Abstract
Although sex chromosomes have been the focus of a great deal of scientific scrutiny, most interest has centred on understanding the evolution and relative importance of X and Z chromosomes. By contrast, the sex-limited W and Y chromosomes have received far less attention, both because of their generally degenerate nature and the difficulty in studying non-recombining and often highly heterochromatic genomic regions. However, recent theory and empirical evidence suggest that the W and Y chromosomes play a far more important role in sex-specific fitness traits than would be expected based on their size alone, and this importance may explain the persistence of some Y and W chromosomes in the face of powerful degradative forces. In addition to their role in fertility and fecundity, the sex-limited nature of these genomic regions results in unique evolutionary forces acting on Y and W chromosomes, implicating them as potentially major contributors to sexual selection and speciation. Recent empirical studies have borne out these predictions and revealed that some W and Y chromosomes play a vital role in key sex-specific evolutionary processes.
Keywords: Y chromosome, W chromosome, Sexual selection, Gene acquisition, Gene conversion, Non-homologous recombination
Although they generally represent a restricted proportion of any given genome, sex chromosomes are fascinating because of their unique evolutionary properties. The two main diploid varieties of sex chromosome systems are classed by whether the female or male is the heterogametic sex, and although male heterogamety seems to be more common overall than female heterogamety, both types of sex chromosome systems have arisen numerous independent times in animals as well as plants (Charlesworth and Mank 2010; Bachtrog et al. 2011), offering numerous potential clades for the study of sex chromosome evolution.
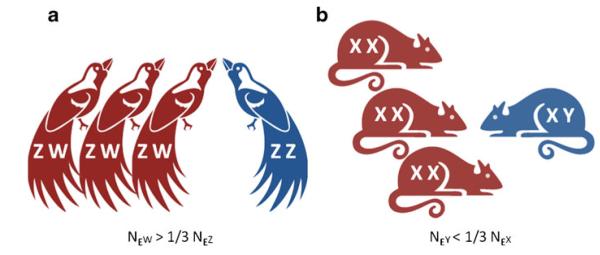
Within male and female heterogametic systems, there are four types of sex chromosomes. In female heterogamety, the homogametic sex (males) carry two Z chromosomes, and the heterogametic sex (females) has one Z and one W. In male heterogamety, females (the homogametic sex) have two X chromosomes and males (the heterogametic sex) carry one X and one Y chromosome. Therefore, X and Z chromosomes are similar in that they are present in both sexes, albeit in unequal amounts. By contrast, W and Y chromosomes are sex-limited, with the Y present solely in males and the W restricted to females. This sex limitation greatly reduces their number, and for every reproducing male–female pair, there is only one W or Y chromosome for every three Z or X chromosomes (Fig. 1). I will therefore refer to the W and Y as minor sex chromosomes, and will use the term major sex chromosomes in reference to the X and Z. These terms are based on abundance and do not refer in any way to size (discussed below) or relative effect. In fact, the effect of the minor sex chromosomes is, in many ways, anything but minor.
Fig. 1.
Most sex chromosomes come in either of two primary types. a Animals such as birds, lepidopterans and plants such as wild strawberry (Spigler et al. 2008) show a pattern of female heterogamety, with a major Z chromosome and minor W chromosome. The W chromosome is limited to females, and therefore the effective population size of the W (NEW) is less than NEZ. b In male heterogamety, which is present in animals such and mammals and salmon (Woram et al. 2003), as well as plants including Silene (Guttman and Charlesworth 1998), the X chromosome is present in both sexes while the Y is restricted to males. NEY is less than NEX
Although minor sex chromosomes are sometimes far smaller than their major chromosome partner, this is not always, or even often, the case. In some recent sex chromosomes, the minor sex chromosomes are actually larger than the major chromosome, as is the case in papaya (Liu et al. 2004). However, major and minor sex chromosome pairs are more often largely homomorphic, differing little in overall size and gene content. This is true for most origins of sex chromosomes in many fish (Devlin and Nagahama 2002; Mank et al. 2006), plants (Matsunaga and Kawano 2001; Ming et al. 2007), reptiles (Ezaz et al. 2009; Janes et al. 2010) and amphibians (Nakamura 2009; Stöck et al. 2011a). Even in clades that share a common and orthologous pair of sex chromosomes, there can be large variation among species in the degree of difference between the major and minor sex chromosomes. For example, snakes share a homologous set of ZW sex chromosomes, and although the Z and W are highly differentiated in some species, they are largely identical in pythons (Matsubara et al. 2006). The avian Z and W chromosomes are similarly highly differentiated in most birds (Stiglec et al. 2007); however, the ratite Z and W are nearly identical and the tinamou Z and W are only partially differentiated (Ogawa et al. 1998; Tsuda et al. 2007). Interestingly, the avian Z and W chromosomes seem to have differentiated multiple independent times, rather than once in the ancestor of modern birds, as the paleognath (ratites and tinamous) and neognath (all other modern birds) divergence of the Z and W are convergent but unrelated (Mank and Ellegren 2007; Suh et al. 2011). The fact that degeneration does not always correlate with age suggests sex chromosome differentiation is not an inevitable process. Although the exact reasons for this are not known, the role of haploid selection may explain some cases of old but preserved minor sex chromosome. In organisms where the haploid phase is considerable, such as plants, a degenerated Y or W chromosome would leave half of the gametophytes lacking significant numbers of genes (Chibalina and Filatov 2011). Haploid selection does not explain the persistence of the ratite and python W chromosomes, however, as the haploid phase does not differ between these taxa and their relatives with more degenerate sex chromosomes.
Even though highly differentiated minor sex chromosomes are relatively rare among plants and animals, they are particularly interesting due to their advanced state, and derived minor sex chromosomes share several key properties with each other. The first key feature of many minor sex chromosomes is their reduced gene content compared to the major sex chromosome. For example, just 27 distinct proteins are encoded by the male-specific region of the human Y chromosome (Skaletsky et al. 2003), compared to >1,000 found on the human X (Ross et al. 2005). Even large karyotypic minor sex chromosomes often carry very few genes. Although the Drosophila melanogaster Y chromosome is karyotypically quite large, it is completely distinct from the paired X chromosome and carries just 12 known genes (Koerich et al. 2008), a difference of two orders of magnitude from the X chromosome. In addition to the general reduction in gene content, minor sex chromosomes are often littered with repetitive sequence (Skaletsky et al. 2003; Stiglec et al. 2007), providing little evidence of functional utility at the same time as complicating complete sequencing and assembly. Finally, the minor sex chromosomes are often transcriptionally inactivated in some fashion (Turner 2007; Royo et al. 2010). The reduced gene content, heterochromatic nature and transcriptional silencing have all combined to create a perception that the minor sex chromosomes have little functional contribution beyond initial sex determination; however, as discussed below, minor sex chromosomes in many animals play an important role in functions beyond sex determination itself.
