Abstract
Heparin is the most widely used pharmaceutical to control blood coagulation in modern medicine. A health crisis that took place in 2008 led to a demand for production of heparin from non-animal sources. Chinese hamster ovary (CHO) cells, commonly used mammalian host cells for production of foreign pharmaceutical proteins in the biopharmaceutical industry, are capable of producing heparan sulfate (HS), a related polysaccharide naturally. Since heparin and HS share the same biosynthetic pathway, we hypothesized that heparin could be produced in CHO cells by metabolic engineering. Based on the expression of endogenous enzymes in the HS / heparin pathways of CHO-S cells, human N-deacetylase/N-sulfotransferase (NDST2) and mouse heparan sulfate 3-O-sulfotransferase 1 (Hs3st1) genes were transfected sequentially into CHO host cells growing in suspension culture. Transfectants were screened using quantitative RT-PCR and Western blotting. Out of 120 clones expressing NDST2 and Hs3st1, 2 clones, Dual-3 and Dual-29, were selected for further analysis. An antithrombin III (ATIII) binding assay using flow cytometry, designed to recognize a key sugar structure characteristic of heparin, indicated that Hs3st1 transfection was capable of increasing ATIII binding. An anti-factor Xa assay, which affords a measure of anticoagulant activity, showed a significant increase in activity in the dual-expressing cell lines. Disaccharide analysis of the engineered HS showed a substantial increase in N-sulfo groups, but did not show a pattern consistent with pharmacological heparin, suggesting that further balancing the expression of transgenes with the expression levels of endogenous enzymes involved in HS / heparin biosynthesis might be necessary.
Keywords: heparin, Chinese hamster ovary cells, anticoagulant, LC-MS, flow cytometry
1. Introduction
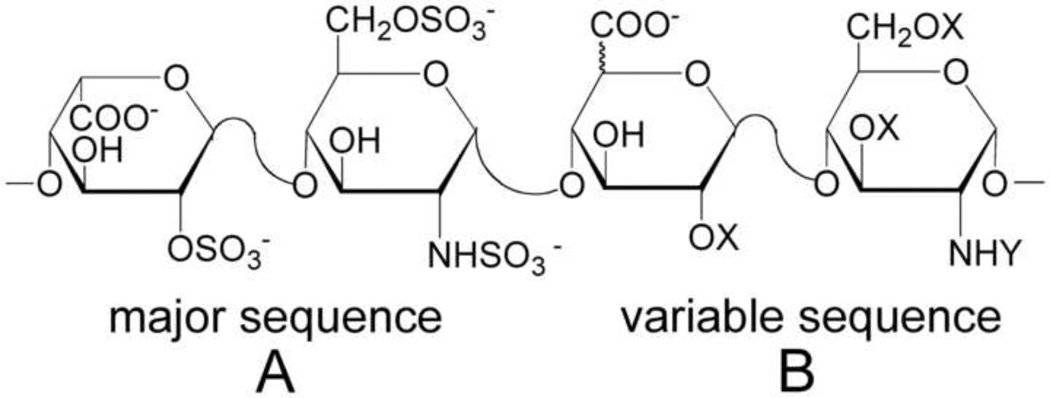
Heparin is a highly sulfated, linear glycosaminoglycan (GAG) which consists of repeated disaccharides, l-iduronic acid (IdoA) or d-glucuronic acid (GlcA) and N-acetyl-d-glucosamine (GlcNAc). Heparin chains are polydisperse (molecular weight: 5,000 – 40,000) and contain significant sequence heterogeneity (Ahsan et al., 1995). Fully sulfated heparin chains are composed of uniform, repeating sequences of trisulfated disaccharides whereas heparin chains having intermediate level of sulfation (2.5 sulfo groups per disaccharide) are composed of long segments of fully sulfated sequences with intervening undersulfated domains (Fig. 1). Heparin is widely used clinically as an anticoagulant, particularly for surgery and kidney dialysis. However, in early 2008, there was a marked increase in serious adverse events associated with heparin therapy, with thousands of patients who showed severe symptoms such as rash, angioedema, hypotension, and tachycardia, and nearly 100 associated deaths in the United States alone. After thorough investigation, it was discovered that the heparin injected into patients had been contaminated by oversulfated GAGs (Guerrini et al., 2008; Pan et al., 2010), highlighting the potential risks of contamination from the current methods of heparin production.
Fig. 1.
The structures of the major (A) and variable (B) repeating disaccharides comprising heparin, where X = SO3− or H and Y = SO3− or COCH3.
Currently, heparin is extracted from animal tissues such as porcine intestines or bovine lungs. In addition to possible contamination, lot-to-lot variation is also a limitation in heparin extraction because factors such as animal-care conditions affect heparin quality and composition. In addition, the supply of animal tissues is limited, while the demand for heparin is increasing. Moreover, there is a risk of the presence of infectious agents (e.g. prions or viruses) in heparin prepared from animal tissues. Therefore, it would be preferable to produce heparin under conditions where the quality and quantity of heparin could be controlled. Thus, the situation for heparin production today is akin to the production of biological proteins and peptides such as insulin before the advent of recombinant DNA technology.
Chinese hamster ovary (CHO) cells, the most widely used cells for therapeutic protein production, are good candidates for production of a bioengineered heparin. They express many of the enzymes involved in glycosylation; they are relatively safe from biological contamination such as viruses, and they can be adapted to suspension culture and easily scaled up. More importantly, CHO cells produce heparan sulfate (HS), a less sulfated GAG that has same basic disaccharide units as heparin, which is typically made by connective tissue type (serosal) mast cells. HS plays many important roles in physiological and pathophysiological processes including angiogenesis (Fuster and Wang, 2010), development (Hacker et al., 2005; Yamaguchi et al., 2010), cell adhesion and proliferation (Tumova et al., 2000), inflammation (Wang et al., 2005), and viral and bacterial infection (Spear et al., 1992) through its interaction with chemokines, cytokines, and growth factor receptors (Bishop et al., 2007). HS has considerably lower anticoagulant activity than heparin (Marcum and Rosenberg, 1987), but both are mediated through the interaction of a unique pentasaccharide motif (an antithrombin (ATIII) binding site), with ATIII, a serine-protease inhibitor (Capila and Linhardt, 2002; Muszbek et al., 2010).
Extensive studies by Lindahl and co-workers have shown that heparin and HS are biosynthesized through a similar pathway. HS/heparin biosynthesis begins in the endoplasmic reticulum with the transfer of xylose to a specific serine residue of the core protein (Robinson et al., 1978). Addition of two galactose molecules followed by glucuronic acid gives rise to the common linkage tetrasaccharide, which is modified by a unique glucosaminyltransferase, Extl3. The chains are then polymerized by the sequential addition of d-glucuronic acid (GlcA) and N-acetyl-d-glucosamine (GlcNAc), catalyzed by a copolymerase complex consisting of exostosin 1 (Ext1) and exostosin 2 (Ext2) (Esko and Selleck, 2002). As the chain polymerizes, a series of modifications takes place starting with N-deacetylation and N-sulfonation of GlcNAc residues, followed by epimerization of GlcA to IdoA, and finally, the introduction of O-sulfo groups at different positions of the glucosamine (GlcN) and uronic acid residues. Complete or nearly complete modification of this nascent GAG chain results in a highly N-sulfo, O-sulfo, IdoA-rich GAG commonly referred to as heparin, which serves as the source of material for further fractionation to generate pharmaceutical heparin. HS is characterized by partial modification of the chains resulting in O-sulfo poor and GlcNAc, GlcA-rich chains (Casu, 1985; Nairn et al., 2007). Many of the enzymes involved in HS biosynthesis have multiple isozymes, which have tissue-specific and developmentally regulated expression (Bai et al., 1999). All of the known enzymes involved in the HS pathway have been cloned and the genes coding for them have been identified from a variety of organisms, including some from Chinese hamster. The expression pattern and activity level of these isozymes presumably determines which particular form of HS will be synthesized.
