Abstract
Evolutionary relationships are typically inferred from molecular sequence data using a statistical model of the evolutionary process. When the model accurately reflects the underlying process, probabilistic phylogenetic methods recover the correct relationships with high accuracy. There is ample evidence, however, that models commonly used today do not adequately reflect real-world evolutionary dynamics. Virtually all contemporary models assume that relatively fast-evolving sites are fast across the entire tree, whereas slower sites always evolve at relatively slower rates. Many molecular sequences, however, exhibit site-specific changes in evolutionary rates, called “heterotachy.” Here we examine the accuracy of 2 phylogenetic methods for incorporating heterotachy, the mixed branch length model—which incorporates site-specific rate changes by summing likelihoods over multiple sets of branch lengths on the same tree—and the covarion model, which uses a hidden Markov process to allow sites to switch between variable and invariable as they evolve. Under a variety of simple heterogeneous simulation conditions, the mixed model was dramatically more accurate than homotachous models, which were subject to topological biases as well as biases in branch length estimates. When data were simulated with strong versions of the types of heterotachy observed in real molecular sequences, the mixed branch length model was more accurate than homotachous techniques. Analyses of empirical data sets confirmed that the mixed branch length model can improve phylogenetic accuracy under conditions that cause homotachous models to fail. In contrast, the covarion model did not improve phylogenetic accuracy compared with homotachous models and was sometimes substantially less accurate. We conclude that a mixed branch length approach, although not the solution to all phylogenetic errors, is a valuable strategy for improving the accuracy of inferred trees.
Keywords: heterotachy, covarion, phylogenetics, maximum likelihood, mixed model, evolutionary heterogeneity, mixed branch length
Introduction
The evolutionary process is complex and dynamic. Selection pressures can vary as organisms diversify. Even when selection is relatively constant at the organismal level, evolutionary constraints acting at particular sites in a molecule may be variable, because the sites subject to specific functional constraints change over evolutionary time (Fitch and Markowitz 1970). As a result, some fast-evolving sites can become slow-evolving (and vice versa) in different lineages (Lopez et al. 2002). Such evolutionary dynamics are largely ignored by existing “homotachous” evolutionary models—including those allowing among-site rate variation—which assume that fast-evolving sites are fast across the entire tree, whereas more constrained sites are always slow-evolving (see Yang 1996a, and fig. 1A and B). It has been shown that homotachous models are inadequate to capture the shifting dynamics of molecular evolution for protein-coding sequences (Fitch and Markowitz 1970; Fitch 1971, 1976; Miyamoto and Fitch 1995; Germot and Philippe 1999; Gaucher et al. 2001; Gu 2001, 2003; Philippe and Lopez 2001; Huelsenbeck 2002; Lopez et al. 2002; Susko et al. 2002; Ané et al. 2005; Lockhart et al. 2005), RNA molecules (Lockhart et al. 1998; Steel et al. 2000; Galtier 2001; Brown 2005; Baele et al. 2006), and promoter regions (Taylor et al. 2006).
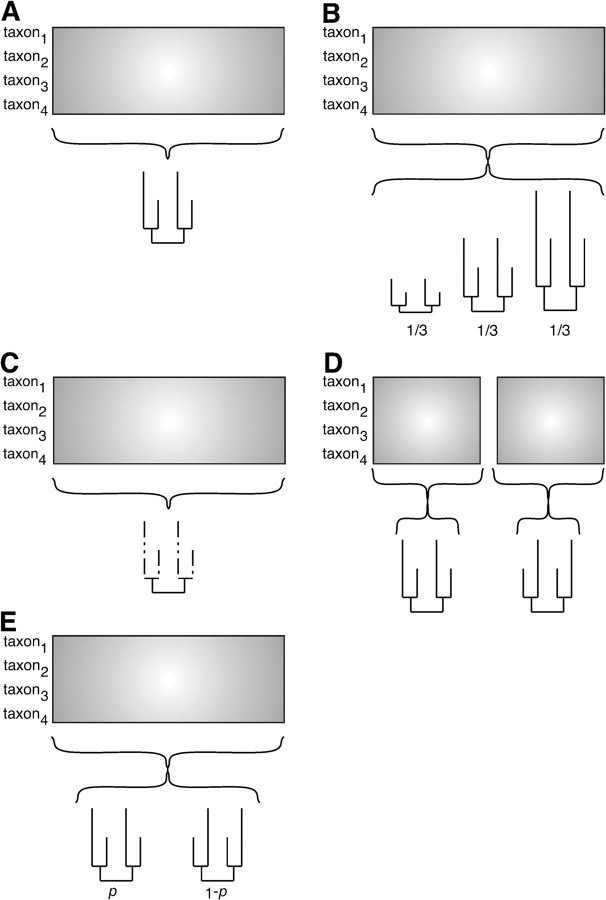
FIG. 1.—
Comparison of homotachous and heterotachous models. (A) In a simple homotachous model, the likelihood of the model is calculated using a single set of branch lengths. (B) A homotachous discrete gamma model of among-site rate variation calculates the likelihood as a weighted sum over multiple branch length classes, but the ratio of each branch length to the others is the same in all classes. (C) A covarion model uses a hidden Markov process allowing sites to switch between variable (solid lines) and invariable (dotted lines) as they evolve. (D) A partitioned model divides sites into categories a priori; the total likelihood is the product over all partitions. (E) A mixed branch length model calculates likelihoods at each site as a weighted sum over multiple independent branch length sets; weights and branch lengths are inferred from the data.
The prevalence of heterotachy—site-specific evolutionary rates that change across the tree (Lopez et al. 2002)—has important implications for phylogenetics. Theoretical arguments suggest that some forms of heterotachy might produce biased inferences of phylogenies or result in lack of resolution when homotachous models are used (Chang 1996; Siddall and Kluge 1999; Štefankovič and Vigoda 2006). Simulation studies have confirmed these predictions, revealing that some forms of heterotachy—but not all (Penny et al. 2001)—can impair the accuracy of homotachous model-based methods (Kolaczkowski and Thornton 2004; Gadagkar and Kumar 2005; Gaucher and Miyamoto 2005; Philippe et al. 2005; Spencer et al. 2005; Susko et al. 2005; Ruano-Rubio and Fares 2007). Analyses of empirical sequence data suggest that heterotachy may be an important cause of real-world phylogenetic error. For example, site-specific rate shifts are at least partially responsible for the failure of homotachous models to recover the correct Microsporidia + Fungi (MF) phylogeny from elongation factor 1α (EF1α) data (Hirt et al. 1999; Inagaki et al. 2003, 2004). Heterotachy seems to be a contributing factor in a variety of other phylogenetic artifacts, as well (Philippe, Lartillot, and Brinkmann 2005; Rodriquez-Ezpeleta et al. 2007).
Two types of statistical models of heterotachy have been developed. First, based on early observations that different sites in a molecular sequence may be invariant in different lineages (Fitch and Markowitz 1970; Fitch, 1971, 1976), the “covarion” model uses a hidden Markov process that allows sites to switch between variable and invariable as they evolve (Tuffley and Steel 1998, see fig. 1C). The simple variant–invariant covarion model has been generalized to allow sites to switch among multiple evolutionary rates (Galtier 2001; Wang et al. 2007). Covarion models provide an improved statistical fit to empirical data compared with homotachous models (Miyamoto and Fitch 1995; Galtier 2001; Huelsenbeck 2002; Wang et al. 2007), but it is unknown whether this improved fit translates into improved phylogenetic accuracy. Evidence suggests that the covarion model does not accurately match the way site-specific evolutionary rates change (Germot and Philippe 1999; Steel et al. 2000; Lopez et al. 2002; Lockhart et al. 2005). Particularly, covarion models assume that 1) the proportion of sites in each rate category is constant across the entire tree and 2) the rate at which sites switch evolutionary rates is proportional to the expected number of substitutions per site. These conditions are unlikely to hold in real molecular sequence data.
