nirS mutant deficient in NO production was shown to have a reduced amount of type III secretion system-dependent exotoxin production compared to wild type. This NO-dependent exotoxin induction was confirmed by the addition of exogenous NO donors [58].
One of the important factors that determines the cellular response to NO is the concentration of NO. High levels of NO are toxic to the cell, but low levels of NO could function as a signal. How NO concentration is modulated in the cell during denitrification is not fully understood, but one possibility is denitrification control by the AHL and PQS molecules because they affect NIR and NOR activity to a different extent. For example, rhl quorum-sensing mutants increase NO production due to the imbalance of NIR and NOR activity, and this induces cell death under denitrifying conditions [30]. PQS upregulates NO-producing NIR enzyme activity, while it downregulates the NO-reducing NOR enzyme activity, suggesting that PQS would induce NO accumulation [36].
7. Biofilm Formation under Denitrifying Conditions
A study by Yawata et al. [59] showed that filamentous cells emerge extensively in biofilms formed under denitrifying conditions. This was followed by the study by Yoon et al., which indicated that the filamentation is due to a defect in cell division, and filamentation of the cells is correlated with biofilm formation [60]. It was also suggested in this study that the filamentation is induced by NO [60]. Filamentation of cells has long been characterised as a trait of cell morphology under stressful conditions, such as UV light exposure, antibiotic treatment, and nutrient deprivation. In contrast, it is known that filamentation of the cell is a part of cell differentiation in bacteria, such as Proteus mirabilis [61]. Currently, it is not known whether the filamentation in denitrifying biofilms is a response to NO toxicity or an adaptive response to denitrifying conditions or whether there is a specific signalling pathway that initialises cell filamentation in P. aeruginosa. Nevertheless, morphological changes may impact cell physiology and behaviours and provide advantages for survival under certain circumstances [62].
Most of the studies investigating biofilm formation in P. aeruginosa have been performed under aerobic conditions. The morphological change in cells under denitrifying conditions causes us to question how the biofilm development process differs under aerobic conditions. Studies using a selected number of mutants have indicated that the same gene may have different effects on biofilm formation under aerobic and anaerobic conditions [30]. These studies suggest the possibility that the biofilm development procedure differs under denitrifying conditions.
8. Perspectives
As our knowledge of energy metabolism under anaerobic conditions expands, new questions arise with respect to the physiology of the cells under those conditions. Under denitrifying conditions, the cells undergo morphological changes [59, 60], and cell physiology is likely to change dynamically with the shift in respiration systems [42]. Understanding the physiology under denitrifying conditions in P. aeruignosa is also important from clinical perspectives because anaerobiosis leads to the alteration in antibiotic tolerance, the mechanism of which is not fully understood [45]. Attenuation of the AHL and PQS signalling systems and different biofilm development processes raise further questions about the social behaviours of the bacteria under anaerobic conditions. It will be interesting to investigate whether additional signals or systems that coordinate group behaviours under anaerobic conditions may exist.
Acknowledgments
The authors thank Core Research for Evolutional Science and Technology (CREST) and the Ministry of Education, Culture, Sports, and Technology of Japan for financial support.
