Abstract
Objective
The Fc receptor like 3 (FCRL3) molecule, involved in controlling B cell signalling, may contribute to the autoimmune disease process. Recently a genome wide screen detected association of neighbouring gene FCRL5 with Graves’ disease (GD). To determine whether FCRL5 represents a further independent B cell signaling GD susceptibility loci we screened 12 tag SNPs, capturing all known common variation within FCRL5, in 5192 UK Caucasian GD index cases and controls.
Design
A case control association study investigating twelve tag SNPs within FCRL5 which captured the majority of known common variation within this gene region.
Patients
A dataset comprising 2504 UK Caucasian GD patients and 2688 geographically matched controls taken from the 1958 British Birth cohort.
Measurements
We used the chi-squared test and haplotype analysis to investigate association between the tag SNPs and GD before performing regression analysis to determine if association at FCRL5 was independent of the known FCRL3 association.
Results
Three of the FCRL5 tag SNPs, rs6667109, rs3811035 and rs6692977 showed association with GD (P=0.015-0.001, OR=1.15-1.16). Logistic regression performed on all FCRL5 and, previously screened, FCRL3 tag SNPs revealed that association with FCRL5 was secondary to linkage disequilibrium with the FCRL3, rs11264798 and rs10489678 SNPs.
Conclusions
FCRL5 does not appear to be exerting an independent effect on the development of GD in the UK. Fine mapping of the entire FCRL region is required to determine the exact location of the etiological variant/s present.
Keywords: Linkage disequilibrium, FCRL3, FCRL5, Graves’ disease, genome wide screening
Introduction
The Fc receptor like (FCRL) gene family, which encode products that play a key role in controlling B cell signalling1, 2, has been shown to be associated with several autoimmune diseases (AIDs). Originally, single nucleotide polymorphism (SNP) screening of chromosome 1q23 in a Japanese rheumatoid arthritis (RA) dataset narrowed down association to within two linkage disequilibrium (LD) blocks, which encoded five FCRL family members, FCRL1-FCRL5 and lymphocyte antigen CD5-like (CD5L)3. Association was further refined to four SNPs within FCRL3 with functional studies revealing that the rs7528684 SNP affected NF-κB binding to FCRL3, thereby causing variation in FCRL3 expression on B cells, with the potential to impact on B cell regulation3. Further functional work has confirmed an inhibitory role for FCRL3 in B cell signalling, with the disease risk genotype causing variation in inhibitory signalling, which in turn could lead to a breakdown in B cell tolerance and autoreactive B cell activity4. Association of the rs7528684 SNP was also replicated in Japanese Graves’ disease (GD) (P=1.7×10−5, OR=1.79) and Systemic lupus erythematosus (SLE) (P=0.0017) subjects3.
Replication of FCRL3 within many AIDs including RA, SLE and type 1 diabetes (T1D) has been inconsistent1, 5. However in GD we replicated association of rs7528684 (and the other three SNPs which we later found to be in complete linkage disequilibrium (LD) with rs7528684) in a UK Caucasian GD cohort of 983 GD patients and 733 controls, although the effect was smaller than that seen in the Japanese GD dataset (P=0.024, OR=1.17)3, 6. Association of additional FCRL3 SNPs was also detected in a smaller independent GD dataset consisting of 625 GD patients and 490 controls (OR=1.50 and 1.25, respectively)7 and within a 15,000 non-synonymous SNP genome wide screen performed by the Wellcome Trust Case Control Consortium (WTCCC) within 900 GD cases and 1500 controls8. Seven tag SNPs employed to screen the whole FCRL3 region for association with GD in a Caucasian collection of 2280 GD and 2616 controls8, revealed association with disease of rs3761959 (P=9.4×10−3, OR=1.16) (which tags previously associated SNPs rs7522061 and rs7528684 with r2>0.80) and another more strongly associated SNP rs11264798 (P=1.6×10−5, OR=1.22)8.
The WTCCC non-synonymous SNP screen also reported association of three SNPs within FCRL5 (P=1.3×10−4-2.8×10−4) with GD, located within the same region as FCRL3 (approximately 125kb away)8. However, the typing of rs6667109, which tagged all three associated FCRL5 SNPs, in our GD dataset of 2280 cases and 2616 controls showed only a trend towards association with GD (P=0.077)8. The aim of this study, therefore, was to use tag SNPs to screen all common variation within FCRL5 in our UK Caucasian GD population to determine if other tag SNPs within FCRL5 are more strongly associated with GD and whether association is independent of the previously detected FCRL3 associations.
