SYNOPSIS
Lafora progressive myoclonus epilepsy (Lafora disease; LD) is a fatal autosomal recessive neurodegenerative disorder caused by loss-of-function mutations in either the EPM2A gene, encoding the dual specificity phosphatase laforin, or the EPM2B gene, encoding the E3-ubiquitin ligase malin. Previously, we and others showed that laforin and malin form a functional complex that regulates multiple aspects of glycogen metabolism, and that the interaction between laforin and malin is enhanced by conditions activating AMP-activated protein kinase (AMPK). Here, we demonstrate that laforin is a phosphoprotein, as indicated by two-dimensional electrophoresis, and we identify Ser25 as the residue involved in this modification. We also show that Ser25 is phosphorylated both in vitro and in vivo by AMPK. Lastly, we demonstrate that this residue plays a critical role for both the phosphatase activity and the ability of laforin to interact with itself and with previously established binding partners. Our data suggest that phosphorylation of laforin-Ser25 by AMPK provides a mechanism to modulate the interaction between laforin and malin. Regulation of this complex is necessary to maintain normal glycogen metabolism. Importantly, Ser25 is mutated in some Lafora disease patients (S25P), and our results begin to elucidate the mechanism of disease in these patients.
Keywords: Laforin, AMPK, phosphorylation, alanine scanning mutagenesis, protein-protein interaction, glucan-phosphatase
INTRODUCTION
Lafora progressive myoclonus epilepsy (LD, OMIM 254780) is a fatal autosomal recessive neurodegenerative disorder characterized by the presence of progressive neurological deterioration, myoclonus and epilepsy (see [1] and [2] for review). LD initially manifests during adolescence with generalized tonic-clonic seizures, myoclonus, absences, drop attacks and visual hallucinations. As the disease proceeds, patients enter into a vegetative state and eventually die, usually within the first decade from onset of the first symptoms ([1], [3]). Mutations causing LD have been identified in two genes, EPM2A ([4], [5]) and EPM2B (NHLRC1) [6], and there is evidence for a third locus [7].
EPM2A encodes laforin, a dual specificity phosphatase with a functional carbohydrate binding domain at the N-terminus ([8], [9]). EPM2B encodes malin, an E3-ubiquitin ligase with a RING finger domain at the N-terminus and six NHL domains involved in protein-protein interactions in the C-terminal region ([6], [10], [11]). A hallmark of LD is the accumulation of insoluble glucans (i.e. carbohydrates) called Lafora bodies (LBs) ([12], [13]). LBs form in the cytoplasm of cells from most tissues. LBs, like normal glycogen, are composed of glucose residues joined by α-1,4-glycosidic linkages with branches occurring via α-1,6-glycosidic linkages (reviewed in [2]). However, the branches are less frequent in LBs compared to glycogen, making LBs water insoluble. It has been recently described that laforin directly interacts with malin, forming a laforin-malin complex, and that laforin recruits specific substrates to be ubiquitinated by malin ([10], [11], [14]). Given the reoccurring theme of glycogen and carbohydrates in LD, it is not surprising that some substrates of the laforin-malin complex are integrally involved in regulating glycogen biosynthesis. The laforin-malin complex ubiquitinates the muscle isoform of glycogen synthase (MGS) [15], the glycogen debranching enzyme (GDE/AGL) [16], and the glycogen targeting subunit of type 1 protein phosphatase (PP1) called protein targeting to glycogen (R5/PTG) ([14], [17]). While multiple groups have corroborated these findings, there is dissent concerning they are in vivo substrates of the malin-laforin complex ([18], [19]).
Despite these important genetic findings, the physiological roles of laforin and malin are only beginning to be understood and it is still unclear which cellular processes, altered by the absence of these proteins, give rise to the devastating disorder of LD. More recently, an alternative function for laforin in glycogen homeostasis has been described ([20], [21], [22]). In this case, laforin acts as a phosphatase of glycogen and it has been proposed that this function might be necessary for the maintenance of normal cellular glycogen ([20], [22], [23]).
We have recently demonstrated that the activity of the laforin-malin complex is modulated by the AMP-activated protein kinase (AMPK), but no modified residue was identified and the mechanism driving this modulation is currently unknown [14]. However, it is clear that activated AMPK increases the interaction between laforin and malin, and the increased interaction enhances the degradation of R5/PTG by the laforinmalin complex [14]. The degradation of R5/PTG inhibits the glycogenic activity of R5/PTG and down-regulates glycogen production. Cumulatively, these results highlight the importance of the laforin-malin complex in regulating glycogen biosynthesis, and these molecular events are consistent with the accumulation of the glycogen-like intracellular LBs. However, despite these findings, it is currently unclear whether the accumulation of Lafora bodies is the cause of the disease or is a secondary determinant of a primarily established metabolic alteration.
As mentioned above, laforin is a dual specificity phosphatase and as such contains a DSP domain (DSPD) at the C-terminus. Accordingly, recombinant laforin is able to dephosphorylate in vitro artificial substrates such as p-nitrophenylphosphate and 3-O-methyl fluorescein phosphate (OMFP; a more sensitive substrate of dual specificity phosphatases) ([24], [25], [26]). In addition to the capacity of laforin to dephosphorylate glycogen (see above), it has been suggested that laforin dephosphorylates proteinaceous substrates such as glycogen synthase kinase 3β (GSK3β) ([27], [28]), although this is a controversial issue ([20], [21]).
In this work, we present evidence that laforin is a phosphoprotein that is phosphorylated at residue Ser25 by AMPK. In addition, we demonstrate that this residue plays critical roles in laforin function, affecting phosphatase activity and the ability of laforin to interact with itself and with previously established binding partners. Since Ser25 is mutated in some Lafora disease patients (S25P), our results begin to elucidate the mechanism of disease in this population of patients.
EXPERIMENTAL
Cell models, culture conditions and genetic methods
Escherichia coli DH5α was used as the host strain for plasmid constructions. E. coli BL21 (RIL) was used for protein production. They were grown in LB (1% peptone,0.5% yeast extract, 1% NaCl, pH 7.5) medium supplemented with 50 mg/L ampicillin. Yeast strains used in this work were FY250 (MATα his3Δ200 leu2Δ1 trp1Δ63 ura3-52) and CTY10-5d (MATa ade2 his3 leu2 trp1 gal4 gal80 URA3::lexAop-lacZ; gift from R. Sternglanz, State University of New York, Stony Brook, USA). Yeast transformation was carried out using the lithium acetate protocol [29]. Yeast cultures were grown in synthetic complete (SC) medium lacking the corresponding supplements to maintain selection for plasmids [30].
Human embryonic kidney (HEK293) cells, hamster ovary (CHO) cells and control and double AMPKα1α2 KO mouse embryonic fibroblasts (MEFs, kindly provided by Dr. Benoit Viollet, Institut Cochin, Université Paris Descartes, Paris, France) were grown in DMEM (Lonza) supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mM glutamine, 10% inactivated fetal bovine serum (FBS, GIBCO). 1.5×106 cells were plated onto 60mm-diameter culture dishes the day before transfection. Cells were transfected with 1 μg of each plasmid using Lipofectamine 2000 (Invitrogen). In [32P]-orthophosphate labelling experiments, we grew cells to subconfluency, transfected the cells and twenty four hours later the cells were washed with phosphate-free DMEM. Then, cells were then incubated in DMEM-free media for 0.5 hours, 0.25mCi/ml [32P]-orthophosphate and 10μM MG132 were added, the cells were incubated for 5 hours, and denatured immunoprecipitations were preformed as previously described [11].
Plasmids
Plasmids pEG202-laforin S25A, T45A, T83A, S98A, T111A, T133A, T136A, T142A, T143A, S158A, S168A, T177A, T187A, T194I, S202/3A, T213A, T216A, T233A, S237/T238A, S273/T274A, S326A, S327A and S330A, were obtained by site directed mutagenesis using plasmid pEG202-laforin [14] as template, the Quick Change kit (Stratagene) and the corresponding mutagenic oligonucleotides. These mutagenic nucleotides contained the corresponding 21 nucleotides upstream the codon to be mutagenized and the downstream 21 nucleotides (45 nucleotides in total), based on the human laforin cDNA. Nucleotides in the mutated codon were replaced by the appropriated nucleotides to code for Ala (only in the case of T194I, nucleotides were changed to code for Ile). Similarly we constructed plasmids pEG202-laforin S25D, S168D, T187D and T194D. In this case nucleotides in the mutated codon were changed to code for Asp. The sequences of all these mutagenic oligonucleotides are available upon request. All mutants were sequenced to ensure that additional mutations were not introduced during the mutagenesis procedure. Plasmids pACT2-laforin WT, S25A, S25D and T194I were obtained by digesting the corresponding pEG202-laforin plasmids with BamHI and SalI and cloning the corresponding fragment into pACT2, digested with the same set of restriction enzymes.