References
- 1.Arai H. Regulation and function of versatile aerobic and anaerobic respiratory metabolism in Pseudomonas aeruginosa . Frontiers in Microbiology. 2011;2(103) doi: 10.3389/fmicb.2011.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zumft WG. Cell biology and molecular basis of denitrification? Microbiology and Molecular Biology Reviews. 1997;61(4):533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Worlitzsch D, Tarran R, Ulrich M, et al. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. Journal of Clinical Investigation. 2002;109(3):317–325. doi: 10.1172/JCI13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanderson K, Wescombe L, Kirov SM, Champion A, Reid DW. Bacterial cyanogenesis occurs in the cystic fibrosis lung. European Respiratory Journal. 2008;32(2):329–333. doi: 10.1183/09031936.00152407. [DOI] [PubMed] [Google Scholar]
- 5.Beckmann C, Brittnacher M, Ernst R, Mayer-Hamblett N, Miller SI, Burns JL. Use of phage display to identify potential Pseudomonas aeruginosa gene products relevant to early cystic fibrosis airway infections. Infection and Immunity. 2005;73(1):444–452. doi: 10.1128/IAI.73.1.444-452.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Son MS, Matthews WJ, Kang Y, Nguyen DT, Hoang TT. In vivo evidence of Pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infection and Immunity. 2007;75(11):5313–5324. doi: 10.1128/IAI.01807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zimmermann A, Reimmann C, Galimand M, Haas D. Anaerobic growth and cyanide synthesis of Pseudomonas aeruginosa depend on anr, a regulatory gene homologous with fnr of Escherichia coli. Molecular Microbiology. 1991;5(6):1483–1490. doi: 10.1111/j.1365-2958.1991.tb00794.x. [DOI] [PubMed] [Google Scholar]
- 8.Yoon SS, Karabulut AC, Lipscomb JD, et al. Two-pronged survival strategy for the major cystic fibrosis pathogen, Pseudomonas aeruginosa, lacking the capacity to degrade nitric oxide during anaerobic respiration. EMBO Journal. 2007;26(15):3662–3672. doi: 10.1038/sj.emboj.7601787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Comolli JC, Donohue TJ. Differences in two Pseudomonas aeruginosa cbb3 cytochrome oxidases. Molecular Microbiology. 2004;51(4):1193–1203. doi: 10.1046/j.1365-2958.2003.03904.x. [DOI] [PubMed] [Google Scholar]
- 10.Kawakami T, Kuroki M, Ishii M, Igarashi Y, Arai H. Differential expression of multiple terminal oxidases for aerobic respiration in Pseudomonas aeruginosa . Environmental Microbiology. 2010;12(6):1399–1412. doi: 10.1111/j.1462-2920.2009.02109.x. [DOI] [PubMed] [Google Scholar]
- 11.Gamper M, Zimmermann A, Haas D. Anaerobic regulation of transcription initiation in the arcDABC operon of Pseudomonas aeruginosa . Journal of Bacteriology. 1991;173(15):4742–4750. doi: 10.1128/jb.173.15.4742-4750.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schreiber K, Krieger R, Benkert B, et al. The anaerobic regulatory network required for Pseudomonas aeruginosa nitrate respiration. Journal of Bacteriology. 2007;189(11):4310–4314. doi: 10.1128/JB.00240-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arai H, Kodama T, Igarashi Y. Cascade regulation of the two CRP/FNR-related transcriptional regulators (ANR and DNR) and the denitrification enzymes in Pseudomonas aeruginosa . Molecular Microbiology. 1997;25(6):1141–1148. doi: 10.1046/j.1365-2958.1997.5431906.x. [DOI] [PubMed] [Google Scholar]
- 14.Benkert B, Quäck N, Schreiber K, Jaensch L, Jahn D, Schobert M. Nitrate-responsive NarX-NarL represses arginine-mediated induction of the Pseudomonas aeruginosa arginine fermentation arcDABC operon. Microbiology. 2008;154(10):3053–3060. doi: 10.1099/mic.0.2008/018929-0. [DOI] [PubMed] [Google Scholar]
