Abstract
Carotenoid cleavage dioxygenases (CCDs) are a group of enzymes that catalyze the oxidative cleavage steps from carotenoids to various carotenoid cleavage products. Some ccd genes have been identified and encoded enzymes functionally characterized in many higher plants, but little in cyanobacteria. We performed a comparative analysis of ccd sequences and explored their distribution, classification, phylogeny, evolution, and structure among 37 cyanobacteria. Totally 61 putative ccd sequences were identified, which are abundant in Acaryochloris marina MBIC 11017, filamentous N2-fixing cyanobacteria, and unicellular cyanobacterial Cyanothece. According to phylogenetic trees of 16S rDNA and CCD, nced and ccd8 genes occur later than the divergence of ccd7, apco, and ccd1. All CCD enzymes share conserved basic structure domains constituted by a single loop formed with seven β-strands and one helix. In this paper, a general framework of sequence-function-evolution connection for the ccd has been revealed, which may provide new insight for functional investigation.
1. Introduction
Cyanobacteria, also known as blue-green algae and blue-green bacteria, are among the earliest branching groups on earth, dating back 2.5–3.5 billion years, based on the fossil evidence [1]. They may be unicellular or filamentous and can be found in almost every conceivable environment, such as marine and freshwater habitats, soil, rocks, and plants [2, 3]. With the capacity of oxygenic photosynthesis similar to the process found in higher plants, cyanobacteria constitute a group of species diverse not only in ecological habitat, but also in genome size and the number of gene, indicating the significance of comparative genome research. The genome size varied from 1.6 Mb (Prochlorococcus sp. MIT9301) to 9.0 Mb (Nostoc punctiforme PCC 73102), and the number of gene ranged from 1,756 (Prochlorococcus marinus MED4) to 8,462 (Acaryochloris marina MBIC11017) [4–6].
A lot of information on the evolutionary history of cyanobacteria has strongly supported an underlying meaning of comparative genome research. Three major clades are observed in cyanobacteria phylogenetic tree (Figure 1). The unicellular cyanobacteria (Prochlorococcus and Synechococcus) from ocean form the first monophyletic group (BS: 98%). They maintain the smallest genome size and account for significant biomass and primary production of marine biosphere [7]. Two Synechococcus elongatus PCC (6301 and 7942) are found at the base of this monophyletic group. Three thermophilic cyanobacteria (Synechococcus sp. JA-2-3B′a (2-13), Thermosynechococcus elongatus BP-1, and Synechococcus sp. JA-3-3Ab) and a thylakoid-lacking marine cyanobacterium (Gloeobacter sp. PCC 7421) [8] together compose the second monophyletic group (BS: 60%). In the last monophyletic group (BS: 64%), the filamentous N2-fixing cyanobacteria (Nostoc sp. PCC 7120, A. variabilis ATCC 29413, N. punctiforme ATCC 29133, A. platensis NIES-39, and T. erythraeum IMS101) build the sister group (BS: 75%) with freshwater unicellular cyanobacteria (Cyanothece sp. ATCC 51142, 8801, 7424, bloom forming Synechocystis sp. PCC 6803, and toxic bloom Microcystis aeruginosa NIES-843). An animal-cyanobacterial symbiont (Acaryochloris marina MBIC11017) and Cyanothece sp. PCC 7425 build the basal branch of this monophyletic group (see Figure 1 and Supplementary Material Table 1 available online at doi: 10.1155/2012/164690).
Figure 1.
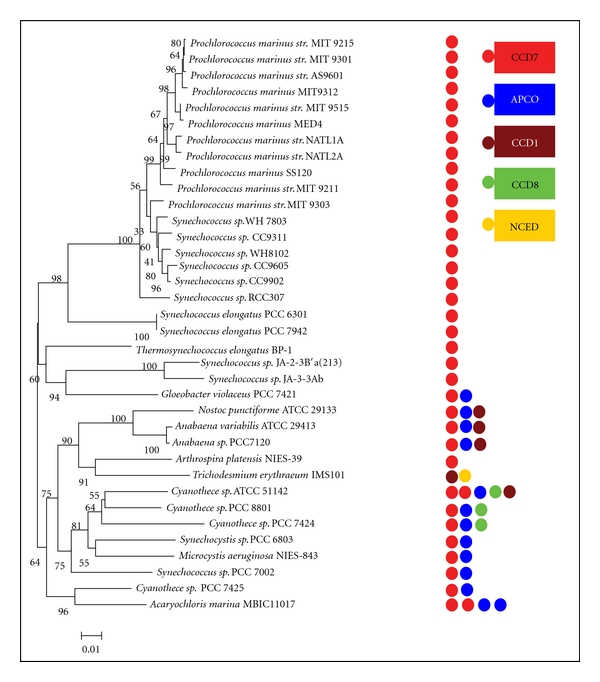
Phylogenetic tree of the sequenced cyanobacterial strains and the distribution of ccd sequences genes. Left: 37 fully sequenced cyanobacteria strains were performed based on 16 s rRNA as described in the methods section. An identical topology was obtained with two different methods (ML, NJ) and different models applied (for ML, generalized time reversible (GTR); for NJ, Kimura 2-parameter). Numbers at the node indicated bootstrap values (%) for 1,000 replicates. Right: the distribution of ccd genes across 37 cyanobacterial strains. Circular boxes represented the different ccd sequences that were labeled in the distinct color. The numbers of ccd sequences were presented by the numbers of circular boxes.
The first identified gene encoding carotenoid cleavage dioxygenase was the Vp14 maize gene which was required for the formation of the abscisic acid (ABA) precursor xanthoxin, representing the rate-limiting step in ABA biosynthesis [9, 10]. Subsequently, homology-based analysis of the existence in all taxa was carried out in silico, which allowed the elucidation of the biosynthesis of several carotenoid-derived compounds, such as retinal in animals [11, 12] and fungi [13], the pigments bixin [14], saffron [15], and neurosporaxanthin [16]. On the basis of the identified substrates and presumed mechanism of catalysis, these enzymes are referred to as carotenoid cleavage dioxygenases (CCDs).
Carotenoid cleavage dioxygenases (CCDs) are a group of enzymes that catalyze the oxidative cleavage steps from carotenoids to various carotenoid cleavage products (apocarotenoids). These apocarotenoids function as signalling molecules with diverse functions including the ubiquitous chromophore retinal, plant hormone abscisic acid, and strigolactones [17, 18]. Other apocarotenoids with unknown functions in plants with high economic value are bixin in Bixa orellana and saffron in Crocus sativus [19]. These enzymes are present in all taxa [17, 20] and exhibit a high degree of regio- and stereospecificity for certain double bond positions as opposed to their frequent promiscuity towards substrates [21]. CCD enzymes have been widely studied in higher plants. In Arabidopsis thaliana, the CCD family includes nine members forming the basis for CCD classification (NCED, CCD1, CCD4, CCD7, and CCD8) in plants [22, 23]. The members of CCD family, Cmccd4a, Cmccd4b, Cmnced3a, and Cmnced3b have, been identified and functionally investigated in Chrysanthemum morifolium [24]. Among CCD family, NCED subfamily involves in abscisic acid (ABA) biosynthesis [10, 17, 23]. The majority of CCD has been shown to reside in plastids, whereas the only exception is CCD1 and its orthologous enzymes in higher plants, which act in the cytoplast to generate C13 and C14 apocarotenoids. Known pathways of carotenoid cleavage leading to various apocarotenoids have been discussed [17, 20, 21, 25, 26]. According to endosymbiotic theory, chloroplasts in plants and eukaryotic algae evolved from cyanobacterial ancestors via endosymbiosis.
It is reasonable to speculate that the CCD enzymes are widely separated in cyanobacteria as well. At present, many ccd genes have been cloned and functionally characterized in vitro from some sequenced cyanobacteria strains, including Synechocystis sp. PCC 6803 [27] and Nostoc sp. PCC 7120 [28–30]. With the completion of genome sequencing of several cyanobacterial species, modifications and supplements are needed.