Materials and Methods
Subjects
A dataset consisting of 2504 unrelated white Caucasian patients with GD from the UK National Autoimmune Thyroid Collection was screened. Patients were recruited from specialist clinics in Birmingham, Bournemouth, Cambridge, Cardiff, Exeter, Leeds, Sheffield and Newcastle in the UK. Clinical definition of GD was as previously described9. A total of 2688 geographically matched white Caucasian control subjects were also obtained from the 1958 British Birth cohort. All patients gave informed written consent and the project was approved by the local ethics committee.
Tag SNP selection and genotyping
Genotyping data for the 39.14 kb FCRL5 region (Phase 2, NCBI Build 36, Caucasian (CEU) population) was downloaded from the International Haplotype Mapping Project website (http://www.hapmap.org). Our analysis of Hapmap genotyping data identified 40 SNPs with minor allele frequencies of ≥5% in the CEU population present within this region. The Tagger pairwise function within Hapmap was used to assign tag SNPs. Twelve tag SNPs: rs6667109, rs12036228, rs6692977, rs6427383, rs3811033, rs3811035, rs3811036, rs3843307, rs17416676, rs3900770, rs3845586 and rs1332714, were chosen to capture the majority of the common variation within this gene and the surrounding area with a minimum r2 of 0.80 (for information on the SNPs which they tag see Supplementary Table 1). The rs6667109 SNP was previously typed as part of the WTCCC genome screen8, in which we used a smaller subsection of our current case control dataset8. We have subsequently increased our dataset by an additional 224 GD cases and 72 controls. As all of our current dataset is being used for this study we wanted to bring the number of GD samples and controls investigated for rs6667109 in line with those investigated for all the other FCRL5 tag SNPs, so screened rs6667109 in the additional 224 GD cases and 72 controls not previously typed.
Assays for all of the above SNPs were purchased from Applied Biosystems, Warrington, UK and genotyped on an ABI7900HT using Taqman® genotyping technologies (Applied Biosystems, UK).
Statistical and haplotype analyses
All SNPs were in Hardy Weinberg equilibrium in both the cases and controls (P=0.92-0.05). When investigating prospective power in our dataset we had >80% power to exclude an OR>1.13-1.24 at α = 0.05 for our selected tag SNPs (see Supplementary Table 1 for the power associated with each individual tag SNP). Allelic and genotypic analysis of case-control data was performed using the χ2-test within the MINITAB statistical package (MINITAB Release 15.1.2, © 1972-2007, Minitab Inc., State College, PA, USA). Odds ratios (OR) with 95% confidence intervals (CI) were calculated by the method of Woolf with Haldane’s modification for small numbers, where appropriate10. Linkage disequilibrium (LD) between tag SNPs was analyzed using the pairwise LD measure D′ and r2 and haplotype blocks constructed using the solid spine haplotype algorithm within the computer program Haploview Version 3.2 (http://www.broad.mit.edu/mpg/haploview). Logistic regression and imputation analysis was performed using the PLINK software package11.
Results
FCRL5 genotyping results
Out of the twelve tag SNPs screened, the rs6667109 SNP, which when previously typed in a smaller sub-section of our case control dataset only showed a trend towards association with GD8, was weakly associated with GD in our larger cohort of 2504 GD index cases and 2688 controls (P=0.010, OR=1.16 [95% CI=1.04-1.30]) (see Table 1). Two additional SNPs, rs3811035 (P=0.001, OR=1.15 [95% CI=1.06-1.25]) and rs6692977 (P=0.015, OR=1.11 [95% CI=1.02-1.21]), were also associated with GD in our larger dataset. A minor difference in allele frequency between index cases and controls was also observed with SNPs rs3843307 (P=0.037) and rs3845586 (P=0.019). None of the remaining eight tag SNPs showed any association with GD (P=0.988-0.063) see Table 1.
Table 1.