- 15.Giardina G, Rinaldo S, Johnson KA, Di Matteo A, Brunori M, Cutruzzolà F. NO sensing in Pseudomonas aeruginosa: structure of the Transcriptional Regulator DNR. Journal of Molecular Biology. 2008;378(5):1002–1015. doi: 10.1016/j.jmb.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 16.Pesci EC, Pearson JP, Seed PC, Iglewski BH. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa . Journal of Bacteriology. 1997;179(10):3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearson JP, Gray KM, Passador L, et al. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(1):197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Passador L, Cook JM, Gambello MJ, Rust L, Iglewski BH. Expression of Pseudomonas aeruginosa virulence genes requires cell-to- cell communication. Science. 1993;260(5111):1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 19.Pearson JP, Passador L, Iglewski BH, Greenberg EP. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa . Proceedings of the National Academy of Sciences of the United States of America. 1995;92(5):1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ochsner UA, Reiser J. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa . Proceedings of the National Academy of Sciences of the United States of America. 1995;92(14):6424–6428. doi: 10.1073/pnas.92.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gambello MJ, Iglewski BH. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. Journal of Bacteriology. 1991;173(9):3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ochsner UA, Koch AK, Fiechter A, Reiser J. Isolation and characterization of a regulatory gene affecting rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa . Journal of Bacteriology. 1994;176(7):2044–2054. doi: 10.1128/jb.176.7.2044-2054.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chugani SA, Whiteley M, Lee KM, D’Argenio D, Manoil C, Greenberg EP. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa . Proceedings of the National Academy of Sciences of the United States of America. 2001;98(5):2752–2757. doi: 10.1073/pnas.051624298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juhas M, Wiehlmann L, Huber B, et al. Global regulation of quorum sensing and virulence by VqsR in Pseudomonas aeruginosa . Microbiology. 2004;150(4):831–841. doi: 10.1099/mic.0.26906-0. [DOI] [PubMed] [Google Scholar]
- 25.Gallagher LA, McKnight SL, Kuznetsova MS, Pesci EC, Manoil C. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa . Journal of Bacteriology. 2002;184(23):6472–6480. doi: 10.1128/JB.184.23.6472-6480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diggle SP, Lumjiaktase P, Dipilato F, et al. Functional genetic analysis reveals a 2-Alkyl-4-quinolone signaling system in the human pathogen Burkholderia pseudomallei and related bacteria. Chemistry and Biology. 2006;13(7):701–710. doi: 10.1016/j.chembiol.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 27.Xiao G, Déziel E, He J, et al. MvfR, a key Pseudomonas aeruginosa pathogenicity LTTR-class regulatory protein, has dual ligands. Molecular Microbiology. 2006;62(6):1689–1699. doi: 10.1111/j.1365-2958.2006.05462.x. [DOI] [PubMed] [Google Scholar]
- 28.Mashburn LM, Whiteley M. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature. 2005;437(7057):422–425. doi: 10.1038/nature03925. [DOI] [PubMed] [Google Scholar]
- 29.Tashiro Y, Ichikawa S, Shimizu M, et al. Variation of physiochemical properties and cell association activity of membrane vesicles with growth phase in Pseudomonas aeruginosa . Applied and Environmental Microbiology. 2010;76(11):3732–3739. doi: 10.1128/AEM.02794-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoon SS, Hennigan RF, Hilliard GM, et al. Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Developmental Cell. 2002;3(4):593–603. doi: 10.1016/s1534-5807(02)00295-2. [DOI] [PubMed] [Google Scholar]