Although they are in many ways comparable to each other, W and Y chromosomes differ in one key regard, namely that the W is female-limited and the Y is present only in males. This difference makes contrasts between the evolutionary properties of the W and Y chromosome particularly revealing, as such contrasts can be used to confirm general principles. For example, the sex-limited nature of highly derived minor sex chromosomes means that any gene content not directly involved in sex determination should still function in a sex-specific manner: the Y chromosomes should play an important role in males and W chromosomes should harbour loci important to female function. This has largely been confirmed in Drosophila and mammals, where the Y chromosomes encode genes key to male fertility (Carvalho et al. 2000; Lange et al. 2009), as well as birds, where W-linked genes are expressed in the ovary (Hori et al. 2000). Additionally, sex-specific demographic forces, including mating system and sexual selection (Bachtrog et al. 2011), should act differently on W and Y chromosomes. For example, sexual selection generally acts more powerfully on males (Andersson 1994), resulting in a higher variance in male reproductive success. This means that sex-specific selection pressures in most natural systems should be stronger on Y than W chromosomes, except for the few cases of sex-role reversal (Clutton-Brock 2007). At the same time that sexual selection exerts more powerful selective forces on Y chromosomes, it also lowers the effective population size of Y chromosomes (NEY) compared to W chromosomes (NEW). As shown in Fig. 2, when the reproductive potential of males exceeds that of females, NEY is reduced compared to strictly monogamous mating systems, and NEY < NEW. This reduction in NEY means that genetic drift is a more powerful force on the Y than the W in most cases of sexual selection.
Fig. 2.
Mating system and effective population size of sex chromosomes. By altering the proportion of males and females that contribute to the next generation, or by increasing the variance in reproductive success of one sex compared to the other, the ratio of minor to major sex chromosomes is altered. For male heterogamety, increased variance in male mating success due to sexual selection reduces NEY relative to NEX (a). For female heterogamety (b) mating system results in increased NEW relative to NEZ
Comparing and contrasting the minor sex chromosomes to the remainder of the genome, as well as the different types of minor sex chromosomes to each other, can be used to study several important genetic and evolutionary processes, including the direction and power of sex-specific selection, the strength and consequences of genetic drift and methods to counter it, and the constraints placed on genes by sexual and pleiotropic conflicts. Despite this utility, the minor sex chromosomes have proved to be somewhat more difficult to study than the remainder of the genome, and only recently have some of the necessary resources become available to identify the exact nature and role of Y and W chromosomes. It is my intent here to summarise recent developments regarding the evolution of minor sex chromosome with the aim of highlighting the surprisingly dynamic nature of these restricted genomic regions.
Are minor sex chromosomes degenerates?
For sex chromosomes that descend from an ancestral autosome pair (Charlesworth 1991), the suppression of recombination between the sex determining gene and nearby sexually antagonistic loci allows the major and minor sex chromosomes to diverge from one another, and at the same time initiates several degenerative processes that act on sex-limited portions of the W and Y chromosomes (Rice 1987). Recombination suppression can occur through inversions on the sex chromosomes which prevent recombination between the major and minor sex chromosome in a specific region (Charlesworth and Charlesworth 2000), and sequential inversions result in distinct evolutionary strata along the sex chromosomes (Lahn and Page 1999; Bergero et al. 2007; Nam and Ellegren 2008). Alternatively, sex chromosomes can arise via complete achiasmy in the heterogametic sex (Burt et al. 1991; Berset-Brandli et al. 2008), in which case strata are not observed. Regardless of the mechanism, because the sex-limited portions of the minor sex chromosomes by definition do not recombine with the homologous portions of the major sex chromosomes, all the loci in this region are locked into complete linkage disequilibrium. As a result, minor sex chromosome haplotypes cannot be broken up and selection acts on the net effect of the linkage group as a whole rather than each locus independently. This process, Hill–Robertson interference, results therefore in reduced efficacy of selection at any single locus (Charlesworth and Charlesworth 2000; Berlin et al. 2007; Bachtrog 2008). Additionally, the linkage disequilibrium of the minor sex chromosome can result in hitchhiking, a process where selection for beneficial mutations also fixes deleterious alleles at linked loci. As an effectively haploid asexual portion of the genome, the sex-limited portions of the minor sex chromosome also suffer from Muller’s ratchet (Bachtrog 2008; Kaiser and Charlesworth 2010), whereby stochastic variations in inheritance can lead to the accumulation of genetic load. Finally, the efficacy of selection acting on sex-limited genes is further limited by the reduced NE of minor sex chromosomes (Bachtrog and Charlesworth 2002; Haddrill et al. 2007), as shown in Figs. 1 and 2.
The extensive gene loss from the minor sex chromosomes following the suppression of recombination is the most striking aspect of minor sex chromosome degeneration. For those major and minor sex chromosome pairs that share a common autosomal ancestor, highly derived minor sex chromosomes often carry a small fraction of the genes observed on their major sex chromosome partner. Just a handful of X chromosome orthologues remain on the human Y (Skaletsky et al. 2003), and the same is true of the orthologues shared between the chicken Z and W (Nam and Ellegren 2008). Although this is partly due to gene traffic off major sex chromosomes to autosomes (Potrzebowski et al. 2008; Morkovsky et al. 2010; Zhang et al. 2010a, b), the vast majority of the pattern is due to gene loss from the minor sex chromosomes. The rate of gene loss is best illustrated by neo-Y chromosome in Drosophila miranda, where up to half of the neo-X chromosome orthologues have been pseudogenised on the neo-Y (Bachtrog 2008).
Taken together, the seemingly inexorable forces of Hill–Robertson interference, genetic hitchhiking, Muller’s ratchet and genetic drift act in concert to degrade the minor sex chromosome. This often produces a picture of the W and Y as decrepit and frail, a perception with substantial support. For instance, Caenorhabditis elegans lacks a Y chromosome entirely, leaving males with an unpaired X chromosome. Other sex chromosome systems lacking minor sex chromosomes exist (Lecher et al. 1995; Hawthorne 2001; Doan and Paliulis 2009; Ross et al. 2010), suggesting that the minor sex chromosomes can degenerate to the point of disappearance (Steinemann and Steinemann 2005; Graves 2006). These cases of sex chromosome systems completely lacking minor sex chromosomes further supports the assumption that the Y and W are, in the best cases, entirely expendable, and in the worst cases, in terminal decline.
There are however numerous lines of evidence that, when taken together, reveal a remarkable level of dynamicism in some minor sex chromosomes that act to counter degeneration. First of all, minor sex chromosomes need not degenerate in the first place, as evidenced by the fact that not all homomorphic sex chromosomes are evolutionarily young. Recombination patterns in many disparate animals are defined by phenotypic rather than genotypic sex (Inoue et al. 1983; Wallace et al. 1997; Lynn et al. 2005; Campos-Ramos et al. 2009; Matsuba et al. 2010); therefore, species with a low incidence of sex-reversed individuals will see a rare but steady rate of crossing over between major and minor sex chromosomes. This acts to refresh the minor sex chromosome (Perrin 2009) and can result in ancient yet largely homomorphic sex chromosomes (Stöck et al. 2011b). This is probably not the case in the examples of homomorphic sex chromosomes in ratites or pythons mentioned earlier, but can explain why most sex chromosomes in fish, frogs and some other poikilothermic animals are largely undifferentiated when at least some of them are ancient (Stöck et al. 2011b).
For heteromorphic sex chromosomes, presumably in species that lack fertile sex-reversed individuals, intra-chromosomal recombination can act to counter further minor sex chromosome degeneration. For example, there are numerous palindromes present on the primate Y (Rozen et al. 2003; Lange et al. 2009; Hughes et al. 2010) and avian W (Davis et al. 2010) chromosomes. These palindromes foster intra-chromosomal recombination, as the arms of the Y or W chromosome recombine with each other, leading to both gene conversion and copy number variation for Y- or W-linked genes (Backstrom et al. 2005; Lange et al. 2009). Gene conversion is a mechanism to correct mutated copies, although conversion can work in the other direction as well when a functional copy of a gene is converted by a mutated copy. In addition, copy number variations provide the differences in expression level within populations that selection can then act on in order to purge less fit individuals (Lange et al. 2009). Although non-homologous recombination is usually thought of as a maladaptive, or at least neutral, process, in the case of the minor sex chromosomes it seems to be a mechanism by which the minor sex chromosome can effectively counter the degenerative forces of Hill–Robertson interference and Muller’s ratchet (Marais et al. 2010).