Mixed models to represent heterotachy have also been described (Kolaczkowski and Thornton 2004; Spencer et al. 2005). Mixture modeling is a general statistical approach for incorporating complex heterogeneous processes (McLachlan and Peel 2000). Under a mixed model, the probability of the data is calculated for a variety of simple submodels and then combined to give the probability of the data under the mixed model. Kolaczkowski and Thornton (2004) first suggested that heterotachy could be modeled using multiple sets of branch lengths on the same topology. Likelihoods for each site are calculated as a weighted sum over all sets of branch lengths. Spencer et al. (2005) improved the model by inferring weights from the data as free parameters (see fig. 1E). The mixed branch length model is different from a partitioned model (Yang 1996b; Ronquist and Huelsenbeck 2003) because a partitioned model assigns sites to specified categories a priori (fig. 1D). A partitioned model is useful when biochemical information is sufficient to accurately classify sites into rate categories prior to analysis, whereas a mixed model does not require such prior knowledge.
Unlike the covarion model—which assumes a specific stationary process generates variation in evolutionary rates—the mixed branch length model is a general model of heterotachy that does not make any strong assumptions about the process generating rate variation. Any distribution of evolutionary rates across sites and lineages can be described by allowing different sites to evolve along different branch lengths. The flexibility of the mixed model allows it to fit a variety of patterns of heterotachy.
Little is known about whether the mixed branch length model improves the accuracy of phylogenetic inference. First, simulation studies have shown that the mixed model can perform well on a single, challenging form of heterotachy when the correct number of branch length categories is known in advance (Kolaczkowski and Thornton 2004; Spencer et al. 2005), but its accuracy on other forms of heterotachy has not been assessed. Second, the number of branch length classes required to adequately describe the data is never known in practice; the accuracy with which the number of branch length classes and the parameters of the mixed model can be estimated from sequence data has not been investigated. Third, although theoretical analyses suggest that the mixed model may fail to recover the correct tree under some simplified conditions, even if infinite data were available (Allman and Rhodes 2006; Štefankovič and Vigoda 2006; Matsen and Steel 2007), the relevance of these findings to complex phylogenetic problems is unknown. Finally, the fit of the mixed branch length model to empirical data—and its ability to recover the correct phylogeny under realistic conditions—is not known.
Here we report on the implementation and performance analysis of a general mixed branch length software package for analyzing both nucleotide and protein data (available at http://phylo.uoregon.edu/software/m3l). We introduce a simulated annealing algorithm to estimate maximum likelihood (ML) values of model parameters and tree topology and infer the best-fit number of branch length categories using the Akaike information criterion (AIC) and the Bayesian information criterion (BIC). We use simulation experiments and analyze empirical sequence data to examine the effects of heterotachy on phylogenetic inference and evaluate the ability of the mixed branch length and covarion models to improve phylogenetic accuracy.
Materials and Methods
Phylogenetic Analyses
Mixed branch length model analyses were conducted in a ML framework using novel software (available at http://phylo.uoregon.edu/software/m3l). The mixed branch length model calculates likelihoods using multiple independent sets of branch lengths on the tree (Kolaczkowski and Thornton 2004; Spencer et al. 2005). We have implemented the model as formulated by Spencer et al. (2005). The likelihood of tree t given data X = (x1, x2,…, xm) and branch length sets b = (b1, b2,…, bn) is given by
where each ρi is estimated from the data and P(xk|t, bi) is the probability of the data given branch lengths bi. We used the JC69 model for analyses of simulated data and the Jones, Taylor, Thornton + gamma (4-category discrete gamma approximation) model for analyses of empirical protein sequences. Simulated annealing (Kirkpatrick et al. 1983) was used to optimize tree topology and all model parameters. The annealing schedule used a geometric descent of 1,000 temperatures starting from 1.0 and ending at 10−5. At each temperature, 1,000 parameter changes were attempted, with acceptance based on the Metropolis criterion. For 4-taxon simulations, we performed exhaustive topology searches, optimizing parameters separately on each possible tree. For larger phylogenies, heuristic tree searches were performed using simulated annealing, with topology rearrangements including tree bisection-reconnection, subtree pruning-regrafting, and nearest neighbor interchange. The best-fit number of branch length classes (n) was selected using either AIC (Akaike 1974) or BIC (Schwartz 1978).
Sequence alignments were also analyzed using homotachous ML (which includes models of among-site rate variation such as the gamma model and the proportion of invariable sites model), Bayesian Markov Chain Monte Carlo (BMCMC), and unweighted maximum parsimony (MP). MP and homotachous ML analyses of nucleotide data were conducted using exhaustive topology search in PAUP* 4.0b10 (Swofford 2002). For homotachous ML and BMCMC analyses, the best-fit substitution model was selected by a chi-square hierarchical likelihood ratio test (α = 0.05) assuming the Neighbor-Joining topology, implemented in Modeltest 3.7 (Posada and Crandall 1998). Use of alternative homotachous models did not substantially affect our results (see supplementary fig. S2, Supplementary Material online). Bayesian analyses were conducted using MrBayes 3.1 (Ronquist and Huelsenbeck 2003). Two independent runs of 4 chains were executed until the average standard deviation in clade probabilities dropped below 0.01; the first 5,000 generations were discarded as burn-in. Topology priors were equal for each resolved tree, branch length priors were uniform on (0, 10), and the default priors were used for other model parameters.
To determine the specific effects of various forms of heterotachy on phylogenetic accuracy, we also performed analyses using the true ML model (MLtrue), which correctly partitions sites into branch length categories a priori and estimates branch lengths separately for each category.
Simplified Branch Length Heterogeneity
We simulated data sets of length 5,000 nucleotides (nt) using the JC69 model under 4 simplified types of 4-taxon branch length heterogeneity (see fig. 2): 1) Felsenstein zone heterotachy (FZH), 2) inverse-Felsenstein zone heterotachy (IFZH), 3) single long-branch heterotachy (SLBH), and 4) signal–noise heterotachy (SNH). Both FZH and IFZH partition sites into 2 branch length categories, with equal numbers of sites in each category. Long branches (0.75 expected substitutions/site) lead to 2 terminal lineages, whereas short branches (0.05) lead to the other 2 terminal lineages, but the lineages with long terminal branches are different in different branch length categories. In the case of FZH, the long terminal branches are not sister to one another, whereas long branches lead to sister taxa in IFZH. SNH partitions sites into 2 categories; in the first (80% of sites), sequences evolve with long terminal branch lengths (0.75) and a zero-length internal branch. In the other category, terminal branch lengths are short (0.05), and the internal branch length varies between 0.0 and 0.4. SLBH consists of 4 branch length categories with equal numbers of sites; in each category, a single lineage has a long terminal branch (0.75), whereas all other terminal branches are short (0.05). In each case, the internal branch length (which is the same in all categories) varied between 0.0 and 0.4. Two hundred replicate sequence alignments were simulated under each set of evolutionary conditions.
FIG. 2.—

Mixed branch length model improves phylogenetic accuracy under idealized heterotachous conditions. Sequences of 5,000 nt each were simulated using the tree at left in each panel, with long terminal branch lengths 0.75 substitutions/site and short terminals 0.05. Left graph in each panel plots the proportion of correct inferences made using homotachous, heterotachous, and the true partitioned model (see key at lower right) against the internal branch length of the true tree. Right graphs plot internal (left) and terminal (right) branch lengths estimated by maximum likelihood using true, mixed branch length, and homotachous models against the true internal branch length on which sequences were simulated; dotted lines indicate perfect correspondence between estimated and true lengths. (A) Felsenstein-zone heterotachy. (B) Inverse Felsenstein-zone heterotachy. Inset bar graph shows the proportion of replicates from which each method recovered each possible resolved topology when data were generated with an internal branch length of zero. (C) Signal-noise heterotachy. (D) Single long-branch heterotachy. Bottom left panel shows per-site likelihood calculated on an ideal infinite data set (see Materials and Methods) using homotachous ML plotted against increasing internal branch length; sequence was generated under SLBH conditions with a true internal branch length of 0.01. Bottom right panel shows accuracy when >95% support is required to resolve the phylogeny.
Phylogenetic analyses were conducted as described above. Accuracy was determined by calculating the proportion of replicates for which the correct phylogeny was uniquely recovered. The internal branch length at which 50% of inferred trees were correct (BL50) was estimated for each method using nonlinear regression (Kolaczkowski and Thornton 2004), and the accuracy of different methods was compared by comparing BL50 estimates using a 2-way t-test. Bias was examined by simulating sequences under heterotachous conditions but with a zero-length internal branch. The proportion of replicates falsely resolved with support >0.95 was measured using nonparametric bootstrapping (1,000 replicates) for ML and MP and posterior probabilities for BMCMC.