- 31.Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. Journal of Bacteriology. 2003;185(7):2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toyofuku M, Nomura N, Fujii T, et al. Quorum sensing regulates denitrification in Pseudomonas aeruginosa PAO1. Journal of Bacteriology. 2007;189(13):4969–4972. doi: 10.1128/JB.00289-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.D’Argenio DA, Wu M, Hoffman LR, et al. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Molecular Microbiology. 2007;64(2):512–533. doi: 10.1111/j.1365-2958.2007.05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoffman LR, Kulasekara HD, Emerson J, et al. Pseudomonas aeruginosa lasR mutants are associated with cystic fibrosis lung disease progression. Journal of Cystic Fibrosis. 2009;8(1):66–70. doi: 10.1016/j.jcf.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffman LR, Richardson AR, Houston LS, et al. Nutrient availability as a mechanism for selection of antibiotic tolerant Pseudomonas aeruginosa within the CF airway. PLoS Pathogens. 2010;6(1) doi: 10.1371/journal.ppat.1000712. Article ID e1000712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toyofuku M, Nomura N, Kuno E, Tashiro Y, Nakajima T, Uchiyama H. Influence of the Pseudomonas quinolone signal on denitrification in Pseudomonas aeruginosa . Journal of Bacteriology. 2008;190(24):7947–7956. doi: 10.1128/JB.00968-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rampioni G, Pustelny C, Fletcher MP, et al. Transcriptomic analysis reveals a global alkyl-quinolone-independent regulatory role for PqsE in facilitating the environmental adaptation of Pseudomonas aeruginosa to plant and animal hosts. Environmental Microbiology. 2010;12(6):1659–1673. doi: 10.1111/j.1462-2920.2010.02214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Diggle SP, Matthijs S, Wright VJ, et al. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chemistry and Biology. 2007;14(1):87–96. doi: 10.1016/j.chembiol.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 39.Bredenbruch F, Geffers R, Nimtz M, Buer J, Häussler S. The Pseudomonas aeruginosa quinolone signal (PQS) has an iron-chelating activity. Environmental Microbiology. 2006;8(8):1318–1329. doi: 10.1111/j.1462-2920.2006.01025.x. [DOI] [PubMed] [Google Scholar]
- 40.Häussler S, Becker T. The pseudomonas quinolone signal (PQS) balances life and death in Pseudomonas aeruginosa populations. PLoS Pathogens. 2008;4(9) doi: 10.1371/journal.ppat.1000166. Article ID e1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toyofuku M, Nakajima-Kambe T, Uchiyama H, Nomura N. The effect of a cell-to-cell communication molecule, Pseudomonas quinolone signal (PQS), produced by p. aeruginosa on other bacterial species. Microbes and Environments. 2010;25(1):1–7. doi: 10.1264/jsme2.me09156. [DOI] [PubMed] [Google Scholar]
- 42.Filiatrault MJ, Wagner VE, Bushnell D, Haidaris CG, Iglewski BH, Passador L. Effect of anaerobiosis and nitrate on gene expression in Pseudomonas aeruginosa . Infection and Immunity. 2005;73(6):3764–3772. doi: 10.1128/IAI.73.6.3764-3772.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee KM, Yoon MY, Park Y, Lee JH, Yoon SS. Anaerobiosis-induced loss of cytotoxicity is due to inactivation of quorum sensing in Pseudomonas aeruginosa . Infection and Immunity. 2011;79(7):2792–2800. doi: 10.1128/IAI.01361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schertzer JW, Brown SA, Whiteley M. Oxygen levels rapidly modulate Pseudomonas aeruginosa social behaviours via substrate limitation of PqsH. Molecular Microbiology. 2010;77(6):1527–1538. doi: 10.1111/j.1365-2958.2010.07303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borriello G, Werner E, Roe F, Kim AM, Ehrlich GD, Stewart PS. Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrobial Agents and Chemotherapy. 2004;48(7):2659–2664. doi: 10.1128/AAC.48.7.2659-2664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams P, Cámara M. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: a tale of regulatory networks and multifunctional signal molecules. Current Opinion in Microbiology. 2009;12(2):182–191. doi: 10.1016/j.mib.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 47.Gusarov I, Shatalin K, Starodubtseva M, Nudler E. Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science. 2009;325(5946):1380–1384. doi: 10.1126/science.1175439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shatalin K, Gusarov I, Avetissova E, et al. Bacillus anthracis-derived nitric oxide is essential for pathogen virulence and survival in macrophages. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(3):1009–1013. doi: 10.1073/pnas.0710950105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arai H, Mizutani M, Igarashi Y. Transcriptional regulation of the nos genes for nitrous oxide reductase in Pseudomonas aeruginosa . Microbiology. 2003;149(1):29–36. doi: 10.1099/mic.0.25936-0. [DOI] [PubMed] [Google Scholar]