Allelic and genotypic association of FCRL5 tag SNPs within a large UK Caucasian Graves’ disease cohort
| FCRL5 | Genotype /Allele |
Graves’ disease (%) |
Control subjects (%) |
Chi-squared | P | OR | 95% CI | ||
|---|---|---|---|---|---|---|---|---|---|
| rs3845586 | A | 2842 | (60.4) | 3125 | (62.7) | 5.461 | 0.019 | 1.10 | 1.02-1.20 |
| G | 1866 | (39.6) | 1861 | (37.3) | |||||
| AA | 853 | (36.2) | 973 | (39.0) | 5.519 | 0.063 | |||
| AG | 1136 | (48.3) | 1179 | (47.3) | |||||
| GG | 365 | (15.5) | 341 | (13.7) | |||||
| rs3843307 | G | 3079 | (66.1) | 3355 | (68.1) | 4.372 | 0.037 | 1.09 | 1.01-1.119 |
| A | 1581 | (33.9) | 1573 | (31.9) | |||||
| GG | 1004 | (43.1) | 1131 | (45.9) | 4.491 | 0.106 | |||
| GA | 1071 | (46.0) | 1093 | (44.4) | |||||
| AA | 255 | (10.9) | 240 | (9.7) | |||||
| rs3900770 | A | 4490 | (94.2) | 4738 | (94.2) | 0.037 | 0.847 | 0.98 | 0.83-1.17 |
| G | 274 | (5.8) | 294 | (5.8) | |||||
| AA | 2115 | (88.8) | 2232 | (88.7) | 0.380 | 0.827 | |||
| AG | 260 | (10.9) | 274 | (10.9) | |||||
| GG | 7 | (0.3) | 10 | (0.4) | |||||
| rs3811036 | T | 4390 | (92.5) | 4505 | (92.1) | 0.569 | 0.451 | 0.94 | 0.81-1.10 |
| G | 356 | (7.5) | 387 | (7.9) | |||||
| TT | 2030 | (85.5) | 2078 | (85.0) | 1.112 | 0.574 | |||
| TG | 330 | (13.9) | 349 | (14.2) | |||||
| GG | 13 | (0.6) | 19 | (0.8) | |||||
| rs3811035 | A | 2733 | (61.4) | 3242 | (64.7) | 11.009 | 0.001 | 1.15 | 1.06-1.25 |
| G | 1719 | (38.6) | 1770 | (35.3) | |||||
| AA | 830 | (37.3) | 1030 | (41.1) | 11.610 | 0.003 | |||
| AG | 1073 | (48.2) | 1182 | (47.2) | |||||
| GG | 323 | (14.5) | 294 | (11.7) | |||||
| rs6692977 | C | 2883 | (63.8) | 3257 | (66.3) | 5.886 | 0.015 | 1.11 | 1.02-1.21 |
| T | 1629 | (36.1) | 1657 | (33.7) | |||||
| CC | 930 | (41.2) | 1076 | (43.8) | 6.485 | 0.039 | |||
| CT | 1023 | (45.3) | 1105 | (45.0) | |||||
| TT | 303 | (13.4) | 276 | (11.2) | |||||
| rs3811033 | G | 3718 | (81.6) | 4130 | (82.8) | 2.055 | 0.152 | 1.08 | 0.97-1.20 |
| A | 836 | (18.4) | 860 | (17.2) | |||||
| GG | 1524 | (66.9) | 1702 | (68.2) | 3.824 | 0.148 | |||
| GA | 670 | (29.4) | 726 | (29.1) | |||||
| AA | 83 | (3.7) | 67 | (2.7) | |||||
| rs1332714 | T | 3132 | (70.3) | 3431 | (70.1) | 0.026 | 0.872 | 0.99 | 0.91-1.08 |
| G | 1324 | (29.7) | 1461 | (29.9) | |||||
| TT | 1117 | (50.1) | 1221 | (49.9) | 0.025 | 0.988 | |||
