SUMMARY
Centromeres are essential chromosomal regions required for kinetochore assembly and chromosome segregation. The composition and organization of centromeric nucleosomes containing the essential histone H3 variant CENP-A (CID in Drosophila) is a fundamental, unresolved issue. Using immunoprecipitation of CID mononucleosomes and cysteine crosslinking, we demonstrate that centromeric nucleosomes contain CID dimers in vivo. Furthermore, CID dimerization and centromeric targeting require a residue implicated in formation of the four helix bundle, which mediates intra-nucleosomal H3 dimerization and nucleosome integrity. Taken together, our findings suggest that CID nucleosomes are octameric in vivo and that CID dimerization is essential for correct centromere assembly.
Keywords: chromatin, centromere, CENP-A, CID, nucleosome
INTRODUCTION
The centromere is a unique chromosomal domain responsible for kinetochore formation and faithful chromosome segregation during cell division. Centromere identity and propagation are determined epigenetically in most eukaryotes. CENP-A is the centromere-specific histone H3 variant (CID in flies), which is required for proper localization of nearly all other centromere and kinetochore proteins (Allshire and Karpen, 2008). Furthermore, CID mislocalization leads to ectopic centromere formation (Olszak et al., 2011). These results place CENP-A at or near the top of the centromere assembly pathway.
Knowledge of CENP-A nucleosome structure in vivo is essential to understanding the mechanisms responsible for CENP-A replenishment and maintenance. Canonical nucleosomes contain a (H3/H4)2 tetramer and two H2A/H2B dimers. However, recent investigations present contradictory views of CENP-A nucleosome composition and structure. Studies using immunoprecipitation (IP) of digested bulk chromatin or in vitro assembly have suggested an octameric structure for CENP-A nucleosomes, where two copies of CENP-A take the place of H3 (Foltz et al., 2006; Sekulic et al., 2010; Shelby et al., 1997; Tachiwana et al., 2011; Yoda et al., 2000). Of note was the demonstration that Cse4 (S. cerevisiae CENP-A) nucleosomes are histone octamers in vivo, since they contain Cse4 dimers and H2A/H2B (Camahort et al., 2009). However, another study suggested a Cse4 complex consisting of Cse4/H4 tetramers and the Cse4 assembly factor Scm3 in place of H2A/H2B dimers (Mizuguchi et al., 2007). Finally, it was reported that Drosophila CID and human CENP-A are in ‘hemisomes’ containing only one copy each of CID/CENP-A, H4, H2A and H2B (Dalal et al., 2007; Dimitriadis et al., 2010). At this time, it is unclear whether these discrepancies reflect biologically relevant differences among organisms or cell cycle stages, or result from technical differences (Black and Cleveland, 2011).
Analysis of the CENP-A domains have shed some light on this controversy. The CATD (CENP-A Targeting Domain) comprising loop I and helix αII of the CENP-A histone fold domain (HFD) is required for centromeric targeting in yeast, flies, and humans, and for interaction with HJURP/Scm3-like CENP-A assembly factors (Black et al., 2004; Hu et al., 2011; Vermaak et al., 2002; Zhou et al., 2011). In addition, CENP-A shares significant homology with H3 in the four helix bundle structure formed between the C-terminal halves of αII and αIII helices and required for intra-nucleosomal H3 dimerization (Luger et al., 1997). This region in CENP-A is required for in vitro formation of crystallized (CENP-A/H4)2 tetramers (Sekulic et al., 2010) and octameric CENP-A nucleosomes (Tachiwana et al., 2011). In Cse4, it is required for in vitro nucleosome reconstitution, centromere localization in vivo, and cell viability (Camahort et al., 2009), but not for the interaction with Scm3 (Zhou et al., 2011). These findings strongly suggest that Cse4 nucleosomes are octameric in vivo, and that the four helix bundle mediates Cse4 dimerization and centromere localization. Whether these properties are conserved in CENP-A nucleosomes from other organisms, especially those with epigenetically-regulated ‘regional’ centromeres, has not been addressed.
In this study, we use a combination of biochemical, molecular and cell biological approaches to elucidate the composition of Drosophila centromeric CID nucleosomes in vivo. We show that purified CID mononucleosomes contain two copies of CID, consistent with an octameric structure, and provide evidence that the four helix bundle is required for CID dimerization and centromere localization.