Recently, 37 genomes of unicellular and filamentous cyanobacteria have been available, which facilitate cyanobacterial systemic analysis for metacaspases family [31], serine/threonine protein kinases [32], restriction modification system [33], and carotenoids biosynthesis [34]. These genome-sequencing projects undoubtedly bring great convenience to the searching for novel ccd genes by bioinformatic tools. In this study, five characterized ccd genes from Arabidopsis thaliana and Zea mays were selected as queries to search for cyanobacterial ccd genes. A BLASTP-plus-phylogeny reconstruction approach was employed to analyze CCD protein sequences in cyanobacteria, emphasizing an overall view on their distribution, classifications, phylogeny, evolution, and structure. Better understanding of cyanobacterial CCD enzymes may provide deeper insights into the evolution and the functional investigation of CCD enzymes in all organisms.
2. Materials and Methods
2.1. Computational Search for Novel ccd Genes
37 species of cyanobacteria, including Prochlorococcus, Synechococcus, Synechocystis, Gloeobacter, Cyanothece, Microcystis, Trichodesmium, Acaryochloris, Anabaena, and Nostoc (http://genome.kazusa.or.jp/cyanobase/), were used in this analysis. The ccd genes from higher plants Arabidopsis thaliana, Tomato, and Zea may (Table 1) were obtained from National Center for Biotechnology Information (NCBI) and were used to construct a query protein set. Each protein in this query data set was used to search for the potential novel sequences in all cyanobacterial species from whole genome sequences available, by using the basic local alignment search tool (BLASTP) with E-value < 1E−10 [35–37]. Proteins found by this method that fit the criteria for genuine CCD enzymes were added to the query set for another round of BLASTP searches. The searches were iterated until convergence.
Table 1.
Carotenoid cleavage dioxygenases protein sequences from distinct organisms in this paper.
| Name | Species | Accession number | Type |
|---|---|---|---|
| NCED | Arabidopsis thaliana | NP_189064.1 | Streptophyta |
| NCED | Solanum lycopersicum | CAB10168.1 | Streptophyta |
| NCED | Phaeodactylum tricornutum CCAP 1055/1 | XP_002177588.1 | Heterokontophyta |
| CCD1 | Zea mays | AAZ22348.1 | Streptophyta |
| CCD4 | Arabidopsis thaliana | sp|O49675.1 | Streptophyta |
| CCD7 | Arabidopsis thaliana | AEC10494.1 | Streptophyta |
| CCD7 | Solanum lycopersicum | ACY39882.1 | Streptophyta |
| CCD8 | Arabidopsis thaliana | NP_195007.2 | Streptophyta |
| RPE | Ostreococcus tauri | XP_003080965.1 | Chlorophyta |
| RPE | Ajellomyces dermatitidis ATCC 18188 | EGE78156.1 | Fungi |
| RPE | Pyrenophora tritici-repentis Pt-1C-BFP | EDU50284.1 | Fungi |
| CO | Mycobacterium vanbaalenii PYR-1 | YP_951059.1 | Bacteria |
| PP | Chlamydomonas reinhardtii | EDP06596.1 | Chlorophyta |
| LSD | Ostreococcus tauri | CAL53034.1 | Chlorophyta |
| APCO | Ostreococcus tauri | CAL50095.1 | Chlorophyta |
2.2. Multiple Sequence Alignment and Phylogenetic Analysis
All protein sequences identified by BLASTP were aligned using ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/) [38]. The final alignment was further refined after excluding the relatively poorly conserved regions at the protein ends and consisted of sequences spanning the conserved domains. 16S rDNA and CCD phylogenetic trees were constructed by the NJ (Neighbor-Joining) method, using the program MEGA 4.0 (http://www.megasoftware.net/) [39] and ML (Maximum-Likelihood) method, using the program PHYML (http://www.atgc-montpellier.fr/phyml/binaries.php/) [40], with bootstrap support values deriving from 1000 randomized and replicated datasets. The Le and Gascuel evolutionary model [41] was selected for the protein phylogenies assuming an estimated proportion of invariant sites and a gamma correction. Graphical representation and edition of the phylogenetic tree were performed with TreeDyn (v198.3) [42]. In the phylogenetic analysis of CCD, CCD enzymes of fungi were used as outgroup to root the tree.
2.3. Motif Scanning and Structure Domain Analysis
To identify conserved motifs, Multiple Expectation Maximization for Motif Elicitation (MEME) version 2.2 [43] was employed with a set of parameters as follows: number of repetitions-any, maximum number of motifs-100, number of sites (≥5 and ≤50), and optimum motif width set to ≥6 and ≤200 [44, 45]. The Simple Modular Architecture Research Tool (SMART) (http://smart.embl-heidelberg.de/) and Conserved Domains Database (CDD) (http://www.ncbi.nlm.nih.gov/cdd/) were applied to predict the structure domains of these CCD proteins sequences, relying on hidden Markov models and Reverse Position-Specific BLAST separately [46, 47].
2.4. Tertiary Structure Prediction
To well understand the evolution of certain enzyme, protein structure was analyzed using homology modeling. The protein sequences of CCD from Prochlorococcus marinus MIT9312 (PMT9312_0282), Nostoc punctiforme ATCC 29133 (Npun_F0298), Cyanothece sp. PCC 7424 (PCC7424_5321), and Anabaena sp. PCC 7120 (all1106) were submitted to the protein model server: SWISS-MODEL Web server [48, 49] (http://swissmodel.expasy.org/) though Automated Model, respectively. All the manipulations were performed using Pdb-Viewer [50, 51].
3. Results
3.1. Identification of Putative CCD Proteins
The 37 completed cyanobacteria were used in this research, and the detail information about key features of these cyanobacterial species was summarized in Supplementary material Table 1. The phylogenetic tree based on 16S rDNA was showed in Figure 1. The candidate genes identified and the distribution across cyanobacterial strains in this study were listed in Table 2 and Figure 1. Totally 61 putative ccd genes were predicted and annotated from 37 completed cyanobacterial genomes using BLASTP programs with the query sequences. 50 of these genes were originally annotated as retinal pigment epithelial membrane protein (RPE), lignostilbene-α, β-dioxygenase (LSD), or carotenoid oxygenase (CO). The remaining 11 proteins were accepted as CCD enzymes in this research, including 5 hypothetical proteins, 4 undefined proteins, 1 apocarotenoid 15,15′-dioxygenase (APCO), and 1 similar to neoxanthin cleavage enzyme. Interestingly, RPE65 (retinal pigment epithelium 65-kDa protein) shows a significant degree of sequence homology to the CCD family. Thus, many CCD enzymes were originally annotated RPE in cyanobacteria.
Table 2.
List of putative ccd genes identified across 37 cyanobacterial in this paper.