Wild type CHO-K1 cells produce simple, less sulfated HS as they lack certain biosynthetic enzymes or exploit different isozymes in comparison with mast cells, which are the native site of heparin production. Moreover, CHO cells produce a lower amount of HS compared to other cultured cells such as fibroblasts and some tumor cell lines (Zhang et al., 2006). CHO cells reportedly express two out of four N-deacetylase/N-sulfotransferases (Ndst), two out of three heparan sulfate 6-O-sulfotransferases (Hs6st) and none of seven heparan sulfate 3-O-sulfotransferases (Hs3st) (Zhang et al., 2006). CHO cells have been widely used to study different aspects of glycosylation. Mutants lacking certain biosynthetic enzymes were used to determine the components of the HS biosynthetic pathway and their order within the pathway (Zhang et al., 2006). However, CHO cells lack or weakly express Hs3sts and do not produce HS that binds or activates ATIII. Introduction of Hs3st1 or Hs3st5 into CHO cells bestowed ATIII binding activity upon HS produced in the cells (Zhang et al., 2001a; Zhang et al., 2001b; Duncan et al., 2004), indicating that they have the capacity to generate the ATIII-precursor sequence.
Although many glycosylation-related enzymes related to GAG pathways have been engineered into or genetically inactivated in CHO cells to determine their functions (Bame and Esko, 1989; Bai and Esko, 1996; Zhang et al., 2001a; Zhang et al., 2001b), there have been no published efforts to produce heparin in CHO cells. Since heparin and HS share a common biosynthetic pathway, we hypothesized that CHO cells could be metabolically engineered to produce heparin. In this work, we evaluated the expression levels of metabolic enzymes and isozymes involved in the biosynthetic pathway of HS / heparin. We then employed genetic engineering to construct stable cell lines that expressed the relevant enzymes. Finally, the structure and activity of the engineered HS was characterized.
2. Materials and Methods
All chemicals and media were purchased from Sigma-Aldrich (St. Louis, MO), Invitrogen (Carlsbad, CA) or Thermo Fisher Scientific (Waltham, MA) and used without modification. CHO-S cells (Invitrogen) and dual NDST2 and Hs3st1 expressing cell lines were routinely seeded at 2 × 105 cells/ml in 125ml polycarbonate Erlenmeyer flasks (Corning, Corning, NY) containing 30 ml of culture medium (described in Section 2.2.) and cultured on orbital shakers agitated at 125 rpm in a humidified 37 °C incubator with 5% CO2.
2.1. Evaluation of the expression of HS / heparin biosynthetic enzymes by RT-PCR
2.1.1. Isolation of rat mast cells
Peritoneal rat mast cells were isolated from ~6-week old adult rats based on a modification of widely used methods for mast cell isolation and staining (Gustafson and Pihl, 1967; Nemeth and Rohlich, 1980; Martynova et al., 2005). After rats were sacrificed by CO2 fixation and decapitation, their abdomens were skinned, followed by injection of ~60 ml of sterile phosphate buffered saline (PBS). After a gentle massage of the abdomen, the injected buffer was collected and centrifuged at 120 × g for 10 min. The pellets were re-suspended in 0.5 ml of PBS and layered on the top of a preformed 88% Percoll gradient and centrifuged at 450 × g for 15 min. The mast-cell-containing fraction, which occupied the lower part of the tube, was collected. The presence of the mast cells was confirmed by staining with 0.005% Ruthenium red. The 88% Percoll gradient was prepared by dilution of a 100% Percoll gradient with 1X RPMI-1640. The 100% Percoll gradient was made by adding 10 ml of 10X RPMI-1640 containing 200 mM HEPES/ 1M NaOH to 90 mL of commercial Percoll solution (GE Healthcare, Piscataway, NJ). The tubes were then centrifuged at 12,000 × g for 30 min at 4 °C to make the gradient.
2.1.2. RNA extraction, RT-PCR reaction
Total RNA was extracted from the mast cells isolated from 6 rats and from 4 × 106 CHO-S cells using RNeasy mini kits (Qiagen, Germantown, MD) according to the manufacturer‘s instructions. The amount of RNA was assessed by NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). 60–90 ng of total RNA per sample was used in reverse transcription and polymerase chain reactions using a SuperScript III Platinum One-Step RT-PCR kit (Invitrogen). A list of primers used is given in Supplementary Table S1. RT-PCR products were separated and visualized by agarose gel electrophoresis using ethidium bromide.
2.2. Transfection and selection of NDST2 and Hs3st1 expressing cell lines
The human NDST2 gene (GenBank: BC035711.1, Thermo Fisher Scientific) was amplified using a primer pair, which includes BamHI and NotI restriction enzyme sites (forward primer: 5`-GAGCTCGGATCCACTATGCTCCAGTTGTGGAAGGTG-3`; reverse primer: 5`-CTCGAGCGGCCGCTCAGCCCAGACTGGAATGCTG-3`) and inserted into the pcDNA3.1/Neo expression vector (Invitrogen). The mouse Hs3st1 gene, kindly provided by Professor Jian Liu at University of North Carolina (Xu et al., 2008), was cut by EcoRI and XhoI restriction enzymes and inserted into the pcDNA3.1/Zeo expression vector (Invitrogen). CHO-S cells (2 × 106 cells) were transfected with the NDST2 gene using a Nucleofector® II (Lonza, Basel, Switzerland) according to the manufacturer’s instructions (kit V, program U-024). The transfected cells were seeded at 6.7 × 105 cells/ml and incubated at 37 °C and 5% CO2 in static 6-well plate cultures (Corning) for 24 hours after transfection. Next, the cells (104 cells/ml) were seeded into ClonaCell®-TCS Medium (STEMCELL Technologies, Vancouver, Canada) supplemented with 1mg/ml of Geneticin® (Invitrogen) and grown at 37 °C and 5% CO2 for two weeks. Selected NDST2 expressing cell clones were then transfected with the Hs3st1 gene and inoculated into semi-solid medium supplemented with 1 mg/ml of Geneticin® and 500 µg/ml of Zeocin™ (Invitrogen) in the same manner as for the development of NDST2 expressing cell lines. The host CHO-S cell line and dual NDST2 and Hs3st1 expressing cell lines were maintained in CD CHO medium (Invitrogen) supplemented with 8 mM GlutaMAX™ (Invitrogen) and 15 ml of hypoxanthine/thymidine solution per 500 ml of medium (HT, Mediatech, Manassas, VA). In addition, 1 mg/ml of Geneticin® and 500 µg/ml of Zeocin™ were added to the medium for dual-expressing cell lines.
2.3. Screening of NDST2 / Hs3st1 transfected cell lines by RT-PCR and immunoblotting
RT-PCR was conducted as described above for wild-type CHO-S cells. For total protein extraction, exponentially growing cells were lysed in Nonidet-P40 lysis buffer (Boston Bioproducts, Ashland, MA) on ice for 30 min in the presence of a cocktail of protease and phosphatase inhibitors, (Thermo Fisher Scientific) which contained AEBSF, aprotinin, bestatin, E-64, leupeptin, and pepstatin A. Protein concentrations were determined using BCA assay (Thermo Fisher Scientific). 40 µg of total protein was loaded and separated on 4–20% polyacrylamide gels (Thermo Fisher Scientific) at 150 V. Tris-Hepes-SDS buffer was used as the running buffer. Proteins were transferred onto a PVDF membrane (Bio-Rad Laboratories, Hercules, CA), probed with relevant primary antibodies (described below), and then detected using the appropriate HRP-conjugated secondary antibody and chemiluminescent (Super Signal West Pico ECL substrate, Thermo Fisher Scientific) exposure on high performance chemiluminescence film (Amersham Hyperfilm ECL, GE Healthcare). The primary and secondary antibodies used are the following: rabbit anti-gamma-tubulin (T3320, Sigma-Aldrich); goat anti-Ndst2 (sc-16764), goat anti-Hs3st1 (sc-104313), goat anti-Hs6st1 (sc-109943), rabbit anti-Hs6st3 (sc-84308, Santa Cruz Biotechnology, Santa Cruz, CA); mouse anti-Glce (glucuronyl C5-epimerase, H00026035-B01P, Abnova, Taipei City, Taiwan); goat anti-rabbit HRP-conjugated (31460), goat anti-mouse HRP-conjugated (31430, Thermo Fisher Scientific); donkey anti-goat HRP-conjugated (sc-2020, Santa Cruz Biotechnology).