Plasmids pCMVmyc-laforin WT, S25A, S25D and T194I were obtained by digesting the corresponding pACT2-laforin plasmids with SfiI and BglII and subcloning the corresponding fragment into pCMVmyc (BD Biosciences), digested with the same set of restriction enzymes. Plasmids pWSGST-laforin WT, S25A and S25D were obtained by digesting the corresponding pEG202-laforin plasmids with BamHI/SalI and subcloning the fragments into the pWSGST vector [31].
Other plasmids used in this study were: pACT2-malin, pACT2-R5/PTG, pACT2-AMPKα2 [14]. Plasmid pGEX4T1-VHR was a generous gift of Dr. Rafael Pulido (Centro de Investigacion Principe Felipe, Valencia, Spain). Plasmid pEGFP-N1 was from BD-Biosciences. pEGB2 AMPKα1 wild type, -kinase dead (KD, AMPKα1 D157A), and -constitutively active (CA, AMPKα1 residues 1-312) were kind gifts from Dr. Reuben Shaw (Salk Institute, San Diego, USA) [32].
Yeast two hybrid analyses
Yeast CTY10.5d strain was co-transformed with combinations of pACT2-malin, pACT2-R5/PTG and pACT2-AMPKα2 and different pEG202-laforin mutant plasmids. Transformants were grown in selective SC medium (4% glucose) and shifted to low glucose conditions (0.05% glucose) for 3 hours when indicated. Then, β-galactosidase activity was assayed in permeabilized cells and expressed in Miller Units as in [33].
Expression of recombinant proteins
E.coli transformants harboring the pGEX4T1-VHR plasmid were grown in 500 ml of LB/ampicillin. Transformants were grown at 37°C until the absorbance at 600 nm reached a value of around 0.3. IPTG was then added to a final concentration of 0.1 mM, and cultures were maintained overnight at 25 °C. Cells were harvested and resuspended in 20 ml of sonication buffer [50 mM HEPES-NaOH pH 7.0, 150 mM NaCl, 10% glycerol, 0.1% Triton X-100, 2 mM DTT, 2 mM PMSF and complete protease inhibitor cocktail (Roche)]. Cells were disrupted by sonication and the fusion proteins purified by passing the extracts through columns containing 1 ml bed volume of glutathionesepharose (GE Healthcare). GST-fusion proteins were eluted from the column with 50 mM glutathione. Samples were stored at -80°C. FY250 yeast cells were transformed with plasmids pWSGST-laforin WT, S25A and S25D. Yeast extracts were prepared as described in [34]. Extraction buffer was 50 mM Tris/HCl (pH 7.5), 150 mM NaCl, 0.1% Triton X-100, 1 mM dithiothreitol and 10% glycerol, and contained 2 mM PMSF and a complete protease inhibitor cocktail (Roche). GST-fusion proteins were purified and eluted as described above.
Immunodetection
HEK293 cells were transfected with the corresponding plasmids. Twenty-four hours after transfection, cells were scraped on ice in lysis buffer [10 mM TrisHCl pH 8; 150 mM NaCl, 15 mM EDTA; 0.6 M sucrose, 0.5% nonidet P-40 (NP-40), complete protease inhibitor cocktail (Roche), 1 mM PMSF, 50 mM NaF and 5 mM Na2P2O7]. Cells were lysed by repeated passage through a 25-gauge needle. Twenty-five μg of total protein from the soluble fraction of cell lysates were analyzed by SDS-PAGE and western blotting using appropriate antibodies: anti-myc, anti-tubulin (Sigma) and anti-GFP (Immunokontact). Yeast extracts were prepared as described previously [34]; samples (15 μg) were separated by SDS-PAGE and analyzed by western blotting using the corresponding anti-LexA (Santa Cruz Biotechnology), anti-HA (Sigma) and anti-GST (GE Healthcare) antibodies.
Non-denaturing gel electrophoresis
Cell extracts were freshly prepared in SDS- and DTT-free loading buffer (125 mM Tris-HCl, 20% Glycerol), samples were not heated, and then analysed by regular SDS-PAGE and western blotting.
2D-electrophoresis
Mammalian HEK293 cells were transfected with pCMVmyc-laforin (wild type, S25D and T194I) plasmids and 24 hours after transfection, extracts were prepared in urea lysis buffer (9.7 M urea, 4% CHAPS, 20 mM DTT). Myc-laforin derivatives were analyzed by two-dimensional electrophoresis (2DE) using an IPGphor (Amersham Bioscience) instrument. For the first dimension (IEF), 50 μg of total protein (in 100 μl of 9.7 M urea, 4% CHAPS, 20 mM DTT and 0.5 % IPG buffer) were loaded on a 7 cm IPG strip (pH range 4-7) using the following focusing conditions: 500 V for 30 min, 1000 V for 30 min and 5000 V for 80 min. Electrophoretic separation (second dimension) was performed using SDS-PAGE. When indicated, extracts were also prepared in λ-phosphatase buffer containing 2 mM MnCl2. Fifty micrograms of crude extract was treated at 30°C for 30 min with 50 Units of λ-phosphatase (New England Biolabs). Reactions were stopped by adding two volumes of urea lysis buffer and analyzed by 2-D electrophoresis as above. The same protocol was used to analyze mouse embryonic fibroblasts transfected with pCMVmyc-laforin.
In vitro kinase assays
Kinase assays were performed with 5.0 ng of recombinant AMPK (Sigma) incubated with 100 ng of recombinant non-tagged human laforin (Hs-laforin-NT) in kinase buffer (25 mM MOPS, pH 7.2, 12.5 mM glycerol, 25 mM MgCl2, 5 mM EDTA, 2 mM EGTA, and 0.25 mM DTT) along with 0.5 mM AMP and 1 μCi of [γ-32P]ATP (Perkin Elmer). After 15 minutes at 30°C, SDS-PAGE buffer was added to each sample at 4°C to stop the reaction. Samples were separated by SDS-PAGE, followed by transfer to PVDF, and autoradiography.
Identification of phosphorylation sites
In vitro kinase assays were performed as above in the absence of [γ-32P]ATP. After purification of Hs-laforin by affinity chromatography, SDS-PAGE, and Coomassie blue staining, bands corresponding to Hs-laforin were excised from the gel. The gel slices were reduced with dithiothreitol and alkylated with iodoacetamide. Following in-gel trypsin and chymotrypsin digestion, the peptides were analyzed by automated microcaplillary liquid chromatography-tandem mass spectrometry. Fused-silica capillaries (100 μm i.d.) were pulled using a P-2000 CO2 laser puller (Sutter Instruments, Novato, CA) to a 5 μm i.d. tip and packed with 10 cm of 5 μm Magic C18 material (Agilent, Santa Clara, CA) using a pressure bomb. This column was then placed in-line with a Dionex 3000 HPLC equipped with an autosampler. The column was equilibrated in buffer A, and the peptide mixture was loaded onto the column using the autosampler. The HPLC separation at a flow rate of 300 nl/min was provided by a gradient between Buffer A and Buffer B (98% acetonitrile, 0.1% formic acid). The HPLC gradient was held constant at 100% buffer A for 5 min after peptide loading followed by a 30-min gradient from 5% buffer B to 40% buffer B. Then, the gradient was switched from 40% to 80% buffer B over 5 min and held constant for 3 min. Finally, the gradient was changed from 80% buffer B to 100% buffer A over 1 min, and then held constant at 100% buffer A for 15 more minutes. The application of a 1.8 kV distal voltage electrosprayed the eluted peptides directly into a Thermo LTQ ion trap mass spectrometer equipped with a custom nanoLC electrospray ionization source. Full masses (MS) spectra were recorded on the peptides over a 400-2000 m/z range, followed by five tandem mass (MS/MS) events sequentially generated in a datadependent manner on the first, second, third, fourth and fifth most intense ions selected from the full MS spectrum (at 35% collision energy). Mass spectrometer scan functions and HPLC solvent gradients were controlled by the Xcalibur data system (ThermoFinnigan, San Jose, CA). MS/MS spectra were extracted from the RAW file with ReAdW.exe (http://sourceforge.net/projects/sashimi). The resulting mzXML file contains all the data for all MS/MS spectra and can be read by the subsequent analysis software. The MS/MS data was searched with Inspect [35] against a database containing laforin in addition to an E.coli database plus common contaminants, with modifications: +16 on Methionine, +57 on Cysteine, +80 on Serine, Threonine, and Tyrosine. Only peptides with at least a p-value of 0.01 were analyzed further. Phosphorylated peptides were manually verified.