| Species | Gene | Length | Annotation | Proposed function |
|---|---|---|---|---|
| P. marinus MED4 | 0280 | 507 | CCD7 | |
| P. marinus MIT9312 | 0282 | 494 | RPE | CCD7 |
| P. marinus MIT9313 | 1879 | 507 | CCD7 | |
| P. marinus SS120 | 0312 | 496 | CCD7 | |
| P. marinus str. AS9601 | 03031 | 494 | RPE | CCD7 |
| P. marinus str. MIT 9211 | 03071 | 495 | RPE | CCD7 |
| P. marinus str. MIT 9215 | 03051 | 494 | RPE | CCD7 |
| P. marinus str. MIT 9301 | 03041 | 494 | RPE | CCD7 |
| P. marinus str. MIT 9303 | 25111 | 507 | RPE | CCD7 |
| P. marinus str. MIT 9515 | 03131 | 495 | RPE | CCD7 |
| P. marinus str. NATL1A | 03601 | 497 | RPE | CCD7 |
| P. marinus str. NATL2A | 1646 | 497 | LSD | CCD7 |
| A. platensis NIES-39 | 06860 | 492 | LSD | CCD7 |
| S. elongates PCC 6301 | 1315d | 493 | LSD | CCD7 |
| S. elongates PCC 7942 | 0196 | 493 | BCD | CCD7 |
| S. sp. CC9311 | 0261 | 504 | RPE | CCD7 |
| S. sp. CC9605 | 0221 | 488 | LSD | CCD7 |
| S. sp. CC9902 | 0248 | 489 | LSD | CCD7 |
| S. sp. JA-2-3B′a (2-13) | 0263 | 482 | LSD | CCD7 |
| S. sp. JA-3-3Ab | 1068 | 482 | LSD | CCD7 |
| S. sp. PCC 7002 | A1425 | 492 | LSD | CCD7 |
| S. sp. PCC 7002 | A2200 | 484 | RPE | APCO |
| S. sp. RCC307 | 2292 | 486 | LSD | CCD7 |
| S. sp. WH 7803 | 0270 | 495 | LSD | CCD7 |
| S. sp. WH8102 | 0227 | 489 | CCD7 | |
| S. sp. PCC 6803 | 1541 | 490 | HP | CCD7 |
| S. sp. PCC 6803 | 1648 | 480 | HP | APCO |
| T. elongatus BP-1 | 0015 | 487 | LSD | CCD7 |
| T. IMS101 | 3794 | 464 | CO | CCD1 |
| T. IMS101 | 3212 | 489 | CO | NCED |
| G. violaceus PCC 7421 | 3689 | 482 | LSD | CCD7 |
| G. violaceus PCC 7421 | 2774 | 475 | HP | APCO |
| M. aeruginosa NIES-843 | 09040 | 515 | CO | CCD7 |
| M. aeruginosa NIES-843 | 60600 | 490 | HP | APCO |
| N. punctiforme ATCC 29133 | 2869 | 460 | CO | CCD1 |
| N. punctiforme ATCC 29133 | 0298 | 498 | CO | CCD7 |
| N. punctiforme ATCC 29133 | 0776 | 491 | CO | APCO |
| C. sp. PCC 8801 | 0352 | 487 | CO | CCD8 |
| C. sp. PCC 8801 | 4340 | 495 | CO | CCD7 |
| C. sp. PCC 8801 | 0015 | 468 | CO | APCO |
| C. sp. PCC 7424 | 4841 | 464 | CO | CCD1 |
| C. sp. PCC 7424 | 0935 | 488 | CO | CCD8 |
| C. sp. PCC 7424 | 0449 | 491 | CO | CCD7 |
| C. sp. PCC 7424 | 5321 | 477 | CO | APCO |
| C. sp. PCC 7425 | 4222 | 501 | CO | CCD7 |
| C. sp. PCC 7425 | 3964 | 486 | CO | APCO |
| C. sp. ATCC 51142 | 2215 | 459 | RPE | CCD1 |
| C. sp. ATCC 51142 | 3977 | 486 | RPE | CCD8 |
| C. sp. ATCC 51142 | 2665 | 494 | LSD | CCD7 |
| C. sp. ATCC 51142 | 4927 | 481 | CO | CCD7 |
| C. sp. ATCC 51142 | 2515 | 468 | CO | APCO |
| A. marina MBIC11017 | 0661 | 488 | LSD | CCD7 |
| A. marina MBIC11017 | 3102 | 483 | RPE | CCD7 |
| A. marina MBIC11017 | 1272 | 479 | RPE | APCO |
| A. marina MBIC11017 | 0925 | 485 | RPE | APCO |
| A. sp. PCC 7120 | 1106 | 475 | NCE | CCD1 |
| A. sp. PCC 7120 | 4284 | 497 | LSD | CCD7 |
| A. sp. PCC 7120 | 4895 | 472 | HP | APCO |
| A. variabilis ATCC 29413 | 2047 | 765 | RPE | CCD1 |
| A. variabilis ATCC 29413 | 3710 | 463 | RPE | CCD1 |
| A. variabilis ATCC 29413 | 1236 | 510 | RPE | CCD7 |
Amid diverse cyanobacterial genomes, the number of ccd gene varies from 1 to 5. Within unicellular cyanobacteria, N2-fixing Cyanothece sp. ATCC 51142 has 5 ccd genes, much more than other species. An animal-cyanobacterial symbiont (Acaryochloris marina MBIC11017) has 4 ccd genes. All of the Synechococcus and Prochlorococcus marinus strains have only one ccd gene except that Synechococcus sp. PCC 700 has two ccd genes. Two unicellular strains inhabit in freshwater (Synechocystis sp. PCC 6803 and Microcystis aeruginosa NIES-843) and one marine cyanobacteria (Gloeobacter violaceus PCC 7421) each contains two ccd genes. The Cyanothece group has two or more ccd genes, including Cyanothece sp. PCC 7425 (2), Cyanothece sp. PCC 8801 (3), and Cyanothece sp. PCC 7424 (4). Compared to unicellular cyanobacteria, filamentous N2-fixing cyanobacteria have more ccd gene (3 for Anabaena sp. PCC 7120, Anabaena variabilis ATCC 29413, and Nostoc punctiforme ATCC 29133, 2 for Trichodesmium erythraeum IMS101). However, Arthrospira platensis NIES-39 contains only one ccd gene.
3.2. Phylogenetic Analysis of ccd Genes in Cyanobacteria
Considering the confusion created by unspecific annotation with possibly separate evolutionary histories, the translated CCD proteins from cyanobacteria and the characterized CCD enzymes from bacteria, eukaryotic algae, higher plants, and fungi were used to perform a phylogenetic analysis. The tree topology matches our Neighbor-Joining tree (data not shown), and the high branch support values coincide with high neighbor joining bootstrap values, suggesting that the CCD phylogeny is robust in different tree reconstruction methods. Therefore, only the ML trees were displayed in this paper. The CCD phylogenetic tree was rooted in the CCD of fungi. Observation of the tree revealed that all CCD enzymes fell into four clades (Figure 2): clade 1: CCD7, clade 2: APCO, clade 3: CCD1/NCED/CCD4, and clade 4: CCD8.
Figure 2.
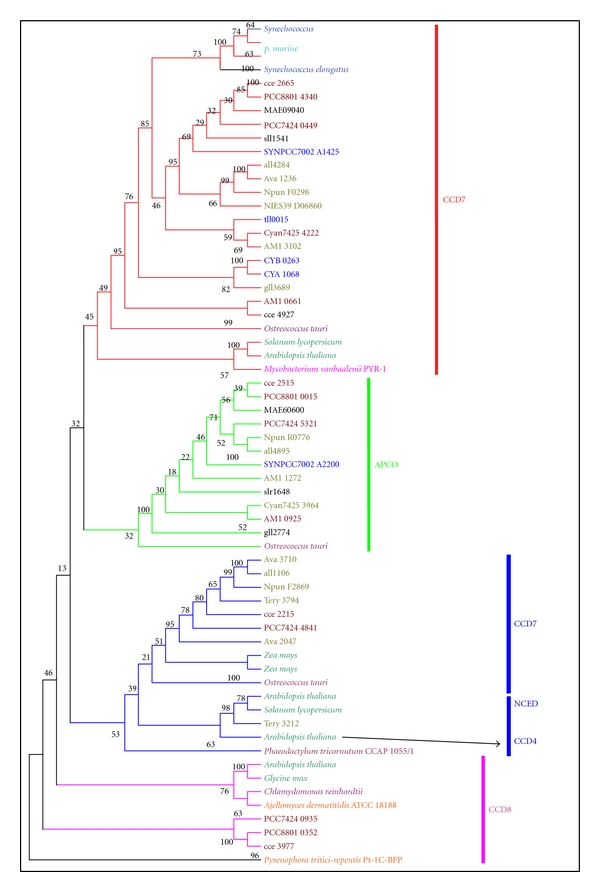
Maximum-likelihood tree of CCD enzymes of cyanobacteria, bacteria, eukaryotic algae, fungi, and higher plants. The model of LG+I+G was applied to construct ML tree using PHYML as described in the methods section. Numbers at the node indicated bootstrap values (%) for 1,000 replicates. Cyanobacterial CCD enzymes IDs and the strain names are as in Table 2 and Figure 1. Red line: CCD7, green: APCO, blue: CCD1, CCD4, and NCED, pink: CCD8. Species belonged to similar clade were represented by colored boxes. Orange: fungi, wheat: bacteria, purple: eukaryotic algae, barium: higher plants, cyan: cyanobacterial Prochlorococcus marinus, blue: Thermosynechococcus elongates, Synechococcus elongates, and Synechococcus, ruby: cyanobacterial Cyanothece, and smudge: filamentous cyanobacteria. Unlabeled species included Synechocystis sp. PCC 6803 (sll1541 and slr1648), Gloeobacter violaceus PCC 7421 (gll3689 and gll2774), Microcystis aeruginosa NIES-843 (MAE09040 and MAE60600).