RESULTS
Characterization of CID chromatin by salt extraction and sucrose gradient ultracentrifugation
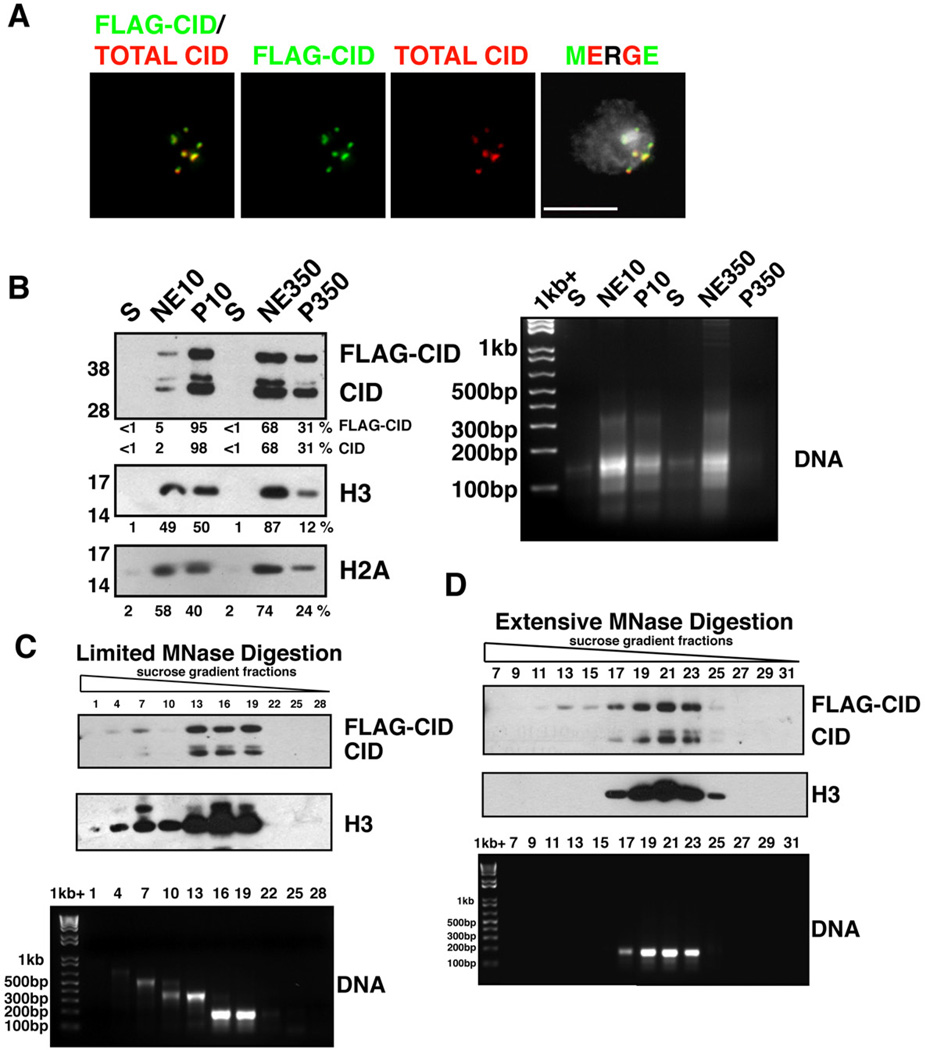
To study the composition of centromeric chromatin, we generated a Drosophila S2 cell line constitutively expressing N-terminally FLAG-tagged CID controlled by a copia promoter (FLAGCID). Immunofluorescence (IF) analysis showed that FLAG-CID properly localizes to centromeres, with a pattern identical to endogenous CID (Figure 1A). Western blots demonstrated that FLAG-CID comprises about 2/3 of total CID in chromatin (Figure 1B). FLAG-CID IP from micrococcal nuclease (MNase)-digested bulk chromatin containing mono- to oligo- nucleosomes revealed significant enrichment for endogenous CID, centromeric CAL1, and H2A, relative to IPs from untagged controls (Figure S1). Thus, FLAG-CID is closely associated with endogenous CID and other known centromeric chromatin components.
Figure 1. Characterization of CID chromatin.
- FLAG-CID (green) and total CID (red) colocalize at centromeres in a stable S2 cell line. FLAG-CID and total CID were stained by α-FLAG and α-CID antibodies, respectively. DNA staining by DAPI (grey). Scale bar = 5 µm.
- (left) FLAG-CID and endogenous CID are more resistant to salt extraction than H3. After MNase digestion, soluble nucleoplasmic fractions (S), chromatin extracted with either 10 mM or 350 mM salt (NE10 and NE350, respectively), and insoluble nuclear pellets (P10 and P350, respectively) were analyzed by Western blots for CID, H3 and H2A. (right) DNA agarose gel with the same samples.
- (top) FLAG-CID and endogenous CID co-fractionate with peaks of H3 in sucrose gradient fractions after limited MNase digestion. (bottom) DNA agarose gel shows sizes consistent with mainly mono-, di- or tri-nucleosomes.
- (Top) FLAG-CID and endogenous CID co-migrate with H3 in sucrose gradient fractions after extensive MNase digestion. (bottom) DNA agarose gel from the same samples.
Also see Figure S1.
We investigated the solubility of CID chromatin by digesting S2 nuclei with MNase and successively extracting chromatin with increasing salt concentrations (Figure 1B). Using a low salt buffer (10 mM NaCl), more than 95% of CID nucleosomes remain insoluble, whereas ~50% of H3 nucleosomes are soluble. In addition, high salt (350 mM NaCl) extracts ~90% of H3 nucleosomes, and ~70% of CID nucleosomes. Thus, CID chromatin is less soluble than H3 chromatin.
We next used linear sucrose gradient ultracentrifugation to characterize CID nucleosomes (Figures 1C and 1D). Limited MNase digestion converted bulk chromatin into mono- and oligonucleosomes, while extensive digestion converted most chromatin to a single peak of ~150 bp DNA consistent with mononucleosomes. Under both conditions, fractions containing H3 mononucleosomes or oligonucleosomes are also enriched for CID and FLAG-CID. Importantly, no CID or FLAG-CID was observed in fractions of higher sucrose density, where hemisomes would be expected to reside. We conclude that H3 and CID nucleosomes exhibit similar sedimentation velocity in ultracentrifugation. These results are inconsistent with hemisomes, which would contain fewer histones and shorter DNA than H3 nucleosomes.