To assess the asymptotic performance of ML with infinite data, ideal pseudodata with no stochastic error were analyzed. We calculated the expected frequency of each character state pattern (f(x)) under SLBH conditions with an internal branch length of 0.01. These state pattern frequencies are the frequencies that would occur if infinite sequence data were available. We implemented an algorithm to calculate likelihoods under a homotachous model directly from this vector of expected pattern frequencies, producing a per-site likelihood equivalent to that which would be obtained from infinite data. The per-site likelihood of tree t given state pattern x is calculated by raising the probability of the pattern, given the tree, to the frequency with which that pattern is expected to occur: L(t|x) = P(x|t)f(x). The total per-site likelihood of the tree is the product of this partial likelihood over all possible state patterns. We calculated the likelihoods of internal branch lengths between 0.0 and 0.01 expected substitutions/site, with other branch lengths optimized using ML.
We also examined the accuracy with which different phylogenetic models estimated branch lengths from finite data. For each set of simulation conditions, we calculated the expected or mean set of branch lengths across sites using homotachous ML, the mixed branch length model, and the correct ML model (MLtrue). The single set of inferred branch lengths are the expected lengths across sites for homotachous ML. For the mixed model and MLtrue, expected branch lengths over sites were calculated by multiplying each site-specific branch length by the weight associated with that length and then summing over all weighted site-specific branch lengths. For the mixed model, weights are estimated from the data, whereas weights are correctly assigned a priori for MLtrue. In the case of terminal branches, we report the average expected branch length over all 4 terminals.
Types of Heterotachy Observed in Molecular Evolution
To simulate stationary covarion dynamics, we simulated sequence data using the covarion model described by Tuffley and Steel (1998). We used a 4-taxon Felsenstein zone phylogeny (see fig. 3A) with nonsister long (0.75 expected substitutions/site) and short (0.05) terminal branch lengths to generate 5,000-nt sequences using the JC69 model. A hidden Markov process was used to allow sites to continuously switch between variable and invariable states, with the rate of switching varying from 0.2 to 2.0 switches/substitution. The internal branch length varied from 0.0 to 0.4. Note that the covarion model implemented in MrBayes is the true model in this case.
FIG. 3.—
Mixed branch length model improves phylogenetic accuracy under simulated conditions derived from observations of heterotachy in empirical sequence data. Proportion of replicate data sets from which the correct tree was uniquely recovered by each method is plotted against increasing internal branch length; sequences of 5,000 nt were simulated using the trees at left, with long terminal branch lengths of 0.75 expected substitutions/site. (A) Sequences were simulated along a single set of branch lengths using a stationary covarion process in which sites switch between variable and invariable states as they evolve. Series are not shown for the true model (which is the same as the covarion model in this case) and the mixed branch length model (which is the same as the homotachous model). (B) Data were simulated under a model of covarion rate shifts in which some invariable sites become released from selection in nonsister lineages; other sites that were previously variable then become invariable to compensate for the relaxation of selective constraint at correlated sites. The proportion of correct inferences using each method is plotted against the length of the internal branch (for variable sites only) on the true tree. (C) Sequence data were simulated with convergent proportions of invariable sites in nonsister lineages.
To simulate correlated rate shifts, we generated sequence data using an evolutionary process in which groups of sites exhibit correlated changes in evolutionary rates in nonsister lineages (see fig. 3B). Sequences were simulated on a 4-taxon ((AB),(CD)) phylogeny, with sites divided into 4 classes as follows: 25% of sites were invariable throughout the tree; 25% of sites were invariable in lineages B and D but released from selection—indicated by long branches (0.75)—in lineages A and C; 25% of sites were variable in lineages B and D (terminal branch lengths 0.75) but constrained in lineages A and C to compensate for loss of evolutionary constraints in the previously described class of sites. The remaining sites (25%) were variable in all lineages (terminal branch length 0.75). We varied the internal branch length for variable sites from 0.0 to 0.8.
To simulate changing proportions of invariable sites in different lineages, we divided sites into 3 different rate classes. Twenty-five percent of sites were invariable throughout the entire ((AB),(CD)) tree; 25% were invariable only in lineages A and C; and 50% were always variable (see fig. 3C). Terminal branch lengths for variable sites were 0.75, and the internal branch length varied between 0.0 and 0.8.
Empirical Sequence Data
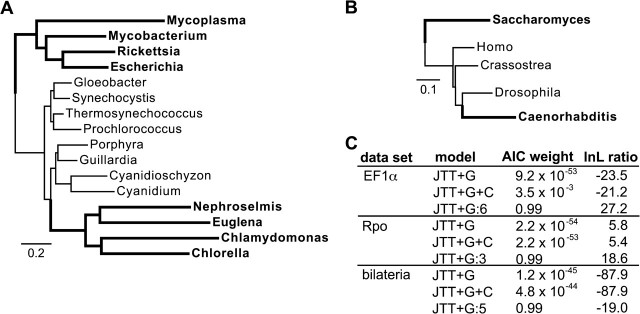
We analyzed the Micro* data set of Inagaki et al. (2004) (349 sites, 24 taxa) using homotachous ML (JTT + gamma model), MP, BMCMC (JTT + gamma + covarion), and the mixed branch length model using JTT + gamma with a variable number of branch length categories. ML analyses were conducted using 4 gamma rate categories, with branch lengths and shape parameter optimized using simulated annealing. BMCMC analyses were conducted using MrBayes v3.1 (Ronquist and Huelsenbeck 2003) as described above. ML scores for the covarion model were calculated using software provided by Zhou et al. (2007), with the same parameters as used in their original study. We calculated the best-fit number of branch length classes for the mixed branch length model using AIC. For each number of branch length classes (from 1 to 7), we inferred the ML phylogeny using simulated annealing; the likelihoods obtained were used to calculate AIC scores for each model, and the number of classes with the lowest AIC score was selected as the best-fit model. We calculated the likelihood ratio of the correct MF tree to the artifactual Microsporidia + Archaebacteria (MA) tree and assessed the support for the most likely hypothesis using the approximately unbiased (AU) test (Shimodaira 2002) implemented in CONSEL v0.1i (Shimodaira and Hasegawa 2001).
We calculated the weight of evidence in favor of the model selected by AIC using Akaike weights (see Posada and Buckley 2004). We calculated the difference in AIC score between each model i and the model selected by AIC: ΔAICi = AICi − AICselected. The Akaike weight in favor of model i (wi) is
Using the ML topology inferred under the model selected by AIC, we calculated the posterior probability that each site evolved according to each set of inferred branch lengths. The posterior probability of branch length set bi given site x was calculated by multiplying the proportion of sites expected to evolve under branch length set bi (ρi) by the likelihood obtained for that branch length set (P(x|t, bi)) and dividing by the total likelihood summed over all branch length sets:
For post hoc partitioned analysis, we used posterior probability cutoffs of 0.7, 0.8, 0.9, 0.95, and 0.99 to classify sites into categories. A site x was assigned to a particular class i if the posterior probability of that class (P(bi|x, t)) was greater than the cutoff. In each case, the likelihood ratio MF/MA was calculated using the JTT + gamma model, with branch lengths optimized independently for each class of sites. We also performed a partitioned analysis in which each site was assigned to the class with the highest posterior probability.
We compressed the original RNA polymerase (Rpo) alignment of Lockhart et al. (2005) using Gblocks to remove ambiguously aligned regions (Castresana 2000). We used a minimum number of conserved sequences of 8, a minimum number of flanking sequences of 12, a maximum contiguous nonconserved region length of 20, a minimum block length of 5, and allowing gaps with half the total number of taxa. This resulted in an alignment of 1,773 amino acids (aa). This alignment was analyzed using the JTT + G4 mixed branch length model with 1–7 branch length classes. For each number of classes, the ML tree was inferred using simulated annealing. We calculated the best-fit number of classes using AIC and estimated the weight of evidence in favor of the best-fit model using Akaike weights as described above. We calculated the support in favor of the correct red + green algae tree using the likelihood ratio of the best tree with red+green algae versus the best tree with the alternative nonphytosynthetic bacteria + green algae topology (fig. 7A).