As shown in Figures 1 and 2, the first clade, ccd7, was composed of almost all the cyanobacterial strains (in this paper), except for filamentous nitrogen fixation Trichodesmium erythraeum IMS101. The ccd7-homologous genes from two unicellular marine cyanobacteria Synechococcus (except for Synechococcus sp. PCC 7002) and Prochlorococcus constituted the first subfamily. Amino acid sequence identity of genes from this subfamily ranged from 59% to 98% (Supplementary Material Table 2). In the second subfamily, ccd7-homologous genes from two hot-spring habitat cyanobacteria (Synechococcus sp. JA-3-3Ab and Synechococcus sp. JA-2-3B′a (2-13)) and a thylakoid-lacking cyanobacterium (Gloeobacter sp. PCC 7421) clustered into a group. Other cyanobacterial ccd7 genes clustered into the last group, including filamentous cyanobacteria and unicellular cyanobacterial Cyanothece. Moreover, ccd7-homologous genes were discovered among eukaryotic microalgae (Ostreococcus tauri), bacteria (Mycobacterium vanbaalenii PYR-1), and higher plants (Solanum lycopersicum and Arabidopsis thaliana). All of these CCD7s are orthologues because of obvious evolutionary relationships with high bootstrap value.
All the apco-homologous genes clustered into clade 2, including filamentous cyanobacteria (Anabaena variabilis ATCC 29413, Anabaena sp. PCC 7120, and Acaryochloris marina MBIC11017) and unicellular cyanobacteria Cyanothece. In addition, apco-homologous genes from Gloeobacter violaceus PCC 7421, Microcystis aeruginosa NIES-843, Synechocystis sp. PCC 6803, and Synechococcus sp. PCC 7002 were also assembled into this group. Amino acid sequence identity of genes from this clade ranged from 43% to 76% (Supplementary Material Table 3). It is interesting that two copies of apco-homologous genes existed in animal-cyanobacterial symbiont (Acaryochloris marina MBIC11017). Moreover, ccd7-homologous gene was discovered in eukaryotic algae (Ostreococcus tauri) while was absent in higher plants. Results from BLASTP and phylogenetic tree suggest that a close evolutionary relationship exists between CCD7 enzyme and APCO enzyme, between CCD7 enzyme and APCO enzyme.
CCD1 enzyme from filamentous cyanobacteria, algae, as well as higher plant (Zea mays) stays in a sister group relation with NCED and CCD4 enzyme from higher plant. They build the third monophyletic group with NCED enzymes from Phaeodactylum tricornutum as the basal branch of this clade. Amino acid sequence identity of genes from CCD1, NCED, and CCD8 was summarized in Supplementary Material Table 4. This group included all filamentous cyanobacteria, except for A. platensis NIES-39. Surprisingly, the ccd1-homologous gene was absent in all unicellular cyanobacteria except for Cyanothece ATCC51142, suggesting this organism might acquire this gene by horizontal gene transfer. It is interesting that the protein sequence of Tery_3212 from Trichodesmium erythraeum IMS101 is highly similar with NCED from Arabidopsis thaliana (99%). It is worth considering that CCD4 enzyme from Arabidopsis thaliana form a monophyletic with NCED enzyme, indicating that they may origin from common ancestor and acquire different function under natural selection during the evolution. CCD8 enzymes are widely distributed in eukaryote (fungi, eukaryotic algae, and higher plants), whereas they are missing in all cyanobacteria except for Cyanothece (PCC 7424, 8801 and ATCC 51142), suggesting these cyanobacteria obtain this gene by horizontal gene transfer or produce it later under the natural selection during the evolution.
3.3. Conserved Motifs
According to CDD and SMART domain analyses, 8 protein sequences which were originally annotated as hypothesis protein or unidentified belonged to cyanobacterial CCD enzymes categories as well. All the protein sequences belonged to Pfam: RPE65 (IPR004294), which included β-carotene-15,15′-monooxygenase (BCDO1; EC 1.14.99.36), β-carotene-9′,10′-dioxygenase (BCDO2/CCD7), 9-cis-epoxycarotenoid dioxygenase (NCED), β-apocarotenoid-15,15′-oxygenase (APCO), and retinal pigment epithelial membrane protein (RPE). However, domain analyses failed to further distinguish them. To facilitate the classification of different types of CCD enzymes, conserved motifs were identified by MEME tool and multiple sequence alignments with ClustalW. There were two typical glycine-rich (GX7GX5GP and HHPFDGDGMI) motifs and one leucine-rich (LALWEAX2PX4PX2PX4PX2L) motif existed in all cyanobacterial CCD proteins (Figure 3). Moreover, CCD7 from Synechococcus and Prochlorococcus organisms shared 59–98% similarities (Supplementary material Table 2) and possessed conserved patterns (EAFSAHPX2D, NPLPFX2GX2GAAQCLXS). CCD7 from other cyanobacteria organisms shared 54–95% similarities (Supplementary Material Table 5) and possessed conserved patterns (APXGX2GEPXFXPXP and LX2HXPYXLHG). In addition, RGXFGX4GG, FXFIHDFX2TX5F, and FVFHHXNA motifs also existed in CCD7 enzymes from distinct organisms (Figures 3(a) and 3(b)). It was interesting that protein sequences of CCD7 and CCD1 have the same PX3PX2FH box in the N-terminal (Figures 3(b) and 3(d)). All these results suggested that all ccd genes have originated from a common ancestor and exhibited different functions under distinct natural selection during the evolution.
Figure 3.
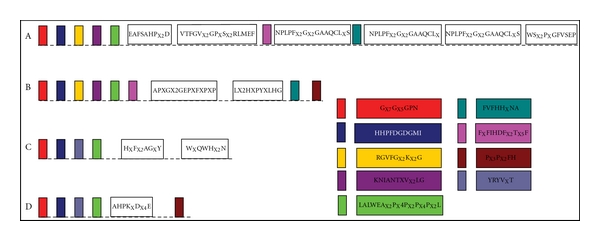
The conserved motifs of the cyanobacterial CCD enzymes. Schematic representation of motifs identified in cyanobacterial CCD enzymes using MEME motif search tool and ClustalW. The results were edited by hand. Length of box did not correspond to length of motif. Boxes represented the same or similar motifs that were labeled in the same color. (a) CCD7 from Synechococcus and Prochlorococcus marinus, (b) CCD7 from other cyanobacterial strains, (c) APCO, and (d) CCD1 and NCED.
3.4. Structure of CCD Enzymes
To understand the evolution of cyanobacterial CCD enzymes, protein structures of CCD7 from Prochlorococcus MIT9312 (PMT9312_0282) and Nostoc punctiforme ATCC 29133 (Npun_F0298), APCO from Cyanothece sp. PCC 7424 (PCC7424_5321), and CCD1 from Anabaena sp. PCC 7120 (all1106) were analyzed using homology modeling as described material and methods. A comparable analysis for the tertiary structure of different type of CCD-encoding ORFs from cyanobacteria revealed that a single loop formed with seven β-strands and one helix has conserved in four models, which may be related to binding domain (Figure 4).
Figure 4.