FLAG-CID mononucleosomes contain stoichiometric amount of endogenous CID
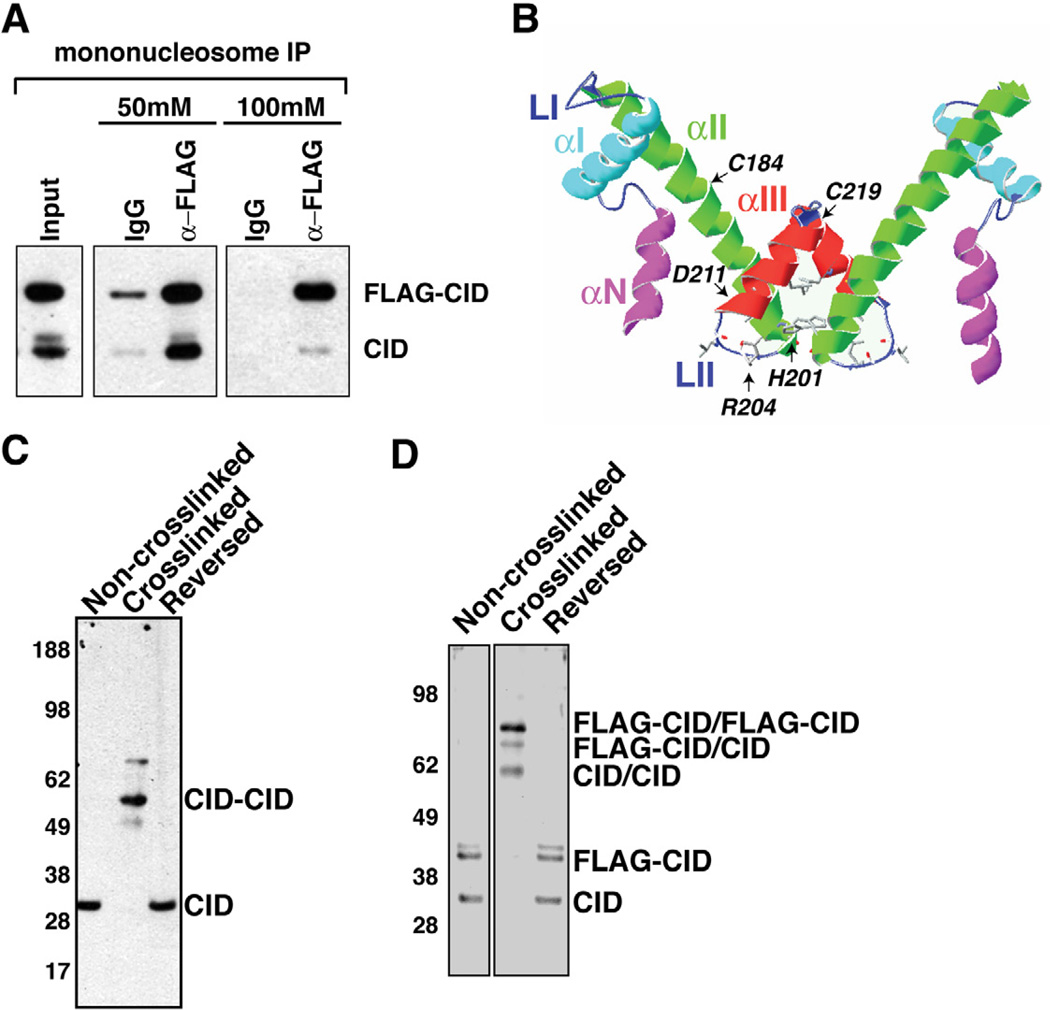
To examine the stoichiometry of CID nucleosomes in more detail, we immunoprecipitated FLAG-CID nucleosomes without crosslinking from either oligonucleosome or mononucleosome fractions. IPs from oligonucleosome preparations, which were conducted with stringent washes (350 mM NaCl), resulted in detection of both FLAG-CID and endogenous CID (Figure S2A), suggesting that tagged and native CID are physically close in chromatin containing only a few nucleosomes. In contrast, mononucleosome IPs detected only sub-stoichiometric amounts of endogenous CID with FLAG-CID after 300 mM NaCl washes, despite the presence of H2A and H2B (Figure S2B). This could suggest that only half nucleosomes are recovered, or that FLAG-CID does not assemble with endogenous CID. However, ~85% of endogenous CID was co-depleted by the α-FLAG beads (Figure S2C), indicating that endogenous CID interacts with FLAG-CID but is subsequently lost during stringent washes.
These results raised the possibility that CID nucleosome stability is sensitive to high salt conditions, as observed for variant nucleosomes containing both H3.3 and H2A.Z (Jin and Felsenfeld, 2007). Therefore, we performed FLAG-CID IP on mononucleosome fractions with increasing stringency of washes from 50 mM to 100 mM NaCl. Analysis of endogenous CID recovery from α-FLAG IP after each wash revealed that approximately stoichiometric amounts of endogenous CID co-precipitated with FLAG-CID after 50 mM NaCl, relative to their ratio in the input (Figure 2A, 50 mM). In contrast, endogenous CID levels were reduced relative to FLAG-CID after 100 mM NaCl washes (Figure 2A, 100 mM). These observations suggest that each CID nucleosome contains two CID molecules in vivo, in agreement with an octameric structure, though the integrity of solubilized CID nucleosomes is sensitive to high salt.