FIG. 7.—
Mixed branch length model increases support for correct phylogenetic relationships. We analyzed 3 empirical data sets using both homotachous and mixed branch length models. (A) The correct 16-taxon Rpo phylogeny; the incorrect tree groups the green algal plastids with nonphotosynthetic bacteria (shown in bold) (B) The correct bilaterian phylogeny; the incorrect tree places the taxa in bold together. (C) We calculated the log-likelihood ratio (lnL) of the correct versus incorrect phylogenies. Positive lnLs indicate support for the correct tree, whereas negative values indicate support for the wrong tree. AIC weights indicate the inferred support for each model. The correct and incorrect trees for EF1α are shown in figure 4. Models used were homotoachous with gamma-distributed among-site rate variation (JTT+G), covarion (+C), or the mixed branch length model (:n, where n is the best-fit number of branch-length categories using AIC).
We analyzed a multigene data set from Philippe, Lartillot, and Brinkmann (2005) using the same approach as with the Rpo data. To reduce the computational burden of working with long sequences, we selected a reduced 5-taxon data set (fig. 7B) and removed columns containing gaps or missing characters from the alignment, resulting in a sequence length of 16,791 aa. We inferred the best-fit model and the ML topology as for the Rpo data. In this case, we calculated the likelihood ratio of the best tree with nematodes + insects (the well-corroborated tree) versus the best tree with nematodes + fungi.
Results
To determine the effects of heterotachy on phylogenetic inference, we conducted 3 different kinds of analyses, each designed to address a different question. First, to assess how specific forms of heterotachy affect phylogenetic accuracy, we simulated sequences under very challenging conditions in which sites evolve on various combinations of heterogeneous branch lengths. Second, to examine more empirically relevant forms of heterotachy under controlled conditions, we simulated sequences under strong versions of the types of heterotachy observed in real data sets. Finally, to determine the potential of the mixed branch length and covarion models for addressing real phylogenetic problems, we analyzed empirical sequence data known to have evolved heterotachously on well-known phylogenies.
Under each condition, we asked 2 questions: 1) how different forms of unincorporated heterotachy affect the performance of homotachous models and 2) whether evolutionary models incorporating heterotachy produce more accurate phylogenies (see fig. 1 for a diagram of the models used in this study). Two heterotachous models were used: 1) a Bayesian implementation of the covarion model (Tuffley and Steel 1998) and 2) a ML implementation of the mixed branch length model (Kolaczkowski and Thornton 2004; Spencer et al. 2005), with the number of branch length classes estimated from the data using AIC (Akaike [1974]) and BIC (Schwartz [1978]), 2 widely used methods of statistical model selection (Posada and Buckley 2004).
Simulations of Simplified Branch Length Heterogeneity
To elucidate the types of problems that different forms of heterotachy might cause and the ability of heterotachous models to address these problems, we examined data sets generated using 4 types of challenging, stereotyped branch length combinations. We compared the phylogenetic accuracy of the mixed branch length and covarion models of heterotachy with that of the best-fit homotachous model by plotting the fraction of correct inferences using each method against increasing phylogenetic signal (internal branch length). To reveal the specific effects of heterotachy, we compared the accuracy of each method with that of the true partitioned model (MLtrue), which correctly assigns sites to branch length categories a priori and estimates separate branch lengths within each category. We also examined the accuracy of MP.
Under all conditions studied, unincorporated heterotachy substantially reduced the accuracy of homotachous models. The mixed branch length model was significantly more accurate, recovering the correct tree with less phylogenetic signal and producing more accurate estimates of expected branch lengths across sites (fig. 2). Although the mixed model generally exhibited reduced statistical power compared with MLtrue (see supplementary fig. S1, Supplementary Material online), the mixed model was not biased under any of these conditions. In contrast, the covarion and homotachous models were subject to strong topological biases, loss of statistical power, and inference of hard polytomies, depending on the specific pattern of heterotachy in the data.
Under the first set of conditions—sequences generated on a tree with 2 long and 2 short terminal branches—both homotachous and covarion models were severely biased in favor of the long-branch attraction tree (fig. 2A and B). As with classical long-branch attraction, the direction of bias depended on which taxa had long branches. As previously observed (Kolaczkowski and Thornton 2004), when long terminals were not sister to one another, the bias favored an incorrect tree (fig. 2A). When sister taxa had long branches, the bias favored the correct phylogeny, as indicated by strong support for this tree even when the internal branch length was zero (fig. 2B). In contrast, the mixed branch length model was unbiased, producing inferences of topology and estimates of branch lengths much more similar to those obtained using MLtrue.
Under the second set of conditions—sites with strong phylogenetic signal simulated together with randomized noisy sites—homotachous and covarion models both exhibited a severe reduction in statistical power to resolve the correct phylogeny compared with MLtrue (fig. 2C). The mixed branch length model was more accurate; the performance improvement was small but statistically significant (P < 0.001). Branch length estimates were much more accurate using the mixed model compared with homotachous ML under these conditions.
Under the third set of conditions—in which sites are released from selection in different lineages—homotachous ML incorrectly estimated a zero-length internal branch on the most likely topology, inferring a hard polytomy (fig. 2D). This polytomous tree was recovered even when sequences of effectively infinite length were analyzed, indicating that homotachous ML is statistically inconsistent under these conditions. The mixed branch length model, in contrast, was not biased and recovered the correct phylogeny significantly more often. Although homotachous BMCMC was also unbiased, trees inferred using BMCMC were very weakly supported; when strong support was required to resolve the phylogeny, the accuracy of BMCMC was reduced to that of homotachous ML. The covarion model did not improve performance (see supplementary text, section 1, Supplementary Material online).
Across all conditions, AIC selected the correct number of branch length classes for mixed model analysis more often than it selected a too-simple model, and it never overestimated model complexity (see supplementary table S1, Supplementary Material online). In contrast, BIC favored an underparameterized model under some conditions.
Simulations of Types of Heterotachy Observed in Molecular Evolution
Studies of heterotachy have revealed 3 important features. First, a stationary covarion model of evolution generally fits empirical data better than homotachous models (Miyamoto and Fitch 1995; Galtier 2001; Huelsenbeck 2002). Second, different sites in the sequence may be invariable in different lineages (Fitch and Markowitz 1970; Fitch 1971, 1976). Third, the proportion of invariable sites has been observed to vary among lineages (Germot and Philippe 1999; Steel et al. 2000; Lockhart et al. 2005). To examine the potential effects of these types of heterotachy on phylogenetic inference, we simulated sequences under 3 simplified models: 1) a stationary covarion model in which every site may continuously switch between variable and invariable at a constant rate as evolution proceeds, 2) a nonstationary “correlated rate shift” model in which groups of sites exhibit periodic correlated changes in evolutionary rates, and 3) a model in which the proportion of invariable sites differs among lineages. In each case, we simulated sequences along a Felsenstein zone tree with long-branch nonsister lineages (see fig. 3), using challenging conditions and strong heterotachy. Although not necessarily indicative of the levels of heterotachy observed in empirical data sets, these simulations allow us to test for heterotachy-induced topological biases using purposefully difficult conditions of the types likely to be encountered when analyzing real data.
For the stationary covarion process, we simulated sequence data using the model of Tuffley and Steel (1998). Under these conditions, homotachous ML was unbiased and recovered the correct tree with high accuracy (fig. 3A; supplementary fig. S3, Supplementary Material online). The accuracy of the covarion model was the same as that of the simpler homotachous model. Model selection criteria did not support multiple branch length categories under these conditions.
To simulate correlated rate shifts, we partitioned sites on the ((AB),(CD)) phylogeny into 50% invariable and 50% variable. In lineages A and C, half the invariable sites are released from selection and become variable; a corresponding number of previously variable sites become invariable in the same lineages (fig. 3B). Under these conditions, homotachous models were strongly biased, and the covarion model performed even more poorly. In contrast, the mixed model was substantially more accurate than homotachous models and was unbiased, performing almost as well as MLtrue.