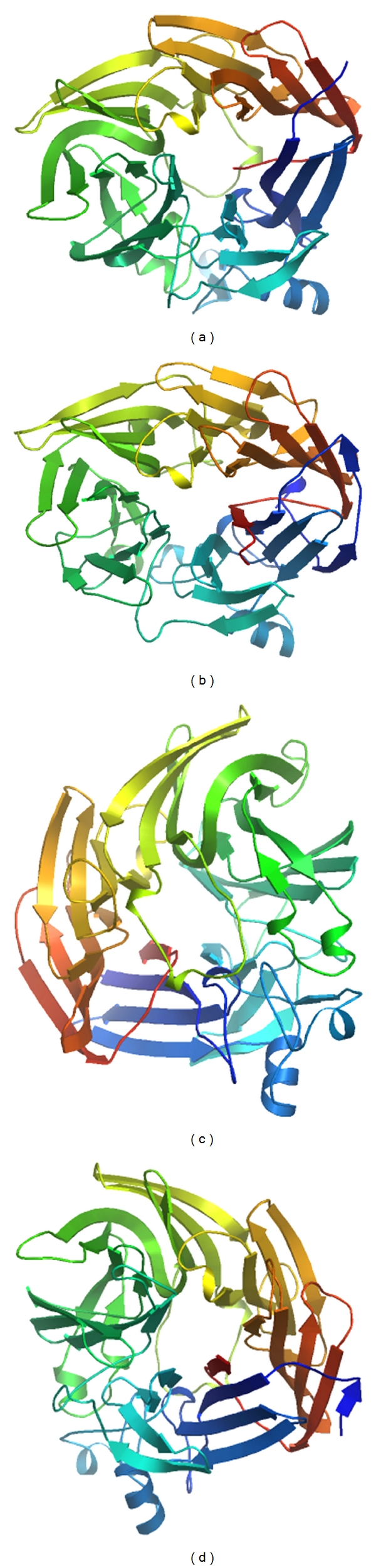
Model of structure of CCD enzymes from cyanobacteria strains. (a) The structure model of CCD7 from Prochlorococcus MIT9312 (PMT9312_0282), (b) the structure model of CCD7 from Nostoc punctiforme ATCC 29133 (Npun_F0298), (c) the structure model of APCO from Cyanothece sp. PCC 7424 (PCC7424_5321), (d) the structure model of CCD1 from Anabaena sp. PCC 7120 (all1106).
CCD7-encoding ORFs from different cyanobacterial organisms had conserved structure, which included α-helix (1), four-antiparallel β strand (4), two-, three-, and five-antiparallel β strand (1, 1, and 1), respectively. The additional sheets in the N- and C- terminal are the only difference (Figures 5(a) and 5(b)). As showed in Figures 5(c) and 5(d), five-antiparallel β strand existing in CCD7 was lacking in that of the APCO and CCD1. However, two α-helixes were present in CCD1-encoding ORFs. We supposed all the CCD-encoding ORFs in a given lineage may evolve through gene duplication that happened under natural selection during the evolution.
Figure 5.
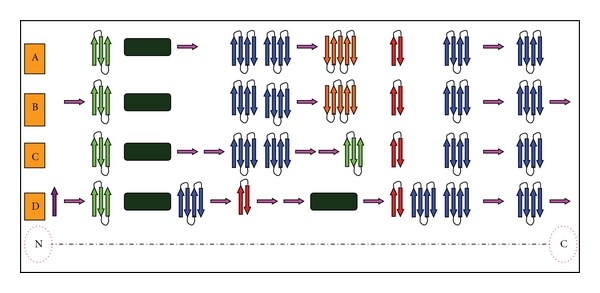
Sketch of the chain-fold of CCD enzyme showing the seven-bladed β-sheet as the basic motif. Distinct types of β-sheets were presented by colored arrowheads. Bottle green boxes stand for α-helix. (a) The second structure of CCD7 enzyme from Prochlorococcus MIT9312 (PMT9312_0282), (b) CCD7 from Nostoc punctiforme ATCC 29133 (Npun_F0298), (c) APCO from Cyanothece sp. PCC 7424 (PCC7424_5321), (d) CCD1 from Anabaena sp. PCC 7120 (all1106).
4. Discussion
The distribution of putative CCD encoding open reading frames (ORFs) in cyanobacteria is an integrated function of the genome sizes and the ecophysiological properties. In large extent, most cyanobacterial strains possess proportionate numbers of putative ccd genes with genome sizes, except for a few particular cases, for example, Arthrospira platensis NIES-39 and Gloeobacter violaceus PCC 7421. All of Prochlorococcus and most Synechococcus (except for Synechococcus sp. PCC 7002) strains, which live in the oligotrophic open ocean and have smaller genome sizes, maintain one CCD encoding ORF. Gene duplication event or larger genome size than other Synechococcus species may be responsible for two ccd genes existed in Synechococcus sp. PCC 7002. Metacaspases were found to be absent in all Prochlorococcus and marine Synechococcus strains, except Synechococcus sp. PCC 7002 [31]. Compared to unicellular cyanobacteria with smaller genome sizes, CCD-encoding ORFs are abundant in filamentous with larger genome sizes. On the other hand, filamentous heterocystous cyanobacteria in response to the absence of combined nitrogen and exhibiting ecological properties including broad symbiotic competence with plants and fungi are responsible for containing more putative CCD-encoding ORFs even after allowing for their larger genome sizes [32, 33]. Moreover, according to our results, we speculated that the diverse distributions of ccd genes may reflect various environmental selective pressures. Unicellular Cyanothece that inhabits in freshwater has two or more CCD encoding ORFs, which is beyond the scaling effect of genome size. Similar phenomenon occurred in Synechocystis sp. PCC 6803 and Microcystis aeruginosa NIES-843. Considering the similar genome size, environmental selective pressure may take responsibility for this difference. This parallel pattern of distribution was provided by other cyanobacterial systemic analysis. For example, metacaspases systerm, Serine/threonine kinases, and restriction modification system in cyanobacteria indicate remarkable reduction of these proteins and environmental stress response systems in the ocean [31–33]. Gene lost is revealed to facilitate these cyanobacteria (Prochlorococcus) to acclimatize to the oligotrophic environment. The major driving force was supposed to be “a selective process sustaining the adaptation of these cyanobacteria,” which was discussed by Dufresne et al. (2005) in detail [52].
It is well accepted that there are NCED, CCD1, CCD4, CCD7, and CCD8 subfamily in higher plants [21]. Among them, NCED plays key role in cleavage of the 11,12 double bond of 9-cis violaxanthin or 9′-cis neoxanthin, which results in the formation of abscisic acid (C15) [10]. Our results showed that nced-homologous genes were absent in all cyanobacteria except for Trichodesmium erythraeum ISM 101, suggesting they did not contain ABA. These results were consistence with Pryce (1972), who supposed that, as distinct from higher plants, algae and liverworts did not contain ABA and its functions were fulfilled by lunularic acid [53]. Surprisingly, this hormone has been found in green microalgae (Chlorella sp., Dunaliella salina, and Haematococcus pluvialis) [54, 55] and also in the thalli of brown macrophytes from the genus Ascophyllum and some species of Laminaria [56].
Converting C40 trans-carotenoids to C27 apocarotenoids and the following step C27 to C18 is catalyzed by the CCD7 and CCD8 enzymes, respectively. They involve in the biosynthesis of strigolactones (C18) [57–59]. The recombinant CCD7 protein of Arabidopsis exhibited a specific 9′-10′(but not 9-10/9′-10′) cleavage activity in vitro converting β-carotene to the C27 compound β-apo-10′-carotenal and the C13 compound β-ionone. The C18 compound was formed by a secondary cleavage of the C27 apocarotenoid generated by AtCCD7 [59]. These two types of CCDs have transit peptides indicative of their action in plastids [60]. In this paper, ccd7-homologous genes were present in all cyanobacteria except for Trichodesmium erythraeum ISM 101 and ccd8-homologous genes were discovered in Cyanothece. Our results strongly support a cyanobacterial origin of CCD7 proteins in eukaryotic algae and higher plants (Figure 2). However, further investigation is necessary to explain whether a cyanobacterial origin of CCD8 proteins.