In instances where the majority of the minor sex chromosome is sex specific, gene loss from the minor sex chromosomes seems to slow considerably over time and may even halt at a certain point. Rather than thinking of gene loss from minor sex chromosomes as a linear process with a natural endpoint of total extinction, the process appears to be asymptotic, as gene loss appears to slow down, and even cease, once the minor sex chromosome coding content has shrunk to a certain point (Bachtrog 2008; Kaiser and Charlesworth 2010). This is partially due to the fact that fewer active genes present fewer targets for mutational silencing, and partially because the remaining genes are likely subject to particularly strong purifying selection that acts to maintain coding content.
Minor sex chromosomes also accumulate new material or even entirely new segments. The Y chromosomes in the Drosophila clade illustrate this most clearly. Although the X chromosome is highly conserved across Drosophila species (Drosophila 12 Genomes Consortium 2007), the coding compliments of the Y chromosomes of several Drosophilids are largely non-overlapping, and this seems to be due to gene gain rather than gene decay, as the acquisition rate is roughly 10 times higher than the rate of gene loss (Koerich et al. 2008). Divergence is also the case for the human and chimpanzee Y chromosomes, which differ quite remarkably in both gene content and structure (Hughes et al. 2010). Interestingly, the difference is primarily due to divergence of the chimpanzee Y, as the gorilla and human Y chromosomes are rather more conserved (Goto et al. 2009), suggesting that the chimp mating system has caused a shift in sex-specific selection pressures on chimpan-zee males, and this may have played a role in altering Y chromosome divergence rate (Wilson and Makova 2009). Birds also show evidence of rapid turnover of W chromosome gene content and repetitive elements (Stiglec et al. 2007), and given the range of avian mating systems that includes polyandry (Jenni 1974), strict monogamy (Griffith et al. 2002) and polygyny (Slagsvold and Lifjeld 1994), this might be an ideal clade with which to test the role of mating system in minor chromosome divergence.
From these diverse lines of evidence, it seems that far from disappearing, some minor sex chromosomes seem to be thriving in the face of the myriad of degenerative forces acting against them. This survival and persistence implies that the minor sex chromosomes must play a vital role for selection to have been strong enough to preserve them.
What sorts of genes remain on and relocate to minor sex chromosomes?
In speculating about why minor sex chromosomes persist in the face of strong degradative forces, it can be useful to change the question of how minor sex chromosomes degenerate to ask what sorts of genes escape degeneration, and what sorts of genes relocate to the minor sex chromosomes and succeed there. Loci on minor sex chromosomes are subject to different selection pressures compared to genes in the remainder of the genome, primarily because the minor sex chromosomes are present and expressed in only one sex. This carries with it a host of evolutionary implications (detailed below), but for immediate purposes, sex limitation means Y chromosomes are selected only in males resulting in male-specific functions for most Y chromosome loci. Similarly, genes on W chromosomes are only selected in females; therefore, they nearly always fulfil female-specific roles.
In some cases, genes on minor sex chromosomes have a clear sex-specific function: namely they are responsible for determining sex. For example, SRY on the Y chromosome is responsible for initiating the male-sex determination cascade in most therian mammals (Sekido and Lovell-Badge 2009), and a gene called DM-W on the W chromosome of Xenopus is required for ovary initiation in females (Yoshimoto et al. 2008). Other examples of sex determination genes on minor sex chromosomes are common (Matsuda et al. 2002; Peichel et al. 2004; Graves 2008), and this vital yet restricted role might explain the persistence of some minor sex chromosomes, as sex determination presumably results in strong preservative selection. This explanation does not apply to all minor heteromorphic sex chromosomes, however, as sex determination in many other taxa is based on the dose of the major sex chromosome (Smith et al. 2009; Williams and Carroll 2009). For those sex chromosome systems that are based on X or Z chromosome gene dose, the minor sex chromosomes are not required for sex determination. Without some other vital role to play, these minor sex chromosomes would be expected to simply disintegrate and disappear, leaving X0 or Z0 species such as C. elegans. The fact that the minor sex chromosomes persist in many systems where sex is based on major chromosome dose, such as Drosophila and birds, suggests that they play some additional and vital role other than simply determining sex.
Therefore, it is not surprising that there are different classes of genes residing on minor sex chromosomes aside from primary sex determining loci. The most commonly studied class are major and minor orthologues that originated before the sex chromosomes diverged. Many sex chromosomes descend from an ancestral autosome (Charlesworth 1991), and in these cases, there are often X–Y or Z–W orthologous loci that show clear shared ancestry in animals (Lahn and Page 1999; Nam and Ellegren 2008) and plants (Bergero et al. 2007). It should be noted that not all minor sex chromosomes are orthologous to their respective major chromosome, as the Drosophila Y is not related to the X, and instead derives from a supernumery B chromosome (Carvalho et al. 2009); therefore, X and Y loci in Drosophila are not expected to show any evidence of shared ancestry. However, for instances where the major and minor sex chromosomes share a common autosomal ancestor, the orthologous genes on minor sex chromosomes may be maintained in order to balance gene dose in the heterogametic sex (Mank et al. 2011), but more commonly, the minor chromosome copy takes on a related but sex-specific function (Skaletsky et al. 2003). HINTW on the W chromosome of chicken is one such example. This gene has an orthologue on the Z chromosome (HINTZ), yet HINTW has taken on a different role from HINTZ, and the expression of the gene is key to ovary development (Hori et al. 2000), yet the W does not determine sex in birds (Smith et al. 2009).
Other minor sex chromosome loci do not derive from the ancestral autosome, but rather have transposed onto the W or Y after it became a sex chromosome. This can happen for individual loci, or it can happen in a wholesale manner when large genomic regions fuse to the minor sex chromosome or start segregating with the major sex chromosome (neo-sex chromosomes), as seen in the neo-sex chromosomes of D. miranda (Bachtrog and Charlesworth 2002), the black muntjac (Zhou et al. 2008) as well as a large portion of the Y in placental mammals that fused to the sex chromosomes after the split with marsupials (Waters et al. 2001). Duplications and translocations to the minor chromosome will be favoured and maintained for certain types of genes, namely genes that confer a benefit to the heterogametic sex, especially if there is a cost to the homogametic sex (Connallon and Clark 2010). This selective advantage is due to the fact that relocating to the minor chromosome maintains the benefit to the heterogametic sex at the same time as it eliminates sexual conflict with the homogametic sex. As there is strong evidence for sexual conflict currently or previously affecting a substantial proportion of coding regions (Chapman et al. 2003; Mank and Ellegren 2009; Harano et al. 2010; Innocenti and Morrow 2010), there are potentially many loci for which translocation to the minor chromosome would be selectively favoured. This may explain many of the cases of gene acquisition by minor sex chromosomes (Koerich et al. 2008), as well as the presence of entire regions that are not orthologous to the respective major chromosome, as is the case for the entire Drosophila Y (Carvalho et al. 2009). Of course, sexually antagonistic loci cannot relocate to the minor sex chromosome with the intention of resolving conflict; rather, these factors determine whether a chance event will be selectively favoured and, if so, maintained.
