Abstract
Host plant shifts by phytophagous insects play a key role in insect evolution and plant ecology. Such shifts often involve major behavioral changes as the insects must acquire an attraction and/or lose the repulsion to the new host plant's odor and taste. The evolution of chemotactic behavior may be due, in part, to gene expression changes in the peripheral sensory system. To test this hypothesis, we compared gene expression in the olfactory organs of Drosophila sechellia, a narrow ecological specialist that feeds on the fruit of Morinda citrifolia, with its close relatives Drosophila simulans and Drosophila melanogaster, which feed on a wide variety of decaying plant matter. Using whole-genome microarrays and quantitative polymerase chain reaction, we surveyed the entire repertoire of Drosophila odorant receptors (ORs) and odorant-binding proteins (OBPs) expressed in the antennae. We found that the evolution of OR and OBP expression was accelerated in D. sechellia compared both with the genome average in that species and with the rate of OR and OBP evolution in the other species. However, some of the gene expression changes that correlate with D. sechellia’s increased sensitivity to Morinda odorants may predate its divergence from D. simulans. Interspecific divergence of olfactory gene expression cannot be fully explained by changes in the relative abundance of different sensilla as some ORs and OBPs have evolved independently of other genes expressed in the same sensilla. A number of OR and OBP genes are upregulated in D. sechellia compared with its generalist relatives. These genes include Or22a, which likely responds to a key odorant of M. citrifolia, and several genes that are yet to be characterized in detail. Increased expression of these genes in D. sechellia may have contributed to the evolution of its unique chemotactic behavior.
Keywords: olfactory receptors, Drosophila sechellia, gene expression, microarrays, regulatory evolution, host plant preferences
Introduction
The adaptation of phytophagous insects to host plant chemistry influences both insect evolution and plant community structure (Becerra 1997, 2007; Renwick 2001). Host plant shifts are particularly important in insect speciation and ecological diversification, leading, for example, to the origin of new agricultural pests (Hawthorne and Via 2001; Dres and Mallet 2002; Nosil et al. 2002; Linn et al. 2003). Changes in chemotactic behavior are an integral part of these shifts as the insect must acquire an attraction and/or overcome the repulsion to the new food source.
Drosophila use both taste and smell to find food, mates, and appropriate environments for their offspring. Olfactory receptors (ORs) are important for locating suitable food sources at a distance; once at a food source or oviposition site, both gustatory and olfactory systems contribute to the fly's behavior (Takamura and Fuyama 1980; Hallem et al. 2006; Ebbs and Amrein 2007). Here, we examine the evolution of olfactory system by contrasting gene expression in Drosophila sechellia, a narrow host plant specialist, with that of its generalist relatives Drosophila simulans and Drosophila melanogaster.
Physiology and Biochemistry of Taste and Smell in Drosophila
The role of the Drosophila “nose” is played by the third antennal segment, which carries several hundred olfactory sensilla, and the maxillary palp, which carries ∼60 sensilla (Shanbhag et al. 1999). These sensilla have 3 major morphological types—basiconic, trichoid, and coeloconic—each of which can be further subdivided into several subtypes based on innervation patterns and neurophysiological responses. For example, the large basiconic sensilla on the antennae are classified into ab1, ab2, and ab3 subtypes (Shanbhag et al. 1999; Stocker 2001; Couto et al. 2005). Each sensillum is innervated by 1–4 olfactory neurons, which project into the antennal lobe of the brain (Couto et al. 2005; Fishilevich and Vosshall 2005). The sensitivity of each chemoreceptory neuron to a range of volatile compounds is determined by transmembrane OR proteins. Most neurons in the antennal sensory organs express a single OR, although some types of neurons express 2 or 3 (Vosshall et al. 1999; Dobritsa et al. 2003; Yao et al. 2005). Each OR responds to a subset of volatile compounds in a concentration-dependent manner, with different ORs having distinct but overlapping response spectra (de Bruyne and Warr 2005; Hallem et al. 2006; Smith 2007). Most ORs are sensitive to alcohols, ketones, fatty acids, esters, and other volatile compounds present in the fruit (Yao et al. 2005; Hallem and Carlson 2006), whereas a few are responsible for the perception of fly pheromones (Kurtovic et al. 2007; van der Goes van Naters and Carlson 2007).
Contact chemoreception (i.e., taste) is mediated by gustatory organs located on the legs, proboscis, wings, and genitalia (Dunipace et al. 2001; de Bruyne and Warr 2005; Hallem et al. 2006; Smith 2007). These organs express a different set of transmembrane proteins called gustatory receptors (GRs) (Scott et al. 2001; Thorne et al. 2005; Ebbs and Amrein 2007). Both olfactory and gustatory organs also express odorant-binding proteins (OBPs), which are secreted proteins that function in the lymph fluid that fills the sensilla lumen (Galindo and Smith 2001; Graham and Davies 2002; Hekmat-Scafe et al. 2002). One of their functions may be to chaperone the ligands through the lymph to the neuron dendrites where they can be bound by an OR or GR. Other hypothesized functions of OBPs include concentration of odorant molecules in the sensillum lymph, removal or detoxification of odorants, and coinitiation of signal transduction through OR and GR receptors (Rutzler and Zwiebel 2005; Hallem et al. 2006). In general, little is known about the binding specificity of OBPs. The only Drosophila OBP characterized in detail, Obp76a (also called lush), is essential for the sensitivity of olfactory neurons to cis-vaccenyl acetate, which acts as a sex and aggregation pheromone in D. melanogaster (Xu et al. 2005; Ha and Smith 2006).
Host Plant Choice in Drosophila
Drosophila exploit a wide variety of food sources, including fungi, flowers, cambium and sap fluxes of woody plants, cacti, and both fleshy and dry fruits (Markow and O'Grady 2005). In the D. melanogaster species group, most species are ecological generalists that feed on decaying fruit and other decaying vegetation. However, a few species in this group have evolved into narrow specialists restricted to a small number of host plants, such as Eucalyptus and Eugenia for Drosophila flavohirta, Pandanus for Drosophila erecta, and Morinda citrifolia for D. sechellia (Lachaise et al. 1988; McEvey et al. 1989).
Drosophila sechellia, in particular, is a promising model for understanding the genetic and molecular mechanisms underlying the evolution of host plant preferences. Whereas its closest relatives—D. melanogaster, D. simulans, and Drosophila mauritiana—are generalists, D. sechellia almost exclusively uses relatively unspoiled ripe fruit of Morinda (Lachaise et al. 1988). Drosophila sechellia preference for this plant typifies the complex interplay of smell and taste in fly behavior. Field and laboratory studies have shown that volatile compounds present in the ripe Morinda fruit are sufficient to attract D. sechellia and repel D. simulans and D. melanogaster over large distances, suggesting that long-range host plant choice is mediated by ORs (Louis and David 1986; R'Kha et al. 1991; Higa and Fuyama 1993; Amlou et al. 1998; Legal et al. 1999). Part of this attraction is likely due to the octanoic and hexanoic acids that are abundant in Morinda, although other medium-chain fatty acids and derived compounds may also play a role (Amlou et al. 1998; Legal et al. 1999). Once on its host, contact-mediated gustatory responses become important, especially during oviposition. Recent work has implicated Obp57d/e, expressed in leg gustatory organs, in behavioral differences between D. sechellia and its relatives. Replacement of D. melanogaster Obp57d/e with the D. sechellia ortholog causes transgenic flies to stop avoiding oviposition on media containing compounds from Morinda fruit (Matsuo et al. 2007).
Although not as extreme as D. sechellia, both D. melanogaster and D. simulans also exhibit species-specific food and oviposition site preferences (David et al. 2004). Even within D. melanogaster, there is evidence for considerable variation in odor-driven food preference (van Delden and Kamping 1990; Possidente et al. 1999).
Evolution of the Olfactory System in D. sechellia and its relatives
Morphology of the third antennal segment and the distribution of morphologically distinguishable sensilla are well conserved among the close relatives of D. melanogaster (the melanogaster species subgroup), although D. sechellia exhibits subtle changes in the relative numbers and fine structure of some sensilla (Stensmyr et al. 2003; Dekker et al. 2006). However, neurophysiological recordings have shown that D. sechellia is unique in this subgroup in not having any basiconic sensilla with the ab2 response spectrum, with a concomitant increase in the number of ab3 and a decrease in the number of ab1 sensilla (Stensmyr et al. 2003; Dekker et al. 2006). In addition, the dominant ligand for the ab3 receptors has shifted from ethyl hexanoate in D. melanogaster and most other species to methyl hexanoate in D. sechellia and D. simulans. Drosophila sechellia ab3 receptors show increased sensitivity to hexanoate esters compared with D. melanogaster. These changes may have been a key factor in the ecological specialization of D. sechellia as hexanoates are major components of the Morinda fruit headspace—the volatile odors around the fruit (Dekker et al. 2006).