The formation of C13 and C14 from C27 is catalyzed by CCD1 or CCD4 enzymes in the higher plant [61–63]. The recombinant CCD1 or CCD4 enzymes from several plants have been shown to preferentially catalyze a single-step symmetrical cleavage at the 9-10/9′-10′ double bonds of various C40 carotenoids [17, 60, 64, 65]. Interestingly, the CsCCD4 recombinant enzymes produced considerably more β-ionone than CsCCD1, implying that the CCD4 enzymes may have higher enzyme activities [66]. In planta, CCD1 and CCD4 differ in their subcellular location being cytosolic and plastidial, respectively. Surprisingly, ccd1-homologous genes were discovered in N2-fixing cyanobacteria whereas ccd4-homologous gene was absent in all cyanobacteria (Figure 1). This relationship between the cyanobacterial, phototrophic eukaryotic algae, and higher plants strongly supports a cyanobacterial origin of CCD1 proteins (Figure 2). This conclusion has been demonstrated by Scherzinger and Al-Babili [29], who found that the NSC1 (NosCCD) enzyme is an ortholog of plant CCD1 enzymes [29]. Moreover, in planta, CCD1 and/or CCD4 might exhibit additional cleavage specificities including 7-8/7′-8′ cleavage in a certain subcellular environment or guided by a specific cofactor to provide the altered cleavage activity required for generation of crocetin and picrocrocin precursors. Recombinant CCD1 has been shown to exhibit 5-6/5′-6′ or 7-8/7′-8′ activities in addition to 9-10/9′-10′ under certain artificial conditions in vitro but only on acyclic or monocyclic substrates [67].
Apart from the numerous enzymes acting on C40 carotenoids, there are some special CCD enzymes converting only β-apocarotenals but not of bicyclic β-carotene. This has led to its classification as an apocarotenoid 15-15′-oxygenase (APCO). For example, APCO (apocarotenoid cleavage oxygenase) from the cyanobacterium Synechocystis sp. PCC 6803 cleaves the 15-15′double bond of various all-trans-apocarotenoids [20]. In this paper, apco-homologous genes were discovered by BLASTP analysis in some cyanobacteria and eukaryotic algae, which may be responsible for the production of apocarotenoids-derived volatile compounds in these organisms. For example, Microcystis aeruginosa blooms liberate β-ionone [20, 25]. Moreover, it was worthy consideration that apco-homologous genes were absent in higher plant and were evolutional closed with ccd7 genes. Kiefer et al. [68] identified a new type of APCO, which catalyses the asymmetric cleavage of β-carotenoid at the C9′-C10′ position and produces the volatile compound β-ionone (C13) and apo-10′-carotenal (C27) [68].
Considering the sites (chloroplast) of C40 carotenoids biosynthesis and the location of each CCD enzyme, some hypothesis were summarized: (1) the additional function of cleavage C40 carotenoids suggests that the ccd1 and ccd7 genes may origin in cyanobacteria via eukaryotic algae towards higher plants; (2) the ccd1 gene transfers to cytosol by endosymbiotic gene transfer during the evolution in higher plants; (3) the CCD1, CCD7, and APCO enzymes occur earlier than other members of CCDs (CCD4, CCD8, and NCED) enzymes in cyanobacteria; (4) the apco-homologous gene was absent because of gene lost, whereas the ccd4-, ccd8-, and nced-homologous genes were present because of gene duplication in higher plant. These hypothesis were supported by one of our results that ccd7-, apco-, and ccd1-homologous genes were widespread in cyanobacterial strains, while ccd8- and nced-homologous genes only existed in some species, and ccd4-homologous genes were absent in all cyanobacteria (Figures 1 and 2).
Another interesting result was the highly similar tertiary structure for different types of CCD enzymes from distinct cyanobacterial strains (Figures 4 and 5). Indeed, sequence alignments within the CCD family clearly show that the most highly conserved regions are within the β-strands forming the propeller as demonstrated in Figures 3, 4, and 5. This strongly suggests that all members of the CCDs family have the same β propeller chain-fold and may therefore be modeled along the lines of the other CCD structures and share several same characteristics. Schwartz et al. [10], Redmond et al. [12], and Kiefer et al. [68] found that members of the CCD family share several characteristics: first, they require a Fe2+ for catalytic activity [10, 12, 68]; second, they contain four conserved histidines that are thought to coordinate iron binding; third, they contain a conserved peptide sequence at their carboxyl terminus that minimally constitutes a signature sequence for the family.
At present, the only established structure of a CCDs family member is apocarotenoids oxygenase (APCO) from Synechocystis PCC 6803 [69], and the mechanisms of choosing and modulating substrate specifically have been discussed [20, 70]. As observed in numerous β propeller structures, the active center of APCO is located near the propeller axis on the top side. The individual CCO family members choose and modulate substrate by changing length hand sequence of the connecting loops, while retaining the rigid propeller scaffold for stability [71–73]. More analysis is necessary to further delineate the mechanisms of choosing substrate exactly for each CCD family member. However, such an analysis is beyond the scope of this paper.
5. Conclusion
CCD enzymes play significant role in forming of carotenoids cleavage products, which are essential to several prokaryotes, eukaryotic algae, fungi, and higher plants. A total of 61 putative ccd genes have been identified across 37 species of cyanobacteria. The ccd7-, ccd1-, and apco-homologous genes are widespread in cyanobacteria and ccd8-homologous genes only exist in a few species. The distribution of ccd genes in unicellular and filamentous cyanobacteria relies on the genome size and ecological habitat. According to BLASTP results and phylogenetic tree of 16 s rDNA and CCD, it seems that CCD7, APCO, and CCD1 enzymes appeared earlier than other members of CCD enzymes. A slight difference exists between distinct types of CCD enzymes by motif scanning, while the secondary and tertiary structures are highly similar through homology modeling. All CCD enzymes share conserved basic configuration, which is constituted by a single loop formed with seven β-strands and one helix. This paper may provide new insight for the evolutional and functional investigation of CCD enzyme in cyanobacteria.
Supplementary Material
Supplementary Table 1: List of kea features and genomes information from cyanobacterial strains analyzed in this paper. Note: HL, high light adaptation; LL, low light adaptation; GS, genome size.
Supplementary Table 2: The percentage of amino acid sequence similarity for CCD7 proteins from Synechococcus and Prochlorococcus.
Supplementary Table 3: The percentage of amino acid sequence similarity for APCO proteins from distinct organisms.
Supplementary Table 4: The percentage of amino acid sequence similarity for CCD1, NCED, and CCD8 proteins from distinct organisms.
Supplementary Table 5: The percentage of amino acid sequence similarity for CCD7 proteins from distinct organisms (except Synechococcus and Prochlorococcus)
Acknowledgments
The author thank Professor Fangqing Zhao (Beijing Institutes of Life Science, Chinese Academy of Sciences) for carefully modifying the draft. This work was supported by the National Natural Science Foundation of China (40876082), Chinese Academy of Sciences, international partnership program “Typical environment processes and their effects on resources,” and Outstanding Young Scholars Fellowship of Shandong Province (Molecular Phycology JQ200914).
Abbreviations
- CCD:
Carotenoid cleavage dioxygenase
- CCD (1, 4, 7, and 8):
Carotenoid cleavage dioxygenase (1, 4, 7, and 8)
- NCED:
Nine-cis-epoxycarotenoid dioxygenase
- APCO:
Apocarotenoid 15,15′ oxygenase
- RPE:
Retinal pigment epithelial membrane protein
- CO:
Carotenoid oxygenase
- LSD:
Lignostilbene-α, β-dioxygenase
- HP:
Hypothetical proteins
- UP:
Undefined proteins
- NCE:
Neoxanthin cleavage enzyme
- BS:
Bootstrap value.