Regardless of whether they are ancestral genes with new sex-specific functions, or new additions to the minor sex chromosomes, the genes on the Y and W that are not directly associated with determining sex are typically directly tied to either fertility or other reproductive traits. For example, the Y chromosome harbours the majority of the variation for male attractiveness in guppies (Postma et al. 2011), male fitness in D. melanogaster (Chippindale and Rice 2001; Lemos et al. 2008) and even a disproportionately large amount of variation for dimorphic traits in the plant Silene latifolia (Scotti and Delph 2006). Work in my laboratory indicates that female fertility is similarly tied to the expression of several W-linked genes chickens (Mogadam et al. in preparation). In some of these examples, it is clear that a large part of the general fitness effects of the minor sex chromosome is due to epistatic regulation by the minor sex chromosome of the other genomic regions (Chippindale and Rice 2001; Lemos et al. 2008), and other studies cannot rule this out (Postma et al. 2011). Epistasis is clearly not the only mechanism connecting the minor sex chromosome to fitness, however, as specific loci on the Y have been directly tied to fertility in humans (Lange et al. 2009) as well as Drosophila (Gepner and Hays 1993; Carvalho et al. 2000; Ding et al. 2010), and W-linked genes seem key to female fecundity as well in birds (Hori et al. 2000). Regardless of the mechanism by which minor sex chromosomes influence fertility, it is unlikely that ancestral autosomes that gave rise to a minor sex chromosome would harbour such a large fraction of the loci related to sex-specific function, so it is reasonable to conclude that this characteristic is a product of the sex-specific selection pressures arising from sex chromosome divergence.
Why do some minor sex chromosomes persist?
The sex-specific nature of the minor sex chromosomes gives some clues about why these sex chromosomes can persist, namely that they do something vitally important in the sex to which they are confined. This suggests that purifying selection, already potentially a powerful force on the minor sex chromosomes due to their haploid nature, is potentially even stronger still in maintaining genes essential to fertility. Sex-specific selection pressures are therefore expected to be extremely strong on minor sex chromosomes (Connallon and Clark 2010), possibly strong enough to maintain heteromorphic minor sex chromosomes, or at least some portion of them, and prevent complete degeneration. Put another way, because Y chromosome genes are essential to male fitness, the chromosome is subject to extremely strong male-specific selection pressures which in turn may help maintain the vital Y chromosome coding content associated with male fertility against the degenerative forces acting against the chromosome. Female-specific selection would similarly be expected to maintain W-linked genes with key roles in female fertility.
Interestingly, loci on minor sex chromosomes may be more able to respond to sex-specific selection pressures, as they are freed up from other constraints imposed by the other sex. The vast majority of autosomal, X-linked and Z-linked genes are expressed in both females and males (though many of these genes are expressed at different levels in the sexes), and selection in one sex may fail to produce a change due to constraints acting in the other sex (Fig. 3). This type of intra-locus sexual conflict is common (Bonduriansky and Chenoweth 2009), and if unresolved, it can impose a severe burden on populations (Connallon et al. 2010). Sexual conflict can be resolved in some cases via genetic mechanisms such as sex-biased gene expression (Connallon and Knowles 2005) or genetic imprinting (Day and Bonduriansky 2004); however, these opportunistic regulatory changes take time to evolve and must be in place before constrained genes can respond to sex-specific selection pressures.
Fig. 3.
Traits encoded by genes on minor sex chromosomes are not subject to intra-locus sexual conflict, and therefore are more able to respond to sex-specific selection. Autosomal genes are expressed in both males and females, and male-specific selection (blue arrow) may be counter to female-specific selection (red arrow), resulting in unresolved conflict over optimal trait values (a). This is resolved for genes on minor sex chromosomes, as male-specific selection on Y-linked traits (b) is unconstrained by expression in females, and similarly, female-specific selection on W-linked traits (c) is unconstrained by expression in males. In these latter two panels, the trait responds to sex-specific selection by moving to male (Y chromosome) or female (W chromosome) optimum
The sex limitation of the minor sex chromosomes eliminates the possibility of sexual conflict, potentially leaving the loci on W and Y chromosomes more responsive to female- or male-specific selection pressures as response is not dependent upon a regulatory mechanism to resolve constraints in the other sex. This suggests a somewhat circular process that may act as a feedback loop, as minor sex chromosomes are subject to stronger sex-specific selection pressure and the loci encoded on them are able to respond more quickly to it (Postma et al. 2011). This sex-specific nature also explains why some gene duplications to the minor are selectively favoured—for some sexually antagonistic genes, relocation to the minor sex chromosome resolves conflict with the other sex.
Sex-specific selection pressures may also help explain the multitude of examples of sex chromosome turnover. Existing sex chromosome systems can be supplanted directly by newly emergent and non-homologous sex chromosomes, as documented in salmon (Woram et al. 2003), sticklebacks (Ross et al. 2009) and rice fish (Matsuda et al. 2002), and emergent but still homomorphic sex chromosomes can explain the loss of master sex determining genes in other animals (Just et al. 1995). These turnover events likely represent changes directly from existing sex chromosome systems rather than complete losses of sex chromosomes followed by gains. The former case is supported by the many examples of populations polymorphic for sex chromosomes (Orzack et al. 1980; Ogata et al. 2003, 2008); the latter would presumably cause extinction before new sex chromosomes could emerge. Theoretical work suggests that turnover of sex chromosomes can be driven by sex-specific selection pressures that are sexually antagonistic (van Doorn and Kirkpatrick 2007, 2010), and there is empirical support for this from cichlids (Roberts et al. 2009). Therefore, it is possible that even as sex-specific selection pressures drive minor sex chromosome evolution, they also cause complete replacement of sex chromosome systems.
Evolutionary implications of the minor sex chromosomes
Although there are often genes on minor sex chromosomes that directly encode fitness traits as described above, it is also often the case that the minor sex chromosome interacts epistatically with the remainder of the genome regarding some fitness traits (Chippindale and Rice 2001; Lemos et al. 2008; Ding et al. 2010). This suggests that the limited coding content of minor sex chromosomes has a long reach. It is also interesting to speculate whether the role of minor sex chromosome in fitness and fertility, combined with the rapid rate of change in minor sex chromosome coding content and structure (Koerich et al. 2008; Hughes et al. 2010), means that the minor sex chromosomes play an important role in Haldane’s Rule. Haldane’s Rule, which states that ‘when in the F1 offspring of two different animal races one sex is absent, rare or sterile, that sex is the heterozygous sex’ (Haldane 1922), has traditionally been linked to changes on major sex chromosomes (Masly and Presgraves 2007; Presgraves 2008). However, given the clear fertility role of many minor sex chromosomes, as well as the evidence suggesting that epistatic interactions between the minor sex chromosome and the remainder of the genome greatly influence fertility, it seems reasonable that the minor sex chromosome evolution and turnover could explain at least the sterility aspects of Haldane’s Rule (Sackton et al. 2011). This would be more likely for minor sex chromosomes that are highly differentiated from their major sex chromosome pair and could be due to epistatic interactions with any part of the remaining genome. However, given the faster rate of evolution generally observed on major sex chromosomes (Mank et al. 2010) and the Large-X effect (or Large-Z) observed in speciation (Presgraves 2003, 2008; Masly and Presgraves 2007), it is possible that the main source of epistatic incompatibilities may be between the major and minor sex chromosomes.
Beyond a potential role in Haldane’s Rule, there is evidence that minor sex chromosomes may play a direct role in sexual selection and, via this route, in speciation due to mating preferences. In addition to the role in sex-specific fertility and fecundity, minor sex chromosomes have been shown to harbour loci directly tied to some sexually selected traits involved in mating preferences, such as courtship behaviour in Anopheles mosquitoes (Fraccaro et al. 1977) and sexually selected colouration in guppies (Winge 1922, 1927). Sexual selection due to mating preference is predicted to drive speciation events (Lande 1981; van Doorn et al. 2009), and it is reasonable that if minor sex chromosomes do indeed generally play an important role in encoding sexually selected traits, and sexually selected traits contribute to speciation, then minor sex chromosomes may be important in speciation by sexual selection.