| TG | 898 | (40.3) | 989 | (40.4) | |||||
| GG | 213 | (9.6) | 236 | (9.7) | |||||
| rs6427383 | A | 3539 | (77.8) | 3968 | (79.1) | 2.486 | 0.115 | 0.99 | 0.91-1.08 |
| T | 1011 | (22.2) | 1048 | (20.9) | |||||
| AA | 1382 | (60.7) | 1575 | (62.8) | 2.452 | 0.294 | |||
| AT | 775 | (34.1) | 818 | (32.6) | |||||
| TT | 118 | (5.2) | 115 | (4.6) | |||||
| rs17416676 | G | 3128 | (70.6) | 3475 | (71.2) | 0.370 | 0.543 | 1.03 | 0.94-1.12 |
| A | 1304 | (29.4) | 1409 | (28.8) | |||||
| GG | 1107 | (50.0) | 1256 | (51.4) | 1.589 | 0.452 | |||
| AG | 914 | (41.2) | 963 | (39.5) | |||||
| AA | 195 | (8.8) | 223 | (9.1) | |||||
| rs6667109 * | T | 3445 | (82.5) | 3912 | (84.6) | 6.601 | 0.010 | 1.16 | 1.04-1.30 |
| A | 729 | (17.5) | 714 | (15.4) | |||||
| TT | 1428 | (68.4) | 1660 | (71.8) | 6.465 | 0.039 | |||
| TA | 589 | (28.2) | 592 | (25.6) | |||||
| AA | 70 | (3.4) | 61 | (2.6) | |||||
| rs12036228 | C | 3639 | (77.7) | 3981 | (78.9) | 1.858 | 0.173 | 1.07 | 0.97-1.18 |
| T | 1043 | (22.3) | 1067 | (21.1) | |||||
| CC | 1420 | (60.7) | 1572 | (62.3) | 1.930 | 0.381 | |||
| CT | 799 | (34.1) | 837 | (33.2) | |||||
| TT | 122 | (5.2) | 115 | (4.5) | |||||
All SNPs are shown within their forward oriental (5′-3′). All SNPs which showed association at both the allelic and genotypic level are highlighted in bold. All P values are shown uncorrected. χ2 = Chi-squared, OR = Odds Ratio, 95% CI = 95% Confidence Intervals.
This SNP was previously typed in a subsection of our GD case control dataset as part of the WTCCC genome screen8 and so to bring the numbers of GD samples and controls in line with those investigated for all the other FCRL5 tag SNPs we screened an additional 224 GD cases and 72 controls.
Haplotype analysis between FCRL5 and FCRL3
Although tag SNPs are selected to take into account known LD within a gene region, tag SNPs information is only based on reference data within Hapmap generated from screening 90 CEPH individuals which provides us with a generalized overview of the LD within the region. The additional data obtained from typing larger datasets allows us to carefully dissect local haplotype structures which may not be evident in the smaller reference dataset from which tag SNPs are chosen. As FCRL3 is within 125kb of FCRL5 and as seven FCRL3 tag SNPs (representing all known common variation within this gene region) were also screened in our GD dataset8, we performed a haplotype analysis of FCRL3 and FCRL5 tag SNPs (see Figure 1).