Carbohydrate binding assay
Crude extracts from yeast cell cultures (400 ml) expressing GST-laforin (WT, S25A and S25D) were obtained as above. Extracts (5 ml) were passed through columns containing 150 μl bed volume of amylose-sepharose (GE Healthcare). Flow-through was collected and columns were washed twice with 5 ml of extraction buffer, collecting the second wash volume. Bound proteins were eluted with 50 μl of SDS-PAGE sample buffer. Collected samples (30 μl) were analysed by SDS-PAGE and western blotting using anti-GST (GE Healthcare) antibodies.
Phosphatase activity
In vitro phosphatase assays were performed using 4 μg of GST-fusion proteins diluted in phosphatase buffer (0.1 M Tris-HCl, 40 mM NaCl, 10 mM DTT), in the presence of 0.5 mM 3-O-methyl fluorescein phosphate (OMFP, Sigma). Reactions were carried out at 37° C in a final volume of 200 μl in 96-well ELISA plates. Phosphatase activity was measured for 3 hours as absorbance at 490 nm. One Unit of phosphatase activity is defined as the one unit of change in the absorbance at 490 nm per min of assay.
Molecular modeling
HHpred search [36] and InterPro domain scan [37] were used to determine the best available template for modeling the CBD and DSP of laforin. The top three hits from each were aligned with the CBD of laforin using PROfile Multiple Alignment with predicted Local Structure 3D (PROMALS) [38]. A homology model for the CBD of laforin was generated using the ESyPred3D server [39] with the PDB file 1CYG (Geobacillus stearothermophilus cyclodextrin glycosyltransferase; 29 % identity) as the template. Similarly, the PDB file 1WRM (Homo sapiens DUSP22 phosphatase; 13.2 % identity) was used to generate a homology model for the laforin DSP domain. Cyclodextrin moieties were docked after superimposition of laforin CBD structural model with the Aspergillus niger glucoamylase CBD structure (PDB: 1AC0). Images were generated with PyMOL (http://www.pymol.org).
Statistical analyses
Values are given as means ± SD of at least three independent experiments. When indicated, differences between groups were analyzed by two-tailed student’s t-tests. The significance has been considered at * p<0.05 and ** p<0.01, as indicated in each case.
RESULTS
Mutation of Ser/Thr residues to Ala influences the interaction of laforin with malin and with other established binding partners
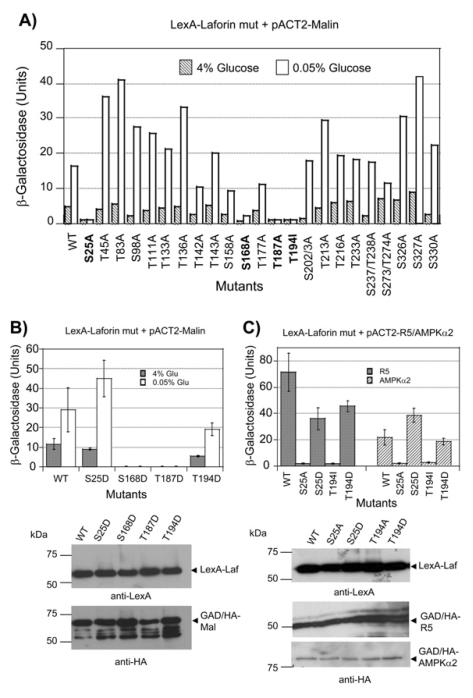
We and others have described that laforin interacts physically with malin ([10], [11], [14]). We have also recently described that the interaction between laforin and malin is enhanced by conditions that activate AMP-activated protein kinase (AMPK, a sensor of energy status), such as low glucose conditions. In addition, we demonstrated that laforin but not malin directly interacted with different subunits of the AMPK complex [14]. To test whether phosphorylation of laforin is responsible for the enhanced interaction between laforin and malin, we substituted each of the 26 Ser/Thr residues present in laforin with Ala residues and analyzed by yeast two-hybrid the strength of the laforinmalin interaction. As reported previously, the two-hybrid interaction between wild type laforin and malin increased under low glucose conditions (Fig. 1A), possibly because of the action of activated endogenous SNF1 complex (the yeast AMPK orthologue) [14]. We then analyzed the interaction between mutated laforin and malin and noticed that some Ser/Thr to Ala substitutions did not greatly affect the glucose regulated interaction between laforin and malin, whereas others improved the interaction between these two proteins under conditions of low glucose (i.e. T45A, T83A, T136A, S327A, etc.). However, only four mutations abolished the interaction between laforin and malin, in both high and low glucose conditions, namely S25A, S168A, T187A and T194I (in the latter case, the Thr residue was substituted by Ile to mimic a mutation recovered from Lafora patients [40]). Western blot analyses confirmed that all the mutated forms were expressed at similar levels (not shown).
Fig. 1.
Alanine scanning mutagenesis of laforin and its effect on interaction with malin. A) pEG202-laforin plasmids containing the described mutations were introduced into CTY10-5d yeast strain in combination with plasmid pACT2-malin. Transformants were grown in 4% glucose containing medium until they reach exponential phase. Then, an aliquot of the cultures was shifted to low glucose conditions (0.05% glucose) for 3 hours. Cells from both growth conditions were used to determine the β-galactosidase activity, as an indication of two-hybrid interaction (see Experimental section). Results are the mean values of four to six different transformants, with a standard deviation of less than 15% in each case. B) The two-hybrid interaction between the indicated plasmids was measured as in (A); bars indicate standard deviation. Yeast crude extract (15 μg) from the different transformants were analyzed by western blotting using anti-LexA and anti-HA antibodies. Molecular size standards are in the left. C) The twohybrid interaction between the corresponding protein fusions was measured as above in cells growing in 4% glucose; bars indicate standard deviation. Yeast crude extract (15 μg) from the different transformants were analyzed by western blotting using anti-LexA and anti-HA antibodies.
We reasoned that if the phosphorylation of one of these residues was responsible for the enhanced interaction between laforin and malin under low glucose conditions, then, substitution of this residue to the phosphomimetic residue Asp should recover the interaction between laforin and malin. Thus, we constructed the corresponding Ser/Thr to Asp mutants and observed that in the case of the S168D and T187D mutants the interaction with malin was still impaired, suggesting that these two residues may play important roles in the conformation of the protein and changes to either Ala or Asp could severely affect the overall structure of the protein. Alternatively, in the case of S25D and T194D the interaction with malin was recovered, although this recovery was lower in the T194I mutant (Fig. 1B; western blot analyses confirmed that the mutated forms were expressed at similar levels as wild type). In the case of S25D mutant, the interaction with malin under low glucose conditions was larger than in wild type, suggesting that a negative charge on this residue allowed a better regulation of the interaction between laforin and malin by glucose.
In addition to interacting with malin, we have also described that laforin interacts with the PP1 regulatory subunit R5/PTG and with subunits of the AMPK complex [14]. Therefore, we investigated if mutations in Ser25 and Thr194 affected the interaction with these partners. We observed that the S25A and T194I mutants did not interact with either R5/PTG or AMPKα2, whereas the S25D and T194D forms allowed the interaction with these components (Fig. 1C; western blot analyses confirmed that the mutated forms were expressed at similar levels as wild type).
Laforin is a phosphoprotein and is phosphorylated on residue Ser25
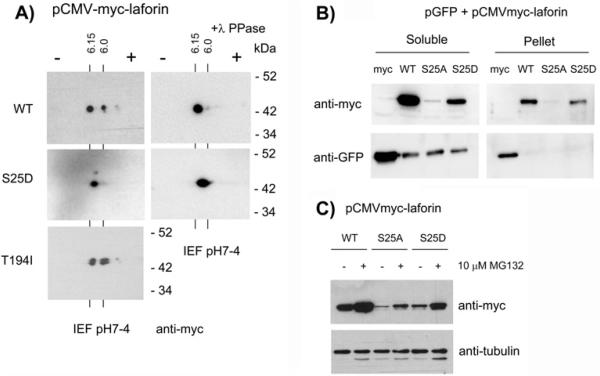
The results described above indicated that the substitution of Ser/Thr residues (S25, T194) by non-phosphorylatable residues (Ala and Ile, respectively), prevented the interaction of laforin with different binding partners. These results suggested that either the presence of a polar amino acid (Ser/Thr) or the phosphorylation of these residues could be essential for the interaction of laforin with its corresponding partners. To discern between the two, we expressed the mutated forms in mammalian cells (HEK293) and analyzed their in vivo phosphorylation status by two-dimensional electrophoresis (2DE). As shown in Fig. 2A (upper left panel), wild type laforin migrates as two major species on the 2DE gels, indicative of phosphoprotein species. Furthermore, treatment of the sample with λ-phosphatase eliminated the spot that moved faster towards the positive pole (right spot), suggesting that this form is due to phosphorylation (Fig. 2A, upper right panel). The isoelectric point of the left spot was in agreement with the calculated pI for the myc-laforin fusion protein (pI: 6.15) and the isoelectric point of the right spot (pI: 6.00) was compatible with the acquisition of one phosphate group.