To assess the potential effects of changes in lineage-specific proportions of invariable sites, we simulated data on the ((AB),(CD)) phylogeny, with lineages A and C having 50% invariable sites, whereas lineages B and D had only 25% (fig. 3C). The mixed branch length model was more accurate than other methods under these conditions, whereas homotachous models were strongly biased in favor of the long-branch attraction topology. The covarion model was significantly less accurate than homotachous models.
These results show that mixed branch length analysis can improve the quality of inferred phylogenies under a variety of conditions when sequences evolve heterotachously. In contrast, the covarion model was less accurate than simpler homotachous models in some cases and was no more accurate than homotachous models even when it precisely matched the true evolutionary conditions.
Empirical Sequence Analysis
Although simulations can establish the potential impacts of heterotachy on phylogenetic accuracy, the true test of any method is how accurately it can reconstruct correct evolutionary relationships from real sequence data. To determine whether the mixed branch length model can improve the accuracy of phylogenies inferred from empirical sequences, we analyzed 3 data sets in which heterotachy is thought to cause phylogenetic error.
First, we analyzed the EF1α data set of Inagaki et al. (2004). Previous analyses have shown that when the Eukaryote phylogeny is inferred from these data using a homotachous evolutionary model, the Microsporidia are artifactually grouped with the Archaebacterial outgroup (the MA tree) rather than correctly with Fungi (MF, see fig. 4A). Prior analyses also show that systematic removal of sites exhibiting strong rate changes across the Archaebacteria/Eukaryote split reduces support for the incorrect placement of Microsporidia, suggesting that heterotachy may be at least partially responsible for this phylogenetic artifact (Inagaki et al. 2004).
FIG. 4.—
Mixed branch length model recovers the correct MF grouping from EF1α sequence data. (A) Correct MF tree is shown at left, and incorrect MA tree is shown at right, with branch lengths inferred by ML using the JTT + gamma model. (B) The difference in AIC scores between each model and the model with minimal AIC is plotted for the JTT + gamma model with 1–7 branch length classes. The tree topology was estimated by ML separately for each model. Arrow indicates the model with minimal AIC score, which is the model selected by AIC. (C) The log-likelihood ratio of the MF tree to the MA tree is plotted for models with increasing number of branch length classes, with negative lnL ratios indicating support for the incorrect MA tree and positive values indicating support for the correct MF tree. The significance of support for the best tree in each case was assessed using the AU test; p-values assuming each model are shown at right. Arrow indicates the model selected by AIC.
To analyze the EF1 data set using the mixed branch length model, we used an unconstrained topology search based on simulated annealing (see Materials and Methods) to infer the ML phylogeny assuming mixed models with 1–7 branch length classes. The best-fit number of classes—and resulting topology inference—was determined using AIC and BIC. AIC gave very strong support for branch length heterogeneity, selecting 6 as the best-fit number of branch length classes with Akaike weight >0.99 (fig. 4B; supplementary table S2, Supplementary Material online). BIC selected the covarion model with strong support (BIC weight >0.99).
The mixed model selected by AIC strongly supported the correct MF tree over the artifactual MA phylogeny (P = 0.021, fig. 4C). Whenever the number of branch length classes was underestimated, support shifted in favor of the MA tree. Overestimating the complexity of the model reduced support for the correct phylogeny but did not favor the incorrect tree. The covarion model, which was preferred by BIC, recovered the incorrect MA tree and gave negligible support (posterior probability <0.05) for the correct MF tree.
Concerns have been raised that AIC may systematically overestimate model complexity (Hurvich and Tsai 1989; Alfaro and Huelsenbeck 2006); BIC can be biased in favor of a too-simple model (Weakliem 1999). To determine the accuracy of AIC and BIC in this case, we simulated protein sequence data of the same length as the original data (349 aa) using the JTT + gamma model with 4 branch length classes—a model simpler than the one inferred by AIC—and parameter values estimated from the original data (see supplementary fig. S4, Supplementary Material online). We found that AIC was slightly conservative, selecting the correct number of branch length classes in 75% of trials; the number of classes was underestimated as 2 in the remaining 25% and was never overestimated. In contrast, BIC was strongly biased, selecting a 2-category model from 93% of replicates and a 1-category model in the remaining cases. These results show that an AIC/mixed model approach can improve phylogenetic accuracy in real data analysis. BIC and the covarion model were inferior strategies under these conditions.
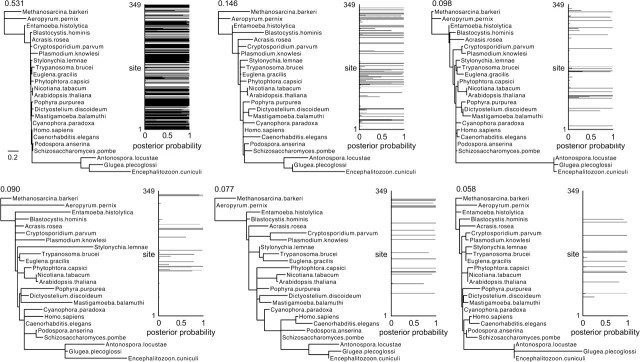
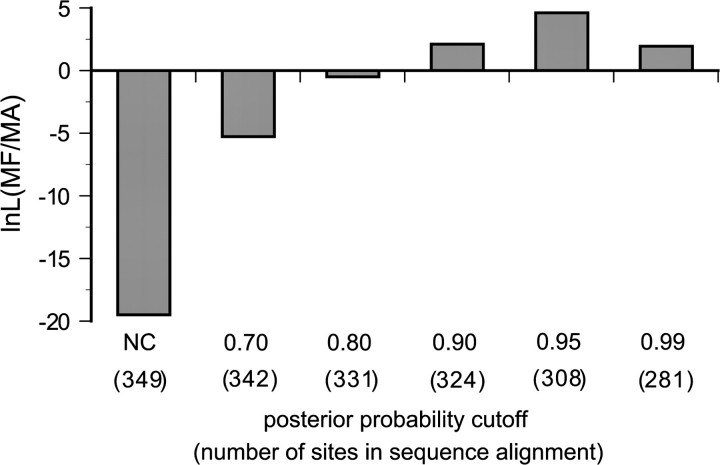
To determine if incorporating heterotachy is responsible for the improved phylogenetic accuracy of the mixed model, we performed partitioned analyses, with partitions inferred using the ML tree assuming a 6-category mixed model. We calculated the posterior probability of each branch length class for each site in the data set (see Materials and Methods). Most of the sites were decisively categorized with high posterior probability (fig. 5): 93% of sites were unambiguously categorized with posterior probability greater than 0.9; 88% of sites were categorized with posterior probability greater than 0.95; and 81% of sites were categorized with greater than 0.99 posterior probability. We used a variety of posterior probability cutoffs to generate strongly supported partitions; sites with posterior probability less than the cutoff were excluded (fig. 6). We found that using a high posterior probability cutoff to classify sites on the MF tree resulted in support for the correct phylogeny, indicating that partitioning sites based on mixed model analysis is sufficient to recover the correct tree. These results are consistent with the hypothesis that the mixed model is capturing an important aspect of EF1α evolution; however, it is impossible to rule out heterogeneity in other aspects of the evolutionary process—such as shifts in relative transition rates—as contributing to the improved performance of the mixed model.
FIG. 5.—
Mixed model analysis of EF1α data partitions sites into branch length categories. We plot the posterior probability that each site in the alignment evolved according to each set of branch lengths inferred using a 6-category mixed branch length model (inferred branch lengths shown to the left of each graph). The number above each tree indicates the inferred proportion of sites expected to evolve according to those branch lengths. The tree topology is the same as the MF tree in figure 4A; the Microsporidia clade has been placed at the bottom for space.
FIG. 6.—
Partitioning sites based on mixed branch length analysis recovers the correct MF tree from EF1α data. The log-likelihood ratio of the MF tree to the MA tree is plotted for a partitioned analysis, with sites categorized into groups based on posterior probabilities calculated from a 6-category mixed branch length analysis. Support for the correct MF tree or the incorrect MA tree is indicated by positive or negative lnL values, respectively. NC indicates that no cutoff was used; each site was placed in the category having the highest posterior probability.