Conclusions
Taken together, the recent evidence summarised above indicates that some minor sex chromosomes persist despite strong deleterious forces, possibly because the strong preservative forces result from the essential role these regions play in sex-specific fitness. However, the evidence is piecemeal and tenuous at this point, and it is difficult to know if the importance and persistence of some minor sex chromosomes represent a general trend or simply are anecdotal examples to counter the contradictory examples of minor sex chromosome loss. A generalised understanding will require both comparative sequence data from a large number of species as well as evolutionary studies focused on the explicit functional role of the minor sex chromosome. Neither of these are easy tasks.
Complete sequences of minor chromosomes often require arduous laboratory efforts beyond even that needed for high coverage genome sequencing efforts (Bachtrog 2008; Koerich et al. 2008; Hughes et al. 2010). However, next-generation sequencing allows for relatively quick and efficient identification of minor sex chromosome exomes (Portela et al. 2010). This methodology could accelerate studies of minor sex chromosome coding content and expression levels across populations and related species, although without non-coding sequence, it is difficult to identify cis- versus trans-regulatory changes. Exome and expression data in conjunction with pedigrees (Postma et al. 2011) and fitness assays (Lemos et al. 2008) can also help reveal the importance of the minor sex chromosomes. All these tools implemented together may hopefully reveal general trends in minor sex chromosome evolution, and either confirm or refute the small but mighty nature of these regions of the genome.
Acknowledgements
Peter Harrison, Hooman Moghadam, Jen Perry, Marie Pointer and Alison Wright provided helpful comments and suggestions, as did three anonymous reviewers. Support is gratefully acknowledged from the BBSRC and the ERC 7th Framework Programme (grant agreement no. 260233).
Abbreviations
- DMRT1
Doublesex and mab-3 related transcription factor 1—a conserved gene in the vertebrate sex determination pathway that is also key in male development in Drosophila and Caenorhabditis elegans
- DM-W
Doublesex mab-3 related gene on the W— a gene showing similarity to parts of DMRT1 on the Xenopus W chromosome that plays a role in ovary development (Yoshimoto et al. 2008)
- HINTW
Histidine triad nucleotide binding protein W—also called Wpkci, W-linked protein kinase C inhibitor (Hori et al. 2000)
- HINTZ
Histidine triad nucleotide binding protein Z—the Z chromosome gametolog to HINTW
- NE
Effective population size
- NEW
The effective population size of the W chromosome
- NEX
The effective population size of the X chromosome
- NEY
The effective population size of the Y chromosome
- NEZ
The effective population size of the Z chromosome
- SRY
Sex-determining region Y—the sex determining gene on the eutherian Y chromosome
References
- Andersson M. Sexual selection. Princeton University Press; Princeton: 1994. [Google Scholar]
- Bachtrog D. The temporal dynamics of processes underlying Y chromosome degeneration. Genetics. 2008;179:1513–1525. doi: 10.1534/genetics.107.084012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D, Charlesworth B. Reduced adaptation of a non-recombining neo-Y chromosome. Nature. 2002;416:323–326. doi: 10.1038/416323a. [DOI] [PubMed] [Google Scholar]
- Bachtrog D, Kirkpatrick M, Mank JE, McDaniel SF, Pires JC, Rice WR, Valenzuela N. Are all sex chromosomes created equal? Trends Genet. 2011;27:350–357. doi: 10.1016/j.tig.2011.05.005. [DOI] [PubMed] [Google Scholar]
- Backstrom N, Ceplitis H, Berlin S, Ellegren H. Gene conversion drives the evolution of HINTW, an ampliconic gene on the female-specific avian W chromosome. Mol Biol Evol. 2005;22:1992–1999. doi: 10.1093/molbev/msi198. [DOI] [PubMed] [Google Scholar]
- Bergero R, Forrest A, Kamau E, Charlesworth D. Evolutionary strata on the X chromosomes of the dioecious plant Silene latifolia: evidence from new sex-linked genes. Genetics. 2007;175:1945–1954. doi: 10.1534/genetics.106.070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin S, Tomaras D, Charlesworth B. Low mitochondrial variability in birds may indicate Hill–Robertson effects on the W chromosome. Heredity. 2007;99:389–396. doi: 10.1038/sj.hdy.6801014. [DOI] [PubMed] [Google Scholar]
- Berset-Brandli L, Jaquiery J, Broquet T, Ulrich Y, Perrin N. Extreme heterochiasmy and nascent sex chromosomes in European tree frogs. Proc Biol Sci. 2008;275:1577–1585. doi: 10.1098/rspb.2008.0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonduriansky R, Chenoweth SF. Intralocus sexual conflict. Trends Ecol Evol. 2009;24:280–288. doi: 10.1016/j.tree.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Burt A, Bell G, Harvey PH. Sex differences in recombination. J Evol Biol. 1991;4:259–277. [Google Scholar]
- Campos-Ramos R, Harvey SC, Penman DJ. Sex-specific differences in the synaptonemal complex in the genus Oreochromis (Cichlidae) Genetica. 2009;135:325–332. doi: 10.1007/s10709-008-9280-8. [DOI] [PubMed] [Google Scholar]
- Carvalho AB, Lazzaro BP, Clark AG. Y chromosomal fertility factors kl-2 and kl-3 of Drosophila melanogaster encode dynein heavy chain polypeptides. Proc Natl Acad Sci USA. 2000;97:13239–13244. doi: 10.1073/pnas.230438397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho AB, Koerich LB, Clark AG. Origin and evolution of Y chromosomes: Drosophila tales. Trends Genet. 2009;25:270–277. doi: 10.1016/j.tig.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman T, Arnqvist G, Bangham J, Rowe L. Sexual conflict. Trends Ecol Evol. 2003;18:41–47. [Google Scholar]
- Charlesworth B. The evolution of sex chromosomes. Science. 1991;251:1030–1033. doi: 10.1126/science.1998119. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Philos Trans R Soc Lond B Biol Sci. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Mank JE. The birds and the bees and the flowers and the trees: lessons from the genetic mapping of sex determination in plants and animals. Genetics. 2010;186:9–31. doi: 10.1534/genetics.110.117697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibalina MV, Filatov DA. Plant Y chromosome degeneration is retarded by haploid purifying selection. Curr Biol. 2011;21:1475–1479. doi: 10.1016/j.cub.2011.07.045. [DOI] [PubMed] [Google Scholar]
- Chippindale AK, Rice WR. Y chromosome polymorphism is a strong determinant of male fitness in Drosophila melanogaster. Proc Natl Acad Sci USA. 2001;98:5677–5682. doi: 10.1073/pnas.101456898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clutton-Brock T. Sexual selection in males and females. Science. 2007;318:1882–1885. doi: 10.1126/science.1133311. [DOI] [PubMed] [Google Scholar]