Fig. 2.
Laforin is phosphorylated at residue Ser25. A) Cell extracts from HEK293 cells transfected with plasmids pCMVmyc-laforin (WT, S25D and T194I) were analyzed by 2D-electrophoresis and western blotting using anti-myc monoclonal antibodies. Cell extracts were also treated with λ-phosphatase as described in Experimental section. The calculated pI for each spot is indicated. Molecular size standards are indicated in the right. B) Cell extracts from HEK293 cells co-transfected with plasmids pCMVmyclaforin (WT, S25A and S25D) and pEGFP-N1 (expressing GFP), were prepared for immunodetection as described in Experimental section. In this case, lysis buffer contained 0.5% TritonX100 instead of 0.5% nonidet P40. Extracts were centrifuged at 13,000 rpm for 10 min at 4°C to separate soluble from pellet fractions. These fractions were treated with SDS-PAGE sample buffer and analyzed by western blotting using anti-myc and anti-GFP antibodies. C) HEK293 cells co-transfected with plasmids pCMVmyc-laforin (WT, S25A and S25D) were treated or not with 10 μM MG132 (a proteasome inhibitor) for 8 hours. Then, crude extracts were obtained and analyzed by western blotting using anti-myc and anti-tubulin (loading control).
We attempted to analyze the phosphorylation pattern of a laforin-S25A mutant but the steady state levels of this protein were greatly diminished (Fig. 2B). Cotransfection experiments using a plasmid expressing GFP under the same promoter as the laforin constructs indicated that the low levels of the laforin-S25A protein were not due to different transfection efficiency, since similar levels of GFP were observed in cells co-transfected with laforin or laforin-S25A constructs. In addition, laforin-S25A was not in the pellet after a high speed spin, indicating that it did not form insoluble aggregates (Fig. 2B). These results indicate that the stability of the laforin-S25A form could be greatly reduced, likely due to enhanced proteolytic turnover. In agreement with this proposal we observed an accumulation of laforin-S25A protein when the cells were treated with the proteasome inhibitor MG132 (Fig. 2C). Therefore, although we cannot discard the possibility that the S25A mutation could affect the stability of the corresponding mRNA when expressed in mammalian cells, our results suggest that the laforin-S25A form is very unstable and gets rapidly degraded by the cellular proteolytic system. The reduced levels of laforin-S25A were only observed when the constructs were expressed in mammalian cells, but not when they were expressed in yeast (see Fig. 1).
Since laforin-S25D was expressed in mammalian cells (although at lower levels than wild type), we used this form in the 2DE experiments. We observed that laforin-S25D migrated as a single spot and treatment of the extracts with λ-phosphatase did not produce any shift in its mobility, indicating that this form was not phosphorylated in vivo (Fig. 2A, middle panel). Due to the negative charge of the new Asp residue, this form migrated between pI 6.15 and 6.0. Conversely, laforin-T194I showed the presence of two major spots that migrated at similar isoelectric points as those that we observed for wild type laforin (Fig. 2A, lower panel) (the observed doublet in each spot could be due to partial degradation of the sample). Cumulatively, these results indicate that laforin is a phosphoprotein in vivo and that Ser25 is the residue that is phosphorylated. The lack of interaction between laforin-S25A with malin, R5/PTG and AMPK is likely due to either the absence of Ser25 phosphorylation or most likely to an altered conformation of laforin-S25A mutant. On the contrary, the decreased interaction of laforin-T194I with these proteins is likely due to a conformational change due to the presence of a non-polar Ile194 residue and not to a lack of phosphorylation of this mutant.
Mutation of Ser25 to either Ala or Asp maintains binding to carbohydrates but reduces phosphatase activity
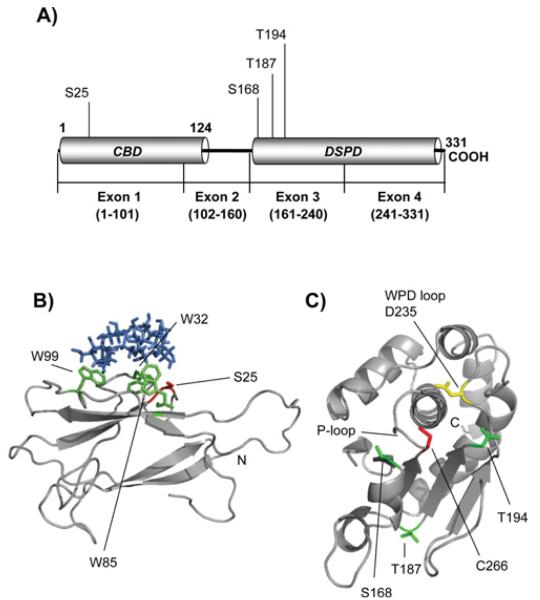
Since no crystal structure of laforin is yet available and in order to map the position of the Ser25 residue in the laforin molecule, we modelled the structure of the carbohydrate binding domain of laforin (residues 1-116) (Fig. 3A and 3B). We first searched the published CBD crystal structures to identify the best template for homology modelling. We identified the Geobacillus stearothermophilus cyclodextrin glycosyltransferase (PDB: 1CYG) as having the maximum identity with laforin (29%) and generated a homology model using 1CYG as the template. The homology model suggests that the laforin CBD folds into the characteristic two β-sheets fold, each consisting of three to six antiparallel β-strands and with the N- and C-termini pointing towards opposite ends of the longest axis of the molecule (Fig. 3A and 3B) [41]. Conserved aromatic residues involved in carbohydrate binding were readily observable in the laforin CBD structure: W32, W85 and W99. In the model, these residues form a compact, rigid and surfaceexposed hydrophobic site containing inter-ring spacing appropriate for binding to α(1,4)-linked glucoses, as is the case for glycoamylase [41]. These results are similar to those previously reported when the laforin CBD was modelled based on the crystal structure of Bacillus circulans cyclodextrin glycosyltransferase (PDB: 2DIJ, [9]). In both cases, Ser25 is located at the beginning of a hairpin loop, close to the carbohydrate binding area (Fig. 3B).
Fig. 3.
Structure models of laforin CBD and DSPD domains. A) Diagram of the position of the CBD and DSPD domain in the primary structure of human laforin. The positions of residues described in this work are also indicated. B) Laforin CBD domain (residues 1-116) was in silico modelled, as described in Experimental section, using the crystal structure of Geobacillus stearothermophilus cyclodextrin glycosyltransferase (PDB: 1CYG) as a template. The positions of the Trp residues involved in carbohydrate binding (W32, W85 and W99) are indicated in green and the position of residue Ser25 is marked in red. β-cyclodextrin is coloured in blue. The N-terminus is also indicated. C) The laforin dual specificity phosphatase domain (residues 157-326) was in silico modelled, as described in Experimental procedures, using the crystal structure of human DUSP22 (PDB: 1WRM) as a template. The position of the characteristic P-loop, containing the catalytic C266 residue (in red), and the WPD-loop, containing the conserved D235 (in yellow), is indicated. The positions of other residues described in this study (S168, T187 and T194; in green) and the C-terminus are also indicated.
We also modelled the laforin dual specificity phosphatase domain (DSPD, residues 157 to 326) by comparing it with that of the human DUSP22 phosphatase (PDB: 1WRM; [42]) (Fig. 3C). Again, this in silico approach suggested that laforin DSPD folded into the characteristic fold consisting of four to five β-sheets surrounded by α-helices [43]. In this structure, the characteristic P-loop (phosphate binding-loop), containing the catalytic C266 residue and the WPD-loop (TPD235 in laforin) with the conserved Asp residue (D235) pointing towards the catalytic grove, were clearly observed (Fig. 3C). In this structure we were also able to map the position of residues S168, T187 and T194 whose mutation to Ala (Ile in the case of T194) resulted in impairment of interaction of laforin with different binding partners (see above).