Olfactory and gustatory systems of D. sechellia have also undergone considerable changes at the molecular level. Six OR and 13 GR genes have been inactivated by loss-of-function mutations in this species—a rate of gene loss much higher than in its generalist relatives. Coding sequence evolution in the remaining OR and GR genes has also been accelerated in D. sechellia compared with D. simulans (McBride 2007; Nozawa and Nei 2007; Tunstall et al. 2007). Interestingly, genes inactivated in D. sechellia include Or22b, one of the receptor proteins expressed in the hexanoate-sensitive ab3 sensilla. These gene losses may have contributed to the host plant shift in D. sechellia, although for any given gene it is unclear whether its loss was a cause or a consequence of ecological specialization. As D. sechellia is an island endemic with a small effective population size, the relative roles of selective and demographic factors in gene loss are not always clear.
In the evolution of an ecological specialist from a generalist ancestor, some genes and receptors may be rendered superfluous, whereas others may take on increasingly prominent roles. Although most work to date has focused on the former (McBride 2007; Nozawa and Nei 2007; Tunstall et al. 2007), it is necessary to elucidate both sides of this process to understand the molecular mechanisms of an ecological shift. Accelerated evolution of many ORs in D. sechellia may in part reflect functional changes increasing or decreasing their sensitivity to ecologically relevant odorants. However, changes in the relative abundance of different receptor proteins in olfactory neurons may also affect their response spectra and contribute to the unique chemotactic behavior of this species. In this report, we examine the divergence of gene expression in the olfactory system between D. sechellia and its generalist relatives D. simulans and D. melanogaster, focusing particularly on OR and OBP genes. We hypothesized that D. sechellia may have experienced “gain-of-function” expression changes, in addition to the loss-of-function changes already described. Indeed, we find that a number of genes are strongly upregulated in D. sechellia compared with its relatives, suggesting that they could be involved in the evolution of host plant preferences. At the same time, changes in some genes implicated in response to key Morinda odorants may predate the divergence of D. sechellia from its generalist relatives.
Materials and Methods
Microarray Experiments
Gene expression was compared among 4 genotypes: D. sechellia isofemale line 81, multifemale D. simulans strain from Winters, CA, and 2 isogenic strains of D. melanogaster, WI58 and WI89 (Yang and Nuzhdin 2003). Males and females were analyzed separately. For each genotype and sex, we used 3 independent biological replicates of 32–35 individuals each, for a total of 24 samples. All dissections were performed between 1 and 2 h after eclosion. From each fly, the third antennal segment and arista were removed with microsurgical scissors (Storz Ophthalmic Instruments, San Dimas, CA) and placed immediately in Trizol (Invitrogen, Carlsbad, CA) on ice. Dissected samples were frozen in Trizol at −80 °C until RNA extraction.
RNA extraction and quality control, cDNA synthesis, and linear amplification and labeling of complementary RNA were performed as described (Barmina et al. 2005). Each replicate RNA sample isolated from the antennae contained 500–1,250 ng of total RNA. A total of 75 ng from each sample was amplified to obtain 122–172 μg of biotin-labeled complementary RNA. A total of 15 μg from each labeled sample were hybridized to Affymetrix Drosophila genome v2.0 microarrays.
Signal intensities were quantified using MAS 5.0 (http://www.affymetrix.com). Microarray probes that did not match perfectly the genome sequences of all 3 species were removed from the analysis (see Supplementary Information online). Normalization was performed by centering natural log-transformed signal intensities to the chip median. Probe sets that produced “absent” calls for all replicates were excluded from further analysis. For each remaining probe set, we fitted the following analysis of variance (ANOVA) model:
where Yjkn is the log-normalized transcript abundance for the jth species (j = melanogaster, simulans, or sechellia), kth sex (k = male or female), and nth replicate (n = 1,2,3). The lj in this model is the effect of species, sk is the effect of sex, and lsjk is the species × sex interaction. Normality of the residuals (ϵjkn) for each probe set was checked using Shapiro–Wilk's test. All effects with P values below the false discovery rate (FDR) of 0.05 (Benjamini and Hochberg 1995) were considered significant (for a review, see Verhoeven et al. 2005).
We first examined each probe set for the effects of species, sex, and species × sex interaction. Genes that showed significant sex or species × sex interaction effects were considered sexually dimorphic. Further analyses were performed separately for sexually dimorphic and monomorphic genes. For the sexually monomorphic genes, we used a reduced ANOVA model Yjn = μ + lj + ϵjn to test for the effect of species in each of 4 separate pairwise comparisons: D. sechellia versus D. simulans, D. sechellia versus D. melanogaster, D. simulans versus D. melanogaster, and D. melanogaster WI58 versus D. melanogaster WI89. Within each genotype, gene expression measurements from both sexes were combined for this analysis. The same 4 pairwise comparisons were performed for the sexually dimorphic genes using the same reduced ANOVA model, but the ANOVA was performed separately for males and females.
Confirmation of Microarray Data by Quantitative Polymerase Chain reaction
To confirm the microarray data, we measured the expression of 34 antennal OR and OBP genes using quantitative rt–polymerase chain reaction (quantitative rt-PCR) as described in the Supplementary Information online. We then compared the microarray and qPCR results for each gene and for each pair of species, for a total of 99 tests (supplementary table 1, Supplementary Material online). The qPCR data were corrected for multiple testing using an FDR level of 0.05. In 75 out of 99 comparisons, qPCR and microarray hybridization showed expression differences in the same direction; most of these comparisons were significant at FDR = 0.05 according to both microarray and qPCR results (supplementary table 1, Supplementary Material online). This level of concordance compares favorably with the results of cross-platform validation experiments in which identical RNA samples were examined on different microarrays (Bammler et al. 2005; Dallas et al. 2005; Morey et al. 2006; Wang et al. 2006; Stafford and Brun 2007) or quantified by microarrays and massively parallel sequencing (Liu et al. 2007). Given the different technical biases of microarray hybridization and qPCR, we have strong evidence that the genes showing concordant expression differences in both analyses have diverged among species and strains. In cases where qPCR and microarray data disagree (∼25% of comparisons), no conclusion can be drawn about the direction and magnitude of change because we do not know which method provides a more accurate estimate of transcript abundance under our experimental conditions.
Reconstruction of Ancestral Gene Expression Levels
Ancestral gene expression states were reconstructed from the microarray data using AncML v 1.0 (Schluter et al. 1998). For each species and strain, gene expression levels were estimated as the least-squares mean of normalized, log-transformed signal intensities across all samples (6 samples each for D. sechellia, D. simulans, and each strain of D. melanogaster). Male and female samples were combined for this analysis. A neutral phylogenetic tree for reconstructing ancestral character states was obtained by selecting from published genome sequences 10 noncoding regions evenly spaced along each of the major chromosome arms (50 loci total, mean length ∼500 bases). Sequences from Drosophila yakuba were used as outgroups to root the trees. Phylogenetic reconstruction was performed for each locus using MrBayes version 3.1.2 (Huelsenbeck and Ronquist 2001) under a GTR + Γ model. We chose this general model rather than estimating a specific model for each locus to ensure that branch lengths were comparable across genes. We did not observe any particular biases among the loci in either tree topology or branch lengths. For ancestral reconstruction, we used the median branch length across all loci for each species so that the tree was as follows: ([{mel58:0.008, mel89:0.008}: 0.0309, {sech:0.0269, sim:0.0168}: 0.021], yak).
Ancestral reconstruction was performed for all transcripts for which we had masked microarray data (N = 11,631). We treated gene expression levels as a continuously varying quantitative trait and estimated the ancestral expression states for the most recent common ancestor (MRCA) of the 2 D. melanogaster lines, the MRCA of D. simulans and D. sechellia, and the MRCA of all 4 taxa. We also calculated the standard errors of the reconstructed expression states. The amount of change in gene expression along a particular branch of the phylogeny was calculated by subtracting the mean expression value of the extant taxon from the corresponding MRCA estimate.
Independent contrasts analysis was performed for OR and OBP genes using the Contrast program in PHYLIP (Felsenstein 2005). For each pair of genes, we estimated the covariance and correlation of expression levels across genotypes. These estimates take into account phylogenetic relationships among species and strains (Felsenstein 1985). The square pairwise correlation matrices were visualized using cell plots in JMP v6 (SAS Institute, Cary, NC), where colors indicate the strength and sign of correlations (supplementary fig. 1, Supplementary Material online).