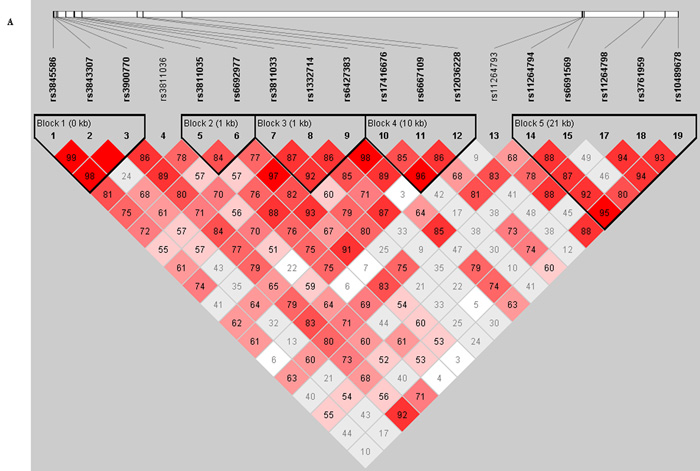
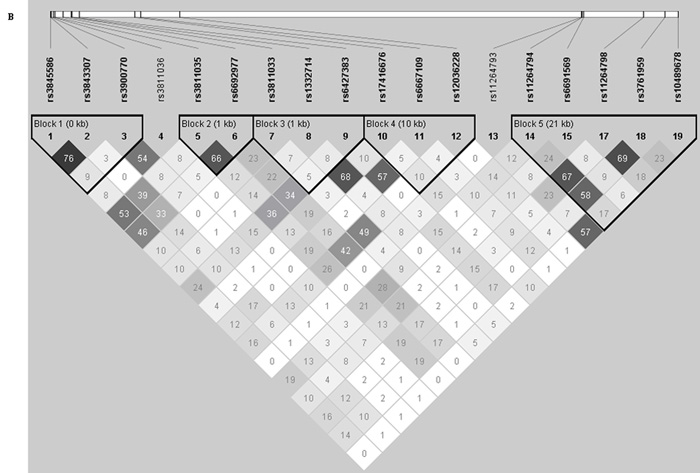
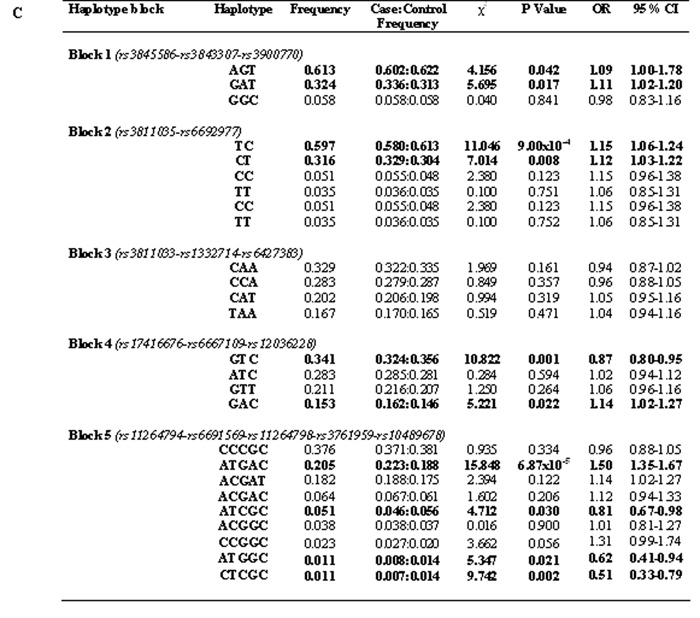
Figure 1. Haplotype analysis between FCRL5 and FCRL3 tag SNPs.
Linkage disequilibrium (LD) between tag SNPs was analyzed using the pairwise LD measure D′ and r2 and haplotype blocks constructed using the solid spine haplotype algorithm within the computer program Haploview Version 3.2 (http://www.broad.mit.edu/mpg/haploview). The solid spine haplotype method searches for a "spine" of strong LD running from one marker to another along the base of the triangle in the LD chart (this would mean that the first and last markers in a block are in strong LD with all intermediate markers but that the intermediate markers are not necessarily in LD with each other). Each diamond either represents the D′ or r2 measure of LD. A: Displays the LD parameter, D′, where the darker shades of red represent greater D′-values. B: Shows the correlation coefficient of LD, r2, darker shades of black represent greater r2 values. C: Shows association of haplotypes produced by Haplotype Block 1, Haplotype Block 2 and Haplotype Block 4 within FCRL5 and Haplotype Block 5 within FCRL3 (highlighted in bold). χ2 = Chi-squared, OR = Odds Ratio, 95% CI = 95% Confidence Intervals.
All P values are represented as uncorrected.
No haplotype blocks were detected between FCRL3 and FCRL5. Four LD blocks were detected within FCRL5. Association of the rs3811035 allele T-rs6692977 allele C in haplotype block 2 showed the strongest association with GD (P=9.00x10−4, OR=1.15 [95% CI=1.06-1.24]). Other smaller associations were also observed in block 2 (P=0.008), block 1 (P=0.042-0.017) and in block 4 which was comprised of the previously associated rs6667109 SNP and the rs17416676 and rs6667109 SNPs (P=0.022-0.001). One haplotype block (block 5) was detected within FCRL3 spanning the previously associated rs11264798 and rs3761959 SNPs and previously non-associated rs112644794, rs6691569 and rs10489678 SNPs. This block produced one common haplotype block strongly associated with GD (P=6.87×10−5) and several rarer protective haplotypes with minor allele frequencies ≤5.1% (P=0.030-0.002) see Figure 1.