- 50.Trunk K, Benkert B, Quäck N, et al. Anaerobic adaptation in Pseudomonas aeruginosa: definition of the Anr and Dnr regulons. Environmental Microbiology. 2010;12(6):1719–1733. doi: 10.1111/j.1462-2920.2010.02252.x. [DOI] [PubMed] [Google Scholar]
- 51.Fetar H, Gilmour C, Klinoski R, Daigle DM, Dean CR, Poole K. mexEF-oprN multidrug efflux operon of Pseudomonas aeruginosa: regulation by the MexT activator in response to nitrosative stress and chloramphenicol. Antimicrobial Agents and Chemotherapy. 2011;55(2):508–514. doi: 10.1128/AAC.00830-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tian ZX, Fargier E, Mac Aogáin M, Adams C, Wang YP, O’Gara F. Transcriptome profiling defines a novel regulon modulated by the LysR-type transcriptional regulator MexT in Pseudomonas aeruginosa . Nucleic Acids Research. 2009;37(22):7546–7559. doi: 10.1093/nar/gkp828. Article ID gkp828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arai H, Hayashi M, Kuroi A, Ishii M, Igarashi Y. Transcriptional regulation of the flavohemoglobin gene for aerobic nitric oxide detoxification by the second nitric oxide-responsive regulator of Pseudomonas aeruginosa . Journal of Bacteriology. 2005;187(12):3960–3968. doi: 10.1128/JB.187.12.3960-3968.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Firoved AM, Wood SR, Ornatowski W, Deretic V, Timmins GS. Microarray analysis and functional characterization of the nitrosative stress response in nonmucoid and mucoid Pseudomonas aeruginosa . Journal of Bacteriology. 2004;186(12):4046–4050. doi: 10.1128/JB.186.12.4046-4050.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barraud N, Hassett DJ, Hwang SH, Rice SA, Kjelleberg S, Webb JS. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa . Journal of Bacteriology. 2006;188(21):7344–7353. doi: 10.1128/JB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barraud N, Schleheck D, Klebensberger J, et al. Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di-GMP levels, and enhanced dispersal. Journal of Bacteriology. 2009;191(23):7333–7342. doi: 10.1128/JB.00975-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Attila C, Ueda A, Wood TK. PA2663 (PpyR) increases biofilm formation in Pseudomonas aeruginosa PAO1 through the psl operon and stimulates virulence and quorum-sensing phenotypes. Applied Microbiology and Biotechnology. 2008;78(2):293–307. doi: 10.1007/s00253-007-1308-y. [DOI] [PubMed] [Google Scholar]
- 58.Van Alst NE, Wellington M, Clark VL, Haidaris CG, Iglewski BH. Nitrite reductase NirS is required for type III secretion system expression and virulence in the human monocyte cell line THP-1 by Pseudomonas aeruginosa . Infection and Immunity. 2009;77(10):4446–4454. doi: 10.1128/IAI.00822-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yawata Y, Nomura N, Uchiyama H. Development of a novel biofilm continuous culture method for simultaneous assessment of architecture and gaseous metabolite production. Applied and Environmental Microbiology. 2008;74(17):5429–5435. doi: 10.1128/AEM.00801-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoon MY, Lee KM, Park Y, Yoon SS. Contribution of cell elongation to the biofilm formation of Pseudomonas aeruginosa during anaerobic respiration. PLoS ONE. 2011;6(1) doi: 10.1371/journal.pone.0016105. Article ID e16105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morgenstein RM, Szostek B, Rather PN. Regulation of gene expression during swarmer cell differentiation in Proteus mirabilis . FEMS Microbiology Reviews. 2010;34(5):753–763. doi: 10.1111/j.1574-6976.2010.00229.x. [DOI] [PubMed] [Google Scholar]
- 62.Justice SS, Hunstad DA, Cegelski L, Hultgren SJ. Morphological plasticity as a bacterial survival strategy. Nature Reviews Microbiology. 2008;6(2):162–168. doi: 10.1038/nrmicro1820. [DOI] [PubMed] [Google Scholar]