References
- 1.Stanier RY, Cohen-Bazire G. Phototrophic prokaryotes: the cyanobacteria. Annual Review of Microbiology. 1977;31:225–274. doi: 10.1146/annurev.mi.31.100177.001301. [DOI] [PubMed] [Google Scholar]
- 2.Xiaoyuan C, Yang Q, Zhao F, et al. Comparative analysis of fatty acid desaturases in cyanobacterial genomes. Comparative and Functional Genomics. 2008;2008 doi: 10.1155/2008/284508. Article ID 284508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scanlan D. Cyanobacteria: ecology, niche adaptation and genomics. Microbiology Today. 2001;28:128–130. [Google Scholar]
- 4.Rocap G, Larimer FW, Lamerdin J, et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003;424(6952):1042–1047. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- 5.Dufresne A, Salanoubat M, Partensky F, et al. Genome sequence of the cyanobacterium Prochlorococcus marinus SS120, a nearly minimal oxyphototrophic genome. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(17):10020–10025. doi: 10.1073/pnas.1733211100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meeks JC, Elhai J, Thiel T, et al. An overview of the Genome of Nostoc punctiforme, a multicellular, symbiotic Cyanobacterium. Photosynthesis Research. 2001;70(1):85–106. doi: 10.1023/A:1013840025518. [DOI] [PubMed] [Google Scholar]
- 7.Partensky F, Hess WR, Vaulot D. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiology and Molecular Biology Reviews. 1999;63(1):106–127. doi: 10.1128/mmbr.63.1.106-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakamura Y, Kaneko T, Sato S, et al. Complete genome structure of gloeobacter violaceus PCC 7421, a cyanobacterium that lacks thylakoids (Supplement) DNA Research. 2003;10(4):181–201. doi: 10.1093/dnares/10.4.181. [DOI] [PubMed] [Google Scholar]
- 9.Tan BC, Schwartz SH, Zeevaart JAD, Mccarty DR. Genetic control of abscisic acid biosynthesis in maize. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(22):12235–12240. doi: 10.1073/pnas.94.22.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartz SH, Tan BC, Gage DA, Zeevaart JAD, McCarty DR. Specific oxidative cleavage of carotenoids by VP14 of maize. Science. 1997;276(5320):1872–1874. doi: 10.1126/science.276.5320.1872. [DOI] [PubMed] [Google Scholar]
- 11.Von Lintig J, Vogt K. Filling the gap in vitamin A research. Molecular identification of an enzyme cleaving β-carotene to retinal. Journal of Biological Chemistry. 2000;275(16):11915–11920. doi: 10.1074/jbc.275.16.11915. [DOI] [PubMed] [Google Scholar]
- 12.Redmond TM, Gentleman S, Duncan T, et al. Identification, expression, and substrate specificity of a mammalian β-carotene 15,15′-dioxygenase. Journal of Biological Chemistry. 2001;276(9):6560–6565. doi: 10.1074/jbc.M009030200. [DOI] [PubMed] [Google Scholar]
- 13.Prado-Cabrero A, Scherzinger D, Avalos J, Al-Babili S. Retinal biosynthesis in fungi: characterization of the carotenoid oxygenase CarX from Fusarium fujikuroi. Eukaryotic Cell. 2007;6(4):650–657. doi: 10.1128/EC.00392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bouvier F, Dogbo O, Camara B. Biosynthesis of the food and cosmetic plant pigment bixin (annatto) Science. 2003;300(5628):2089–2091. doi: 10.1126/science.1085162. [DOI] [PubMed] [Google Scholar]
- 15.Bouvier F, Suire C, Mutterer J, Camara B. Oxidative remodeling of chromoplast carotenoids: identification of the carotenoid dioxygenase CsCCD and CsZCD genes involved in Crocus secondary metabolite biogenesis. Plant Cell. 2003;15(1):47–62. doi: 10.1105/tpc.006536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prado-Cabrero A, Estrada AF, Al-Babili S, Avalos J. Identification and biochemical characterization of a novel carotenoid oxygenase: elucidation of the cleavage step in the Fusarium carotenoid pathway. Molecular Microbiology. 2007;64(2):448–460. doi: 10.1111/j.1365-2958.2007.05665.x. [DOI] [PubMed] [Google Scholar]
- 17.Auldridge ME, McCarty DR, Klee HJ. Plant carotenoid cleavage oxygenases and their apocarotenoid products. Current Opinion in Plant Biology. 2006;9(3):315–321. doi: 10.1016/j.pbi.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 18.Camara B, Bouvier F. Oxidative remodeling of plastid carotenoids. Archives of Biochemistry and Biophysics. 2004;430(1):16–21. doi: 10.1016/j.abb.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 19.Pfander H, Wittwer F. Carotenoid composition in safran (author’s transl) Helvetica Chimica Acta. 1975;58(7):2233–2236. doi: 10.1002/hlca.19750580736. [DOI] [PubMed] [Google Scholar]
- 20.Kloer DP, Schulz GE. Structural and biological aspects of carotenoid cleavage. Cellular and Molecular Life Sciences. 2006;63(19-20):2291–2303. doi: 10.1007/s00018-006-6176-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Floss DS, Walter MH. Role of carotenoid cleavage dioxygenase 1 (CCD1) in apocarotenoid biogenesis revisited. Plant Signaling and Behavior. 2009;4(3):172–175. doi: 10.4161/psb.4.3.7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahrazem O, Trapero A, Gómez MD, Rubio-Moraga A, Gómez-Gómez L. Genomic analysis and gene structure of the plant carotenoid dioxygenase 4 family: a deeper study in Crocus sativus and its allies . Genomics. 2010;96(4):239–250. doi: 10.1016/j.ygeno.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 23.Tan BC, Joseph LM, Deng WT, et al. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant Journal. 2003;35(1):44–56. doi: 10.1046/j.1365-313x.2003.01786.x. [DOI] [PubMed] [Google Scholar]
- 24.Ohmiya A, Kishimoto S, Aida R, Yoshioka S, Sumitomo K. Carotenoid cleavage dioxygenase (CmCCD4a) contributes to white color formation in Chrysanthemum petals. Plant Physiology. 2006;142(3):1193–1201. doi: 10.1104/pp.106.087130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giuliano G, Al-Babili S, Von Lintig J. Carotenoid oxygenases: cleave it or leave it. Trends in Plant Science. 2003;8(4):145–149. doi: 10.1016/S1360-1385(03)00053-0. [DOI] [PubMed] [Google Scholar]
- 26.Alder A, Holdermann I, Beyer P, Al-Babili S. Carotenoid oxygenases involved in plant branching catalyse a highly specific conserved apocarotenoid cleavage reaction. Biochemical Journal. 2008;416(2):289–296. doi: 10.1042/BJ20080568. [DOI] [PubMed] [Google Scholar]
- 27.Ruch S, Beyer P, Ernst H, Al-Babili S. Retinal biosynthesis in Eubacteria: in vitro characterization of a novel carotenoid oxygenase from Synechocystis sp. PCC 6803. Molecular Microbiology. 2005;55(4):1015–1024. doi: 10.1111/j.1365-2958.2004.04460.x. [DOI] [PubMed] [Google Scholar]
- 28.Marasco EK, Vay K, Schmidt-Dannert C. Identification of carotenoid cleavage dioxygenases from Nostoc sp. PCC 7120 with different cleavage activities. Journal of Biological Chemistry. 2006;281(42):31583–31593. doi: 10.1074/jbc.M606299200. [DOI] [PubMed] [Google Scholar]
- 29.Scherzinger D, Al-Babili S. In vitro characterization of a carotenoid cleavage dioxygenase from Nostoc sp. PCC 7120 reveals a novel cleavage pattern, cytosolic localization and induction by highlight. Molecular Microbiology. 2008;69(1):231–244. doi: 10.1111/j.1365-2958.2008.06282.x. [DOI] [PubMed] [Google Scholar]
- 30.Scherzinger D, Ruch S, Kloer DP, Wilde A, Al-Babili S. Retinal is formed from apo-carotenoids in Nostoc sp. PCC7120: in vitro characterization of an apo-carotenoid oxygenase. Biochemical Journal. 2006;398(3):361–369. doi: 10.1042/BJ20060592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang Q, Qin S, Wu QY. Genome-wide comparative analysis of metacaspases in unicellular and filamentous cyanobacteria. BMC Genomics. 2010;11(1, article 198) doi: 10.1186/1471-2164-11-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, Zhao F, Guan X, Yang Y, Liang C, Qin S. Genome-wide survey of putative Serine/Threonine protein kinases in cyanobacteria. BMC Genomics. 2007;8, article 395 doi: 10.1186/1471-2164-8-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao F, Zhang X, Liang C, Wu J, Bao Q, Qin S. Genome-wide analysis of restriction-modification system in unicellular and filamentous cyanobacteria. Physiological Genomics. 2006;24(3):181–190. doi: 10.1152/physiolgenomics.00255.2005. [DOI] [PubMed] [Google Scholar]