Logistic Regression Analysis
Logistic regression analysis was performed to determine if association present within FCRL5 was independent of previously detected associations within neighbouring FCRL3. Logistic regression analysis was performed using the PLINK software package11. When we first entered all of the FCRL3 and FCRL5 tag SNPs into a regression model the FCRL3 rs11264798 SNP was shown to be the most associated SNP within the model (P=1.84x10−7). We found that the rs11264798 SNP improved all other FCRL3 and FCRL5 SNP associations (P=0.004-7.95x10−8). When conditioning for the effects of the rs11264798 SNP, all of the other SNPs except rs10489678 (P=0.001), which previously showed no association with GD, were no longer associated with disease (P=0.984-0.082). When conditioning on both rs11264798 and rs10489678 no other SNP within the model remained associated with GD (P=0.872-0.057), suggesting that association within FCRL5 was the result of LD with the FCRL3 rs11264798 and rs10489678 SNPs.
Interestingly when screened in our case control study the rs10489678 SNP was not associated with GD (see Table 1). There was little evidence for historical recombination between rs10489678 and rs11264798 (D′ = 0.94), but the r2 = 0.19 suggested these SNPs were at least partially independent (see Figure 1). Haplotype analysis revealed that rs10489678 formed part of a haplotype block within FCRL3 containing rs11264798 together with rs11264794, rs6691569 and rs3761959 (see Figure 1 block 5). The most strongly associated haplotype within FCRL3 contains the rs11264798 disease associated allele G and the rs10489678 allele C (P=6.87x10−5) present within 22.3% of the cases and 18.8% of the controls. However when analysing the only haplotype to contain the rs11264798 allele G and the rs10489678 allele T, this haplotype which was present in 18.8% of cases and 17.5% of controls showed no association with GD (P=0.1218) (see Figure 1). Furthermore, several other less common haplotypes with the rs11264798 allele G-rs10489678 allele C were not associated with GD. This suggests that the causal variant could lie exclusively on the most common rs11264798 allele G-rs10489678 allele C haplotype.
Discussion
Genome wide screening of 15,000 non-synonymous SNPs in a UK Caucasian GD cohort not only confirmed association of the previously detected GD susceptibility loci FCRL3 but also detected association of neighbouring FCRL5 with GD8. In our present study we performed tag SNP screening of FCRL5 in an enlarged dataset, confirming association of this gene with GD. Logistic regression analysis performed on FCRL5, and previously typed FCRL3 tag SNPs, revealed that any associations within FCRL5 were the result of LD with FCRL3 rs11264798 and rs10489678 tag SNPs. Although association at FCRL5 was shown to be secondary to association at FCRL3, we are unable to say that FCRL3 contains the etiological variant represented by these associations. Whilst it may be tempting to embark upon a functional analysis of rs11264798 (the most strongly associated FCRL3 tag SNP), haplotype analysis suggests that this may not be the etiological variant, as the high risk G allele is only associated when present with the rs10489678 C allele (not the rs10489678 T allele), suggesting that the etiological variant could be elsewhere within this region.
When trying to determine the Crohn’s disease etiological variant represented by associated markers present within a 1.25-Mb gene desert on chromosome 5, LD was investigated between these markers and a series of SNPs identified by Dixon et al that affected gene expression12. Markers within this 1.25Mb gene desert, which contains no known genes or CpG islands which are a common feature of many gene promoters, were shown to be associated with neighbouring SNPs known to alter Prostaglandin E Receptor 4 (PTGER4) gene expression, thereby suggesting neighbouring PTGER4 as a major potential susceptibility locus for Crohn’s disease within this region of linkage13. As Dixon et al12 identified SNPs which altered gene expression throughout the genome, we investigated whether known gene expression altering SNPs in neighbouring genes could also explain association of rs11264798 or rs10489678 with GD. Although eight functional gene expression altering SNPs were located 200 kb either side of rs11264798 or rs10489678, additional analysis revealed that there was no LD between any of these eight SNPs and either rs11264798 or rs10489678, suggesting that this approach could not provide further insights into the location of the etiological variants within the FCRL region for GD.