2.4. Activity analysis of engineered HS by flow cytometry
2.4.1. Fluorescent labeling of ATIII and fibroblast growth factor-2 (FGF-2)
ATIII and FGF-2 were labeled with amine-reactive 4,4-difluoro-5-phenyl-4-bora-3,4a-diaza-s-indacene-3-propionic acid, succinimidyl ester (BODIPY R6G, SE, Invitrogen) as described previously (Martin et al., 2009). In brief, ATIII or FGF-2 solutions were prepared by dissolving 1 mg of ATIII or FGF-2 in 100 µl of 0.1 M sodium bicarbonate buffer. 10 µl of BODIPY R6G solution was added to the ATIII or FGF-2 solution, and the reaction mixtures were incubated in the dark at 37 °C for 1 hr with continuous stirring. The reactions were stopped by adding 1 ml of sterile PBS and purified with 3000 MWCO spin columns (Millipore, Bedford, MA). The concentrated BODIPY R6G-conjugated FGF-2 or ATIII (2 µl from stock solution) was diluted in 100 µl of PBS containing 10% FBS and stored at −20 °C for up to 14 days until used directly for labeling cells.
2.4.2. Immunofluorescence assays
Flow cytometry experiments were performed as described by Zhang et al. (2006) with BODIPY R6G-conjugated ATIII or FGF-2. A sample containing 106 cells was washed with cold, sterile PBS containing 10% FBS and incubated with BODIPY R6G-conjugated FGF-2 or ATIII at 4 °C for 30 min in the dark. The cells were washed with sterile PBS containing 10% FBS. A minimum of 10,000 cells from each sample was analyzed on a BD LSRII flow cytometer with FITC excitation filters (530 nm, BD Biosciences, Franklin Lakes, NJ). The 488-nm argon-ion laser was used for excitation and the 530/30 nm bandpass emission filter was used to detect fluorescence. The experiment was performed twice with live CHO-S, Dual-3 and Dual-29 cells and once with CHO-S and Dual-3 or CHO-S and Dual-29 cells, respectively. In total, three experiments were performed at three different time points to compare CHO-S with Dual-3 and CHO-S with Dual-29, respectively. Statistical analysis was performed on the normalized mean fluorescence with students t-test comparing CHO-S with Dual-3 or Dual-29 (n=3).
2.5. Anticoagulant-activity analysis of bioengineered HS by anti-Factor Xa assay
The anti-factor Xa anticoagulation activity was based on a chromogenic assay from a published method (Chen et al., 2005) with a heparin anti-FXa assay kit (HemosIL™, Instrumentation Laboratory, Bedford, MA). In brief, the GAGs purified as described below from CHO cell lines or a heparin standard (at various concentrations) were dissolved in Tris-EDTA buffer (50 mM Tris, 7.5 mM EDTA, and 175 mM NaCl, pH 8.4). The reaction mixture, which consisted of 25 µl of ATIII stock solution (1 IU/ml) and 25 µl of the GAGs or heparin, was incubated at 37 °C for 2 min. 25 µl of Factor Xa (13.6 nkat/ml) was added. After incubating at 37 °C for 4 min, 25 µl of chromogenic substrate S-2765 (0.75 mg/ml) was added. The absorbance of the reaction mixture was measured at 405 nm continuously for 10 min in a 96-well plate reader (SpectraMax M5, Molecular Devices, Sunnyvale, CA).
2.6. Disaccharide analysis of engineered HS
2.6.1. Isolation and purification of GAGs from cells and culture media
Disaccharide analysis of engineered HS / heparin was carried out as described previously (Yang et al., 2011). The cell samples were individually subjected to proteolysis at 55°C with 10% (w/v) of actinase E (20 mg/ml in HPLC grade water, Kaken Biochemicals, Tokyo, Japan) for 20 h. After proteolysis, particulates were removed from the resulting solutions by passing each through a 0.22 µm membrane syringe filter. Samples were then concentrated using Microcon YM-10 centrifugal filter units (10 kDa molecular weight cutoff, Millipore) by centrifugation at 12,000 × g and washed with 15 ml of distilled water to remove peptides. The retentate was collected and lyophilized. Samples were dissolved in 0.5 ml of 8 M urea containing 2% CHAPS (pH 8.3). A Vivapure Mini Q H spin column (Viva science, Edgewood, NJ) was prepared by equilibrating with 200 µl of 8 M urea containing 2% CHAPS (pH 8.3). To remove any remaining proteins, the clarified, filtered samples were loaded onto and run through the Vivapure MINI Q H spin columns under centrifugal force (700 × g). The columns were then washed with 200 µl of 8 M urea containing 2% CHAPS at pH 8.3, followed by five washes with 200 µl of 200 mM NaCl. GAGs were released from the spin column by washing three times with 50 µl of 16% NaCl. GAGs were desalted with YM-10 spin columns. The GAGs were lyophilized and stored at room temperature for future use.
2.6.2. Enzymatic digestion and reverse-phase ion-pairing ultra-performance liquid chromatography mass spectrometry (RPIP-UPLC-MS)
The GAGs recovered from cells and media were quantified by microcarbazole assay (Zhang et al., 2009) and then used to prepare a stock solution from which 5 µg of analyte could be removed. Cloning, Escherichia coli expression, and purification of recombinant heparin lyase I (EC 4.2.2.7), heparin lyase II (no EC assigned), and heparin lyase III (EC 4.2.2.8) from Flavobacterium heparinum were performed as described previously (Godavarti et al., 1996; Yoshida et al., 2002; Shaya et al., 2006). Heparin lyase I, II, and III (5mU each, assayed prior to use) in 5 µl of 25 mM Tris 500 mM NaCl, and 300 mM imidazole buffer (pH 7.4) were added to 5 µg of GAG sample in 25 µl of distilled water and incubated at 37 °C for 10 h to completely degrade the GAG sample. The products were recovered by centrifugal filtration using a YM-10 microconcentrator, and the HS / heparin disaccharides were recovered in the flow-through and lyophilized. The digested GAG disaccharides were redissolved in water at a final concentration of 50 to 100 ng/2 µl for LC–MS analysis.
LC–MS analyses were performed on an Agilent 1200 LC/MSD instrument (Agilent Technologies, Wilmington, DE) equipped with a 6300 ion trap and a binary pump followed by a UV detector equipped with a high-pressure cell. The column used was an Acquity UPLC BEH C18 column (2.1 × 150 mm, 1.7 µm, Waters, Milford, MA). Eluent A was water/acetonitrile (85:15, v/v), and eluent B was water/acetonitrile (35:65, v/v). Both eluents contained 12 mM tributylamine (TrBA) and 38 mM NH4OAc with pH adjusted to 6.5 with acetic acid. Disaccharide analysis was performed using a gradient of solution A for 10 min, followed by a linear gradient from 10 to 40 min (0–50% solution B) at a flow rate of 100 µl/min. The column effluent entered the source of the electrospray ionization (ESI)–MS for continuous detection by MS. The electrospray interface was set in negative ionization mode with a skimmer potential of −40.0 V, a capillary exit of −40.0 V, and a source temperature of 350°C to obtain the maximum abundance of the ions in a full-scan spectrum (200–1500 Da). Nitrogen (8 L/min, 40 psi) was used as a drying and nebulizing gas.
Quantification analysis of HS / heparin disaccharides was performed using calibration curves constructed by separation of increasing amounts of unsaturated HS / heparin disaccharide standards (2, 5, 10, 15, 20, 30, 50, and 100 ng per disaccharide). Unsaturated disaccharides standards of HS / heparin (0S: ΔUA-GlcNAc, NS: ΔUA-GlcNS, 6S: ΔUA-GlcNAc6S, 2S: ΔUA2S-GlcNAc, NS2S: ΔUA2S-GlcNS, NS6S: ΔUA-GlcNS6S, 2S6S: ΔUA2S-GlcNAc6S, triS: ΔUA2S-GlcNS6S) were obtained from Iduron (Manchester, UK). Linearity was assessed based on the amount of disaccharide and peak intensity in MS. All analyses were performed in triplicate.