Ancestral character reconstruction relies on a number of assumptions, including that all characters are independent and that each character follows a Brownian motion model—that is, each subsequent change in character value is independent of all previous changes (Felsenstein 1985). The first assumption is almost certainly violated for gene expression, which is controlled by a limited number of transcription factors. The second assumption may be violated, as well, for genes evolving under directional selection. Thus, the results of comparative analysis for individual genes should be interpreted with caution. Finally, because D. sechellia and D. simulans were each represented by only 1 strain in this study, inferences of interspecific divergence may be confounded with intraspecific variation. Despite these qualifications, genome-wide patterns of expression divergence revealed by this analysis may provide valuable insights into the evolutionary history of each species (e.g., Holloway et al. 2007).
Results and Discussion
Extensive Interspecific Differences in Olfactory Gene Expression
A total of 51 out of 60 known OR genes and 38 out of 53 annotated OBPs were detected in RNA samples isolated from the third antennal segment (table 1 and supplementary tables 2 and 3 [Supplementary Material online]). This includes 38 out of 38 ORs and 15 out of 16 OBPs with previously described expression patterns in the antennae (Galindo and Smith 2001; Couto et al. 2005). OR transcripts are present in the third antennal segment at levels similar to the genome-wide distribution, whereas OBP transcripts are more abundant than the genome average (for details, see Supplementary Information online). Thus, our comparative analysis captures the entire or nearly entire set of antennal OR and OBP genes.
Table 1.
Numbers of Genes Showing Expression Differences in the Microarray Analysis
| Number and Percentage of Genes Significant for the Main Effect of Genotype at FDR < 0.05 across All Samples | Number and Percentage of Genes Significant for the Main Effect of Genotype at P < 0.001 in Pairwise Comparisons | |||||
| Gene Class | Total Number of Detected Genes | Drosophila sechellia versus Drosophila simulans | Drosophila sechellia versus Drosophila melanogaster | Drosophila simulans versus D. melanogaster | Between Strains of D. melanogaster | |
| ORs | 51 | 46 (92.0%) | 14 (30.4%) | 19 (41.3%) | 27 (58.7%) | 10 (21.7%) |
| OBPs | 38 | 37 (97.4%) | 19 (50.0%) | 21 (55.3%) | 20 (52.6%) | 3 (7.9%) |
| ORs and OBPs | 89 | 83 (93.3%) | 33 (37.1%) | 40 (44.9%) | 47 (52.8%) | 13 (14.6%) |
| Whole genome | 9,830 | 7,882 (80.2%) | 1,946 (24.7%) | 4,657 (59.1%) | 4,399 (55.8%) | 1,153 (14.6%) |
Gene expression in the antennae has diverged extensively among the 3 species. For the genome as a whole, 80.2% of genes show a significant effect of genotype in the ANOVA at FDR = 0.05 (table 1), indicating that the expression levels of these genes differ significantly across species. In pairwise comparisons, 24.7% of genes differ between D. sechellia and D. simulans, 59.1% differ between D. sechellia and D. melanogaster, 55.8% differ between D. simulans and D. melanogaster, and 14.6% differ between the 2 isogenic strains of D. melanogaster at P < 0.001 (table 1).
Divergence of olfactory gene expression is accelerated relative to genome-wide expression divergence (Fisher's exact test, P = 0.0004). A total of 92% of detected ORs (46 out of 51) and 97.4% of OBPs (37 out of 38) showed a significant effect of genotype at FDR = 0.05. Among these genes, 14 ORs and 19 OBPs differ between D. sechellia and D. simulans, 19 ORs and 21 OBPs differ between D. sechellia and D. melanogaster, 27 ORs and 20 OBPs differ between D. simulans and D. melanogaster, and 10 ORs and 3 OBPs differ between the 2 isogenic strains of D. melanogaster at P < 0.001. Of the 34 OR and OBP genes examined by qPCR (see Materials and Methods), 20 showed significant differences in expression between D. sechellia and D. simulans, 23 differed between D. sechellia and D. melanogaster, 17 differed between D. simulans and D. melanogaster, and 15 differed between the 2 strains of D. melanogaster. Thus, in contrast to electrophysiological analysis (Stensmyr et al. 2003; Dekker et al. 2006), we find that significant changes in the olfactory system are not limited to the specialist D. sechellia and may affect ecologically similar generalist species such as D. simulans and D. melanogaster.
Few genes showed evidence of sex-biased expression in the antennae; however, the proportion of sexually dimorphic genes was higher for olfactory genes than for the genome as a whole. At a lenient significance threshold (FDR = 0.2), 32% of ORs and 11.8% of OBPs were expressed in a sex-biased manner, compared with the genome-wide level of 4.1% (Fisher's exact test, P < 0.0001). At least some of these differences may be attributed to the quantitative differences between males and females in the relative number of different sensilla subtypes (Shanbhag et al. 1999). We found that sexually dimorphic genes showed greater interspecific divergence and that this was true both for olfactory and for other genes. For the genome as a whole, 79.7% of sexually monomorphic genes but 91.9% of sexually dimorphic (at FDR = 0.2) genes showed a significant effect of genotype (Fisher's exact test, P < 0.0001). For the olfactory genes, the corresponding proportions were 92.2% and 100% (table 2). Increased divergence of sex-biased genes was observed in all 3 interspecific comparisons (table 2).
Table 2.
Effect of Sexual Dimorphism on Interspecific Divergence of Gene Expression
| Total Number of Detected Genes | Number and Percentage of Genes Significant for the Main Effect of Genotype at FDR < 0.05 across All Samples | Number and Percentage of Genes Significant for the Main Effect of Genotype at P < 0.001 in Pairwise Comparisons | |||
| Drosophila sechellia versus Drosophila simulans | Drosophila sechellia versus Drosophila melanogaster | Drosophila simulans versus D. melanogaster | |||
| All genes, sexually dimorphic at FDR < 0.2 | 406 | 373 (91.9%) | 111 (27.3%) | 218 (58.4%) | 216 (57.9%) |
| All genes, sexually monomorphic | 9,424 | 7509 (79.7%) | 1835 (19.5%) | 4439 (47.1%) | 4183 (44.4%) |
| OR and OBP genes, sexually dimorphic at FDR < 0.2 | 20 | 20 (100%) | 10 (50%) | 13 (65%) | 14 (70%) |
| OR and OBP genes, sexually monomorphic | 64 | 59 (92.2%) | 20 (33.9%) | 35 (59.3%) | 30 (50.8%) |
Evolution of Olfactory Gene Expression in D. sechellia
Differences in gene expression between 2 extant species can, in principle, be due to evolutionary changes in either, or both, phylogenetic lineages following their divergence from their latest common ancestor. Drosophila sechellia is an island endemic with narrow host plant specificity, suggesting that olfactory gene expression may have undergone greater changes on the D. sechellia branch than on the D. simulans branch. Our analysis confirms this prediction. A total of 37.1% of ORs and OBPs differ between D. sechellia and D. simulans at P < 0.001, compared with 24.7% for the genome as a whole (table 1) (Fisher's exact test, P < 0.0001). In contrast, the corresponding numbers are 44.9% versus 59.1% for the D. sechellia–D. melanogaster comparison, 52.8% versus 55.8% for the D. simulans–D. melanogaster comparison, and 14.6% versus 14.6% for the comparison between 2 strains of D. melanogaster (table 1).
We used a phylogenetic approach to explore the origins of gene expression differences between D. sechellia and D. simulans. For each gene, we reconstructed the mean and standard deviation (SD) of gene expression level in the latest common ancestor of these species in a maximum likelihood framework (see Materials and Methods for a discussion of this approach). We then asked whether the expression of each gene has undergone a greater absolute change in the D. sechellia or in the D. simulans lineage. On average, we expect greater changes in D. sechellia than in D. simulans under any mode of evolution as D. sechellia has the longer branch in the phylogenetic tree used for character reconstruction. Indeed, for the genome as a whole, 74% of genes show greater absolute change in D. sechellia (table 3). Among ORs and OBPs, however, 94% show greater change on the D. sechellia than on the D. simulans branch. This proportion is significantly greater than the genome average (Fisher's exact test, P = 0.0029). For 25 OR and OBP genes, the amount of change on the D. sechellia lineage exceeded 1 SD of the estimated ancestral value and for 15 genes it exceeded 2 SDs. In contrast, the amount of change on the D. simulans branch exceeded 1 SD of the ancestral estimate for only 14 genes and none exceeded 2 SDs (table 3). This analysis suggests that olfactory gene expression has evolved considerably faster on the branch leading to D. sechellia than on the D. simulans branch.