Figure 2. CID nucleosomes contain CID dimers.
- Endogenous CID coimmunoprecipitates with FLAG-CID from mononucleosomes. Inputs consist of mononucleosomes. α-mouse IgG beads were used in control IPs (IgG).
- Schematic display of the HFD of CID using a Cse4 model as reference (Bloom et al., 2006). Helices- αN, -αI, -αII, -αIII, and Loops LI, LII are indicated. CATD is comprised of LI and αII. The putative four helix bundle is comprised of the C terminal halves of αII and αIII.
- Cysteine crosslinking of S2 chromatin. CID monomers migrate at 30 kDa without crosslinking and after crosslink reversal. After crosslinking, the majority of CID migrate at 60 kDa, consistent with the size of dimers.
- Cysteine crosslinking of FLAG-CID chromatin. FLAG-CID and endogenous CID migrate as monomers before crosslinking and after crosslink reversal. After crosslinking, three major bands were detected by CID antibody that correspond to CID/CID, FLAG-CID/CID and FLAG-CID/FLAG-CID, respectively.
Also see Figure S2.
Cysteine crosslinking reveals CID dimers
If CID nucleosomes contain two copies of CID whose interactions are unstable in the N-ChIP experiments, crosslinking of intra-nucleosomal CID molecules should facilitate recovery of CID dimers. The zero-distance sulfhydryl group oxidation agent 1,10, o-phenanthroline-copper has been shown to crosslink intra-nucleosomal histone H3 molecules through cysteine C110, which resides in the four helix bundle structure (Figures 2B, S2D)(Gould et al., 1980).
Although a crystal structure for CID nucleosomes has not been generated, ClustalW alignment shows that CID contains all the residues implicated in H3-H3’ and Cse4-Cse4’ interactions (Camahort et al., 2009; Luger et al., 1997) (Figure S2D). Thus, this region could also form the four helix bundle in an octameric CID nucleosome, and two cysteines (C184 and C219) in αII and αIII helices could mediate oxidative crosslinking (Figure S2D, arrowheads) (Gould et al., 1980).
To test whether CID nucleosomes contain one or two CIDs, extensively digested chromatin was crosslinked with copper phenanthroline. Effective crosslinking was confirmed by H3 dimerization and reversal by 2-mercaptoethanol (Figure S2E). CID in non-crosslinked samples migrated as one band of 30 kDa (Figure 2C). After crosslinking, most CID migrated at an apparent molecular weight of 60.3 kDa, consistent with CID dimers (Figures 2C, S2G), and less than 1.1±0.53% (n=3) of CID was detected as monomers. After crosslink reversal, CID migrated at the monomer size, confirming that the 60.3 kDa CID band observed after crosslinking was due to cysteine oxidation (Figure 2C).
To further test for the presence of CID dimers in vivo, we performed the same experiment using chromatin from cells constitutively expressing FLAG-CID (Figure 2D). We detected two major CID bands in non-crosslinked and reverse-crosslinked chromatin, corresponding to monomeric endogenous CID and FLAG-CID (Figure 3C). After crosslinking, we detected three major CID bands at 60.3, 68.4 and 79.4 kDa, respectively (Figures 3C, S2G). The sizes of these slower migrating CID bands are consistent with dimer formation between two endogenous CIDs, one endogenous CID and one FLAG-CID, or two FLAG-CIDs, respectively. The slower migratingbands (80 kDa and 70 kDa) were also detected by the α-FLAG antibody, validating the formation of FLAG-CID/FLAG-CID and FLAG-CID/CID dimers (Figures S2F, S2G). A weak band at 56 kDa of unknown nature was also detected (Figure S2F). These cysteine crosslinking results provide additional, strong evidence that two CID molecules closely interact within mononucleosomes.
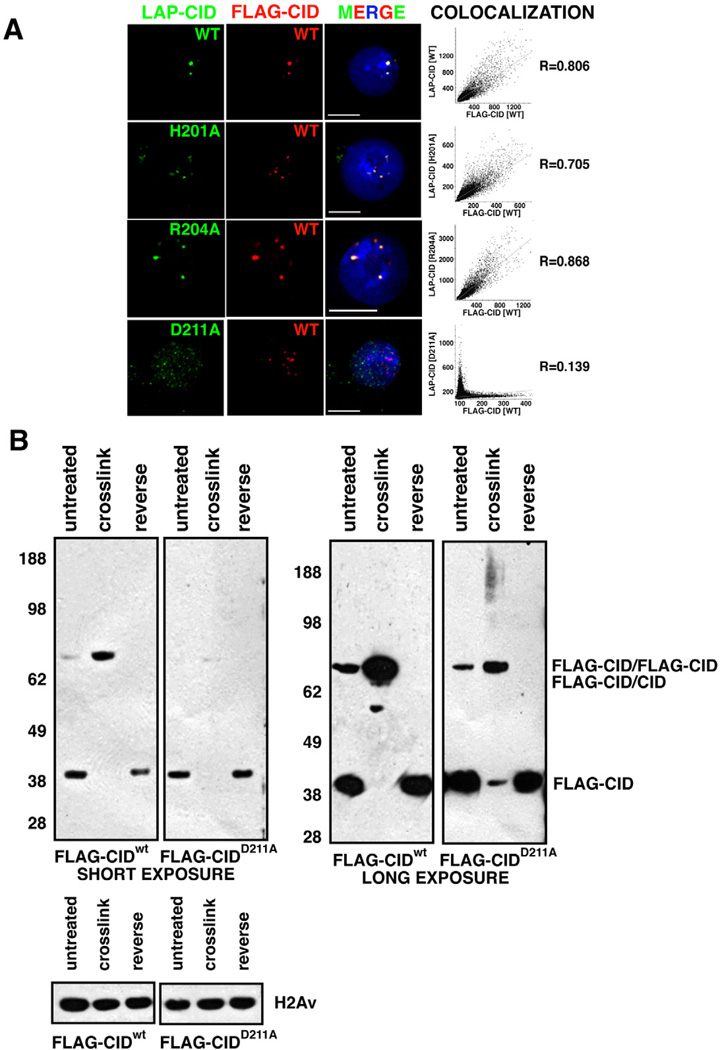
Figure 3. CIDD211A mutation disrupts centromere-specific localization and dimerization of CID.