3. Results
3.1. Evaluation of expression level of enzymes involved in HS / heparin biosynthesis
CHO-S host cells were first analyzed to determine the native expression levels of biosynthetic enzymes required for HS / heparin production and identify essential biosynthetic enzymes that were absent or in low abundance. Rat mast cells, a native producer of heparin, were used as a positive control. As primary mast cells do not divide in culture, they were freshly isolated from the peritoneal cavity of rats and confirmed by ruthenium red staining, a highly cationic stain (Supplementary Fig. S1). Ruthenium red binds to the HS anionic sulfo groups, permitting rapid identification of mast cells. Due to the absence of a CHO-cell or Chinese hamster genomic database at the time these studies were initiated, primer sets for RT-PCR were designed from the homologous regions of corresponding genes from mouse (M. musculus) and long-tailed hamster (C. longicaudatus). The sequences of the primer sets and the expression of the enzymes in CHO-S cells are summarized in Supplementary Table S1 and Supplementary Fig. S2, respectively. The RT-PCR result indicates that Ndst1, Ndst2, C5Epi, Hs2st1, Hs3st1, Hs6st1, and Hs6st3 are clearly expressed in rat mast cells (Table 1 and Supplementary Fig. S2), suggesting the necessity of these enzymes for heparin biosynthesis. RT-PCR and immunoblotting results for CHO-S cells show that Ndst1, Glce, Hs2st1, Hs6st1, Hs6st2, and Hs6st3 are expressed in CHO-S cells, whereas Ndst2 and Hs3st5 expression were not detected (Supplementary Fig. S2), which is consistent with previous studies (Zhang et al., 2006). In addition, expression of Hs3st1 in CHO-S cells was not detected by RT-PCR; however, it was faintly detected by Western blotting (this difference is presumably due to a mismatch between the mouse-derived PCR probes and the unknown CHO sequence). Expression of HS biosynthetic enzymes is summarized in Table 1. The inactivation of Ndst2 in mice has a profound effect on mast-cell heparin (Forsberg et al., 1999), leading us to identify it as one of the critical enzymes for introduction into the CHO-S cells.
Table 1.
Expression of HS / heparin biosynthetic enzymes in rat mast cells and CHO-S cells as determined by RT-PCR and Western blotting.
| Cell type | Ndst1 | Ndst2 | Glce | Hs2st1 | Hs6st1 | Hs6st2 | Hs6st3 | Hs3st1 | Hs3st5 | |
|---|---|---|---|---|---|---|---|---|---|---|
| RT-PCR | Rat mast | + | + | + | + | + | ?a | + | + | − |
| CHO-S | + | − | + | + | + | + | − | − | − | |
| Western blotting | CHO-S | n/ab | − | + | n/ac | + | n/ab | + | + | − |
RT-PCR of Hs6st2 from rat mast cells yielded a band of incorrect size
Western blotting was not performed for Ndst1 or Hs6st2
Endogenous expression of Hs2st1 was not detected by commercial antibodies due to lack of species cross reactivity
3.2. Development of stable NDST2 and Hs3st1 expressing cell lines
Based on the RT-PCR and immunoblotting results, NDST2 and Hs3st1 were selected for metabolic engineering. NDST2 and Hs3st1 genes were engineered sequentially since cotransfection was not successful (data not shown). At 24 h post-transfection, NDST2 transfected cells were inoculated into semi-solid media to form colonies for clonal selection. Out of 60 colonies, 10 stable cell lines were successfully established and screened by RT-PCR (Supplementary Fig. S3) and Western blotting analysis (data not shown). The NDST2-1 cell line showed the highest level of NDST2 transcription and translation among the NDST expressing clones and also sustained these expression levels for at least 30 days (Supplementary Fig. S3); therefore, it was selected for further engineering by expression of Hs3st1.
The transfection of the Hs3st1 gene, clonal selection, and screening were carried out in the same manner as NDST2 transfection. The established dual NDST2 and Hs3st1 expressing cell lines were denoted as Dual-1, Dual-2, etc. (Supplementary Fig. S4). Out of 120 colonies from semi-solid media culture, 40 clones were established and screened by RT-PCR and Western blotting. NDST2 expression was also evaluated to determine if there was a loss of NDST2 gene expression during the 3 months required to develop dual-expressing cell lines. Selected cell lines are shown in Fig. 2. Two highly expressing cell lines, Dual-3 and Dual-29, were selected for characterization of the engineered HS.
Fig. 2.
Gene and protein expression of NDST2 and Hs3st1 among the selected dual-expressing clones. Based on their levels of gene and protein expression, out of 40 NDST2 and Hs3st1 dual-expressing cell lines, Dual-3 and Dual-29 (asterisks) were selected for further characterization.
3.3. Activity and structural analyses of engineered HS
Flow cytometric analysis of ATIII and FGF-2 binding and anticoagulation assays were conducted to determine the activity of engineered HS. ATIII binding is the crucial first criterion for distinguishing HS from HP. Since HS is displayed on the cell membrane, fluorescently labeled ATIII and FGF-2 can bind to the (putative) ATIII binding site on engineered HS and to highly sulfated GAGs, respectively, and be analyzed by detecting the fluorescence. Fig. 3 shows the increase in the fluorescence signal from ATIII and FGF-2 in bioengineered Dual-3 and Dual-29 clones compared to the CHO-S cells, suggesting the biological activity of the introduced NDST2 and Hs3st1 in the clones.
Fig. 3.
Flow cytometric analysis of dual NDST2 and Hs3st1 expressing cell lines. (A) Histogram of binding activity between fluorescently tagged ATIII and dual NDST2 and Hs3st1 expressing cell lines. (B) Normalized binding activity of ATIII and FGF-2. Error bars represent 95% confidence intervals.
Since positive control cells, which have the complete form of heparin on their cell membrane are not available (as mast cells store heparin in intracellular granules), the activity of the engineered HS could not be compared to that of heparin using the ATIII binding assay. Therefore the engineered HS was isolated and analyzed by an anti-factor Xa anticoagulation assay to determine whether the engineered HS showed increased anticoagulant activity. Since cell-membrane-bound HS proteoglycans can be shed through the action of proteases (Bernfield et al., 1999; Bartlett et al., 2007), the engineered HS was also purified from the culture medium and analyzed by the anti-factor Xa assay. As shown in Fig. 4, the HS extracted from Dual-3 and Dual-29 cell pellets shows significantly increased anticoagulation activity (7.5-fold and 6.8-fold, respectively) compared to the HS from CHO-S host cells. However, the anticoagulant activity of the dual-expressing cells is still considerably lower than that of the pharmaceutical heparin. However, the pharmaceutical heparin has been fractionated to obtain high anticoagulant activity, whereas the bioengineered HS used in the activity assay was unfractionated, which may explain some of the difference in activity. The HS purified from culture media of Dual-3 and Dual-29 also shows markedly increased anticoagulation activity (52.9-fold and 97.2-fold, respectively) compared to CHO-S cells.
Fig. 4.
Factor Xa assay of dual NDST2 and Hs3st1 expressing cell lines. Pharmaceutical heparin was used as a positive control. Error bars represent 95% confidence intervals.