Table 3.
Gene Expression Changes on the Drosophila sechellia and Drosophila simulans Branches
| Phylogeny Branch | ||
| Between D. sechellia and MRCA of D. sechellia and D. simulans | Between D. simulans and MRCA of D. sechellia and D. simulans | |
| All genes | ||
| Greater absolute change on branch | 2,944 (73.7%) | 1,051 (26.3%) |
| Expression change on branch exceeds 1 SD of MRCA estimate | 2,881 (72.1%) | 1,848 (46.3%) |
| Expression change on branch exceeds 2 SDs of MRCA estimate | 1,115 (27.9%) | 0 |
| OR and OBP genes | ||
| Greater absolute change on branch | 31 (93.9%) | 2 (6.1%) |
| Expression change on branch exceeds 1 SD of MRCA estimate | 25 (75.8%) | 14 (42.4%) |
| Expression change on branch exceeds 2 SDs of MRCA estimate | 15 (45.5%) | 0 |
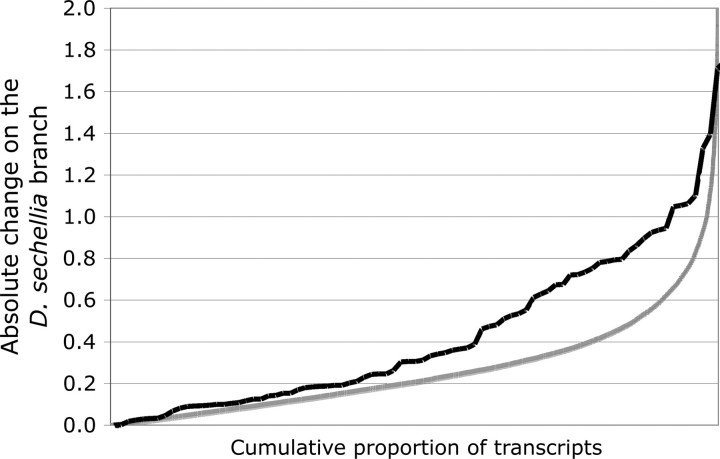
We also compared the absolute amount of change that occurred on the D. sechellia branch between olfactory and nonolfactory genes. This distribution is considerably steeper for olfactory genes, indicating that the evolution of OR and OBP expression has been accelerated in D. sechellia compared with the genome average (fig. 1). A similar though weaker trend is also observed on the D. simulans branch (data not shown), suggesting that olfactory genes may have a general tendency for rapid expression divergence. On average, OBPs have undergone greater changes than ORs (data not shown). These differences remain even after divergence is normalized by mean expression levels, showing that accelerated divergence among some genes is not an artifact of their higher overall expression.
FIG. 1.—
Amount of gene expression change on the Drosophila sechellia branch for all transcripts in the genome (gray line) and for OR and OBP genes (black line). Expression change (y axis) is expressed in standardized units that refer to log-transformed expression levels from the microarray analysis.
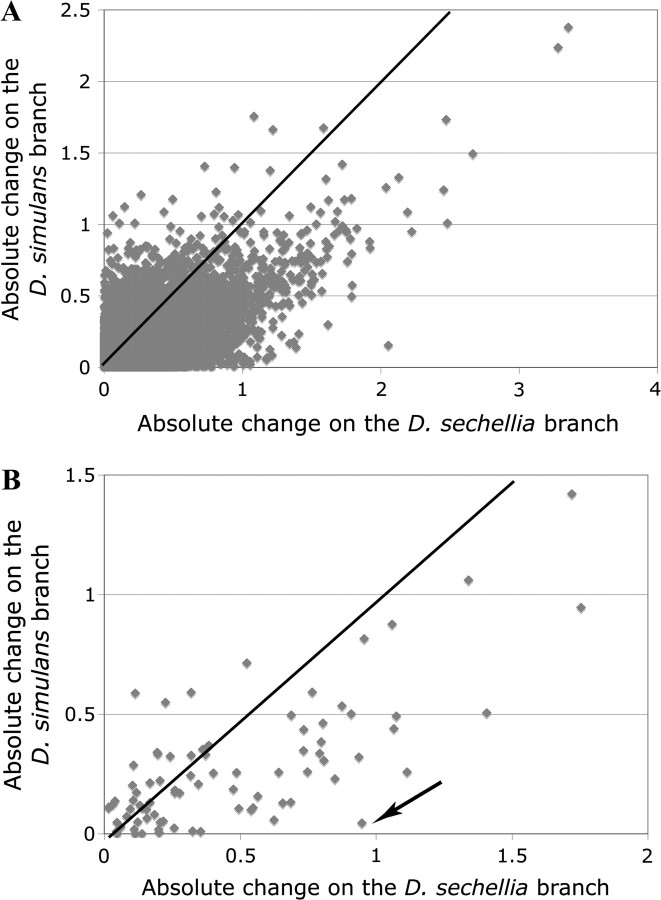
The amount of expression change on the D. sechellia and D. simulans branches is positively correlated both for olfactory genes (r = 0.72, P < 0.0001) and for the genome as a whole (r = 0.54, P < 0.0001) (fig. 2). Because the ancestral values are reconstructed rather than measured independently, positive correlation between branches is expected under most evolutionary scenarios. However, several ORs and OBPs have undergone a large amount of change on the D. sechellia branch but virtually none on the D. simulans branch. Such disparity is unlikely under neutral evolution, suggesting that these genes may be involved in the unique ecological adaptations of this species. The most prominent example of such genes is Or22a (arrow in fig. 2B). The receptor encoded by this gene is sensitive to methyl hexanoate, consistent with an increased sensitivity to this chemical in D. sechellia (Dekker et al. 2006).
FIG. 2.—
Amount of gene expression change on the Drosophila sechellia and Drosophila simulans branches for all transcripts in the genome (A) and for OR and OBP genes (B). Black lines indicate 1:1 diagonals. Expression divergence is expressed as in figure 1. Arrow indicates Or22a.
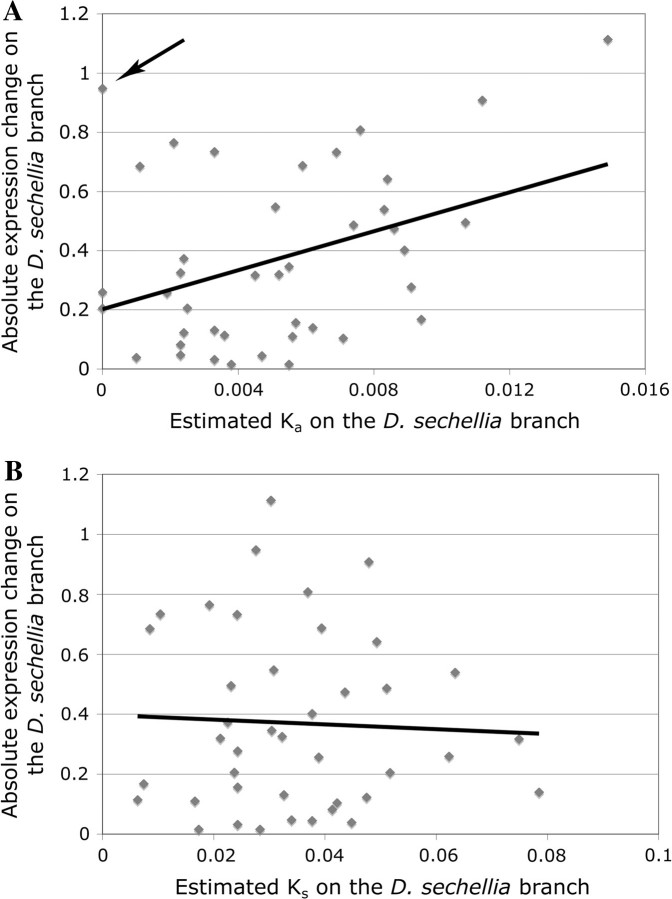
OR expression divergence also correlates with protein sequence evolution. McBride (2007) has estimated lineage-specific nucleotide and amino acid substitution rates for the OR genes in D. sechellia and D. simulans. We compared the estimated amount of change in gene expression on the D. sechellia branch with the rate of amino acid replacement (Ka) on the same branch. The 2 parameters are positively correlated (r = 0.38, P = 0.0071) (fig. 3A). In contrast, we see no correlation between the rate of silent nucleotide substitutions (Ks) and the amount of gene expression change on the D. sechellia branch (r = −0.04, P = 0.598) (fig. 3B). These observations suggest that rapid evolution of OR expression in D. sechellia may have been driven by positive selection, although relaxed purifying selection (e.g., reduced constraint on amino acid composition) is also a plausible explanation. In this analysis as well, Or22a is highly unusual. Expression of this gene has changed dramatically in D. sechellia, even though it did not undergo any changes in protein sequence (arrow in fig. 3A).