- Connallon T, Clark AG. Sex linkage, sex-specific selection, and the role of recombination in the evolution of sexually dimorphic gene exprssion. Evolution. 2010;64:3417–3442. doi: 10.1111/j.1558-5646.2010.01136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon T, Knowles LL. Intergenomic conflict revealed by patterns of sex-biased gene expression. Trends Genet. 2005;21:495–499. doi: 10.1016/j.tig.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Connallon T, Cox RM, Calsbeek R. Fitness consequences of sex-specific selection. Evolution. 2010;64:1671–1682. doi: 10.1111/j.1558-5646.2009.00934.x. [DOI] [PubMed] [Google Scholar]
- Davis JK, Thomas PJ, Program NCS, Thomas JW. A W-linked palindrome and gene conversion in New World sparrows and blackbirds. Chromosom Res. 2010;18:543–553. doi: 10.1007/s10577-010-9134-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day T, Bonduriansky R. Intralocus sexual conflict can drive the evolution of genomic imprinting. Genetics. 2004;167:1537–1546. doi: 10.1534/genetics.103.026211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin RH, Nagahama Y. Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquaculture. 2002;208:191–364. [Google Scholar]
- Ding Y, Zhao L, Yang SA, Jiang Y, Chen YA, Zhao RP, Zhang Y, Zhang GJ, Dong Y, Yu HJ, Zhou Q, Wang W. A young Drosophila duplicate gene plays essential roles in spermatogenesis by regulating several Y-linked male fertility genes. PLoS Genet. 2010;6:e1001255. doi: 10.1371/journal.pgen.1001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan RN, Paliulis LV. Micromanipulation reveals an XO–XX sex determining system in the orb-weaving spider Neoscona arabesca (Walckenaer) Hereditas. 2009;146:180–182. doi: 10.1111/j.1601-5223.2009.02123.x. [DOI] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Ezaz T, Sarre SD, O’Meally D, Graves JAM, Georges A. Sex chromosome evolution in lizards: independent origins and rapid transitions. Cytogenet Genome Res. 2009;127:249–260. doi: 10.1159/000300507. [DOI] [PubMed] [Google Scholar]
- Fraccaro M, Tiepolo L, Laudani U, Marchi A, Jayakar SD. Y chromosome controls mating behavior on Anopheles mosquitos. Nature. 1977;265:326–328. doi: 10.1038/265326a0. [DOI] [PubMed] [Google Scholar]
- Gepner J, Hays TS. A fertility region on the Y chromosome of Drosophila melanogaster encodes a dynein microtubule motor. Proc Natl Acad Sci USA. 1993;90:11132–11136. doi: 10.1073/pnas.90.23.11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto H, Peng L, Makova KD. Evolution of X-degenerate Y chromosome genes in greater apes: conservation of gene content in human and gorilla, but not chimpanzee. J Mol Evol. 2009;68:134–144. doi: 10.1007/s00239-008-9189-y. [DOI] [PubMed] [Google Scholar]
- Graves JAM. Sex chromosome specialization and degeneration in mammals. Cell. 2006;124:901–914. doi: 10.1016/j.cell.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Graves JAM. Weird animal genomes and the evolution of vertebrate sex and sex chromosomes. Annu Rev Genet. 2008;42:565–586. doi: 10.1146/annurev.genet.42.110807.091714. [DOI] [PubMed] [Google Scholar]
- Griffith SC, Owens IPF, Thuman KA. Extra pair paternity in birds: a review of interspecific variation and adaptive function. Mol Ecol. 2002;11:2195–2212. doi: 10.1046/j.1365-294x.2002.01613.x. [DOI] [PubMed] [Google Scholar]
- Guttman DS, Charlesworth D. An X-linked gene with a degenerate Y-linked homologue in a dioecious plant. Nature. 1998;393:263–266. doi: 10.1038/30492. [DOI] [PubMed] [Google Scholar]
- Haddrill PR, Halligan DL, Tomaras D, Charlesworth B. Reduced efficacy of selection in regions of the Drosophila genome that lack crossing over. Genome Biol. 2007;8:R18. doi: 10.1186/gb-2007-8-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane JBS. Sex ratio and unisexual sterility in hybrid animals. J Genet. 1922;12:101–109. [Google Scholar]
- Harano T, Okada K, Nakayama S, Miyatake T, Hosken DJ. Intralocus sexual conflict unresolved by sex-limited trait expression. Curr Biol. 2010;20:2036–2039. doi: 10.1016/j.cub.2010.10.023. [DOI] [PubMed] [Google Scholar]
- Hawthorne DJ. AFLP-based genetic linkage map of the Colorado potato beetle Leptinotarsa decemlineata: sex chromosomes and a pyrethroid-resistance candidate gene. Genetics. 2001;158:695–700. doi: 10.1093/genetics/158.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Asakawa S, Itoh Y, Shimizu N, Mizuno S. Wpkci, encoding an altered form of PKCI, is conserved widely on the avian W chromosome and expressed in early female embryos: implication of its role in female sex determination. Mol Biol Cell. 2000;11:3645–3660. doi: 10.1091/mbc.11.10.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JF, Skaletsky H, Pyntikova T, Graves TA, van Daalen SKM, Minx PJ, Fulton RS, McGrath SD, Locke DP, Friedman C, Trask BJ, Mardis ER, Warren WC, Repping S, Rozen S, Wilson RK, Page DC. Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature. 2010;463:536–539. doi: 10.1038/nature08700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocenti P, Morrow EH. The sexually antagonistic genes of Drosophila melanogaster. PLoS Biology. 2010;8:e1000335. doi: 10.1371/journal.pbio.1000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H, Fukumori Y, Hiroyoshi T. Mapping of autosomal male-determining factors of the housefly, Musca domestica L., by means of sex-reversal. Jpn J Genet. 1983;58:451–461. [Google Scholar]
- Janes DE, Organ CL, Fujita MK, Shedlock AM, Edwards SV. Genome evolution in Reptilia, the sister group of mammals. Annu Rev Genomics Hum Genet. 2010;11:239–264. doi: 10.1146/annurev-genom-082509-141646. [DOI] [PubMed] [Google Scholar]
- Jenni DA. Evolution of polyandry in birds. Amer Zool. 1974;14:129–144. [Google Scholar]
- Just W, Rau W, Vogel W, Akhverdian M, Fredga K, Graves JAM, Lyapunova E. Absence of SRY in species of the vole Ellobius. Nat Genet. 1995;11:117–118. doi: 10.1038/ng1095-117. [DOI] [PubMed] [Google Scholar]
- Kaiser VB, Charlesworth B. Muller’s ratchet and the degeneration of the Drosophila miranda neo-Y chromosome. Genetics. 2010;185:339–348. doi: 10.1534/genetics.109.112789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerich LB, Wang XY, Clark AG, Carvalho AB. Low conservation of gene content in the Drosophila Y chromosome. Nature. 2008;456:949–951. doi: 10.1038/nature07463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- Lande R. Models of speciation by sexual selection on polygenic traits. Proc Natl Acad Sci USA. 1981;78:3721–3725. doi: 10.1073/pnas.78.6.3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange J, Skaletsky H, van Daalen SKM, Embry SL, Korver CM, Brown LG, Oates RD, Silber S, Repping S, Page DC. Isodicentric Y chromosomes and sex disorders as byproducts of homologous recombination that maintains palindromes. Cell. 2009;138:855–869. doi: 10.1016/j.cell.2009.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecher P, Defaye D, Noel P. Chromosomes and nuclear DNA of crustacea. Invertebr Reprod Dev. 1995;27:85–114. [Google Scholar]