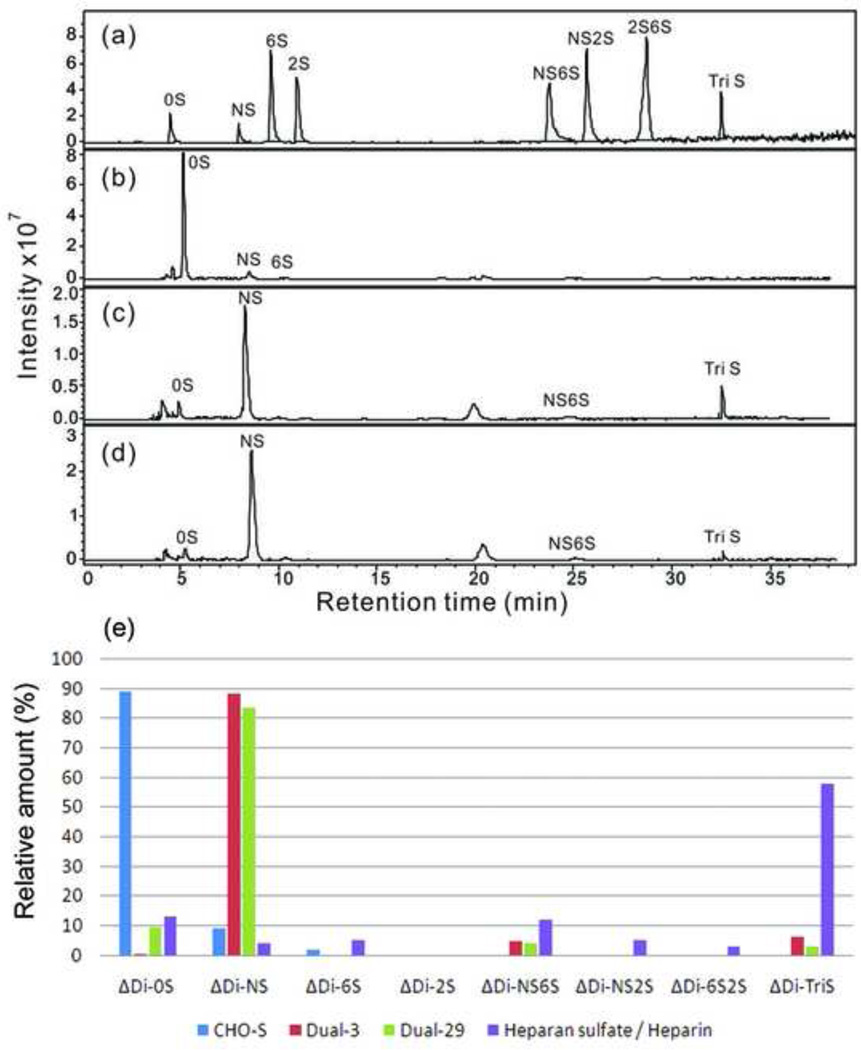
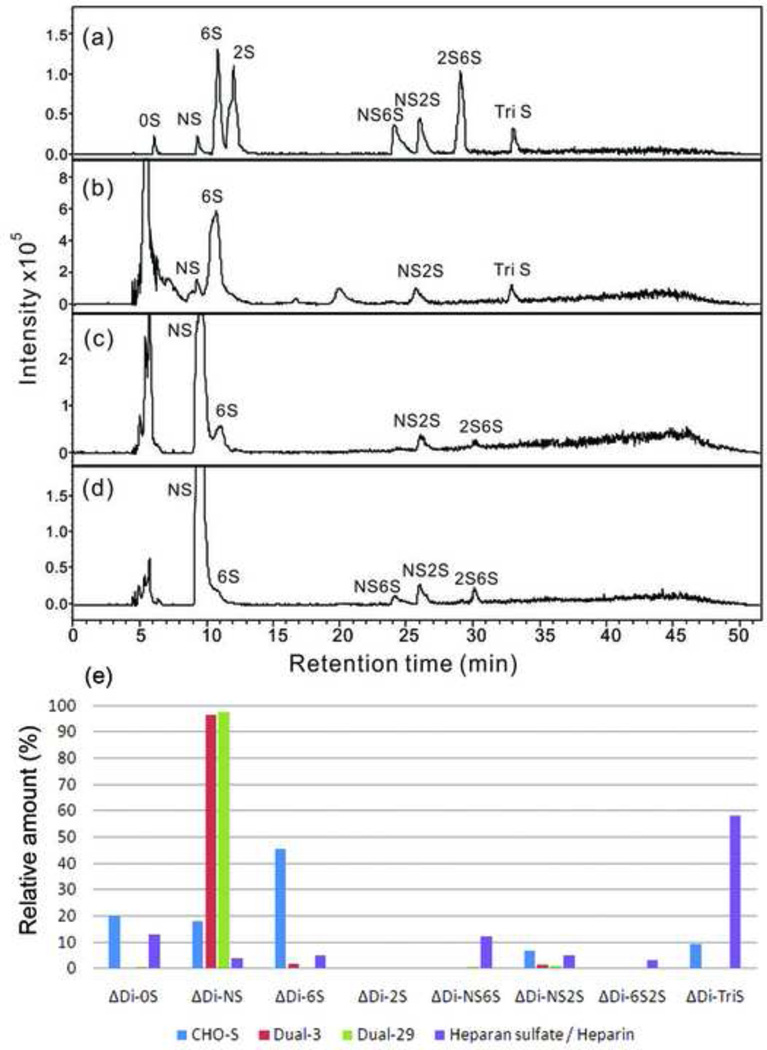
The total GAGs were isolated from cells and culture media using a three-step procedure involving protease digestion, strong anion-exchange chromatography on a spin column, followed by salt release to determine the structure of engineered HS. Purified engineered HS was digested by heparin lyase into disaccharides and analyzed by RPIP-UPLC-MS. The amount of total GAG and HS / heparin were quantified by carbazole assay and LC-MS, respectively. The results are summarized in Table 2. PAGE analysis of the purified GAGs confirmed that they are polymeric in nature (Supplementary Fig. S5). In all samples, the amount of HS / heparin was higher than chondroitin sulfate / dermatan sulfate (CS / DS). In addition, the dual-expressing cell lines showed a significant increase (4.5-fold to 9-fold) in total GAGs secreted into the medium and > 350-fold increase in the amount of HS / heparin relative to the amount of CS / DS. The HS / heparin disaccharide composition of the CHO-S cell line is presented in Fig. 5. The quantitative disaccharide composition of HS / heparin was determined by calibration with disaccharide standards. For the HS / heparin isolated from the CHO-S cells, 0S (88.9%) is the major disaccharide, followed by NS (9.3%) and 6S (2.0%). Interestingly, the HS / heparin disaccharide composition of the culture medium for CHO-S cells shows a substantially different composition, with 6S (45.6%) as the predominant disaccharide, followed by 0S (20.3%), NS (18.1%), TriS (9.2%), and NS2S (6.8%) (Fig. 6). In dual-expressing cell lines, NS (88.3% in Dual-3, 83.7% in Dual-29) was the major disaccharide isolated from the cell pellets, and minor amounts of NS6S and TriS disaccharide were also detected. The major disaccharide from the culture medium for dual-expressing cells was also NS (96.3% in Dual-3 medium, 97.5% in Dual-29 medium). We could not verify the presence of a 3-O-sulfo group containing disaccharide because the action of the Hs3st1 enzyme affords a tetrasaccharide that is not cleavable by heparin lyases (Zhao et al., 2011).
Table 2.
Amount of total GAG and HS / heparin from the cell pellets and media.
| Isolated from cell pellets | Isolated from culture media | |||||
|---|---|---|---|---|---|---|
| CHO-S | Dual-3 | Dual-29 | CHO-S | Dual-3 | Dual-29 | |
| GAGs (µg)a) | 11.4 | 10.3 | 16.9 | 19.3 | 85.2 | 173.2 |
| CS / DS (µg)b) | 2.2 | 1.3 | 1.4 | 0.8 | 0.01 | 0.02 |
| HS / heparin (µg) | 9.2 | 9.0 | 15.5 | 18.5 | 85.2 | 173.2 |
| (HS / heparin) : (CS / DS) | 4.2 : 1 | 6.9 : 1 | 11.1 : 1 | 23.1 : 1 | 8520 : 1 | 8660 : 1 |
Total GAGs were isolated from 5 × 107 cells of each cell line or culture media for the same number of cells.
Chondroitin sulfate / dermatin sulfate comprise the remaining GAGs
Fig. 5.
Disaccharide analysis of HS / heparin from cell pellets by RPIP-UPLC-MS. (a) Extracted ion chromatography (EIC) of heparin disaccharide standards; (b), (c) and (d) EIC of HS / heparin disaccharides of the GAG samples from CHO-S, Dual-3 and Dual-29 cell pellets, respectively. Quantification analysis of heparin/HS disaccharides was performed using calibration curves (e).