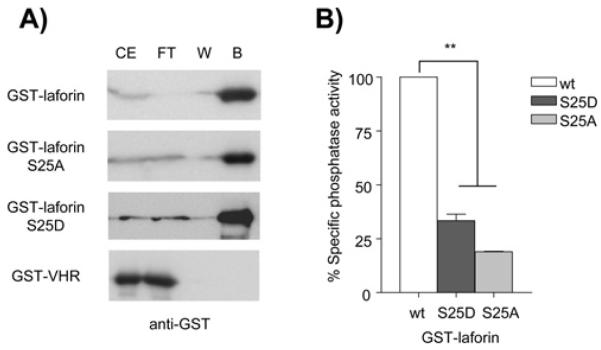
Residue Ser25 is a site mutated in some Lafora disease patients. Since it is close to the carbohydrate binding area, we analyzed whether modifications in this residue could affect carbohydrate binding. With this aim we expressed GST-laforin wild type, S25A and S25D in yeast, as we were unable to produce the laforin-S25A mutant in bacteria. In addition, we expressed a human GST-VHR fusion protein as control. VHR (vaccinia H1-related) is a dual specificity protein phosphatase that lacks a carbohydrate binding domain [44]. As observed in Fig. 4A, GST-laforin wild type, S25A and S25D were all able to bind to amylose-sepharose. On the contrary, GST-VHR did not bind the amylose-sepharose, as expected. These results indicate that changing Ser25 to a small non-polar (Ala) or to an acidic (Asp) amino acid does not affect the capacity of laforin to interact with carbohydrates.
Fig. 4.
Effect of laforin-S25A/D mutations on carbohydrate binding capacity and phosphatase activity. A) Laforin-S25A and S25D mutations do not affect carbohydrate binding capacity of laforin. Yeast crude extracts (CE) containing GST-laforin (WT, S25A and S25D) and GST-VHR fusion proteins were passed through columns containing amylose-sepharose. Flow-through (FT) was collected and columns were washed twice with 5 ml of extraction buffer, collecting the last wash (W). Bound proteins (B) were eluted with 50 μl of SDS-PAGE sample buffer. Collected samples were analysed by SDS-PAGE and western blotting using anti-GST antibodies. A similar volume (30 μl) of CE, FT and the corresponding W was analyzed. B) Effect of S25A and S25D mutations on phosphatase activity of laforin. Phosphatase activity was measured on GST-laforin WT, S25A and S25D proteins purified from yeast. Phosphatase activity was referred to the activity found in the corresponding wild type laforin. Statistical significance was considered at ** p<0.01 (n:3).
We next measured the in vitro phosphatase activity of the laforin mutants using 3-O-methyl fluorescein phosphate (OMFP) as substrate (Fig. 4B). We observed that both mutated laforins showed a dramatic decrease in phosphatase activity, that was more severe in the case of laforin-S25A (20% respect to wild type). These results indicate that the substitution of Ser25 by either a phosphomimetic Asp residue or a nonpolar residue (Ala) appears to impair the phosphatase activity of laforin.
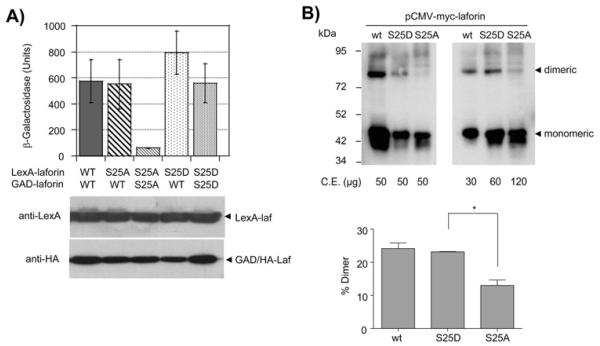
Mutations in Ser25 affect dimerization of laforin
It has been described in the literature that laforin forms dimers and that mutations in the CBD of laforin prevent the dimerization of the protein [45]. In order to determine if dimerization is affected by the Ser25A/D mutants, we measured by two-hybrid analysis the capacity of the different forms to interact with each other. As shown in Fig. 5A, wild type laforin strongly interacted with itself, in agreement with the dimerization reports [45]. However, the laforin S25A mutant was unable to interact with itself, although it could form a stable interaction with wild type laforin. Alternatively, laforin S25D was able to interact with itself and also with wild type laforin. Western blot analyses confirmed that the mutated forms were expressed at similar levels as wild type.
Fig. 5.
Effect of laforin-S25A/D mutations on dimerization capability. A) CTY10.5d yeast cells transformed with the indicated plasmids were used to measure the twohybrid interaction between the indicated proteins, as in legend of Fig. 1. Results are the mean values of four to six different transformants; bars indicate standard deviation. Yeast crude extract (15 μg) from the different transformants were analyzed by western blotting using anti-LexA and anti-HA antibodies. B) Crude extracts from hamster ovary (CHO) cells transfected with plasmids pCMVmyc-laforin (WT, S25D and S25A) were analyzed by non-denaturing electrophoresis and western blotting using anti-myc monoclonal antibodies. Different amounts of the extracts were loaded as indicated. CE, crude extracts. Lower panel shows the quantification of the proportion of dimers in cells expressing the different laforin forms. Values are means of three independent experiments (bars indicate standard deviation, *p<0.05).
Next, we sought to extend our dimerization studies from yeast into mammalian cells. Since the expression of the S25A form in HEK293 cells was very poor (see above), we tested different mammalian cell lines for their capacity to express the S25A mutants and found that hamster ovary (CHO) cells are able to express laforin S25A, although at lower levels than wild type. Extracts from these cells were subjected to nondenaturing electrophoresis (in the absence of SDS and DTT) in order to detect the presence of dimeric forms of laforin. As shown in Fig. 5B, either by loading equal amount of total protein (left panel) or when we adjusted the amount of laforin in the lanes (right panel), we observed the formation of dimeric forms in wild type and S25D forms but an impairment in the formation of these dimers in the S25A mutant (lower panel shows the quantification of the proportion of dimers in the different laforin forms). These results confirmed our previous yeast two-hybrid data (Fig. 5A) and clearly indicated that in mammalian cells, dimerization of S25A mutant is prevented.
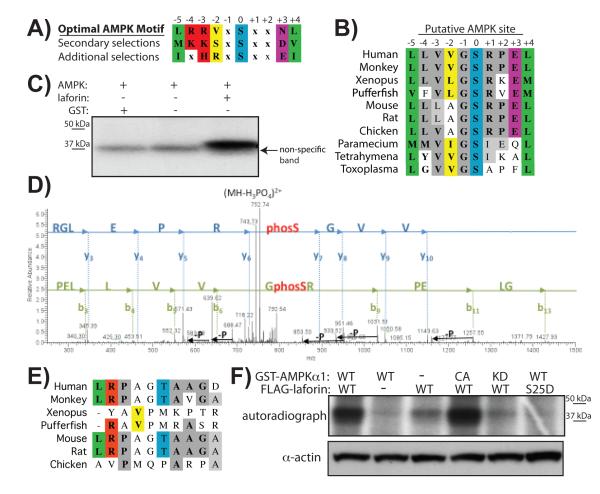
AMPK phosphorylates laforin at Ser25
With the identification of Ser25 as the phosphorylation site, we then set out to determine the kinase that phosphorylates laforin. Given that AMPK and laforin interact with each other [14], we searched the primary sequence of laforin for putative AMPK consensus motifs. It was recently determined that AMPK has preferences at six positions surrounding the phospho-acceptor site, with a consensus of L/M/I, K/R, V/S/L, x, S/T, x, x, N/E, L/M/V/I (Fig. 6A) [46]. We identified laforin Ser25 as a putative AMPK phosphorylation site (Fig. 6B). The residues surrounding laforin Ser25 contain four of the six consensus residues, although no basic residues are present at the positions -4/-3 from the phospho-acceptor. In addition, Ser25 and the surrounding residues are conserved from humans to other vertebrates and even down to protists, which also contain AMPK (Fig. 6B).
Fig. 6.
AMPK phosphorylates laforin at Ser25. A) Optimal AMPK motifs; modified from [46]. B) Evolutionary conservation of a putative AMPK site at laforin-Ser25. C) In vitro phosphorylation of recombinant non-tagged laforin by recombinant AMPK. D) MS/MS identification of the phosphorylation site Ser25 in the laforin tryptic peptide PELLVVGSRPELGR. Individual fragment ions are labelled [y-ions: blue; b-ions: green; –P denotes the neutral loss of H3PO4 (−98 Da)]. E) Evolutionary conservation of laforin-Thr45. F) HEK293 cells were transfected with FLAG-laforin and AMPKα1 wild type, -kinase dead (KD), -constitutively active (CA) forms or empty vector, as indicated. Cells were incubated in [32P]-orthophosphate and MG132 as described in Experimental section. Laforin was immunoprecipitated, proteins separated on a SDSPAGE gel, which was dried and exposed onto film. Actin levels were used as loading control of the amount of proteins in the crude extracts.