To determine whether our results obtained using EF1α sequences can be generalized to other data, we analyzed 2 additional data sets: the 16-taxon plastid/eubacterial Rpo data of Lockhart et al. (2005) and a 5-taxon multigene data set derived from the study of bilaterian phylogeny by Philippe, Lartillot, and Brinkmann (2005). Both data sets have been shown to produce artifactual phylogenies. In the case of the Rpo data, MP incorrectly groups green algal plastids with the nonphotosynthetic bacteria outgroup rather than as a sister group to red algae (fig. 7A). The bilaterian data overwhelmingly support a basal position for nematodes when taxon sampling is poor (fig. 7B). Improving taxon sampling and removing genes with accelerated evolutionary rates shift support in favor of a nematode + insect clade (Philippe, Lartillot, and Brinkmann 2005). For each data set, we identified the best-fit evolutionary model using AIC and inferred the ML tree using simulated annealing. Support in favor of the correct phylogeny versus the incorrect tree was calculated using the log-likelihood ratio (lnL).
The mixed branch length model fits both empirical data sets better than a homotachous model and increases support for the correct phylogeny (fig. 7C). Extremely strong support was observed for choosing a mixed model with 3 and 5 classes, respectively, for the Rpo and bilaterian data. For Rpo, the mixed model improved support for the correct phylogeny from a lnL ratio of 5.8—using a homotachous model—to 18.6 (a >300,000-fold improvement in the likelihood ratio). For the bilaterian data, the mixed model reduced support for the incorrect tree versus the correct phylogeny from −87.9 to −19.0. These results, together with the analysis of EF1α, suggest that the mixed branch length model is likely to be a generally useful strategy for improving phylogenetic accuracy. In some cases—such as the bilaterian example—the mixed model is not sufficient to completely overcome strong topological biases, presumably due to other types of model violations (Lartillot et al. 2007) or inadequate taxon sampling.
Discussion
We have shown that numerous forms of strong heterotachy can cause homotachous models to infer inaccurate phylogenies. These results suggest that phylogenies inferred from molecular data using homotachous models should be interpreted with caution and examined for potential artifacts caused by model misspecification. Because unincorporated heterotachy can introduce strong biases, phylogenetic accuracy is not always improved by increasing the amount of sequence data; under some heterogeneous conditions, model-based techniques infer incorrect trees even when infinite data are available (see also Chang 1996; Kolaczkowski and Thornton 2004; Štefankovič and Vigoda 2006).
We found that the covarion model—the only existing tool for incorporating heterotachy—does not improve phylogenetic accuracy under the conditions we examined, including real sequence data. The failure of the covarion model could be due to 3 potential factors. First, mathematical formulations of the covarion model (Tuffley and Steel 1998) assume that the rate at which sites switch between variable and invariable on any branch is proportional to the rate of character substitution, which is unlikely to be the case for real sequence evolution (Gaucher et al. 2001; Gu 2001, 2003; Susko et al. 2002; Inagaki et al. 2003, 2004). Second, the covarion model assumes that the proportion of invariable sites is constant across lineages, whereas empirical data suggest that different lineages may have different proportions of invariable sites (Germot and Philippe 1999; Steel et al. 2000; Lockhart et al. 2005). Finally, the covarion model does not incorporate correlations in evolutionary rate shifts among different sites, which may be an important feature of molecular evolution (Fitch and Markowitz 1970; Fitch 1971, 1976). Our results show that each of these factors can impair the performance of the covarion model; in some cases, the covarion model's accuracy is even worse than a homotachous model. The covarion model appears to perform well only when it precisely matches the true evolutionary conditions, and even in this case there is no improvement over homotachous models. More general versions of the covarion model that allow sites to switch among multiple evolutionary rates have been developed (Galtier 2001; Wang et al. 2007). These models capture a more subtle stationary rate-switching process than the simple on–off version but do not differ from the simpler covarion model with regard to the 3 factors listed above. For this reason, we predict that these more general covarion models will suffer from the same limitations as the simple on–off model.
The mixed branch length model, in contrast, was dramatically more accurate than both homotachous and covarion models, recovering the correct phylogeny more often and providing better estimates of expected branch lengths across sites under all conditions tested. The mixed model provides a significantly better fit to real sequence data than homotachous models and recovered the correct evolutionary relationships under challenging conditions that cause other methods to fail.
The accuracy of the mixed model depends on the accuracy with which the best-fit number of branch length classes can be estimated. Our analyses suggest that AIC provides a reasonably accurate – albeit slightly conservative – estimate of model complexity when used to select the number of classes for the mixed model. In contrast, BIC was conservatively biased. Previous analyses suggested that AIC may select an overly complex model in some cases but did not address the frequency with which such errors might occur (Zhou et al. 2007). Our results suggest that overfitting errors are likely to be rare. One limitation of our simulation experiments is that the correct evolutionary model was always available; it is not known how AIC or BIC perform when additional forms of heterogeneity not captured by any available model are present in the data. Understanding the properties of model selection procedures such as AIC and BIC in the context of mixed phylogenetic models is an important area for future research.
Theoretical analyses have shown that the mixed model is statistically consistent so long as it is identifiable (Spencer et al. 2005), and that it is identifiable under some conditions (Allman and Rhodes 2006; Štefankovič and Vigoda 2006); however, it may produce inaccurate phylogenies due to nonidentifiability under specific evolutionary conditions using binary data (Matsen and Steel 2007). Our results indicate that the AIC/mixed model strategy is highly accurate on both simulated and empirical data, using both protein and nucleotide sequences: nonidentifiability does not appear to undermine phylogenetic accuracy under the conditions we examined.
The power of the AIC/mixed model to infer accurate phylogenies could depend on the amount of available data as well as the strength and complexity of heterotachy in the data. The mixed model performed extremely well in our simulations, which were done with moderately large data sets (N = 5,000 nt) and a moderate amount of strong heterotachy (2–4 branch length classes). The mixed model also improved phylogenetic accuracy and decisively partitioned sites using smaller empirical data sets (349 and 1,773 aa for EF1α and Rpo, respectively) and, in the case of EF1α, apparently more complex heterotachy. A more detailed understanding of the efficiency of the mixed model on small data sets will require further experiments.
One limitation of the mixed model is that it requires much more computation time than simpler models (see supplementary fig. S5, Supplementary Material online), which could necessitate limiting analyses to smaller data sets. Development and implementation of more efficient optimization algorithms should help overcome this limitation.
In addition to improving phylogenetic accuracy, the mixed branch length model is a potentially useful tool for characterizing the processes that drive molecular sequence evolution, one of the most important standing problems in biology. Branch lengths are often of interest for making inferences about divergence dates, substitution rates, or other aspects of the evolutionary process. We found that homotachous models produce biased estimates of branch lengths when sequences evolve heterotachously. The mixed branch length model produced much more accurate estimates of expected branch lengths across sites and may produce more accurate estimates of other evolutionary parameters, such as the amount of among-site rate variation and the nonsynonymous/synonymous substitution ratio, which could be routinely misestimated due to unincorporated heterogeneity. The ability of the mixed model to decisively partition sites among inferred branch length classes could be used to directly infer site-specific evolutionary properties from sequence data. For example, the identification of sites exhibiting lineage-specific increases in evolutionary rates could be used to predict sites involved in functional shifts. These predictions could then be examined using biochemical and structural approaches (Dean and Thornton 2007).
Current evolutionary models are built from multiple components, typically including a tree topology, branch lengths, a substitution matrix, state frequencies, a proportion of invariable sites, and gamma-distributed rate variation. The evolutionary forces described by these components could be heterogeneous across sites, lineages, or both. A comprehensive approach capable of incorporating a variety of types of heterogeneity would be useful for characterizing the forms of heterogeneity in real data sets. Models incorporating specific types of heterogeneity have been developed (Yang and Roberts 1995; Bruno 1996; Thorne et al. 1996; Galtier and Gouy 1998; Halpern and Bruno 1998; Koshi and Goldstein 1998, 2001; Huelsenbeck and Nielsen 1999; Dimmic et al. 2000; Foster 2004; Lartillot and Philippe 2004; Pagel and Meade 2004; Gowri-Shankar and Rattray 2005; Blanquart and Lartillot 2006); however, these partially heterogeneous models have not been integrated into a coherent framework capable of testing hypotheses about which aspects of the model display significant heterogeneity. Mixed models incorporating multiple types of heterogeneity could provide this general framework, enabling new types of evolutionary and phylogenetic hypotheses to be rigorously examined.