Fig. 6.
Disaccharide analysis of HS / heparin from culture media by RPIP-UPLC-MS. (a) EIC of heparin disaccharide standards; (b), (c) and (d) EIC of HS / heparin disaccharides of the GAG samples from culture media of CHO-S, Dual-3 and Dual-29 cell lines, respectively. Quantification analysis of heparin/HS disaccharides was performed using calibration curves (e).
4. Discussion
Today, production of heparin depends upon extracting GAGs from animal tissues. Despite efforts to synthesize heparin chemically or enzymatically (Kuberan et al., 2003; Noti and Seeberger, 2005; Linhardt et al., 2007; Martin et al., 2009), the sequence diversity and structural complexity of heparin polysaccharides has to date, precluded the development of synthetic laboratory approaches. Some prokaryotic cell strains produce a non-sulfated version of HS / heparin, heparosan, which can be used as a precursor for subsequent heparin synthesis (Wang et al., 2010). However, that will require many steps of enzymatic modification and purification to obtain the relevant sulfated residues and anticoagulant activity. Therefore, a mammalian cell line was chosen as an alternative producer. Producing heparin from mammalian cells is challenging because the production of non-protein macromolecules in mammalian cells has not been investigated intensively, whereas prokaryote strains have been employed to produce many non-protein compounds including polyketides, isoprenoids, bioplastics, and biofuels (Pfeifer et al., 2001; Aldor and Keasling, 2003; Martin et al., 2003; Green, 2011). Since HS / heparin are polysaccharides which are by nature heterogeneous, it is difficult to control their size and sequence, while protein products are precisely transcribed and translated. In addition, increased production of HS / heparin cannot be achieved by genetic amplification in the same manner used for production of most protein therapeutics. It is also necessary to identify the controlling mechanism of the heparin synthesis pathway, which is not fully understood. Moreover, while previous studies have explored CHO cell metabolism (Ahn and Antoniewicz, 2011; Nolan and Lee, 2011) and engineering CHO cells to enhance cell viability or improve recombinant protein glycosylation (Yun et al., 2007; Goh et al., 2010), there are no reports to date using metabolic engineering to produce non-protein products in CHO cells.
In this study, we used rat mast cells, natural producers of heparin, to identify the required expression of HS / heparin biosynthetic enzymes and compared it to that of CHO-S cells. We confirmed that CHO-S cells did not express Ndst2 and showed minimal expression of Hs3st1, which are known to be critical for anticoagulant heparin biosynthesis. Ndst2 plays an important role in the introduction of N-sulfo groups into GlcNAc, which in turn, is important for subsequent sulfonation of the growing HS chain (Sugahara and Kitagawa, 2002). Absence of Ndst2 expression can explain the low level of N-sulfo groups in CHO-S HS. Hs3st1 is responsible for the formation of the unique ATIII binding pentasaccharide, which makes heparin an important anticoagulant therapeutic molecule. We also found that the CHO-S cell line used in this study expresses Hs6st3, which plays a role in refining of the sulfated structure of HS. This has not been reported for other investigated CHO cell lines.
The development of dual NDST2 and Hs3st1 expressing cell lines was confirmed by RT-PCR and immunoblotting. The disaccharide analysis by RPIP-UPLC-MS showed a significant increase in the N-sulfo groups within the disaccharides of the NDST2 and Hs3st1 transfected clones, confirming the activity of NDST2. Further confirmation was provided by flow cytometry, which demonstrated an increase in the binding of fluorescently labeled FGF-2 in clones Dual-3 and Dual-29 compared to CHO-S. The increase in FGF-2 binding is explained by the increased sulfo-group composition of newly synthesized HS, which FGF-2 prefers (Esko and Selleck, 2002). As the available heparin lyases do not cut the HS chain in the 3-O-sulfo group containing sequences, RPIP-UPLC-MS could not be used to confirm the activity of Hs3st1. Flow cytometry confirmed an increase in binding of ATIII to the engineered HS, which showed that Hs3st1 is active. This also was confirmed by anti-factor Xa assay, which affords a measure of HS anticoagulant activity. Quantifying enzyme activity will be necessary to demonstrate the biological activity of the engineered enzymes, but a method is not available at this time.
In the dual-expressing cell lines, the amounts of HS / heparin in the media were increased significantly compared to HS / heparin extracted from the cells, which implies that HS / heparin on the membrane are shed into the medium. Hence, we appear to have increased the metabolic flux through this pathway by metabolic engineering. Increased HS / heparin in the media also supports the hypothesis that bioengineered HS chains will still use the HS core proteins, syndecans and glypicans, for targeting to the outside of the cell. Alternatively, the cells may also express ECM HS proteoglycans. Syndecans are type I transmembrane proteins and glypicans are glycosylphosphatidylinositol-anchored cell-surface proteoglycans. Using these HS core proteins will greatly simplify heparin purification as it will eliminate the necessity of cell lysis to recover the heparin. Overexpression of the core proteins might also be an option to facilitate increased traffic of bioengineered HS / heparin.
From the activity studies, we observed that the engineered HS showed more anticoagulant characteristics compared to normal HS, but there is still room for improvement. In Fig. 5 and Fig. 6, the relative amounts of disaccharide species in dual-expressing cells show unexpected patterns compared to those of pharmaceutical heparin. The major species of dual-expressing cell lines is NS disaccharide, which suggests that the expression level of NDST2 is too high and overwhelms the actions of other enzymes. Despite the fact that enzymes involved in HS biosynthesis have been cloned and expressed, and the relative reaction order in the pathway has been established, many aspects of the biosynthesis such as the type of chain modification, domain placement, and core-protein expression remain poorly understood. The mechanism that controls the HS / heparin biosynthetic pathway is also still unclear, but one widely supported hypothesis is that NDST is involved in the termination of sulfonation in HS / heparin (Esko and Selleck, 2002). Therefore, overexpression of NDST might terminate the sulfonation of HS / heparin before other sulfotransferases were able to act on the HS chain, which means that it may be necessary to balance the expression levels of the transgenes with the endogenous genes. In E. coli or S. cerevisiae, the expression levels of enzymes involved in metabolic pathways have been controlled by gene titration, promoter engineering, or transcriptional regulation (Alper et al., 2005; Pitera et al., 2007; Michalodimitrakis and Isalan, 2009). Our next study will focus on balancing the metabolic engineering to produce an engineered HS identical to the pharmaceutical heparin in terms of structure and activity. This will require understanding the activity of the HS / heparin biosynthetic enzymes in the Golgi apparatus. Assays to establish the activity of GAG biosynthetic enzymes are under development in our laboratory. Although the amount of HS produced by CHO-S cells is not a concern at the moment, increased production to improve the yield of the biosynthesized heparin could be required to meet pharmaceutical needs. We are currently exploring whether adherent CHO cells synthesize greater levels of HS than suspension cells and whether overexpression of Ext1 and Ext2 enzymes, which regulate HS / heparin chain polymerization, will lead to increased HS synthesis. In human embryonic kidney cells, overexpression of Ext1 and Ext2 resulted in increased HS chain length (Busse et al., 2007).
In conclusion, to produce heparin from non-animal sources, stable CHO cell lines expressing both NDST2 and Hs3st1 were established. This is an initial study to produce heparin, a non-protein therapeutic biomolecule, in mammalian cells by metabolic engineering. The engineered HS extracted from those cell lines and culture media showed that the ratio of NS was significantly increased and that ATIII and FGF-2 binding activity were increased compared to CHO-S cells. The disaccharide analysis implied that balancing the expression of enzymes along with modification of other factors such as core proteins might be necessary to achieve a pharmacological heparin.
Supplementary Material
Acknowledgements
This work was funded by a grant from the National Institutes of Health (R01GM090127). Payel Datta was supported in part by a fellowship from Rensselaer Polytechnic Institute. Partial support was provided by grant GM093131 to JDE. The authors would like to thank Professor Richard Stevens (Harvard Medical School) for advice on mast cell purification and Dr. Robert Waniewski for assistance with mast cell isolation.