- Lemos B, Araripe LO, Hartl DL. Polymorphic Y chromosomes harbor cryptic variation with manifold functional consequences. Science. 2008;319:91–93. doi: 10.1126/science.1148861. [DOI] [PubMed] [Google Scholar]
- Liu Z, Moore PH, Ma H, Ackerman CM, Ragiba M, Yu Q, Pearl HM, Kim MS, Charlton JW, Stiles JI, Zee FT, Paterson AH, Ming R. A primitive Y chromosome in papaya marks incipient sex chromosome evolution. Nature. 2004;427:348–353. doi: 10.1038/nature02228. [DOI] [PubMed] [Google Scholar]
- Lynn A, Schrump S, Cherry J, Hassold T, Hunt P. Sex, not genotype, determines recombination levels in mice. Am J Hum Genet. 2005;77:670–675. doi: 10.1086/491718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE, Ellegren H. Parallel divergence and degredation of the avian W sex chromosome. Trends Ecol Evol. 2007;22:389–391. doi: 10.1016/j.tree.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Mank JE, Ellegren H. Sex-linkage of sexually antagonistic genes is predicted by female, but not male, effects in birds. Evolution. 2009;63:1464–1472. doi: 10.1111/j.1558-5646.2009.00618.x. [DOI] [PubMed] [Google Scholar]
- Mank JE, Promislow DEL, Avise JC. Evolution of alternative sex-determining mechanisms in teleost fishes. Biol J Linn Soc. 2006;87:83–93. [Google Scholar]
- Mank JE, Vicoso B, Berlin S, Charlesworth B. Effective population size and the Faster-X effect: empirical evidence and its interpretation. Evolution. 2010;63:663–674. doi: 10.1111/j.1558-5646.2009.00853.x. [DOI] [PubMed] [Google Scholar]
- Mank JE, Hosken DJ, Wedell N. Some inconvenient truths about sex chromosome dosage compensation and the potential role of sexual conflict. Evolution. 2011;65:2133–2144. doi: 10.1111/j.1558-5646.2011.01316.x. [DOI] [PubMed] [Google Scholar]
- Marais GAB, Campos PRA, Gordo I. Can intra-Y gene conversion oppose the degeneration of the human Y chromosome? A simulation study. Genome Biol Evol. 2010;2:347–357. doi: 10.1093/gbe/evq026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masly JP, Presgraves DC. High-resolution genome-wide dissection of the two rules of speciation in Drosophila. PLoS Biology. 2007;5:1890–1898. doi: 10.1371/journal.pbio.0050243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuba C, Alho JS, Merila J. Recombination rate between sex chromosomes depends on phenotypic sex in the common frog. Evolution. 2010;64:3634–3637. doi: 10.1111/j.1558-5646.2010.01076.x. [DOI] [PubMed] [Google Scholar]
- Matsubara K, Tarui H, Toriba M, Yamada K, Nishida-Umehara C, Agata K, Matsuda Y. Evidence for different origin of sex chromosomes in snakes, birds, and mammals and step-wise differentiation of snake sex chromosomes. Proc Natl Acad Sci USA. 2006;103:18190–18195. doi: 10.1073/pnas.0605274103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M, Nagahama Y, Shinomiya A, Sato T, Matsuda C, Kobayashi T, Morrey CE, Shibata N, Asakawa S, Shimizu N, Hori H, Hamaguchi S, Sakaizumi M. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature. 2002;417:559–563. doi: 10.1038/nature751. [DOI] [PubMed] [Google Scholar]
- Matsunaga S, Kawano S. Sex determination by sex chromosomes in dioecious plants. Plant Biol. 2001;3:481–488. [Google Scholar]
- Ming R, Wang JP, Moore PH, Paterson AH. Sex chromosomes in flowering plants. Am J Bot. 2007;94:141–150. doi: 10.3732/ajb.94.2.141. [DOI] [PubMed] [Google Scholar]
- Morkovsky L, Storchova R, Plachy J, Ivanek R, Divina P, Hejnar J. The chicken Z chromosome is enriched for genes with preferential expression in ovarian somatic cells. J Mol Evol. 2010;70:129–136. doi: 10.1007/s00239-009-9315-5. [DOI] [PubMed] [Google Scholar]
- Nakamura M. Sex determination in amphibians. Semin Cell Dev Biol. 2009;20:271–282. doi: 10.1016/j.semcdb.2008.10.003. [DOI] [PubMed] [Google Scholar]
- Nam K, Ellegren H. The chicken (Gallus gallus) Z chromosome contains at least three nonlinear evolutionary strata. Genetics. 2008;180:1131–1136. doi: 10.1534/genetics.108.090324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata M, Ohtani H, Igarashi T, Hasegawa Y, Ichikawa Y, Miura I. Change of the heterogametic sex from male to female in the frog. Genetics. 2003;164:613–620. doi: 10.1093/genetics/164.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata M, Hasegawa Y, Ohtani H, Mineyama M, Miura I. The ZZ/ZW sex-determining mechanism originated twice and independently during evolution of the frog, Rana rugosa. Heredity. 2008;100:92–99. doi: 10.1038/sj.hdy.6801068. [DOI] [PubMed] [Google Scholar]
- Ogawa A, Murata K, Mizuno S. The location of Z- and W-linked marker genes and sequence on the homomorphic sex chromosomes of the ostrich and the emu. Proc Natl Acad Sci USA. 1998;95:4415–4418. doi: 10.1073/pnas.95.8.4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzack SH, Sohn JJ, Kallman KD, Levin SA, Johnston R. Maintenance of the 3 sex chromosome polymorphism in the plaryfish, Xiphophorus maculatus. Evolution. 1980;34:663–672. doi: 10.1111/j.1558-5646.1980.tb04005.x. [DOI] [PubMed] [Google Scholar]
- Peichel CL, Ross JA, Matson CK, Dickson M, Grimwood J, Schmutz J, Myers RM, Mori S, Schluter D, Kingsley DM. The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome. Curr Biol. 2004;14:1416–1424. doi: 10.1016/j.cub.2004.08.030. [DOI] [PubMed] [Google Scholar]
- Perrin N. Sex reversal: a fountain of youth for sex chromosomes? Evolution. 2009;63:3043–3049. doi: 10.1111/j.1558-5646.2009.00837.x. [DOI] [PubMed] [Google Scholar]
- Portela J, Crunau C, Cosseau C, Beltran S, Dantec C, Parrinello H, Boissier J. Whole-genome in silico subtractive hybridization (WISH)—using massing sequencing for the identification of unique and repetitive sex-specific sequences: the example of Schistosoma mansoni. BMC Genomics. 2010;11:387. doi: 10.1186/1471-2164-11-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma E, Spyrou N, Rollins LA, Brooks RC. Sex-dependent selection differentially shapes the genetic variation on and off the guppy Y chromosome. Evolution. 2011;65:2145–2156. doi: 10.1111/j.1558-5646.2011.01314.x. [DOI] [PubMed] [Google Scholar]
- Potrzebowski L, Vinckenbosch N, Marques AC, Chalmel F, Jegou B, Kaessmann H. Chromosomal gene movements reflect the recent origin and biology of therian sex chromosomes. PLoS Biology. 2008;6:709–716. doi: 10.1371/journal.pbio.0060080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC. A fine-scale genetic analysis of hybrid incompatibilities in Drosophila. Genetics. 2003;163:955–972. doi: 10.1093/genetics/163.3.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC. Sex chromosomes and speciation in Drosophila. Trends Genet. 2008;24:336–343. doi: 10.1016/j.tig.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WR. The accumulation of sexually antagonistic genes as a selective agent promoting the evolution of reduced recombination between primitive sex chromosomes. Evolution. 1987;41:911–914. doi: 10.1111/j.1558-5646.1987.tb05864.x. [DOI] [PubMed] [Google Scholar]