As there are numerous other members of the FCRL family encoded within this region, all of which play important roles in B cell signalling1, 2, detailed mapping of the entire region is essential to enable the etiological variant/s to be localized before determining how variation in B cell signalling could be aiding autoimmune onset. To try to determine if other stronger associations than rs11264798 and rs10489678 could exist elsewhere within the FCRL region, in particular within FCRL4 which is located between FCRL3 and FCRL5, we performed imputation analysis within the PLINK software package11 to statistically predict additional SNP genotyping results within the surrounding region. Although a further ninety six SNPs were imputed within the region, including eleven SNPs located within FCRL4, the FCRL3 rs11264798 SNP remained the most associated SNP within this region (see Supplementary Figure 1 for further details).
Interestingly association of the FCRL3 rs7528684 SNP, identified by regression in the Oriental population to be the most associated SNP within this region, was not replicated in many Caucasian AIDs datasets, apart from GD1, 5. Although we detected association of rs3761959, that tagged rs7528684, with GD, logistic regression showed that association of rs3761759 was secondary to the FCRL3 rs11264798 and rs10489678 SNPs. This suggests that rs3761759, and by proxy rs7528684, is not the likely FCRL etiological variant in our Caucasian GD population. Although studies on the rs7528684 SNP in many other AIDs has ruled out association of FCRL3 with these diseases, once the FCRL etiological variant/s have been established for Caucasian GD patients these can then be screened in additional Caucasian AID datasets to confirm whether or not this gene region is associated with other AIDs within the Caucasian population.
This study highlights the well described effects of LD between genes and the problems that this poses when attempting to identify primary aetiological disease causing DNA variants14. Further fine mapping within the entire FCRL region is needed to confirm the imputation results and to enable localisation of the GD etiological DNA variant/s present before embarking upon functional studies to determine how variation in B cell signalling molecules could be contributing to GD.
Supplementary Material
Supplementary Table 1: Tag SNPs present in FCRL5, the SNPs which they capture and the level of OR that can be excluded for these SNPs
Supplementary Figure 1: Imputation analysis of the FCRL region based on FCRL3 and FCRL5 tag SNP data and data available from Hapmap Phase II
Data from FCRL3 and FCRL5 tag SNPs typed in our GD cohort was merged with Hapmap Phase II genotyping data (release 23) on 90 CEPH individuals from 200kb either side of the FCRL3 rs11264798 and rs10489678 SNPs. This data was then used to perform imputation analysis within the PLINK software package11 to statistically predict additional SNP genotyping results within the surrounding region in our case control dataset. A: PLINK imputed an additional 96 SNPs (highlighted in blue) within the surrounding FCRL region and combined with our 19 previously typed tag SNPs (highlighted in red) the results were plotted (−log P value) against their chromosome position with the most associated SNP within the region highlighted. The dotted line represents the cut off for association and any SNPs above this line show association with GD. B: Location of FCRL family members relative to chromosomal location.
Acknowledgements
We would like to thank patients, nurses and doctors for recruiting into the AITD National Collection. We also wish to acknowledge The Society for Endocrinology for awarding MJS a small project grant and the Wellcome Trust for funding this project. We acknowledge use of DNA from the British 1958 Birth Cohort Collection, funded by the Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02.
Footnotes
Conflicts of interest None of the authors have any conflicts of interest to declare in connection with this manuscript.
The authors have no relevant financial or other conflicting interests to declare.