FIG. 3.—
Comparison of gene expression change on the Drosophila sechellia branch with the rate of amino acid replacements (A) and silent nucleotide substitutions (B) in coding gene sequences of OR genes. Black lines indicate linear regressions. Expression divergence is expressed as in figure 1. Arrow indicates Or22a.
We find that OBPs, like the OR genes (McBride 2007; Tunstall et al. 2007), evolve more rapidly in D. sechellia than in D. simulans (supplementary tables 4 and 5, Supplementary Material online). In contrast to OR genes (fig. 3), we find no significant correlation between the rate of expression divergence and amino acid replacement rate among OBP genes (r = 0.10, P = 0.296). Thus, protein sequence and expression divergence of OBPs do not appear to be under the same selective regime as they are for OR genes. Obp50a and Obp56c may be an exception as they show elevated dN/dS ratios in D. sechellia but not in D. simulans (supplementary table 5, Supplementary Material online). Interestingly, Obp50a expression is greatly increased in D. sechellia relative to the other species (see below), suggesting that this gene could be involved in sensory specialization.
Gene Expression Changes in ab2 and ab3 Sensilla
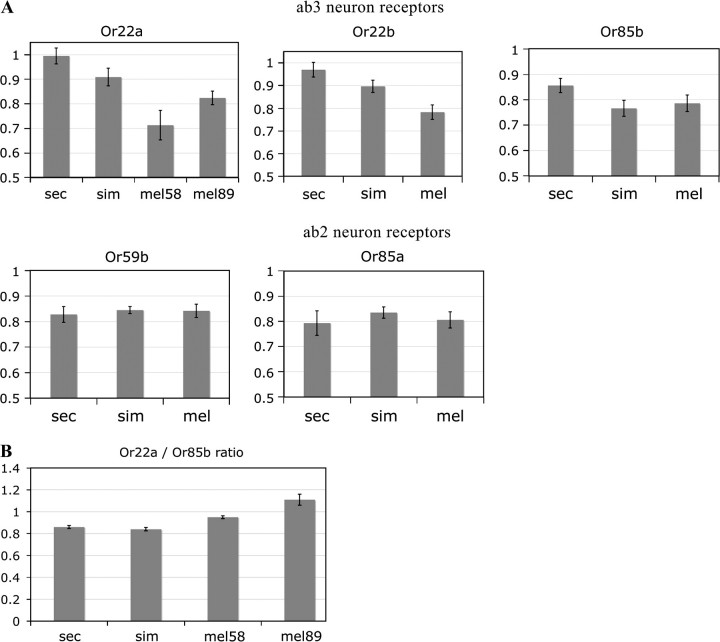
Electrophysiological analysis shows that large basiconic sensilla of the ab2 subtype are present in all species of the melanogaster subgroup except D. sechellia. The loss of ab2 receptors in this species is accompanied by an increase in the number of ab3 receptors (Stensmyr et al. 2003; Dekker et al. 2006). One might expect that the ORs expressed in ab2 neurons (Or59b and Or85a) would not be expressed in D. sechellia, whereas the ORs expressed in ab3 neurons (Or22a, Or22b, and Or85b) would be overexpressed in D. sechellia but expressed at similar levels in D. simulans and D. melanogaster. We find, however, that Or59b and Or85a do not differ significantly among the 3 species (fig. 4A). Thus, there is no strict correspondence between the levels of OR expression, as measured in the entire third antennal segment, and the prevalence of physiologically defined receptor subtypes. In the future, it will be interesting to determine where Or59b and Or85a are expressed in D. sechellia and what functions they perform in this species. One possibility is that they have become expressed in the ab1 sensilla because ab1A neurons in D. sechellia show a strong response to ethyl-3-hydroxybutyrate, a key ligand for Or85a and the ab2B neuron in which it is expressed in D. melanogaster (Hallem et al. 2004; Dekker et al. 2006). Consistent with this explanation, Dekker et al. (2006) found the response spectra of large basiconic sensilla to be considerably more variable in D. sechellia than in D. melanogaster.
FIG. 4.—
Expression of ab2 and ab3 OR genes measured by qPCR. The scale is exponential so that higher bars indicate higher expression. Error bars are SDs based on 4–8 independent biological replicates. (A) Expression levels normalized by cytoplasmic actin (Act5C). (B) Ratio of Or22a to Or85b expression.
The expression of Or22a, Or22b, and Or85b is markedly increased in D. sechellia, but Or22a and Or22b also differ strongly between D. simulans and D. melanogaster, and Or22a shows significant differences between the 2 strains of D. melanogaster, as well (fig. 4A and supplementary table 1 [Supplementary Material online]). This suggests that some evolutionary changes in gene expression in the ab3 sensilla may have occurred prior to the divergence between D. sechellia and D. simulans. Importantly, interspecific divergence in Or22a, Or22b, and Or85b expression cannot be fully explained by the increase in the number of ab3 sensilla in D. sechellia. Or22a and Or22b are upregulated in both D. sechellia and D. simulans compared with D. melanogaster, whereas Or85b is only upregulated in D. sechellia and does not differ significantly between D. simulans and D. melanogaster (fig. 4A). Thus, the ratio of Or22a to Or85b expression is higher in D. sechellia and D. simulans than in D. melanogaster (t-test, P < 10−6) (fig. 4B). This change may have important implications for chemotactic behavior, although it is not immediately clear how to reconcile it with neurophysiological observations. Or22a is highly sensitive to both methyl hexanoate and ethyl hexanoate, whereas Or85b is much more sensitive to methyl hexanoate (Hallem and Carlson 2006) so that increased expression of Or22a relative to Or85b would be expected to increase the ab3 chemoreceptor's response to ethyl hexanoate compared with methyl hexanoate. In fact, the opposite pattern is observed as the ab3 sensilla show increased sensitivity to methyl hexanoate relative to ethyl hexanoate in D. sechellia and D. simulans compared with D. melanogaster and other tested Drosophila species (Stensmyr et al. 2003).
One potential explanation may involve Or22b. This gene has undergone a large deletion resulting in a loss of function in the D. sechellia lineage (McBride 2007) and has experienced positive selection in the D. simulans lineage (Tunstall et al. 2007). Dobritsa et al. (2003) have found that Or22a is sufficient for all olfactory functions of the ab3A neuron in the Canton-S strain of D. melanogaster, whereas Or22b does not rescue or change sensitivity of that neuron and appears nonfunctional. However, both Or22a and Or22b encode functional receptors in a different wild-type strain of D. melanogaster, Oregon-R (Ray et al. 2007). Moreover, the evolutionary conservation of Or22b in D. simulans as well as more distantly related species such as Drosophila pseudoobscura suggests that Or22b is likely to perform some functional role. It is possible that changes in Or22b have contributed to the increased sensitivity to methyl hexanoate in D. sechellia and D. simulans. In any event, it appears that at least some genetic changes that contribute to the distinct sensory profile of D. sechellia may actually have occurred in the common ancestor of D. simulans and D. sechellia. The role of these changes in the adaptation of D. sechellia to Morinda fruit remains to be tested by behavioral experiments.