Abbreviations used
- GAG
glycosaminoglycan
- IdoA
l-iduronic acid
- GlcA
d-glucuronic acid
- GlcNAc
N-acetyl-d-glucosamine
- CHO
Chinese hamster ovary
- HS
heparan sulfate
- ATIII
antithrombin
- Extl3
glucosaminyltransferase
- Ext
exostosin
- Ndst
N-deacetylase/N-sulfotransferase
- Hs6st
heparan sulfate 6-O-sulfotransferase
- Hs3st
heparan sulfate 3-O-sulfotransferase
- PBS
phosphate buffered saline
- Glce
glucuronyl C5-epimerase
- Hs2st
heparan sulfate 2-O-sulfotransferase
- FGF-2
fibroblast growth factor 2
- BODIPY R6G
4,4-difluoro-5-phenyl-4-bora-3,4a-diaza-s-indacene-3-propionic acid
- RPIP
reverse-phase ion-pairing
- UPLC
ultra-performance liquid chromatography
- MS
mass spectrometry
- TrBA
tributylamine
- ESI
electrospray ionization
- CS
chondroitin sulfate
- DS
dermatan sulfate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at
References
- Ahn WS, Antoniewicz MR. Metabolic flux analysis of CHO cells at growth and non-growth phases using isotopic tracers and mass spectrometry. Metab Eng. 2011;13:598–609. doi: 10.1016/j.ymben.2011.07.002. [DOI] [PubMed] [Google Scholar]
- Ahsan A, Jeske W, Hoppensteadt D, Lormeau JC, Wolf H, Fareed J. Molecular profiling and weight determination of heparins and depolymerized heparins. J Pharm Sci. 1995;84:724–727. doi: 10.1002/jps.2600840612. [DOI] [PubMed] [Google Scholar]
- Aldor IS, Keasling JD. Process design for microbial plastic factories: metabolic engineering of polyhydroxyalkanoates. Curr Opin Biotechnol. 2003;14:475–483. doi: 10.1016/j.copbio.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Alper H, Fischer C, Nevoigt E, Stephanopoulos G. Tuning genetic control through promoter engineering. Proc Natl Acad Sci U S A. 2005;102:12678–12683. doi: 10.1073/pnas.0504604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X, Esko JD. An animal cell mutant defective in heparan sulfate hexuronic acid 2-O-sulfation. J Biol Chem. 1996;271:17711–17717. doi: 10.1074/jbc.271.30.17711. [DOI] [PubMed] [Google Scholar]
- Bai X, Wei G, Sinha A, Esko JD. Chinese hamster ovary cell mutants defective in glycosaminoglycan assembly and glucuronosyltransferase I. J Biol Chem. 1999;274:13017–13024. doi: 10.1074/jbc.274.19.13017. [DOI] [PubMed] [Google Scholar]
- Bame KJ, Esko JD. Undersulfated heparan sulfate in a Chinese hamster ovary cell mutant defective in heparan sulfate N-sulfotransferase. J Biol Chem. 1989;264:8059–8065. [PubMed] [Google Scholar]
- Bartlett AH, Hayashida K, Park PW. Molecular and cellular mechanisms of syndecans in tissue injury and inflammation. Mol Cells. 2007;24:153–166. [PubMed] [Google Scholar]
- Bernfield M, Gotte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, Zako M. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- Bishop JR, Schuksz M, Esko JD. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature. 2007;446:1030–1037. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- Busse M, Feta A, Presto J, Wilen M, Gronning M, Kjellen L, Kusche-Gullberg M. Contribution of EXT1, EXT2, and EXTL3 to heparan sulfate chain elongation. J Biol Chem. 2007;282:32802–32810. doi: 10.1074/jbc.M703560200. [DOI] [PubMed] [Google Scholar]
- Capila I, Linhardt RJ. Heparin-protein interactions. Angew Chem Int Ed Engl. 2002;41:391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Casu B. Structure and biological activity of heparin. Adv Carbohydr Chem Biochem. 1985;43:51–134. doi: 10.1016/s0065-2318(08)60067-0. [DOI] [PubMed] [Google Scholar]
- Chen J, Avci FY, Munoz EM, McDowell LM, Chen M, Pedersen LC, Zhang L, Linhardt RJ, Liu J. Enzymatic redesigning of biologically active heparan sulfate. J Biol Chem. 2005;280:42817–42825. doi: 10.1074/jbc.M504338200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan MB, Chen J, Krise JP, Liu J. The biosynthesis of anticoagulant heparan sulfate by the heparan sulfate 3-O-sulfotransferase isoform 5. Biochim Biophys Acta. 2004;1671:34–43. doi: 10.1016/j.bbagen.2003.12.010. [DOI] [PubMed] [Google Scholar]
- Esko JD, Selleck SB. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- Forsberg E, Pejler G, Ringvall M, Lunderius C, Tomasini-Johansson B, Kusche-Gullberg M, Eriksson I, Ledin J, Hellman L, Kjellen L. Abnormal mast cells in mice deficient in a heparin-synthesizing enzyme. Nature. 1999;400:773–776. doi: 10.1038/23488. [DOI] [PubMed] [Google Scholar]
- Fuster MM, Wang L. Endothelial heparan sulfate in angiogenesis. Prog Mol Biol Transl Sci. 2010;93:179–212. doi: 10.1016/S1877-1173(10)93009-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godavarti R, Davis M, Venkataraman G, Cooney C, Langer R, Sasisekharan R. Heparinase III from Flavobacterium heparinum: cloning and recombinant expression in Escherichia coli. Biochem Biophys Res Commun. 1996;225:751–758. doi: 10.1006/bbrc.1996.1246. [DOI] [PubMed] [Google Scholar]
- Goh JS, Zhang P, Chan KF, Lee MM, Lim SF, Song Z. RCA-I-resistant CHO mutant cells have dysfunctional GnT I and expression of normal GnT I in these mutants enhances sialylation of recombinant erythropoietin. Metab Eng. 2010;12:360–368. doi: 10.1016/j.ymben.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Green EM. Fermentative production of butanol--the industrial perspective. Curr Opin Biotechnol. 2011;22:337–343. doi: 10.1016/j.copbio.2011.02.004. [DOI] [PubMed] [Google Scholar]
- Guerrini M, Beccati D, Shriver Z, Naggi A, Viswanathan K, Bisio A, Capila I, Lansing JC, Guglieri S, Fraser B, Al-Hakim A, Gunay NS, Zhang Z, Robinson L, Buhse L, Nasr M, Woodcock J, Langer R, Venkataraman G, Linhardt RJ, Casu B, Torri G, Sasisekharan R. Oversulfated chondroitin sulfate is a contaminant in heparin associated with adverse clinical events. Nat Biotechnol. 2008;26:669–675. doi: 10.1038/nbt1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson GT, Pihl E. Staining of mast cell acid glycosaminoglycans in ultrathin sections by ruthenium red. Nature. 1967;216:697–698. doi: 10.1038/216697a0. [DOI] [PubMed] [Google Scholar]
- Hacker U, Nybakken K, Perrimon N. Heparan sulphate proteoglycans: the sweet side of development. Nat Rev Mol Cell Biol. 2005;6:530–541. doi: 10.1038/nrm1681. [DOI] [PubMed] [Google Scholar]
- Kuberan B, Lech MZ, Beeler DL, Wu ZL, Rosenberg RD. Enzymatic synthesis of antithrombin III-binding heparan sulfate pentasaccharide. Nat Biotechnol. 2003;21:1343–1346. doi: 10.1038/nbt885. [DOI] [PubMed] [Google Scholar]
- Linhardt RJ, Dordick JS, Deangelis PL, Liu J. Enzymatic synthesis of glycosaminoglycan heparin. Semin Thromb Hemost. 2007;33:453–465. doi: 10.1055/s-2007-982076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcum JA, Rosenberg RD. Anticoagulantly active heparan sulfate proteoglycan and the vascular endothelium. Semin Thromb Hemost. 1987;13:464–474. doi: 10.1055/s-2007-1003523. [DOI] [PubMed] [Google Scholar]