- 34.Liang C, Zhao F, Wei W, Wen Z, Qin S. Carotenoid biosynthesis in cyanobacteria: structural and evolutionary scenarios based on comparative genomics. International Journal of Biological Sciences. 2006;2(4):197–207. doi: 10.7150/ijbs.2.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mount DW. Using the basic local alignment search tool (BLAST) CSH Protocols. 2007;2007 doi: 10.1101/pdb.top17. [DOI] [PubMed] [Google Scholar]
- 36.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 37.Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: q new generation of protein database search programs. Nucleic Acids Research. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 40.Guindon S, Gascuel O. A Simple, Fast, and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood. Systematic Biology. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 41.Le SQ, Gascuel O. An improved general amino acid replacement matrix. Molecular Biology and Evolution. 2008;25(7):1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 42.Chevenet F, Brun C, Bañuls AL, Jacq B, Christen R. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics. 2006;7, article 439 doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bailey TL, Elkan C. The value of prior knowledge in discovering motifs with MEME. Proceedings of International Conference on Intelligent Systems for Molecular Biology. 1995;3:21–29. [PubMed] [Google Scholar]
- 44.Chi X, Yang Q, Lu Y, et al. Genome-wide analysis of fatty acid desaturases in Soybean (Glycine max) Plant Molecular Biology Reporter. 2011;29(4):769–783. [Google Scholar]
- 45.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings of International Conference on Intelligent Systems for Molecular Biology. 1994;2:28–36. [PubMed] [Google Scholar]
- 46.Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Research. 2004;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Research. 2009;37(1):D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 49.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Research. 2003;31(13):3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18(15):2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 51.Benkert P, Biasini M, Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27(3):343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dufresne A, Garczarek L, Partensky F. Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biology. 2005;6(2):p. R14. doi: 10.1186/gb-2005-6-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pryce RJ. The occurrence of lunularic and abscisic acids in plants. Phytochemistry. 1972;11(5):1759–1761. [Google Scholar]
- 54.Tominaga N, Takahata M, Tominaga H. Effects of NaCl and KNO3 concentrations on the abscisic acid content of Dunaliella sp. (Chlorophyta) Hydrobiologia. 1993;267(1–3):163–168. [Google Scholar]
- 55.Kobayashi M, Hirai N, Kurimura Y, Ohigashi H, Tsuji Y. Abscisic acid-dependent algal morphogenesis in the unicellular green alga Haematococcus pluvialis . Plant Growth Regulation. 1997;22(2):79–85. [Google Scholar]
- 56.Nimura K, Mizuta H. Inducible effects of abscisic acid on sporophyte discs from Laminaria japonica Areschoug (Laminariales, Phaeophyceae) Journal of Applied Phycology. 2002;14(3):159–163. [Google Scholar]
- 57.Mouchel CF, Leyser O. Novel phytohormones involved in long-range signaling. Current Opinion in Plant Biology. 2007;10(5):473–476. doi: 10.1016/j.pbi.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 58.Booker J, Auldridge M, Wills S, McCarty D, Klee H, Leyser O. MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Current Biology. 2004;14(14):1232–1238. doi: 10.1016/j.cub.2004.06.061. [DOI] [PubMed] [Google Scholar]
- 59.Schwartz SH, Qin X, Loewen MC. The biochemical characterization of two carotenoid cleavage enzymes from Arabidopsis indicates that a carotenoid-derived compound inhibits lateral branching. Journal of Biological Chemistry. 2004;279(45):46940–46945. doi: 10.1074/jbc.M409004200. [DOI] [PubMed] [Google Scholar]
- 60.Auldridge ME, Block A, Vogel JT, et al. Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant Journal. 2006;45(6):982–993. doi: 10.1111/j.1365-313X.2006.02666.x. [DOI] [PubMed] [Google Scholar]
- 61.Maier W, Peipp H, Schmidt J, Wray V, Strack D. Levels of a terpenoid glycoside (Blumenin) and cell wall-bound phenolics in some cereal mycorrhizas. Plant Physiology. 1995;109(2):465–470. doi: 10.1104/pp.109.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Klingner A, Bothe H, Wray V, Marner FJ. Identification of a yellow pigment formed in maize roots upon mycorrhizal colonization. Phytochemistry. 1995;38(1):53–55. [Google Scholar]
- 63.Walter MH, Fester T, Strack D. Arbuscular mycorrhizal fungi induce the non-mevalonate methylerythritol phosphate pathway of isoprenoid biosynthesis correlated with accumulation of the “yellow pigment” and other apocarotenoids. Plant Journal. 2000;21(6):571–578. doi: 10.1046/j.1365-313x.2000.00708.x. [DOI] [PubMed] [Google Scholar]
- 64.Mathieu S, Terrier N, Procureur J, Bigey F, Günata Z. A Carotenoid Cleavage Dioxygenase from Vitis vinifera L.: functional characterization and expression during grape berry development in relation to C13-norisoprenoid accumulation. Journal of Experimental Botany. 2005;56(420):2721–2731. doi: 10.1093/jxb/eri265. [DOI] [PubMed] [Google Scholar]
- 65.Schwartz SH, Qin X, Zeevaart JAD. Characterization of a novel carotenoid cleavage dioxygenase from plants. Journal of Biological Chemistry. 2001;276(27):25208–25211. doi: 10.1074/jbc.M102146200. [DOI] [PubMed] [Google Scholar]
- 66.Rubio A, Rambla JL, Santaella M, et al. Cytosolic and plastoglobule-targeted carotenoid dioxygenases from Crocus sativus are both involved in β-ionone release. Journal of Biological Chemistry. 2008;283(36):24816–24825. doi: 10.1074/jbc.M804000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vogel JT, Tan BC, McCarty DR, Klee HJ. The carotenoid cleavage dioxygenase 1 enzyme has broad substrate specificity, cleaving multiple carotenoids at two different bond positions. Journal of Biological Chemistry. 2008;283(17):11364–11373. doi: 10.1074/jbc.M710106200. [DOI] [PubMed] [Google Scholar]
- 68.Kiefer C, Hessel S, Lampert JM, et al. Identification and characterization of a mammalian enzyme catalyzing the asymmetric oxidative cleavage of orovitamin A. Journal of Biological Chemistry. 2001;276(17):14110–14116. doi: 10.1074/jbc.M011510200. [DOI] [PubMed] [Google Scholar]
- 69.Kloer DP, Ruch S, Al-Babili S, Beyer P, Schulz GE. The structure of a retinal-forming carotenoid oxygenase. Science. 2005;308(5719):267–269. doi: 10.1126/science.1108965. [DOI] [PubMed] [Google Scholar]
- 70.Walter MH, Strack D. Carotenoids and their cleavage products: biosynthesis and functions. Natural Product Reports. 2011;28(4):663–692. doi: 10.1039/c0np00036a. [DOI] [PubMed] [Google Scholar]
- 71.Takahashi Y, Moiseyev G, Chen Y, Ma JX. Identification of conserved histidines and glutamic acid as key residues for isomerohydrolase activity of RPE65, an enzyme of the visual cycle in the retinal pigment epithelium. FEBS Letters. 2005;579(24):5414–5418. doi: 10.1016/j.febslet.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 72.Ala-Laurila P, Donner K, Koskelainen A. Thermal activation and photoactivation of visual pigments. Biophysical Journal. 2004;86(6):3653–3662. doi: 10.1529/biophysj.103.035626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moiseyev G, Takahashi Y, Chen Y, et al. RPE65 is an iron(II)-dependent isomerohydrolase in the retinoid visual cycle. Journal of Biological Chemistry. 2006;281(5):2835–2840. doi: 10.1074/jbc.M508903200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: List of kea features and genomes information from cyanobacterial strains analyzed in this paper. Note: HL, high light adaptation; LL, low light adaptation; GS, genome size.
Supplementary Table 2: The percentage of amino acid sequence similarity for CCD7 proteins from Synechococcus and Prochlorococcus.
Supplementary Table 3: The percentage of amino acid sequence similarity for APCO proteins from distinct organisms.
Supplementary Table 4: The percentage of amino acid sequence similarity for CCD1, NCED, and CCD8 proteins from distinct organisms.
Supplementary Table 5: The percentage of amino acid sequence similarity for CCD7 proteins from distinct organisms (except Synechococcus and Prochlorococcus)