Evolution of other Olfactory Genes
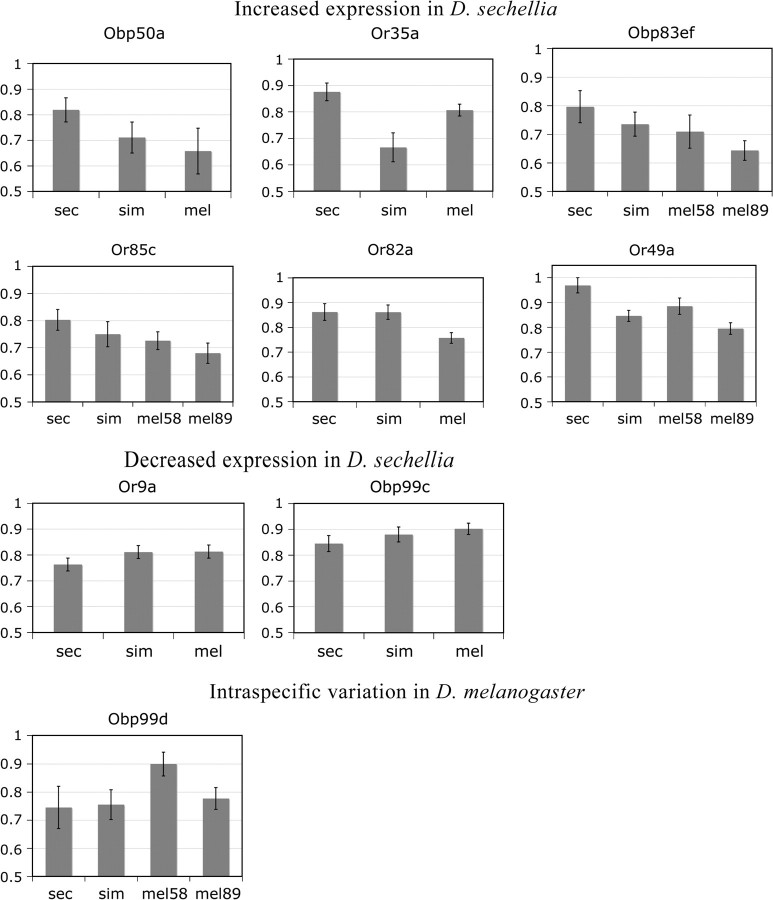
In general, more ORs and OBPs are overexpressed than underexpressed in D. sechellia. Of the genes analyzed by both microarrays and qPCR, only Or9a and Obp99c showed decreased expression in D. sechellia (fig. 5). In contrast, 6 genes (Or35a, Or49a, Or82a, Or85c, Obp50a, and Obp83ef) were upregulated in D. sechellia. All these genes also showed significant differences in expression between D. simulans and D. melanogaster. In most cases (Or85c, Or49a, Obp50a, and Obp83ef), expression level in D. simulans was intermediate between D. sechellia and D. melanogaster (fig. 5). However, Or82a did not differ between D. sechellia and D. simulans, whereas Or35a expression was strongly decreased in D. simulans compared with both D. sechellia and D. melanogaster (fig. 5). Finally, several genes (Or49a, Or85c, Obp83ef, and Obp99d) showed significant differences between the 2 strains of D. melanogaster (fig. 5). Thus, we find evidence for evolutionary changes in olfactory gene expression on all branches of the phylogeny.
FIG. 5.—
Expression of OR and OBP genes measured by qPCR. The scale is exponential so that higher bars indicate higher expression. Error bars are SDs based on 4–8 independent biological replicates. Expression levels were normalized by cytoplasmic actin.
The OR genes upregulated in D. sechellia are expressed in coeloconic (Or35a) and small basiconic (Or82a and Or49a) sensilla (Couto et al. 2005; Yao et al. 2005). Or82a is a narrowly tuned receptor that shows strong response only to geranyl acetate among the tested odorants (Hallem and Carlson 2006). Or35a is a broadly tuned receptor with a spectrum that is generally similar to Or22a and Or85b and includes medium-chain alcohols and aldehydes (e.g., hexanol, hexanal, and their unsaturated equivalents) as well as some short-chain esters (including hexyl acetate, hexenyl acetate, and hexyl butyrate). However, Or35a does not respond to medium-chain esters such as methyl hexanoate or other hexanoate or octanoate esters (Hallem and Carlson 2006). With the exception of Or22a, none of the ORs upregulated in D. sechellia show sensitivity to hexanoic or octanoic acids. This is consistent with the neurophysiological analysis that found no differences between D. melanogaster and D. sechellia in olfactory sensitivity to the hexanoic acid (Dekker et al. 2006), despite its importance in D. sechellia’s chemotaxis and oviposition behavior (Higa and Fuyama 1993; Matsuo et al. 2007). These results suggest that the adaptation of D. sechellia to its host involved changes in its behavioral response to hexanoic and octanoic acids, rather than increased ability to detect these compounds.
Four ORs expressed in trichoid sensilla have been implicated in the reception of pheromones. Or47b and Or88a are sensitive to both male and female cuticular extracts, whereas Or67d and Or65a respond specifically to the male pheromone cis-vaccenyl acetate (Kurtovic et al. 2007; van der Goes van Naters and Carlson 2007). We find that Or47b and Or67d are expressed at a slightly higher level in males than in females, whereas Or88a and Or65a show no evidence of sex-biased expression (supplementary table 1, Supplementary Material online). This observation cannot be fully explained by differences in the abundance of different trichoid sensilla. Or67d is the only OR expressed in the at1 subtype. However, Or47b, Or65a, and Or88a are all expressed in the at4 subtype (Couto et al. 2005). This suggests that Or47b may be regulated by the sex determination pathway independently of other genes expressed in the at4 sensilla and that males may have more copies of Or47b per neuron than females. Interestingly, Or47b shows by far the highest Ka/Ks ratio on the D. sechellia branch of all OR genes (1.28), whereas Or67d, Or88a, and Or65a have medium to low Ka/Ks ratios (0.07–0.19). Thus, it is possible that Or47b has evolved under sexual selection.
No Evidence for Coordinated Evolution of Functionally Similar or Physically Clustered Genes
To test whether OR genes with similar functional specificities show coordinated evolutionary changes in expression, we first estimated pairwise covariances and correlations of gene expression levels using an independent contrasts approach (Felsenstein 1985, 2005). This allowed us to examine the correlations of gene expression across species after accounting for their phylogenetic relationships. Many ORs and OBPs occur in tandem clusters in the genome (Graham and Davies 2002; Hekmat-Scafe et al. 2002). We find that physically clustered genes do not show any consistent tendency to coevolve in expression (supplementary fig. 1, Supplementary Material online). The qPCR analysis confirms this observation. For example, Obp50a differs very strongly in expression between D. sechellia and the other 2 species (supplementary table 1 and fig. 4, Supplementary Material online). In contrast, the adjacent genes Obp50b and Obp50c do not show any interspecific differences (data not shown).
There is also no evidence for coordinated changes in the expression of receptors expressed in the same olfactory organ subtype. For example, Or43b, expressed in the basiconic ab8 sensilla (Couto et al. 2005), shows a negative correlation (−0.7) with the other ab8 gene, Or9a, but a high positive correlation (0.99) with Or47b, which is expressed in the trichoid at4 sensilla (supplementary fig. 1, Supplementary Material online). This indicates that not all interspecific divergences in OR expression can be attributed to changes in the relative numbers of different sensilla subtypes. Rather, increased or decreased expression of at least some OR genes must be due to evolutionary changes either in their cis-regulatory elements or in the transcription factors that control their expression in olfactory neurons. Consistent with this model, the development of different receptors housed in the same sensilla can be controlled by different transcription factors (Ray et al. 2007).
Hallem and Carlson (2006) examined the response of 24 OR genes to each of 110 volatile compounds. Functional similarities among these ORs can be represented as pairwise Euclidean distances in a 110-dimensional space (Hallem and Carlson 2006). We tested whether these distances correlated with the phylogenetic covariances of gene expression levels estimated by the independent contrasts method (supplementary fig. 2, Supplementary Material online). No significant correlation was observed (r = −0.012, rank correlation = −0.002, P = 0.522). Thus, we find no evidence for coordinated evolution of functionally similar ORs.
Some ORs respond to a wide range of chemicals, whereas others respond to only a few (Hallem and Carlson 2006). We asked whether the more narrowly or the more broadly tuned receptors show greater evolutionary changes in expression. Breadth of tuning was quantified as the number of chemicals that produced an excitation response above 100 spikes per second (other cutoffs, ranging from 50 to 150 spikes per second, produced similar results). Gene expression divergence was quantified as the variance of mean expression levels across the 4 genotypes (D. sechellia, D. simulans, and the 2 strains of D. melanogaster). We found a positive correlation between expression divergence and the number of odorants eliciting receptor response (r = 0.46, rank correlation = 0.44, P = 0.011) (supplementary fig. 3A, Supplementary Material online). Separately, we quantified gene expression divergence as the estimated amount of change on the D. sechellia branch. Again, we found a positive correlation between the breadth of tuning and the rate of divergence (r = 0.59, rank correlation = 0.61, P = 0.0012) (supplementary fig. 3B, Supplementary Material online). However, both correlations are due to only 5 rapidly evolving ORs with the broadest functional specificity: Or67a, Or22a, Or85b, Or9a, and Or35a. Aside from these genes, there is not consistent relationship between the breadth of tuning and the rate of expression divergence.
In sum, these 4 analyses show that the patterns of gene expression divergence are not by-products of genomic location, anatomical organization, or broad functional specificity. It is likely that at least some evolutionary changes, particularly the increased expression of several olfactory genes, reflect a specific adaptation of D. sechellia to a new host plant. Behavioral experiments will be needed to test this hypothesis.