To determine if AMPK could phosphorylate laforin in vitro, we incubated recombinant AMPK with recombinant human laforin lacking any epitope tag in the presence of [γ32-P]ATP, separated the proteins by SDS-PAGE, and visualized phosphorylation via autoradiography. Non-tagged laforin was phosphorylated in an AMPK-dependent manner (Fig. 6C), confirming our previous results that indicated AMPK was able to phosphorylate a GST-laforin protein fusion in vitro [14]. To identify the laforin residue(s) phosphorylated by AMPK, we excised bands containing laforin treated with and without AMPK from a Coomassie-stained gel, digested the proteins with trypsin, and analyzed the peptides by nano LC tandem mass spectrometry. Analysis of the tryptic peptides mass spectra revealed Ser25 and Thr45 as the phosphorylation sites (Fig. 6D & Supp. Material Fig. S1). The finding that Ser25 was a major phosphorylation site was in agreement with the data presented thus far. However, we also considered the possibility of AMPK phosphorylating laforin Thr45 and examined the evolutionary conservation of Thr45 and its surrounding residues. The residues around Thr45 of human laforin contain only two of the six residues of the optimal AMPK motif, and these residues, including Thr45 itself, are not conserved in laforin even among vertebrates (Fig. 6E). Since our two-hybrid data indicate that a laforin-T45A does not reduce the interaction between laforin and malin (Fig. 1A) and our 2DE data indicates that Ser25 is the major in vivo phosphorylation site (Fig. 2A), thus, we think that it is unlikely that Thr45 is a physiologically relevant AMPK target and instead we think it is an in vitro artefact.
Next, we investigated if AMPK phosphorylates laforin in tissue culture cells. We co-transfected HEK293 cells with FLAG-laforin and either plasmids expressing AMPKα1 wild type (WT), AMPKα1 kinase dead (KD), AMPKα1 constitutively active (CA) or an empty vector. We grew the cells to sub-confluency, replaced the media with phosphate-free media containing [32P]-orthophosphate and MG132 (a proteasome inhibitor), incubated for 5 hours, and immunoprecipitated laforin using a denatured immunoprecipitation protocol [11] in order to minimize the number of non-laforin proteins that were recovered in the immunoprecipitates. We then separated the proteins in a SDS-PAGE, dried the gel, and exposed the gel to film. The autoradiograph demonstrates that laforin is phosphorylated in cells (Fig. 6F; third lane), and this phosphorylation is enhanced by the overexpression of AMPKα1 (compare the first with the third lane). In addition, phosphorylation of laforin is improved by the overexpression of a constitutively active AMPKα1 form (CA) (Fig. 6F; fourth lane), but no effect on phosphorylation of laforin is observed when overexpressing a kinase dead version of AMPK (KD) (Fig. 6F, fifth lane).
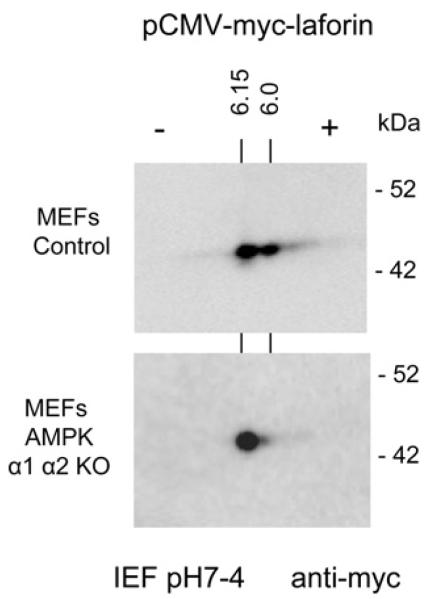
To further prove that AMPK is involved in the in vivo phosphorylation of laforin, we expressed laforin in mouse embryonic fibroblasts (MEFs) from control and double AMPKα1α2 KO mice and subjected cell extracts to 2DE. As shown in Fig. 7, in MEFs from double AMPKα1α2 KO mice laforin migrated as a single spot, indicating that in the absence of AMPK, no other protein kinases are able to phosphorylate laforin. Cumulatively, multiple lines of evidence all confirm, first, that AMPK is able to phosphorylate laforin both in vitro and in vivo and, second, that this modification occurs at residue Ser25.
Fig. 7.
Laforin is not phosphorylated in the absence of AMPK. Mouse embryonic fibroblasts (MEFs) from control and double AMPKα1α2 KO mice were transfected with plasmid pCMVmyc-laforin. Cell extracts were analyzed by 2D-electrophoresis and western blotting using anti-myc monoclonal antibodies, as in legend of Fig. 2. The calculated pI for each spot is indicated. Molecular size standards are indicated in the right.
DISCUSSION
In this work we present evidence that laforin, a dual specificity phosphatase involved in Lafora disease, is a phosphoprotein in vivo and we map the residue involved in this modification to Ser25. In addition, we identify AMPK as the protein kinase that phosphorylates laforin at Ser25, both in vitro and in vivo. These results significantly extend our previous observation that suggested AMPK phosphorylates laforin [14] by identifying the site of phosphorylation.
Laforin-Ser25 is conserved throughout evolution. It is located in the carbohydrate binding domain (CBD) of laforin, close to the surface involved in sugar binding. We show that this residue plays critical roles in the function of laforin, since substitution of Ser25 by Ala, a non-polar amino acid that cannot be phosphorylated, results in loss of interaction of laforin-S25A with multiple known binding partners, such as malin, the PP1 regulatory subunit R5/PTG, and the catalytic subunit of the AMPactivated protein kinase complex (AMPKα2). Our results also indicate that the laforin-S25A mutant exhibits decreased phosphatase activity and decreased dimerization capacity, although this change still allows binding of laforin-S25A to carbohydrates. These results may indicate that substitution of Ser25 by an Ala residue may alter the structure of some part of the protein, resulting in loss of interaction capabilities and loss of catalytic activity. The fact that wild type laforin purified from bacteria (thus, unphosphorylated) has full catalytic activity, suggests again that the cause of the altered phosphatase activity observed in the laforin-S25A mutant is a conformational change and not the absence of phosphorylation. This hypothesis of laforin-S25A undergoing a structural change wouldexplain why the resulting protein is less stable when expressed in mammalian cells. Laforin-S25A would be recognized as an aberrant protein and would be rapidly degraded by the general proteolytic system. It has been described that malin ubiquitinates laforin and targets it for degradation [11]. Our data suggest that degradation of laforin-S25A is likely independent of malin, since no interaction between laforin-S25A and malin could be detected.
Some Lafora disease patients have mutations in Ser25, mutating Ser25 to Pro (S25P). Consistent with our results, a GST-laforin S25P fusion purified from bacteria was able to bind glycogen [47], but this laforin-S25P form did not accumulate in cellculture systems [48]. We therefore suggest that the presence of a non-polar amino acid (either proline or alanine) at residue 25 greatly affects the functionality of the mutated laforin in terms of loss of interaction with binding partners, decreased phosphatase activity, and/or reduced protein stability due to structural changes. In addition, we analyzed the biochemical properties of laforin-S25D, mimicking a constitutive phosphorylated form. We found that this mutant is able to interact with the assayed laforin-binding partners and it is also able to dimerize. In addition, this form is able to bind to carbohydrates. However, its phosphatase activity is greatly reduced. Although speculative, these results suggest that phosphorylation of laforin may reduce its catalytic activity.
Our alanine scanning mutagenesis data identified several Ser/Thr residues whose change to Ala results in increased interaction between laforin and malin. However, since our results indicate that laforin is mainly phosphorylated in one residue (Ser25), we think that the increased interaction in these other mutants is likely due to structural changes and not indicative of phosphorylation events.
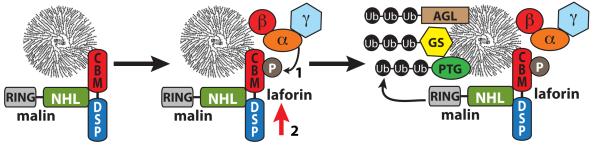
In conclusion, in this work we demonstrate that laforin is a phosphoprotein that is modified at residue Ser25 by AMPK. We have recently reported that the interaction between laforin and malin is enhanced by conditions activating AMPK [14]. Therefore, we propose that phosphorylation of Ser25 by AMPK, although it reduces the catalytic activity of laforin, it enhances the interaction of laforin with malin, improving the formation of a functional laforin-malin complex. Since both laforin and AMPK localize at the glycogen granule because they possess a carbohydrate binding domain, phosphorylation of laforin by AMPK could recruit malin to this site (Fig. 8), triggering the inactivation of glycogenic targets (i.e. R5/PTG, glycogen synthase, and others) and down-regulating glycogen synthesis. If the function of the laforin-malin complex is affected (either because one of the components is not functional or because the interaction between them is prevented), no inactivation of glycogenic targets would take place, resulting in increased glycogen synthesis, which eventually would lead to the appearance of Lafora bodies.
Fig. 8.