- Localization of mutant LAP-CID constructs. FLAG-CIDwt (red) at centromeres colocalizes with LAP-CIDwt (98.5%, n=130)(green), and point mutants LAP-CIDH201A (99.0%, n=96) (green) and LAP-CIDR204A (97.3%, n=112) (green). In contrast, few LAP-CIDD211A (2.63%, n=152) (green) colocalize with FLAG-CIDwt (red), and often associates with non-centromeric regions. Scale bars = 5 µm. The graphs show Pearson’s coefficient of correlation (R) of the individual images.
- FLAG-CIDD211A mutant proteins are defective for dimerization. FLAG-CIDwt was efficiently crosslinked as dimers. In contrast, FLAG-CIDD211A crosslinking converted ~43% into dimmers (short exposure) and 40% into a smear without distinct bands (long exposure), while 17% remained as monomers. Total CIDD211A intensity after crosslinking was calculated as the sum of monomers, dimers and smear. H2Av, which lacks cysteines, was used as loading control.
Also see Figure S3.
A mutation in the CID four helix bundle reduces dimerization and causes CID mislocalization
If CID nucleosomes are octameric and CID dimerization depends on the four helix bundle, then interference with CID-CID’ interactions should disturb stable maintenance and assembly of CID nucleosomes in living cells. The four helix bundle in H3 nucleosomes is stabilized by residue D123, which forms hydrogen bonds with R116 and H113’, respectively (Luger et al., 1997).
We mutated the corresponding CID residues (D211, H201, and R204) potentially involved in dimerization to study their roles in vivo. Each mutation was LAP-tagged at its NH2 terminus and co-transfected with FLAG-CIDwt into S2 cells. As a control, we co-transfected two wild-type CID constructs (LAP-CIDwt and FLAG-CIDwt), and their colocalization at centromeres was demonstrated by IF (Figures 3A, S3A, S3C). Thus, the FLAG-CIDwt distribution can be used to determine if LAP-tagged mutant CID proteins localize to centromeres. Similar to wild-type CID constructs, we observed that the majority of CIDH201A and CIDR204A properly localize to centromeres by IF (Figures 3A, S3C). However, few cells showed centromeric localization for LAP-CIDD211A (Figures 3A, S3). In interphase, LAP-CIDD211A mutant proteins were broadly distributed throughout chromatin and in the cytoplasm. In mitosis, LAP-CIDD211A is spread throughout chromosomes in both cytospin and whole mount IF. This result is significantly different from the specific colocalization of LAP-CIDwt and FLAG-CIDwt to centromeres. We conclude that CID D211, but not the H201 or R204 residues, is required for proper CID localization to centromeres.
We then directly determined whether residue D211 is important for CID dimerization in vivo using cysteine crosslinking. We transiently transfected FLAG-CIDD211A or FLAG-CIDwt control into S2 cells and prepared untreated, crosslinked and reverse-crosslinked chromatin, as conducted previously, from both samples (Figure 3B). For the FLAG-CIDwt control, crosslinking converted nearly 100% FLAG-CIDwt monomers to a single band of the predicted FLAG-CIDwt dimer size. A weak band was also evident consistent with formation of dimers between endogenous CID and FLAG-CIDwt. In contrast, 43% of total FLAG-CIDD211A was detected at the predicted FLAGCIDD211A dimer size after crosslinking, and 17% of FLAG-CIDD211A remained as monomers (RatioDimer:Monomer<3 for FLAG-CIDD211A versus >55 for FLAG-CIDwt). We conclude that D211A inhibits efficient CID dimerization.
Interestingly, combined FLAG-CIDD211A monomers and dimers accounted for 60% of total FLAG-CIDD211A (Figure 3B, compare intensity in crosslinked to reversed). Long exposure revealed a high molecular weight smear accounting for 40% of total FLAG-CIDD211A (Figure 3B). We propose that the smear is formed because inefficient dimerization makes monomeric CIDD211A available for crosslinking with other unknown proteins. We conclude that the CID D211 residue is not only required for proper centromeric localization, but also for efficient dimerization within nucleosomes.