Conclusions
We have carried out a genome-wide survey of interspecific divergence in olfactory gene expression between D. sechellia, an island endemic with narrow host plant specificity, and its generalist relatives D. simulans and D. melanogaster. Our results reveal a number of general evolutionary patterns and suggest several candidate genes that may have contributed to the evolution of host plant preferences.
Evolution of OR and OBP expression was accelerated in D. sechellia compared both with the genome average in that species and with the rate of OR and OBP evolution in the other species. Positive selection may have played a role in the accelerated evolution of OR expression, although relaxed purifying selection is also a plausible explanation in some cases.
Interspecific divergence in olfactory gene expression cannot be fully explained by changes in the relative abundance of different sensilla. Expression of some ORs and OBPs has evolved independently of other genes expressed in the same sensilla.
There is no general tendency for functionally similar or physically clustered genes to coevolve in expression.
A number of OR and OBP genes are upregulated in D. sechellia compared with its generalist relatives. This striking result suggests that the evolution of host plant specialization in D. sechellia likely included gain-of-function changes in addition to the fixation of loss-of-function alleles. Genes with increased expression in D. sechellia include Or22a, which is highly sensitive to methyl hexanoate, as well as several genes that are yet to be characterized in detail.
Some of the gene expression changes that contribute to the distinct neurophysiological phenotype of D. sechellia may predate its divergence from its generalist relative D. simulans.
Supplementary Material
Supplementary tables 1–5 and figures 1–4 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was supported by National Institute of Health (NIH) grant 1R24GM065513-01 to A.K. and L.M.M., National Science Foundation (NSF) grant IOB-0518654 to A.K., NIH grant 1R01GM077618-01A1 to L.M.M., and NSF Grant MCB-0618691 and Funds from the Carolina Center for Genome Sciences to C.D.J. The qPCR equipment was provided by the University of California—Davis Clinical Nutrition Research Unit (DK35747). We would like to thank Ryan Bickel, Deniz Erezyilmaz, Lindy McBride, Carson Miller, and 3 anonymous reviewers for comments that improved the manuscript.
References
- Amlou M, Moreteau B, David JR. Genetic analysis of Drosophila sechellia specialization: oviposition behavior toward the major aliphatic acids of its host plant. Behav Genet. 1998;28:455–464. doi: 10.1023/a:1021689312582. [DOI] [PubMed] [Google Scholar]
- Bammler T, Beyer RP, Bhattacharya S, et al. (64 co-authors) Standardizing global gene expression analysis between laboratories and across platforms. Nat Methods. 2005;2:351–356. doi: 10.1038/nmeth754. [DOI] [PubMed] [Google Scholar]
- Barmina O, Gonzalo M, McIntyre L, Kopp A. Sex- and segment-specific modulation of gene expression profiles in Drosophila. Dev Biol. 2005;288:528–544. doi: 10.1016/j.ydbio.2005.09.052. [DOI] [PubMed] [Google Scholar]
- Becerra JX. Insects on plants: macroevolutionary chemical trends in host use. Science. 1997;276:253–256. doi: 10.1126/science.276.5310.253. [DOI] [PubMed] [Google Scholar]
- Becerra JX. The impact of herbivore-plant coevolution on plant community structure. Proc Natl Acad Sci USA. 2007;104:7483–7488. doi: 10.1073/pnas.0608253104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Method. 1995;57:289–300. [Google Scholar]
- Couto A, Alenius M, Dickson BJ. Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr Biol. 2005;15:1535–1547. doi: 10.1016/j.cub.2005.07.034. [DOI] [PubMed] [Google Scholar]
- Dallas PB, Gottardo NG, Firth MJ, Beesley AH, Hoffmann K, Terry PA, Freitas JR, Boag JM, Cummings AJ, Kees UR. Gene expression levels assessed by oligonucleotide microarray analysis and quantitative real-time RT-PCR—how well do they correlate? BMC Genomics. 2005;6:59. doi: 10.1186/1471-2164-6-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David JR, Allemand R, Capy P, Chakir M, Gibert P, Petavy G, Moreteau B. Comparative life histories and ecophysiology of Drosophila melanogaster and D. simulans. Genetica. 2004;120:151–163. doi: 10.1023/b:gene.0000017638.02813.5a. [DOI] [PubMed] [Google Scholar]
- de Bruyne M, Warr CG. Molecular and cellular organization of insect chemosensory neurons. Bioessays. 2005;28:23–34. doi: 10.1002/bies.20338. [DOI] [PubMed] [Google Scholar]
- Dekker T, Ibba I, Siju KP, Stensmyr MC, Hansson BS. Olfactory shifts parallel superspecialism for toxic fruit in Drosophila melanogaster sibling, D. sechellia. Curr Biol. 2006;16:101–109. doi: 10.1016/j.cub.2005.11.075. [DOI] [PubMed] [Google Scholar]
- Dobritsa AA, van der Goes van Naters W, Warr CG, Steinbrecht RA, Carlson JR. Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron. 2003;37:827–841. doi: 10.1016/s0896-6273(03)00094-1. [DOI] [PubMed] [Google Scholar]
- Dres M, Mallet J. Host races in plant-feeding insects and their importance in sympatric speciation. Philos Trans R Soc Lond B Biol Sci. 2002;357:471–492. doi: 10.1098/rstb.2002.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunipace L, Meister S, McNealy C, Amrein H. Spatially restricted expression of candidate taste receptors in the Drosophila gustatory system. Curr Biol. 2001;11:822–835. doi: 10.1016/s0960-9822(01)00258-5. [DOI] [PubMed] [Google Scholar]
- Ebbs ML, Amrein H. Taste and pheromone perception in the fruit fly Drosophila melanogaster. Pflugers Arch. 2007;454:735–747. doi: 10.1007/s00424-007-0246-y. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125:1–15. [Google Scholar]
- Felsenstein J. PHYLIP (phylogeny inference package) v3.6. Seattle (WA): Department of Genome Sciences, University of Washington; 2005. Distributed by the author. [Google Scholar]
- Fishilevich E, Vosshall LB. Genetic and functional subdivision of the Drosophila antennal lobe. Curr Biol. 2005;15:1548–1553. doi: 10.1016/j.cub.2005.07.066. [DOI] [PubMed] [Google Scholar]
- Galindo K, Smith DP. A large family of divergent Drosophila odorant-binding proteins expressed in gustatory and olfactory sensilla. Genetics. 2001;159:1059–1072. doi: 10.1093/genetics/159.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham LA, Davies PL. The odorant-binding proteins of Drosophila melanogaster: annotation and characterization of a divergent gene family. Gene. 2002;292:43–55. doi: 10.1016/s0378-1119(02)00672-8. [DOI] [PubMed] [Google Scholar]
- Ha TS, Smith DP. A pheromone receptor mediates 11-cis-vaccenyl acetate-induced responses in Drosophila. J Neurosci. 2006;26:8727–8733. doi: 10.1523/JNEUROSCI.0876-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallem EA, Carlson JR. Coding of odors by a receptor repertoire. Cell. 2006;125:143–160. doi: 10.1016/j.cell.2006.01.050. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Dahanukar A, Carlson JR. Insect odor and taste receptors. Annu Rev Entomol. 2006;51:113–135. doi: 10.1146/annurev.ento.51.051705.113646. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Ho MG, Carlson JR. The molecular basis of odor coding in the Drosophila antenna. Cell. 2004;117:965–979. doi: 10.1016/j.cell.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Hawthorne DJ, Via S. Genetic linkage of ecological specialization and reproductive isolation in pea aphids. Nature. 2001;412:904–907. doi: 10.1038/35091062. [DOI] [PubMed] [Google Scholar]
- Hekmat-Scafe DS, Scafe CR, McKinney AJ, Tanouye MA. Genome-wide analysis of the odorant-binding protein gene family in Drosophila melanogaster. Genome Res. 2002;12:1357–1369. doi: 10.1101/gr.239402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa I, Fuyama Y. Genetics of food preference in Drosophila sechellia. I. Responses to food attractants. Genetica. 1993;88:129–136. doi: 10.1007/BF02424469. [DOI] [PubMed] [Google Scholar]