Proposed model of the action of the laforin-malin complex in the regulation of glycogen synthesis. Interaction between laforin and malin is enhanced by phosphorylation of laforin at Ser25 by AMPK (1). A more stable laforin-malin complex results (indicated by the red arrow, 2), resulting in increased ubiquitination of specific substrates, e.g. R5/PTG, glycogen synthase (GS), and glycogen debranching enzyme (AGL/GDE), leading to down-regulation of glycogen synthesis.
Supplementary Material
Figure S1: AMPK phosphorylates laforin at Thr45 in vitro
Non-tagged recombinant laforin was incubated in kinase buffer (see the Experimental section of the main text) in the presence or absence of AMPK, as described in Figure 5 of the main text. Reactions were quenched, proteins separated by SDS/PAGE, the gel was stained with Coomasie Blue, and bands corresponding to non-phosphorylated and phosphorylated laforin were excized and analysed by MS.
ACKNOLEDGMENTS
We want to thank Drs. Rafael Pulido (Centro de Investigacion Principe Felipe, Valencia, Spain) and Reuben Shaw (Salk Institute, La Jolla, CA, USA) for the generous gift of pGEX4T1-VHR and the pEBG2-AMPK plasmids, respectively. We also thank Dr. Benoit Viollet (Institut Cochin, Université Paris Descartes, Paris, France) for control and AMPKα1α2 KO mouse embryonic fibroblasts. We also wish to thank Miguel Heredia (CIBERER, IBV-CSIC, Valencia, Spain) and Maria Elena Fernandez-Sanchez (CIB-CSIC, Madrid, Spain) for their technical assistance in the alanine scanning mutagenesis experiments, Dr. Carolyn Worby for insightful discussions, and the University of Kentucky COBRE Administrative and Proteomics Cores.
FUNDING This work was supported by a grant from the Spanish Ministry of Education and Science (SAF2008-01907) and a grant from Generalitat Valenciana (Prometeo 2009/051) to P.S.; and National Institutes of Health grants R00NS061803, P20RR020171, R01NS070899 and University of Kentucky College of Medicine startup funds to M.S.G.
Abbreviations
- CBD
carbohydrate binding domain
- 2DE
two-dimensional electrophoresis
- DSPD
dual specificity phosphatase domain
- DTT
dithiothreitol
- GFP
green fluorescent protein
- OMFP
3-O-methyl fluorescein phosphate
- PDB
protein data bank
- VHR
vaccinia H1-related phosphatase
REFERENCES
- 1.Delgado-Escueta AV. Advances in lafora progressive myoclonus epilepsy. Curr. Neurol. Neurosci. Rep. 2007;7:428–433. doi: 10.1007/s11910-007-0066-7. [DOI] [PubMed] [Google Scholar]
- 2.Gentry MS, Dixon JE, Worby CA. Lafora disease: insights into neurodegeneration from plant metabolism. Trends Biochem. Sci. 2009;34:628–639. doi: 10.1016/j.tibs.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ganesh S, Puri R, Singh S, Mittal S, Dubey D. Recent advances in the molecular basis of Lafora’s progressive myoclonus epilepsy. J. Hum. Genet. 2006;51:1–8. doi: 10.1007/s10038-005-0321-1. [DOI] [PubMed] [Google Scholar]
- 4.Minassian BA, Lee JR, Herbrick JA, Huizenga J, Soder S, Mungall AJ, Dunham I, Gardner R, Fong CY, Carpenter S, Jardim L, Satishchandra P, Andermann E, Snead OC, 3rd, Lopes-Cendes I, Tsui LC, Delgado-Escueta AV, Rouleau GA, Scherer SW. Mutations in a gene encoding a novel protein tyrosine phosphatase cause progressive myoclonus epilepsy. Nat. Genet. 1998;20:171–174. doi: 10.1038/2470. [DOI] [PubMed] [Google Scholar]
- 5.Serratosa JM, Gomez-Garre P, Gallardo ME, Anta B, de Bernabe DB, Lindhout D, Augustijn PB, Tassinari CA, Malafosse RM, Topcu M, Grid D, Dravet C, Berkovic SF, de Cordoba SR. A novel protein tyrosine phosphatase gene is mutated in progressive myoclonus epilepsy of the Lafora type (EPM2) Hum. Mol. Genet. 1999;8:345–352. doi: 10.1093/hmg/8.2.345. [DOI] [PubMed] [Google Scholar]
- 6.Chan EM, Young EJ, Ianzano L, Munteanu I, Zhao X, Christopoulos CC, Avanzini G, Elia M, Ackerley CA, Jovic NJ, Bohlega S, Andermann E, Rouleau GA, Delgado-Escueta AV, Minassian BA, Scherer SW. Mutations in NHLRC1 cause progressive myoclonus epilepsy. Nat. Genet. 2003;35:125–127. doi: 10.1038/ng1238. [DOI] [PubMed] [Google Scholar]
- 7.Chan EM, Omer S, Ahmed M, Bridges LR, Bennett C, Scherer SW, Minassian BA. Progressive myoclonus epilepsy with polyglucosans (Lafora disease): evidence for a third locus. Neurology. 2004;63:565–567. doi: 10.1212/01.wnl.0000133215.65836.03. [DOI] [PubMed] [Google Scholar]
- 8.Minassian BA, Ianzano L, Meloche M, Andermann E, Rouleau GA, Delgado-Escueta AV, Scherer SW. Mutation spectrum and predicted function of laforin in Lafora’s progressive myoclonus epilepsy. Neurology. 2000;55:341–346. doi: 10.1212/wnl.55.3.341. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Stuckey JA, Wishart MJ, Dixon JE. A unique carbohydrate binding domain targets the lafora disease phosphatase to glycogen. J. Biol. Chem. 2002;277:2377–2380. doi: 10.1074/jbc.C100686200. [DOI] [PubMed] [Google Scholar]
- 10.Lohi H, Ianzano L, Zhao XC, Chan EM, Turnbull J, Scherer SW, Ackerley CA, Minassian BA. Novel glycogen synthase kinase 3 and ubiquitination pathways in progressive myoclonus epilepsy. Hum. Mol. Genet. 2005;14:2727–2736. doi: 10.1093/hmg/ddi306. [DOI] [PubMed] [Google Scholar]
- 11.Gentry MS, Worby CA, Dixon JE. Insights into Lafora disease: malin is an E3 ubiquitin ligase that ubiquitinates and promotes the degradation of laforin. Proc. Natl. Acad. Sci. USA. 2005;102:8501–8506. doi: 10.1073/pnas.0503285102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lafora GR, Glueck B. Beitrag zur histogpathologie der myoklonischen epilepsie. Gesamte Neurol. Psychiatr. 1911;6:1–14. [Google Scholar]
- 13.Collins GH, Cowden RR, Nevis AH. Myoclonus epilepsy with Lafora bodies. An ultrastruc-tural and cytochemical study. Arch. Pathol. 1968;86:239–254. [PubMed] [Google Scholar]
- 14.Solaz-Fuster MC, Gimeno-Alcaniz JV, Ros S, Fernandez-Sanchez ME, Garcia-Fojeda B, Garcia O. Criado, Vilchez D, Dominguez J, Garcia-Rocha M, Sanchez-Piris M, Aguado C, Knecht E, Serratosa J, Guinovart JJ, Sanz P, de Cordoba S. Rodriguez. Regulation of glycogen synthesis by the laforin-malin complex is modulated by the AMP-activated protein kinase pathway. Hum. Mol. Genet. 2008;17:667–678. doi: 10.1093/hmg/ddm339. [DOI] [PubMed] [Google Scholar]
- 15.Vilchez D, Ros S, Cifuentes D, Pujadas L, Valles J, Garcia-Fojeda B, Criado-Garcia O, Fernandez-Sanchez E, Medrano-Fernandez I, Dominguez J, Garcia-Rocha M, Soriano E, de Cordoba S. Rodriguez, Guinovart JJ. Mechanism suppressing glycogen synthesis in neurons and its demise in progressive myoclonus epilepsy. Nat. Neurosci. 2007;10:1407–1413. doi: 10.1038/nn1998. [DOI] [PubMed] [Google Scholar]
- 16.Cheng A, Zhang M, Gentry MS, Worby CA, Dixon JE, Saltiel AR. A role for AGL ubiquitination in the glycogen storage disorders of Lafora and Cori’s disease. Genes Dev. 2007;21:2399–2409. doi: 10.1101/gad.1553207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Worby CA, Gentry MS, Dixon JE. Malin decreases glycogen accumulation by promoting the degradation of protein targeting to glycogen (PTG) J. Biol. Chem. 2008;283:4069–4076. doi: 10.1074/jbc.M708712200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DePaoli-Roach AA, Tagliabracci VS, Segvich DM, Meyer CM, Irimia JM, Roach PJ. Genetic depletion of the malin E3 ubiquitin ligase in mice leads to lafora bodies and the accumulation of insoluble laforin. J. Biol. Chem. 2010;285:25372–25381. doi: 10.1074/jbc.M110.148668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turnbull J, Wang P, Girard JM, Ruggieri A, Wang TJ, Draginov AG, Kameka AP, Pencea N, Zhao X, Ackerley CA, Minassian BA. Glycogen hyperphosphorylation underlies lafora body formation. Ann. Neurol. 2010;68:925–933. doi: 10.1002/ana.22156. [DOI] [PubMed] [Google Scholar]