Supplementary Material
Supplementary text, tables S1 and S2, and figures S1–S5 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank Yuji Inagaki, Peter Lockhart, and Hervé Philippe for making their sequence data available. Hervé Philippe also supplied us with prerelease software. Supported by National Science Foundation DEB-0516530, National Institutes of Health GM62351, NSF IGERT DGE-9972830, and an Alfred Sloan Research Foundation Fellowship to J.W.T.
References
- Akaike H. A new look at the statistical model identification. IEEE Trans Automat Contr. 1974;19:716–723. [Google Scholar]
- Alfaro ME, Huelsenbeck JP. Comparative performance of Bayesian and AIC-based measures of phylogenetic model uncertainty. Syst Biol. 2006;55:89–96. doi: 10.1080/10635150500433565. [DOI] [PubMed] [Google Scholar]
- Allman ES, Rhodes JA. The identifiability of tree topology for phylogenetic models, including covarion and mixture models. J Comput Biol. 2006;15:1101–1113. doi: 10.1089/cmb.2006.13.1101. [DOI] [PubMed] [Google Scholar]
- Ané C, Burleigh JG, McMahon MM, Sanderson MJ. Covarion structure in plastid genome evolution: a new statistical test. Mol Biol Evol. 2005;22:914–924. doi: 10.1093/molbev/msi076. [DOI] [PubMed] [Google Scholar]
- Baele G, Raes J, de Peer YV, Vansteelandt S. An improved method for detecting heterotachy in nucleotide sequences. Mol Biol Evol. 2006;23:1397–1405. doi: 10.1093/molbev/msl006. [DOI] [PubMed] [Google Scholar]
- Blanquart S, Lartillot N. A Bayesian compound stochastic process for modeling nonstationary and nonhomogeneous sequence evolution. Mol Biol Evol. 2006;23:2058–2071. doi: 10.1093/molbev/msl091. [DOI] [PubMed] [Google Scholar]
- Brown RP. Large subunit mitochondrial rRNA secondary structures and site-specific rate variation in two lizard lineages. J Mol Evol. 2005;60:45–56. doi: 10.1007/s00239-004-0119-3. [DOI] [PubMed] [Google Scholar]
- Bruno WJ. Modeling residue usage in aligned protein sequences via maximum likelihood. Mol Biol Evol. 1996;13:1368–1374. doi: 10.1093/oxfordjournals.molbev.a025583. [DOI] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Chang JT. Inconsistency of evolutionary tree topology reconstruction methods when substitution rates vary across characters. Math Biosci. 1996;134:189–215. doi: 10.1016/0025-5564(95)00172-7. [DOI] [PubMed] [Google Scholar]
- Dean AM, Thornton JW. Mechanistic approaches to the study of evolution: the functional synthesis. Nat Rev Genet. 2007;8:675–688. doi: 10.1038/nrg2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmic MW, Mindell DP, Goldstein RA. Modeling evolution at the protein level using an adjustable amino acid fitness model. Pac Symp Biocomput. 2000:18–29. doi: 10.1142/9789814447331_0003. [DOI] [PubMed] [Google Scholar]
- Fitch WM. The nonidentity of invariable positions in the cytochromes c of different species. Biochem Genet. 1971;5:231–241. doi: 10.1007/BF00485794. [DOI] [PubMed] [Google Scholar]
- Fitch WM. The molecular evolution of cytochrome c in eukaryotes. J Mol Evol. 1976;8:13–40. doi: 10.1007/BF01738880. [DOI] [PubMed] [Google Scholar]
- Fitch WM, Markowitz E. An improved method for determining codon variability in a gene and its application to the rate of fixation of mutations in evolution. Biochem Genet. 1970;4:579–593. doi: 10.1007/BF00486096. [DOI] [PubMed] [Google Scholar]
- Foster PG. Modeling compositional heterogeneity. Syst Biol. 2004;53:485–495. doi: 10.1080/10635150490445779. [DOI] [PubMed] [Google Scholar]
- Gadagkar SR, Kumar S. Maximum likelihood outperforms maximum parsimony even when evolutionary rates are heterotachous. Mol Biol Evol. 2005;22:2139–2141. doi: 10.1093/molbev/msi212. [DOI] [PubMed] [Google Scholar]
- Galtier N. Maximum-likelihood phylogenetic analysis under a covarion-like model. Mol Biol Evol. 2001;18:866–873. doi: 10.1093/oxfordjournals.molbev.a003868. [DOI] [PubMed] [Google Scholar]
- Galtier N, Gouy M. Inferring pattern from process: maximum-likelihood implementation of a nonhomogeneous model of DNA sequence evolution for phylogenetic analysis. Mol Biol Evol. 1998;15:871–879. doi: 10.1093/oxfordjournals.molbev.a025991. [DOI] [PubMed] [Google Scholar]
- Gaucher EA, Miyamoto MM. A call for likelihood phylogenetics even when the process of sequence evolution is heterogeneous. Mol Phylogenet Evol. 2005;37:928–931. doi: 10.1016/j.ympev.2005.03.027. [DOI] [PubMed] [Google Scholar]
- Gaucher EA, Miyamoto MM, Benner SA. Function-structure analysis of proteins using covarion-based evolutionary approaches: elongation factors. Proc Natl Acad Sci USA. 2001;19:548–552. doi: 10.1073/pnas.98.2.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germot A, Philippe H. Critical analysis of eukaryotic phylogeny: a case study based on the HSP70 family. J Eukaryot Micorbiol. 1999;46:116–124. doi: 10.1111/j.1550-7408.1999.tb04594.x. [DOI] [PubMed] [Google Scholar]
- Gowri-Shankar V, Rattray M. On the correlation between composition and site-specific evolutionary rate: implications for phylogenetic inference. Mol Biol Evol. 2005;23:352–364. doi: 10.1093/molbev/msj040. [DOI] [PubMed] [Google Scholar]
- Gu X. Maximum-likelihood approach for gene family evolution under functional divergence. Mol Biol Evol. 2001;18:453–464. doi: 10.1093/oxfordjournals.molbev.a003824. [DOI] [PubMed] [Google Scholar]
- Gu X. Functional divergence in protein (family) sequence evolution. Genetica. 2003;118:133–141. [PubMed] [Google Scholar]
- Halpern AL, Bruno WJ. Evolutionary distances for protein-coding sequences: modeling site-specific residue frequencies. Mol Biol Evol. 1998;15:910–917. doi: 10.1093/oxfordjournals.molbev.a025995. [DOI] [PubMed] [Google Scholar]
- Hirt RP, Logsdon JM, Healy B, Dorey MW, Dolittle WF, Embley TM. Microsporidia are related to fungi: evidence from the largest subunit RNA polymerase II and other proteins. Proc Natl Acad Sci USA. 1999;96:580–585. doi: 10.1073/pnas.96.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP. Testing a covariotide model of DNA substitution. Mol Biol Evol. 2002;19:698–707. doi: 10.1093/oxfordjournals.molbev.a004128. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Nielsen R. Variation in the pattern of nucleotide substitution across sites. J Mol Evol. 1999;48:86–93. doi: 10.1007/pl00006448. [DOI] [PubMed] [Google Scholar]
- Hurvich CM, Tsai C-L. Regression and time series model selection in small samples. Biometrika. 1989;76:297–307. [Google Scholar]
- Inagaki Y, Blouin C, Susko E, Roger AJ. Assessing functional divergence in EF-1α and its paralogs in eukaryotes and archaebacteria. Nucleic Acids Res. 2003;31:4227–4237. doi: 10.1093/nar/gkg440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki Y, Susko E, Fast NM, Roger AJ. Covarion shifts cause a long-branch attraction artifact that unites microsporidia and archaebacteria in EF-1α phylogenies. Mol Biol Evol. 2004;21:1340–1349. doi: 10.1093/molbev/msh130. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick S, Gelatt CD, Vecchi MP. Optimization by simulated annealing. Science. 1983;4598:61–680. doi: 10.1126/science.220.4598.671. [DOI] [PubMed] [Google Scholar]