DISCUSSION
CID nucleosome stoichiometry is consistent with an octamer structure
The CENP-A nucleosome is the foundation for centromere structure and function, and its organization has recently been the subject of great debate. Notably, it has been proposed that interphase Drosophila CID nucleosomes are hemisomes (Dalal et al., 2007). We show that the majority of Drosophila CID nucleosomes contain two copies of CID, as well as H2A, H2B (this study) and H4 (Blower et al., 2002). CID nucleosomes co-migrate with octameric H3 nucleosomes in sucrose gradient ultracentrifugation, with no peaks in higher sucrose density fractions where hemisomes would be expected to reside. The possibility that CID hemisomes could exhibit an unusual conformation that results in similar sedimentation velocities as octameric H3 nucleosomes was refuted by the demonstration that endogenous CID and FLAG-CID coimmunoprecipitate stoichiometrically under low salt conditions without crosslinking. The use of purified mononucleosomes provides more conclusive results about the in vivo composition of CENP-A nucleosomes than earlier studies using digested bulk chromatin or in vitro assembled complexes.
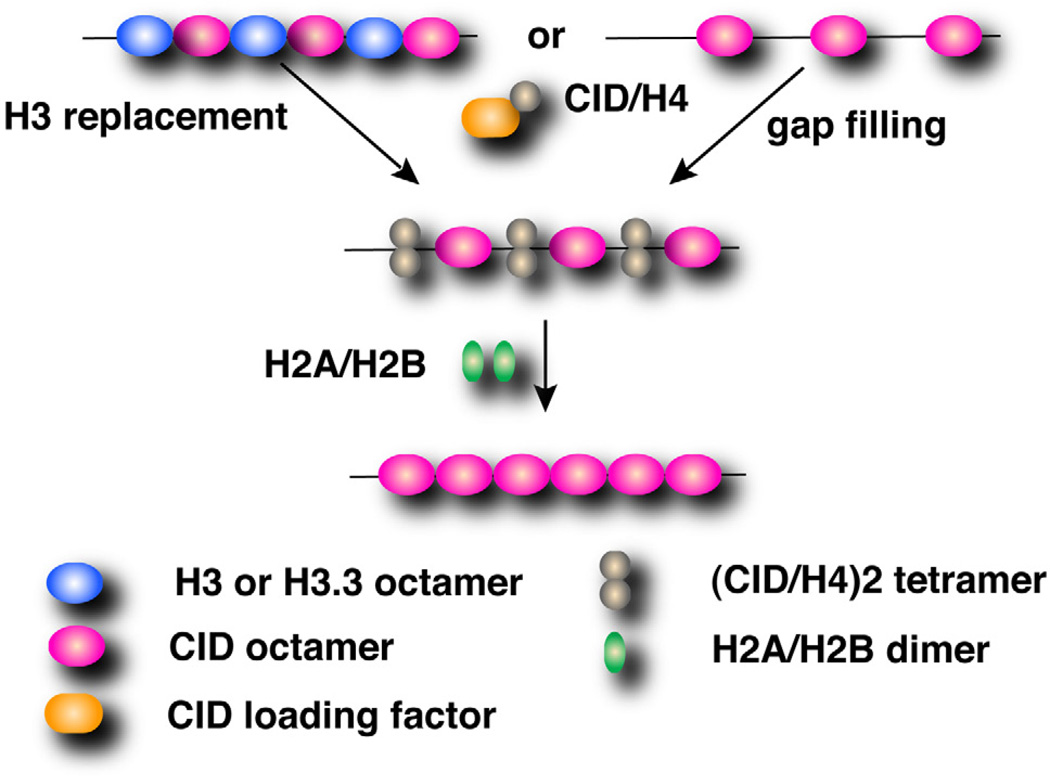
In addition, near complete conversion of CID into a dimeric form with a zero-distance crosslinker contradicts the hemisome model. This reagent has been used to study H3 dimerization in canonical nucleosomes (Gould et al., 1980). The most parsimonious interpretation is that the majority of CID forms dimers during most of the cell cycle. Finally, the CIDD211A mutation reduced dimerization and resulted in broad mislocalization. Thus, a conserved residue important for normal function of the four helix bundle structure in both H3 and Cse4 nucleosomes is essential for dimerization and centromeric localization of CID in vivo. These conclusions are consistent with the structural analyses of in vitro assembled H3 and CENP-A nucleosomes (Luger et al., 1997; Tachiwana et al., 2011), and a Cse4 study in vivo (Camahort et al., 2009). Therefore, we propose that CID dimerization mediated by the putative four helix bundle is essential for assembly and maintenance of octameric CID nucleosomes (Figure 4).
Figure 4. A model for CID deposition incorporating CID dimerization.
In mitosis, two CID/H4 dimers are deposited into centromeres to form an octameric nucleosome, by a hypothetical loading factor recognizing the CATD. CID dimerization through the four helix bundle is required for centromere assembly. Octameric CID nucleosomes are assembled either by replacing H3.3 nucleosomes as observed in human cells (Dunleavy et al., 2011), or de novo into nucleosome gaps generated in S phase (Allshire and Karpen, 2008).