References
- 1.Davis RS. Fc receptor-like molecules. Annu Rev Immunol. 2007;25:525–560. doi: 10.1146/annurev.immunol.25.022106.141541. [DOI] [PubMed] [Google Scholar]
- 2.Ehrhardt GR, Leu CM, Zhang S, Aksu G, Jackson T, Haga C, Hsu JT, Schreeder DM, Davis RS, Cooper MD. Fc receptor-like proteins (FCRL): immunomodulators of B cell function. Adv Exp Med Biol. 2007;596:155–162. doi: 10.1007/0-387-46530-8_14. [DOI] [PubMed] [Google Scholar]
- 3.Kochi Y, Yamada R, Suzuki A, Harley JB, Shirasawa S, Sawada T, Bae SC, Tokuhiro S, Chang X, Sekine A, Takahashi A, Tsunoda T, Ohnishi Y, Kaufman KM, Kang CP, Kang C, Otsubo S, Yumura W, Mimori A, Koike T, Nakamura Y, Sasazuki T, Yamamoto K. A functional variant in FCRL3, encoding Fc receptor-like 3, is associated with rheumatoid arthritis and several autoimmunities. Nat Genet. 2005;37:478–485. doi: 10.1038/ng1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kochi Y, Myouzen K, Yamada R, Suzuki A, Kurosaki T, Nakamura Y, Yamamoto K. FCRL3, an autoimmune susceptibility gene, has inhibitory potential on B-cell receptor-mediated signaling. J Immunol. 2009;183:5502–5510. doi: 10.4049/jimmunol.0901982. [DOI] [PubMed] [Google Scholar]
- 5.Chistiakov DA, Chistiakov AP. Is FCRL3 a new general autoimmunity gene? Hum Immunol. 2007;68:375–383. doi: 10.1016/j.humimm.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 6.Simmonds MJ, Heward JM, Carr-Smith J, Foxall H, Franklyn JA, Gough SC. Contribution of single nucleotide polymorphisms within FCRL3 and MAP3K7IP2 to the pathogenesis of Graves’ disease. J Clin Endocrinol Metab. 2006;91:1056–1061. doi: 10.1210/jc.2005-1634. [DOI] [PubMed] [Google Scholar]
- 7.Owen CJ, Kelly H, Eden JA, Merriman ME, Pearce SH, Merriman TR. Analysis of the Fc receptor-like-3 (FCRL3) locus in Caucasians with autoimmune disorders suggests a complex pattern of disease association. J Clin Endocrinol Metab. 2007;92:1106–1111. doi: 10.1210/jc.2006-2183. [DOI] [PubMed] [Google Scholar]
- 8.WTCCC & TACS Association study of 14,500 nsSNPs in four common diseases identifies variants involved in autoimmunity. Nature Genetics. 2007;39:1329–1337. doi: 10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manji N, Carr-Smith JD, Boelaert K, Allahabadia A, Armitage M, Chatterjee VK, Lazarus JH, Pearce SH, Vaidya B, Gough SC, Franklyn JA. Influences of age, gender, smoking, and family history on autoimmune thyroid disease phenotype. J Clin Endocrinol Metab. 2006;91:4873–4880. doi: 10.1210/jc.2006-1402. [DOI] [PubMed] [Google Scholar]
- 10.Mathews JD. Statistical aspects of immunogenetic association with disease. In: Simons MJ, Tait BD, editors. Detection of immune-associated genetic markers of human disease. Churchill Livingstone, London: 1984. pp. 106–136. [Google Scholar]
- 11.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KCC, Taylor J, Burnett E, Gut I, Farrall M, Lathrop GM, Abecasis GR, Cookson WOC. A genome-wide association study of global gene expression. Nature Genetics. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 13.Libioulle C, Louis E, Hansoul S, Sandor C, Farnir F, Franchimont D, Vermeire S, Dewit O, de Vos M, Dixon A, Demarche B, Gut I, Heath S, Foglio M, Liang LM, Laukens D, Mni M, Zelenika D, Van Gossum A, Rutgeerts P, Belaiche J, Lathrop M, Georges M. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. Plos Genetics. 2007;3 doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarthy MI, Hirschhorn JN. Genome-wide association studies: potential next steps on a genetic journey. Hum Mol Genet. 2008;17:R156–165. doi: 10.1093/hmg/ddn289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: Tag SNPs present in FCRL5, the SNPs which they capture and the level of OR that can be excluded for these SNPs
Supplementary Figure 1: Imputation analysis of the FCRL region based on FCRL3 and FCRL5 tag SNP data and data available from Hapmap Phase II
Data from FCRL3 and FCRL5 tag SNPs typed in our GD cohort was merged with Hapmap Phase II genotyping data (release 23) on 90 CEPH individuals from 200kb either side of the FCRL3 rs11264798 and rs10489678 SNPs. This data was then used to perform imputation analysis within the PLINK software package11 to statistically predict additional SNP genotyping results within the surrounding region in our case control dataset. A: PLINK imputed an additional 96 SNPs (highlighted in blue) within the surrounding FCRL region and combined with our 19 previously typed tag SNPs (highlighted in red) the results were plotted (−log P value) against their chromosome position with the most associated SNP within the region highlighted. The dotted line represents the cut off for association and any SNPs above this line show association with GD. B: Location of FCRL family members relative to chromosomal location.