- Kolaczkowski B, Thornton JW. Performance of maximum parsimony and likelihood phylogenetics when evolution is heterogeneous. Nature. 2004;431:980–984. doi: 10.1038/nature02917. [DOI] [PubMed] [Google Scholar]
- Koshi JM, Goldstein RA. Models of natural mutations including site heterogeneity. Proteins: Struct Funct Genet. 1998;32:289–295. [PubMed] [Google Scholar]
- Koshi JM, Goldstein RA. Analyzing site heterogeneity during protein evolution. Pac Symp Biocomput. 2001:191–202. doi: 10.1142/9789814447362_0020. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Brinkmann H, Philippe H. Suppression of long-branch attraction artifacts in the animal phylogeny using a site-heterogeneous model. BMC Evol Biol. 2007;7(Suppl 1):S4. doi: 10.1186/1471-2148-7-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- Lockhart P, Novis P, Milligan BG, Riden J, Rambaut A, Larkum T. Heterotachy and tree building: a case study with plastids and eubacteria. Mol Biol Evol. 2005;23:40–45. doi: 10.1093/molbev/msj005. [DOI] [PubMed] [Google Scholar]
- Lockhart PJ, Steel MA, Barbrook AC, Huson DH, Charleston MA, Howe CJ. A covariotide model explains apparent phylogenetic structure of oxygenic photosynthetic lineages. Mol Biol Evol. 1998;15:1183–1188. doi: 10.1093/oxfordjournals.molbev.a026025. [DOI] [PubMed] [Google Scholar]
- Lopez P, Casane D, Philippe H. Heterotachy, an important process in protein evolution. Mol Biol Evol. 2002;19:1–7. doi: 10.1093/oxfordjournals.molbev.a003973. [DOI] [PubMed] [Google Scholar]
- Matsen FA, Steel M. Phylogenetic mixtures on a single tree can mimic a tree of another topology. Syst Biol. 2007;56:767–775. doi: 10.1080/10635150701627304. [DOI] [PubMed] [Google Scholar]
- McLachlan G, Peel D. Finite mixture models. John Wiley Sons, Inc; 2000. [Google Scholar]
- Miyamoto MM, Fitch WM. Testing the covarion hypothesis of molecular evolution. Mol Biol Evol. 1995;12:503–513. doi: 10.1093/oxfordjournals.molbev.a040224. [DOI] [PubMed] [Google Scholar]
- Pagel M, Meade A. A phylogenetic mixture model for detecting pattern-heterogeneity in gene sequence or character state data. Syst Biol. 2004;53:571–581. doi: 10.1080/10635150490468675. [DOI] [PubMed] [Google Scholar]
- Penny D, McComish BJ, Charleston MA, Hendy MD. Mathematical elegance with biochemical realism: the covarion model of molecular evolution. J Mol Evol. 2001;53:711–723. doi: 10.1007/s002390010258. [DOI] [PubMed] [Google Scholar]
- Philippe H, Lartillot N, Brinkmann H. Multigene analyses of bilaterian animals corroborate the monophyly of Ecdysozoa, Lophotrochozoa, and Protostomia. Mol Biol Evol. 2005;22:1246–1253. doi: 10.1093/molbev/msi111. [DOI] [PubMed] [Google Scholar]
- Philippe H, Lopez P. On the conservation of protein sequences in evolution. Trends Biochem Sci. 2001;26:414–416. doi: 10.1016/s0968-0004(01)01877-1. [DOI] [PubMed] [Google Scholar]
- Philippe H, Zhou Y, Brinkmann H, Rodrigue N, Delsuc F. Heterotachy and long-branch attraction in phylogenetics. BMC Evol Biol. 2005;5:50. doi: 10.1186/1471-2148-5-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Buckley TR. Model selection and model averaging in phylogenetics: advantages of the AIC and Bayesian approaches over likelihood ratio tests. Syst Biol. 2004;53:793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Rodriquez-Ezpeleta N, Philippe H, Brinkmann H, Becker B, Melkonian M. Phylogenetic analyses of nuclear, mitochondrial, and plastid multigene data sets support the placement of Mesostigma in the Streptophyta. Mol Biol Evol. 2007;24:723–731. doi: 10.1093/molbev/msl200. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Ruano-Rubio V, Fares MA. Artifactual phylogenies caused by correlated distributions of substitution rates among sites and lineages: the good, the bad, and the ugly. Syst Biol. 2007;56:68–82. doi: 10.1080/10635150601175578. [DOI] [PubMed] [Google Scholar]
- Schwartz G. Estimating the dimension of a model. Ann Stat. 1978;6:461–464. [Google Scholar]
- Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- Siddall ME, Kluge AG. Letter to the editor. Cladistics. 1999;15:439–440. doi: 10.1111/j.1096-0031.1999.tb00281.x. [DOI] [PubMed] [Google Scholar]
- Spencer M, Susko E, Roger AJ. Likelihood, parsimony, and heterogeneous evolution. Mol Biol Evol. 2005;22:1161–1164. doi: 10.1093/molbev/msi123. [DOI] [PubMed] [Google Scholar]
- Steel M, Huson D, Lockhart PJ. Invariable sites models and their use in phylogeny reconstruction. Syst Biol. 2000;49:225–232. [PubMed] [Google Scholar]
- Štefankovič D, Vigoda E. Pitfalls of heterogeneous processes for phylogenetic reconstruction. Syst Biol. 2006;56:113–124. doi: 10.1080/10635150701245388. [DOI] [PubMed] [Google Scholar]
- Susko E, Inagaki Y, Field C, Holder ME, Roger AJ. Testing for differences in rates-across-sites distributions in phylogenetic subtrees. Mol Biol Evol. 2002;19:1514–1523. doi: 10.1093/oxfordjournals.molbev.a004214. [DOI] [PubMed] [Google Scholar]
- Susko E, Spencer M, Roger AJ. Biases in phylogenetic estimation can be caused by random sequence segments. J Mol Evol. 2005;61:351–359. doi: 10.1007/s00239-004-0352-9. [DOI] [PubMed] [Google Scholar]
- Swofford DL. Phylogenetic analysis using parsimony (*and other methods) 2002. Sinauer Associates, Sunderland (MA) [Google Scholar]
- Taylor MS, Kai C, Kawai J, Carnici P, Hayashizaki Y, Semple CAM. Heterotachy in mammalian promotor evolution. PLoS Genet. 2006;2:627–639. doi: 10.1371/journal.pgen.0020030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne JL, Goldman N, Jones DT. Combining protein evolution and secondary structure. Mol Biol Evol. 1996;13:666–673. doi: 10.1093/oxfordjournals.molbev.a025627. [DOI] [PubMed] [Google Scholar]
- Tuffley C, Steel M. Modeling the covarion hypothesis of nucleotide substitution. Math Biosci. 1998;147:63–91. doi: 10.1016/s0025-5564(97)00081-3. [DOI] [PubMed] [Google Scholar]
- Wang H-C, Spencer M, Susko E, Roger AJ. Testing for covarion-like evolution in protein sequences. Mol Biol Evol. 2007;24:294–305. doi: 10.1093/molbev/msl155. [DOI] [PubMed] [Google Scholar]
- Weakliem DL. A critique of the Bayesian information criterion for model selection. Sociol Methods Res. 1999;27:359–397. [Google Scholar]
- Yang Z. Among-site rate variatino and its impact on phylogenetic analyses. Trends Ecol Evol. 1996a;11:367–372. doi: 10.1016/0169-5347(96)10041-0. [DOI] [PubMed] [Google Scholar]
- Yang Z. Maximum-likelihood models for combined analyses of multiple sequence data. J Mol Evol. 1996b;42:587–596. doi: 10.1007/BF02352289. [DOI] [PubMed] [Google Scholar]
- Yang Z, Roberts D. On the use of nucleic acid sequences to infer early branchings in the tree of life. Mol Biol Evol. 1995;12:451–458. doi: 10.1093/oxfordjournals.molbev.a040220. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Rodrigue N, Lartillot N, Philippe H. Evaluation of the models handling heterotachy in phylogenetic inference. BMC Evol Biol. 2007;7:206. doi: 10.1186/1471-2148-7-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.