CID dimers could potentially arise from interactions between two hemisomes during crosslinking. However, we have demonstrated that CID dimerizes in mononucleosomes in vivo without crosslinking. Furthermore, we reduced the local CID concentration and minimized potential crosslinking from different nucleosomes by extensive MNase digestion and extraction. We observe that the vast majority, if not all, of soluble CID nucleosomes contain dimers in asynchronous S2 cells, of which over 95% are in interphase and about 50% are in G2. Thus, if CID hemisomes exist, they must be only a small proportion of total CID nucleosomes, or present for a short time in the cell cycle.
Our observations suggest technical reasons for the recovery of hemisomes in previous studies. First, primary amine crosslinking was the major method used in the isolation of CID hemisomes (Dalal et al., 2007). Unlike H3, CID lacks the lysine residue in its HFD that is necessary for efficient primary amine crosslinking (Black and Bassett, 2008). Second, recovery of CID nucleosomes is highly sensitive to salt concentrations in the absence of crosslinking. We suspect that hemisomes were recovered due to dissociation of CID dimers by the higher salt treatment, combined with the absence of residues available for primary amine crosslinking.
Our observation that extracted CID nucleosomes are unstable may stem from loss of specific stabilizing factors during CID nucleosome preparation. It has been reported that CENP-B and CENP-C are absent from human CENP-A mononucleosomes after sucrose gradient ultracentrifugation, but are present in CENP-A oligonucleosomes (Ando et al., 2002). Moreover, partial unwinding of DNA at the nucleosome entry/exit site can disassemble reconstituted human CENP-A nucleosomes (Conde e Silva et al., 2007). Together these findings suggest that instability of CENP-A nucleosomes may be caused by differences in composition and structure compared to H3 nucleosomes.
It is surprising that unstable CENP-A nucleosomes serve as the foundation of a highly stable chromatin domain through cell divisions. However, instability of CID nucleosomes could facilitate elimination of mis-incorporated CID in the absence of limiting stabilizing factors (Hewawasam et al., 2011; Moreno-Moreno et al., 2011; Ranjitkar et al., 2011). Since unwarranted evictions of CID from centromeres would lead to defective kinetochores, the observed lower solubility of CID nucleosomes suggests a stabilizing mechanism important to maintain high concentrations of CID at centromeres. Such a mechanism may involve the constitutive centromere components CAL1 and CENP-C, whose depletion causes loss of CID from centromeres in flies (Erhardt et al., 2008; Goshima et al., 2007).
A conserved CID residue in the four helix bundle is required for CID dimerization and centromere localization
This study strongly supports the idea that CENP-A nucleosomes form through deposition of two CENP-A/H4 molecules. Several studies have emphasized an essential role for the CATD in CENP-A deposition (Black et al., 2004; Foltz et al., 2009; Hu et al., 2011; Vermaak et al., 2002; Zhou et al., 2011), but have not addressed whether and how other regions of CENP-A are also required. We demonstrate dependence of CID dimerization and localization on the conserved D211 residue within the CID four helix bundle. This result indicates that the CATD per se is not sufficient for centromeric CENP-A assembly, and is consistent with the role of the four helix bundle in nucleosome integrity and functions (Camahort et al., 2007; Luger et al., 1997).
Our data suggest that D211 stabilizes the inter- and/or intra-molecular interactions of CID, yet the mislocalization phenotype of CIDD211A may also be interpreted as a result of its failure to interact with an HJURP/Scm3-like assembly factor. The CID-specific assembly factor has not been identified in flies, preventing a direct test of this hypothesis. However, a deletion encompassing the analogous Cse4 residue does not significantly interfere with Cse4-Scm3 interaction (Zhou et al., 2011). This suggests that D211 is unlikely to affect CID assembly factor interactions, and further supports our conclusion that the mislocalization phenotype is due to disruption of the four helix bundle structure and CID dimerization.
In conclusion, we have identified CID dimerization as a key property of CID nucleosomes, and demonstrated its importance to CID localization to centromeres. Our results suggest that CENP-A nucleosomes in a metazoan have identical stoichiometry to that of H3 and Cse4 nucleosomes. These observations lay a foundation for elucidating the epigenetic mechanisms responsible for centromere identity and propagation.
EXPERIMENTAL PROCEDURES
A detailed description of the methods can be found in the Supplemental Information accompanying this manuscript.
Mononucleosome immunoprecipitation
MNase-digested S2 chromatin was centrifuged on a 5–25% sucrose gradient for 22 hrs. FLAG-CID nucleosomes were immunoprecipitated by α-FLAG agarose beads (Sigma). For detecting endogenous CID in FLAG-CID nucleosomes, incubations were performed in a buffer containing 50 mM NaCl, and beads were washed in a buffer containing different salt concentrations (50 mM for low salt or 100 mM NaCl for higher salt) and 0.1% NP-40.
Covalent crosslinking
Extensively digested S2 chromatin was incubated with 0.1 volume of a solution containing 25 mM 1,10,o-phenanthroline and 12.5 mM copper sulphate for crosslinking. Crosslinks were reversed by 1 M 2-mercaptoethanol (Gould et al., 1980).
Statistics
Pearson’s coefficient of correlation for IF images was measured in SoftWorx (Applied Precision, LLC).
Highlights.