- 20.Worby CA, Gentry MS, Dixon JE. Laforin, a dual specificity phosphatase that dephosphorylates complex carbohydrates. J. Biol. Chem. 2006;281:30412–30418. doi: 10.1074/jbc.M606117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tagliabracci VS, Turnbull J, Wang W, Girard JM, Zhao X, Skurat AV, Delgado-Escueta AV, Minassian BA, Depaoli-Roach AA, Roach PJ. Laforin is a glycogen phosphatase, deficiency of which leads to elevated phosphorylation of glycogen in vivo. Proc. Natl. Acad. Sci. USA. 2007;104:19262–19266. doi: 10.1073/pnas.0707952104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gentry MS, Dowen RH, 3rd, Worby CA, Mattoo S, Ecker JR, Dixon JE. The phosphatase laforin crosses evolutionary boundaries and links carbohydrate metabolism to neuronal disease. J. Cell. Biol. 2007;178:477–488. doi: 10.1083/jcb.200704094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tagliabracci VS, Girard JM, Segvich D, Meyer C, Turnbull J, Zhao X, Minassian BA, Depaoli-Roach AA, Roach PJ. Abnormal metabolism of glycogen phosphate as a cause for Lafora disease. J. Biol. Chem. 2008;283:33816–33825. doi: 10.1074/jbc.M807428200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ganesh S, Agarwala KL, Ueda K, Akagi T, Shoda K, Usui T, Hashikawa T, Osada H, Delgado-Escueta AV, Yamakawa K. Laforin, defective in the progressive myoclonus epilepsy of Lafora type, is a dualspecificity phosphatase associated with polyribosomes. Hum. Mol. Genet. 2000;9:2251–2261. doi: 10.1093/oxfordjournals.hmg.a018916. [DOI] [PubMed] [Google Scholar]
- 25.Wang W, Roach PJ. Glycogen and related polysaccharides inhibit the laforin dual-specificity protein phosphatase. Biochem. Biophys. Res. Commun. 2004;325:726–730. doi: 10.1016/j.bbrc.2004.10.083. [DOI] [PubMed] [Google Scholar]
- 26.Girard JM, Le KH, Lederer F. Molecular characterization of laforin, a dual-specificity protein phosphatase implicated in Lafora disease. Biochimie. 2006;88:1961–1971. doi: 10.1016/j.biochi.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Liu Y, Wu C, Zhang H, Zheng X, Zheng Z, Geiger TL, Nuovo GJ, Liu Y, Zheng P. Epm2a suppresses tumor growth in an immunocompromised host by inhibiting Wnt signaling. Cancer Cell. 2006;10:179–190. doi: 10.1016/j.ccr.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 28.Liu R, Wang L, Chen C, Liu Y, Zhou P, Wang Y, Wang X, Turnbull J, Minassian BA, Liu Y, Zheng P. Laforin negatively regulates cell cycle progression through glycogen synthase kinase 3beta-dependent mechanisms. Mol. Cell. Biol. 2008;28:7236–7244. doi: 10.1128/MCB.01334-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rose MD, Winston F, Hieter P. Methods in yeast genetics, a laboratory course manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1990. [Google Scholar]
- 31.Gimeno-Alcaniz JV, Sanz P. Glucose and type 2A protein phosphatase regulate the interaction between catalytic and regulatory subunits of AMP-activated protein kinase. J. Mol. Biol. 2003;333:201–209. doi: 10.1016/j.jmb.2003.08.022. [DOI] [PubMed] [Google Scholar]
- 32.Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, Cantley LC. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc. Natl. Acad. Sci. USA. 2004;101:3329–3335. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ludin K, Jiang R, Carlson M. Glucose-regulated interaction of a regulatory subunit of protein phosphatase 1 with the Snf1 protein kinase in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1998;95:6245–6250. doi: 10.1073/pnas.95.11.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanz P, Alms GR, Haystead TA, Carlson M. Regulatory interactions between the Reg1-Glc7 protein phosphatase and the Snf1 protein kinase. Mol. Cell. Biol. 2000;20:1321–1328. doi: 10.1128/mcb.20.4.1321-1328.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanner S, Shu H, Frank A, Wang LC, Zandi E, Mumby M, Pevzner PA, Bafna V. InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Anal. Chem. 2005;77:4626–4639. doi: 10.1021/ac050102d. [DOI] [PubMed] [Google Scholar]
- 36.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zdobnov EM, Apweiler R. InterProScan--an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 38.Pei J, Tang M, Grishin NV. PROMALS3D web server for accurate multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:W30–34. doi: 10.1093/nar/gkn322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lambert C, Leonard N, De Bolle X, Depiereux E. ESyPred3D: Prediction of proteins 3D structures. Bioinformatics. 2002;18:1250–1256. doi: 10.1093/bioinformatics/18.9.1250. [DOI] [PubMed] [Google Scholar]
- 40.Singh S, Ganesh S. Lafora progressive myoclonus epilepsy: a metaanalysis of reported mutations in the first decade following the discovery of the EPM2A and NHLRC1 genes. Hum. Mut. 2009;30:715–723. doi: 10.1002/humu.20954. [DOI] [PubMed] [Google Scholar]
- 41.Machovic M, Janecek S. Starch-binding domains in the post-genome era. Cell. Mol. Life Sci. 2006;63:2710–2724. doi: 10.1007/s00018-006-6246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yokota T, Nara Y, Kashima A, Matsubara K, Misawa S, Kato R, Sugio S. Crystal structure of human dual specificity phosphatase, JNK stimulatory phosphatase-1, at 1.5 A resolution. Proteins. 2007;66:272–278. doi: 10.1002/prot.21152. [DOI] [PubMed] [Google Scholar]
- 43.Andersen JN, Mortensen OH, Peters GH, Drake PG, Iversen LF, Olsen OH, Jansen PG, Andersen HS, Tonks NK, Moller NP. Structural and evolutionary relationships among protein tyrosine phosphatase domains. Mol. Cell. Biol. 2001;21:7117–7136. doi: 10.1128/MCB.21.21.7117-7136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoyt R, Zhu W, Cerignoli F, Alonso A, Mustelin T, David M. Cutting edge: selective tyrosine dephosphorylation of interferon-activated nuclear STAT5 by the VHR phosphatase. J. Immunol. 2007;179:3402–3406. doi: 10.4049/jimmunol.179.6.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y, Wang Y, Wu C, Liu Y, Zheng P. Dimerization of Laforin Is Required for Its Optimal Phosphatase Activity, Regulation of GSK3beta Phosphorylation, and Wnt Signaling. J. Biol. Chem. 2006;281:34768–34774. doi: 10.1074/jbc.M607778200. [DOI] [PubMed] [Google Scholar]
- 46.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ganesh S, Tsurutani N, Suzuki T, Hoshii Y, Ishihara T, Delgado-Escueta AV, Yamakawa K. The carbohydrate-binding domain of Lafora disease protein targets Lafora polyglucosan bodies. Biochem. Biophys. Res. Commun. 2004;313:1101–1109. doi: 10.1016/j.bbrc.2003.12.043. [DOI] [PubMed] [Google Scholar]
- 48.Ganesh S, Delgado-Escueta AV, Suzuki T, Francheschetti S, Riggio C, Avanzini G, Rabinowicz A, Bohlega S, Bailey J, Alonso ME, Rasmussen A, Thomson AE, Ochoa A, Prado AJ, Medina MT, Yamakawa K. Genotype-phenotype correlations for EPM2A mutations in Lafora’s progressive myoclonus epilepsy: exon 1 mutations associate with an early-onset cognitive deficit subphenotype. Hum. Mol. Genet. 2002;11:1263–1271. doi: 10.1093/hmg/11.11.1263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: AMPK phosphorylates laforin at Thr45 in vitro
Non-tagged recombinant laforin was incubated in kinase buffer (see the Experimental section of the main text) in the presence or absence of AMPK, as described in Figure 5 of the main text. Reactions were quenched, proteins separated by SDS/PAGE, the gel was stained with Coomasie Blue, and bands corresponding to non-phosphorylated and phosphorylated laforin were excized and analysed by MS.