- Holloway AK, Lawniczak MK, Mezey JG, Begun DJ, Jones CD. Adaptive gene expression divergence inferred from population genomics. PLoS Genet. 2007;3:2007–2013. doi: 10.1371/journal.pgen.0030187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Kurtovic A, Widmer A, Dickson BJ. A single class of olfactory neurons mediates behavioural responses to a Drosophila sex pheromone. Nature. 2007;446:542–546. doi: 10.1038/nature05672. [DOI] [PubMed] [Google Scholar]
- Lachaise D, Cariou ML, David JR, Lemeunier F, Tsacas L, Ashburner M. Historical biogeography of the Drosophila melanogaster species subgroup. Evol Biol. 1988;22:159–225. [Google Scholar]
- Legal L, Moulin B, Jallon JM. The relation between structures and toxicity of oxygenated aliphatic compounds homologous to the insecticide octanoic acid and the chemotaxis of two species of Drosophila. Pestic Biochem Physiol. 1999;65:90–101. [Google Scholar]
- Linn C, Jr, Feder JL, Nojima S, Dambroski HR, Berlocher SH, Roelofs W. Fruit odor discrimination and sympatric host race formation in Rhagoletis. Proc Natl Acad Sci USA. 2003;100:11490–11493. doi: 10.1073/pnas.1635049100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Jenssen TK, Trimarchi J, Punzo C, Cepko CL, Ohno-Machado L, Hovig E, Patrick Kuo W. Comparison of hybridization-based and sequencing-based gene expression technologies on biological replicates. BMC Genomics. 2007;8:153. doi: 10.1186/1471-2164-8-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis J, David JR. Ecological specialization in the Drosophila melanogaster species subgroup: a case study of D. sechellia. Acta Oecologica. 1986;7:215–229. [Google Scholar]
- Markow TA, O'Grady PM. Evolutionary genetics of reproductive behavior in Drosophila: connecting the dots. Annu Rev Genet. 2005;39:263–291. doi: 10.1146/annurev.genet.39.073003.112454. [DOI] [PubMed] [Google Scholar]
- Matsuo T, Sugaya S, Yasukawa J, Aigaki T, Fuyama Y. Odorant-binding proteins OBP57d and OBP57e affect taste perception and host-plant preference in Drosophila sechellia. PLoS Biol. 2007;5:e118. doi: 10.1371/journal.pbio.0050118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride CS. Rapid evolution of smell and taste receptor genes during host specialization in Drosophila sechellia. Proc Natl Acad Sci USA. 2007;104:4996–5001. doi: 10.1073/pnas.0608424104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEvey SF, Aulard S, Ralisoa-Randrianasolo O. An Australian drosophilid (Diptera) on Eucalyptus and Eugenia (Myrtaceae) flowers in Madagascar. J Aust Entomol Soc. 1989;28:53–54. [Google Scholar]
- Morey JS, Ryan JC, Van Dolah FM. Microarray validation: factors influencing correlation between oligonucleotide microarrays and real-time PCR. Biol Proced Online. 2006;8:175–193. doi: 10.1251/bpo126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P, Crespi BJ, Sandoval CP. Host-plant adaptation drives the parallel evolution of reproductive isolation. Nature. 2002;417:440–443. doi: 10.1038/417440a. [DOI] [PubMed] [Google Scholar]
- Nozawa M, Nei M. Evolutionary dynamics of olfactory receptor genes in Drosophila species. Proc Natl Acad Sci USA. 2007;104:7122–7127. doi: 10.1073/pnas.0702133104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possidente B, Mustafa M, Collins L. Quantitative genetic variation for oviposition preference with respect to phenylthiocarbamide in Drosophila melanogaster. Behav Genet. 1999;29:193–198. doi: 10.1023/a:1021696019426. [DOI] [PubMed] [Google Scholar]
- R'Kha S, Capy P, David JR. Host-plant specialization in the Drosophila melanogaster species complex: a physiological, behavioral, and genetical analysis. Proc Natl Acad Sci USA. 1991;88:1835–1839. doi: 10.1073/pnas.88.5.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, van Naters WG, Shiraiwa T, Carlson JR. Mechanisms of odor receptor gene choice in Drosophila. Neuron. 2007;53:353–369. doi: 10.1016/j.neuron.2006.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renwick JA. Variable diets and changing taste in plant-insect relationships. J Chem Ecol. 2001;27:1063–1076. doi: 10.1023/a:1010381509601. [DOI] [PubMed] [Google Scholar]
- Rutzler M, Zwiebel LJ. Molecular biology of insect olfaction: recent progress and conceptual models. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2005;191:777–790. doi: 10.1007/s00359-005-0044-y. [DOI] [PubMed] [Google Scholar]
- Schluter D, Price T, Mooers AO, Ludwig D. Likelihood of ancestor states in adaptive radiation. Evolution. 1998;51:1699–1711. doi: 10.1111/j.1558-5646.1997.tb05095.x. [DOI] [PubMed] [Google Scholar]
- Scott K, Brady R, Jr, Cravchik A, Morozov P, Rzhetsky A, Zuker C, Axel R. A chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila. Cell. 2001;104:661–673. doi: 10.1016/s0092-8674(01)00263-x. [DOI] [PubMed] [Google Scholar]
- Shanbhag SR, Muller B, Steinbrecht RA. Atlas of olfactory organs of Drosophila melanogaster. I. Types, external organization, innervation, and distribution of olfactory sensilla. Insect Morphol Embryol. 1999;28:377–397. [Google Scholar]
- Smith DP. Odor and pheromone detection in Drosophila melanogaster. Pflugers Arch. 2007;454:749–758. doi: 10.1007/s00424-006-0190-2. [DOI] [PubMed] [Google Scholar]
- Stafford P, Brun M. Three methods for optimization of cross-laboratory and cross-platform microarray expression data. Nucleic Acids Res. 2007;35:e72. doi: 10.1093/nar/gkl1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stensmyr MC, Dekker T, Hansson BS. Evolution of the olfactory code in the Drosophila melanogaster subgroup. Proc Biol Sci. 2003;270:2333–2340. doi: 10.1098/rspb.2003.2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker RF. Drosophila as a focus in olfactory research: mapping of olfactory sensilla by fine structure, odor specificity, odorant receptor expression, amd central connectivity. Microsc Res Tech. 2001;55:284–296. doi: 10.1002/jemt.1178. [DOI] [PubMed] [Google Scholar]
- Takamura T, Fuyama Y. Behavior genetics of choice of oviposition sites in Drosophila melanogaster. I. Genetic variability and analysis of behavior. Behav Genet. 1980;10:105–120. doi: 10.1007/BF01067322. [DOI] [PubMed] [Google Scholar]
- Thorne N, Bray S, Amrein H. Function and expression of the Drosophila Gr genes in the perception of sweet, bitter and pheromone compounds. Chem Senses. 2005;30:i270–i272. doi: 10.1093/chemse/bjh219. [DOI] [PubMed] [Google Scholar]
- Tunstall NE, Sirey T, Newcomb RD, Warr CG. Selective pressures on Drosophila chemosensory receptor genes. J Mol Evol. 2007;64:628–636. doi: 10.1007/s00239-006-0151-6. [DOI] [PubMed] [Google Scholar]
- van Delden W, Kamping A. Genetic variation for oviposition behavior in Drosophila melanogaster. II. Oviposition preferences and differential survival. Behav Genet. 1990;20:661–673. doi: 10.1007/BF01065877. [DOI] [PubMed] [Google Scholar]
- van der Goes van Naters W, Carlson JR. Receptors and neurons for fly odors in Drosophila. Curr Biol. 2007;17:606–612. doi: 10.1016/j.cub.2007.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven K, Simonsen K, McIntyre LM. Implementing false discovery rate control: increasing your power. Oikos. 2005;108:643–647. [Google Scholar]
- Vosshall LB, Amrein H, Morozov P, Rzhetsky A, Axel R. A spatial map of the olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725–736. doi: 10.1016/s0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- Wang Y, Barbacioru C, Hyland F, Xiao W, Hunkapiller KL, Blake J, Chan F, Gonzalez C, Zhang L, Samaha RR. Large scale real-time PCR validation on gene expression measurements from two commercial long-oligonucleotide microarrays. BMC Genomics. 2006;7:59. doi: 10.1186/1471-2164-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Atkinson R, Jones DN, Smith DP. Drosophila OBP LUSH is required for activity of pheromone-sensitive neurons. Neuron. 2005;45:193–200. doi: 10.1016/j.neuron.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Yang HP, Nuzhdin SV. Fitness costs of Doc expression are insufficient to stabilize its copy number in Drosophila melanogaster. Mol Biol Evol. 2003;20:800–804. doi: 10.1093/molbev/msg087. [DOI] [PubMed] [Google Scholar]
- Yao CA, Ignell R, Carlson JR. Chemosensory coding by neurons in the coeloconic sensilla of the Drosophila antenna. J Neurosci. 2005;25:8359–8367. doi: 10.1523/JNEUROSCI.2432-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.