Drosophila centromeric nucleosomes contain two copies of CID
CID dimerization requires a critical residue for the four helix bundle structure
CID dimerization is essential for normal centromeric localization in vivo
Supplementary Material
ACKNOWLEDGEMENTS
We thank Robert Glaser for α-H2Av and α-H2A antibodies; James Kadonaga and Ming Dong for help with sucrose gradient ultracentrifugation; Robin Allshire for suggesting CID crosslinking; Elaine Dunleavy, Barbara Mellone and Karpen lab members for critical reading and discussions of the manuscript, and Alfred Li for technical help. This work was supported by Susan G. Komen Breast Cancer Foundation (PDF0601222 to W.Z.) and NIH (F32GM086111 to S.C. and GM666272 to G.K).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet. 2008;9:923–937. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando S, Yang H, Nozaki N, Okazaki T, Yoda K. CENP-A, -B, and -C chromatin complex that contains the I-type alpha-satellite array constitutes the prekinetochore in HeLa cells. Mol Cell Biol. 2002;22:2229–2241. doi: 10.1128/MCB.22.7.2229-2241.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Bassett EA. The histone variant CENP-A and centromere specification. Curr Opin Cell Biol. 2008;20:91–100. doi: 10.1016/j.ceb.2007.11.007. [DOI] [PubMed] [Google Scholar]
- Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Jr, Cleveland DW. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- Bloom K, Sharma S, Dokholyan NV. The path of DNA in the kinetochore. Curr Biol. 2006;16:R276–R278. doi: 10.1016/j.cub.2006.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camahort R, Li B, Florens L, Swanson SK, Washburn MP, Gerton JL. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Camahort R, Shivaraju M, Mattingly M, Li B, Nakanishi S, Zhu D, Shilatifard A, Workman JL, Gerton JL. Cse4 is part of an octameric nucleosome in budding yeast. Mol Cell. 2009;35:794–805. doi: 10.1016/j.molcel.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde e Silva N, Black BE, Sivolob A, Filipski J, Cleveland DW, Prunell A. CENP-A-containing nucleosomes: easier disassembly versus exclusive centromeric localization. J Mol Biol. 2007;370:555–573. doi: 10.1016/j.jmb.2007.04.064. [DOI] [PubMed] [Google Scholar]
- Dalal Y, Wang H, Lindsay S, Henikoff S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007;5:e218. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitriadis EK, Weber C, Gill RK, Diekmann S, Dalal Y. Tetrameric organization of vertebrate centromeric nucleosomes. Proc Natl Acad Sci U S A. 2010;107:20317–20322. doi: 10.1073/pnas.1009563107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy E, Almouzni G, Karpen G. H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G1 phase. Nucleus. 2011;2 doi: 10.4161/nucl.2.2.15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt S, Mellone BG, Betts CM, Zhang W, Karpen GH, Straight AF. Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J Cell Biol. 2008;183:805–818. doi: 10.1083/jcb.200806038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, Yates JR, 3rd, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, 3rd, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- Goshima G, Wollman R, Goodwin SS, Zhang N, Scholey JM, Vale RD, Stuurman N. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould HJ, Cowling GJ, Harborne NR, Allan J. An examination of models for chromatin transcription. Nucleic Acids Res. 1980;8:5255–5266. doi: 10.1093/nar/8.22.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewawasam G, Shivaraju M, Mattingly M, Venkatesh S, Martin-Brown S, Florens L, Workman JL, Gerton JL. Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol Cell. 2011;40:444–454. doi: 10.1016/j.molcel.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Liu Y, Wang M, Fang J, Huang H, Yang N, Li Y, Wang J, Yao X, Shi Y, et al. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 2011 doi: 10.1101/gad.2045111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- Moreno-Moreno O, Medina-Giro S, Torras-Llort M, Azorin F. The F Box Protein Partner of Paired Regulates Stability of Drosophila Centromeric Histone H3, CenH3(CID) Curr Biol. 2011;21:1488–1493. doi: 10.1016/j.cub.2011.07.041. [DOI] [PubMed] [Google Scholar]
- Olszak AM, van Essen D, Pereira AJ, Diehl S, Manke T, Maiato H, Saccani S, Heun P. Heterochromatin boundaries are hotspots for de novo kinetochore formation. Nat Cell Biol. 2011 doi: 10.1038/ncb2272. [DOI] [PubMed] [Google Scholar]
- Ranjitkar P, Press MO, Yi X, Baker R, Maccoss MJ, Biggins S. An E3 Ubiquitin Ligase Prevents Ectopic Localization of the Centromeric Histone H3 Variant via the Centromere Targeting Domain. Mol Cell. 2011;40:455–464. doi: 10.1016/j.molcel.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekulic N, Bassett EA, Rogers DJ, Black BE. The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres. Nature. 2010;467:347–351. doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby RD, Vafa O, Sullivan KF. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, Hayashi-Takanaka Y, Oda T, Sato M, Park SY, et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011 doi: 10.1038/nature10258. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- Vermaak D, Hayden HS, Henikoff S. Centromere targeting element within the histone fold domain of Cid. Mol Cell Biol. 2002;22:7553–7561. doi: 10.1128/MCB.22.21.7553-7561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda K, Ando S, Morishita S, Houmura K, Hashimoto K, Takeyasu K, Okazaki T. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc Natl Acad Sci U S A. 2000;97:7266–7271. doi: 10.1073/pnas.130189697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Feng H, Zhou BR, Ghirlando R, Hu K, Zwolak A, Miller Jenkins LM, Xiao H, Tjandra N, Wu C, et al. Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature. 2011;472:234–237. doi: 10.1038/nature09854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.