- Martin JG, Gupta M, Xu Y, Akella S, Liu J, Dordick JS, Linhardt RJ. Toward an artificial Golgi: redesigning the biological activities of heparan sulfate on a digital microfluidic chip. J Am Chem Soc. 2009;131:11041–11048. doi: 10.1021/ja903038d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin VJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol. 2003;21:796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- Martynova MG, Bystrova OA, Moiseeva OM, Evdonin AL, Kondratov KA, Medvedeva ND. The presence of ANP in rat peritoneal mast cells. Cell Res. 2005;15:811–816. doi: 10.1038/sj.cr.7290350. [DOI] [PubMed] [Google Scholar]
- Michalodimitrakis K, Isalan M. Engineering prokaryotic gene circuits. FEMS Microbiol Rev. 2009;33:27–37. doi: 10.1111/j.1574-6976.2008.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muszbek L, Bereczky Z, Kovacs B, Komaromi I. Antithrombin deficiency and its laboratory diagnosis. Clin Chem Lab Med. 2010;48 Suppl 1:S67–S78. doi: 10.1515/CCLM.2010.368. [DOI] [PubMed] [Google Scholar]
- Nairn AV, Kinoshita-Toyoda A, Toyoda H, Xie J, Harris K, Dalton S, Kulik M, Pierce JM, Toida T, Moremen KW, Linhardt RJ. Glycomics of proteoglycan biosynthesis in murine embryonic stem cell differentiation. J Proteome Res. 2007;6:4374–4387. doi: 10.1021/pr070446f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeth A, Rohlich P. Rapid separation of rat peritoneal mast cells with Percoll. Eur J Cell Biol. 1980;20:272–275. [PubMed] [Google Scholar]
- Nolan RP, Lee K. Dynamic model of CHO cell metabolism. Metab Eng. 2011;13:108–124. doi: 10.1016/j.ymben.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Noti C, Seeberger PH. Chemical approaches to define the structure-activity relationship of heparin-like glycosaminoglycans. Chem Biol. 2005;12:731–756. doi: 10.1016/j.chembiol.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Pan J, Qian Y, Zhou X, Pazandak A, Frazier SB, Weiser P, Lu H, Zhang L. Oversulfated chondroitin sulfate is not the sole contaminant in heparin. Nat Biotechnol. 2010;28:203–207. doi: 10.1038/nbt0310-203. author reply 207–211. [DOI] [PubMed] [Google Scholar]
- Pfeifer BA, Admiraal SJ, Gramajo H, Cane DE, Khosla C. Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science. 2001;291:1790–1792. doi: 10.1126/science.1058092. [DOI] [PubMed] [Google Scholar]
- Pitera DJ, Paddon CJ, Newman JD, Keasling JD. Balancing a heterologous mevalonate pathway for improved isoprenoid production in Escherichia coli. Metab Eng. 2007;9:193–207. doi: 10.1016/j.ymben.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Robinson HC, Horner AA, Hook M, Ogren S, Lindahl U. A proteoglycan form of heparin and its degradation to single-chain molecules. J Biol Chem. 1978;253:6687–6693. [PubMed] [Google Scholar]
- Shaya D, Tocilj A, Li Y, Myette J, Venkataraman G, Sasisekharan R, Cygler M. Crystal structure of heparinase II from Pedobacter heparinus and its complex with a disaccharide product. J Biol Chem. 2006;281:15525–15535. doi: 10.1074/jbc.M512055200. [DOI] [PubMed] [Google Scholar]
- Spear PG, Shieh MT, Herold BC, WuDunn D, Koshy TI. Heparan sulfate glycosaminoglycans as primary cell surface receptors for herpes simplex virus. Adv Exp Med Biol. 1992;313:341–353. doi: 10.1007/978-1-4899-2444-5_33. [DOI] [PubMed] [Google Scholar]
- Sugahara K, Kitagawa H. Heparin and heparan sulfate biosynthesis. IUBMB Life. 2002;54:163–175. doi: 10.1080/15216540214928. [DOI] [PubMed] [Google Scholar]
- Tumova S, Woods A, Couchman JR. Heparan sulfate proteoglycans on the cell surface: versatile coordinators of cellular functions. Int J Biochem Cell Biol. 2000;32:269–288. doi: 10.1016/s1357-2725(99)00116-8. [DOI] [PubMed] [Google Scholar]
- Wang L, Fuster M, Sriramarao P, Esko JD. Endothelial heparan sulfate deficiency impairs L-selectin- and chemokine-mediated neutrophil trafficking during inflammatory responses. Nat Immunol. 2005;6:902–910. doi: 10.1038/ni1233. [DOI] [PubMed] [Google Scholar]
- Wang Z, Ly M, Zhang F, Zhong W, Suen A, Hickey AM, Dordick JS, Linhardt RJ. E. coli K5 fermentation and the preparation of heparosan, a bioengineered heparin precursor. Biotechnol Bioeng. 2010;107:964–973. doi: 10.1002/bit.22898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Moon AF, Song D, Pedersen LC, Liu J. Engineering sulfotransferases to modify heparan sulfate. Nat Chem Biol. 2008;4:200–202. doi: 10.1038/nchembio.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Inatani M, Matsumoto Y, Ogawa J, Irie F. Roles of heparan sulfate in mammalian brain development current views based on the findings from Ext1 conditional knockout studies. Prog Mol Biol Transl Sci. 2010;93:133–152. doi: 10.1016/S1877-1173(10)93007-X. [DOI] [PubMed] [Google Scholar]
- Yang B, Weyers A, Baik JY, Sterner E, Sharfstein S, Mousa SA, Zhang F, Dordick JS, Linhardt RJ. Ultra-performance ion-pairing liquid chromatography with on-line electrospray ion trap mass spectrometry for heparin disaccharide analysis. Anal Biochem. 2011;415:59–66. doi: 10.1016/j.ab.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida E, Arakawa S, Matsunaga T, Toriumi S, Tokuyama S, Morikawa K, Tahara Y. Cloning, sequencing, and expression of the gene from bacillus circulans that codes for a heparinase that degrades both heparin and heparan sulfate. Biosci Biotechnol Biochem. 2002;66:1873–1879. doi: 10.1271/bbb.66.1873. [DOI] [PubMed] [Google Scholar]
- Yun CY, Liu S, Lim SF, Wang T, Chung BY, Jiat Teo J, Chuan KH, Soon AS, Goh KS, Song Z. Specific inhibition of caspase-8 and -9 in CHO cells enhances cell viability in batch and fed-batch cultures. Metab Eng. 2007;9:406–418. doi: 10.1016/j.ymben.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Zhang L, Beeler DL, Lawrence R, Lech M, Liu J, Davis JC, Shriver Z, Sasisekharan R, Rosenberg RD. 6-O-sulfotransferase-1 represents a critical enzyme in the anticoagulant heparan sulfate biosynthetic pathway. J Biol Chem. 2001a;276:42311–42321. doi: 10.1074/jbc.M101441200. [DOI] [PubMed] [Google Scholar]
- Zhang L, Lawrence R, Frazier BA, Esko JD. CHO glycosylation mutants: proteoglycans. Methods Enzymol. 2006;416:205–221. doi: 10.1016/S0076-6879(06)16013-9. [DOI] [PubMed] [Google Scholar]
- Zhang L, Lawrence R, Schwartz JJ, Bai X, Wei G, Esko JD, Rosenberg RD. The effect of precursor structures on the action of glucosaminyl 3-O-sulfotransferase-1 and the biosynthesis of anticoagulant heparan sulfate. J Biol Chem. 2001b;276:28806–28813. doi: 10.1074/jbc.M100204200. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Xie J, Liu H, Liu J, Linhardt RJ. Quantification of heparan sulfate disaccharides using ion-pairing reversed-phase microflow high-performance liquid chromatography with electrospray ionization trap mass spectrometry. Anal Chem. 2009;81:4349–4355. doi: 10.1021/ac9001707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Garron ML, Yang B, Xiao Z, Esko JD, Cygler M, Linhardt RJ. Asparagine 405 of heparin lyase II prevents the cleavage of glycosidic linkages proximate to a 3-O-sulfoglucosamine residue. FEBS Lett. 2011;585:2461–2466. doi: 10.1016/j.febslet.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.