- Roberts RB, Ser JR, Kocher TD. Sexual conflict resolved by invasion of a novel sex determiner in Lake Malawi cichlid fishes. Science. 2009;326:998–1001. doi: 10.1126/science.1174705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, Muzny D, Platzer M, Howell GR, Burrows C, Bird CP, Frankish A, Lovell FL, Howe KL, Ashurst JL, Fulton RS, et al. The DNA sequence of the human X chromosome. Nature. 2005;434:325–337. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JA, Urton JR, Boland J, Shapiro MD, Peichel CL. Turnover of sex chromosomes in the stickleback fishes (Gasterosteidae) PLoS Genet. 2009;5:e1000391. doi: 10.1371/journal.pgen.1000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross L, Pen I, Shuker DM. Genomic conflict in scale insects: the causes and consequences of bizarre genetic systems. Biol Rev. 2010;85:807–828. doi: 10.1111/j.1469-185X.2010.00127.x. [DOI] [PubMed] [Google Scholar]
- Royo H, Polikiewicz G, Mahadevaiah SK, Prosser H, Mitchell M, Bradley A, de Rooij DG, Burgoyne PS, Turner JMA. Evidence that meiotic sex chromosome inactivation is essential for male fertility. Curr Biol. 2010;20:2117–2123. doi: 10.1016/j.cub.2010.11.010. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H, Marszalek JD, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Page DC. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature. 2003;423:873–876. doi: 10.1038/nature01723. [DOI] [PubMed] [Google Scholar]
- Sackton TB, Montenegro H, Hartl DL, Lemos B. Interspecific Y chromosome introgression disrupts testis-specific gene expression and male reproductive phenotypes in Drosophila. Proc Natl Acad Sci USA. 2011 doi: 10.1073/pnas.1114690108. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti I, Delph LF. Selective trade-offs and sex-chromosome evolution in Silene latifolia. Evolution. 2006;60:1793–1800. [PubMed] [Google Scholar]
- Sekido R, Lovell-Badge R. Sex determination and SRY: down to a wink and a nudge? Trends Genet. 2009;25:19–29. doi: 10.1016/j.tig.2008.10.008. [DOI] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T, Chinwalla A, Delehaunty A, Delehaunty K, Du H, Fewell G, Fulton L, Fulton R, Graves T, Hou S-F, Latrielle P, Leonard S, Mardis E, Maupin R, McPherson J, Miner T, Nash W, Nguyen C, Ozersky P, Pepin K, Rock S, Rohlfing T, Scott K, Schultz B, Strong C, Tin-Wollam A, Yang S-P, Waterston RH, Wilson RK, Rozen S, Page DC. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Slagsvold T, Lifjeld JT. Polygyny in birds—the role of competition between females and males for parental care. Am Nat. 1994;143:59–94. [Google Scholar]
- Smith CA, Roeszler KN, Ohnesorg T, Cummins DM, Farlie PG, Doran TJ, Sinclair AH. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature. 2009;461:267–271. doi: 10.1038/nature08298. [DOI] [PubMed] [Google Scholar]
- Spigler RB, Lewers KS, Main DS, Ashman TL. Genetic mapping of sex determination in a wild strawberry, Fragaria virginiana, reveals earliest form of sex chromosome. Heredity. 2008;101:507–517. doi: 10.1038/hdy.2008.100. [DOI] [PubMed] [Google Scholar]
- Steinemann S, Steinemann M. Y chromosomes: born to be destroyed. Bioessays. 2005;27:1076–1083. doi: 10.1002/bies.20288. [DOI] [PubMed] [Google Scholar]
- Stiglec R, Ezaz T, Graves JAM. A new look at the evolution of avian sex chromosomes. Cytogenet Genome Res. 2007;117:103–109. doi: 10.1159/000103170. [DOI] [PubMed] [Google Scholar]
- Stöck M, Croll D, Dumas Z, Biollay S, Wang J, Perrin N. A cryptic heterogametic transition revealed by sex-linked DNA markers in Palearctic green toads. J Evol Biol. 2011a;24:1064–1070. doi: 10.1111/j.1420-9101.2011.02239.x. [DOI] [PubMed] [Google Scholar]
- Stöck M, Horn A, Grossen C, Lindtke D, Sermier R, Betto-Colliard C, Dufresnes E, Bonjour E, Dumas Z, Luquet E, Maddalena T, Sousa H Clavero, Martinez-Solano I, Perrin N. Ever-young sex chromosomes in European tree frogs. PLoS Biology. 2011b;9:e1001062. doi: 10.1371/journal.pbio.1001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh A, Kriegs JO, Brosius J, Schmitz J. Retroposon insertions and the chronology of avian sex chromosome evolution. Mol Biol Evol. 2011 doi: 10.1093/molbev/msr147. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Tsuda Y, Nishida-Umehara C, Ishijima J, Yamada K, Matsuda Y. Comparison of the Z and W sex chromosomal architectures in elegant crested tinamou (Eudromia elegans) and ostrich (Struthio camelus) and the process of sex chromosome differentiation in palaeognathous birds. Chromosoma. 2007;116:159–173. doi: 10.1007/s00412-006-0088-y. [DOI] [PubMed] [Google Scholar]
- Turner JMA. Meiotic sex chromosome inactivation. Development. 2007;134:1823–1831. doi: 10.1242/dev.000018. [DOI] [PubMed] [Google Scholar]
- van Doorn GS, Kirkpatrick M. Turnover of sex chromosomes induced by sexual conflict. Nature. 2007;449:909–912. doi: 10.1038/nature06178. [DOI] [PubMed] [Google Scholar]
- van Doorn GS, Kirkpatrick M. Transitions between male and female heterogamety caused by sex-antagonistic selection. Genetics. 2010;186:629–645. doi: 10.1534/genetics.110.118596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn GS, Edelaar P, Weissing FJ. On the origin of species by natural and sexual selection. Science. 2009;326:1704–1707. doi: 10.1126/science.1181661. [DOI] [PubMed] [Google Scholar]
- Wallace H, Wallace BMN, Badawy GMI. Lampbrush chromosomes and chiasmata of sex-reversed crested newts. Chromosoma. 1997;106:526–533. doi: 10.1007/s004120050275. [DOI] [PubMed] [Google Scholar]
- Waters PD, Duffy B, Frost CJ, Delbridge ML, Graves JAM. The human Y chromosome derives largely from a single autosomal region added to the sex chromosomes 80–130 million years ago. Cytogenet Cell Genet. 2001;92:74–79. doi: 10.1159/000056872. [DOI] [PubMed] [Google Scholar]
- Williams TM, Carroll SB. Genetic and molecular insights into the development and evolution of sexual dimorphism. Nat Rev Genet. 2009;10:797–804. doi: 10.1038/nrg2687. [DOI] [PubMed] [Google Scholar]
- Wilson MA, Makova KD. Genomic analyses of sex chromosome evolution. Annu Rev Genom Hum Genet. 2009;10:333–354. doi: 10.1146/annurev-genom-082908-150105. [DOI] [PubMed] [Google Scholar]
- Winge Ö . One-sided masculine and sex-linked inheritance in Lebistes reticulatus. J Genet. 1922;12:145–162. [Google Scholar]
- Winge Ö . The location of eighteen genes in Lebistes reticulatus. J Genet. 1927;18:1–43. [Google Scholar]
- Woram RA, Gharbi K, Sakamoto T, Hoyheim B, Holm LE, Naish K, McGowan C, Ferguson MM, Phillips RB, Stein J, Guyomard R, Cairney M, Taggart JB, Powell R, Davidson W, Danzmann RG. Comparative genome analysis of the primary sex-determining locus in salmonid fishes. Genome Res. 2003;13:272–280. doi: 10.1101/gr.578503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto S, Okada E, Umemoto H, Tamura K, Uno Y, Nishida-Umehara C, Matsuda Y, Takamatsu N, Shiba T, Ito M. A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proc Natl Acad Sci USA. 2008;105:2469–2474. doi: 10.1073/pnas.0712244105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YE, Vibranovski MD, Krinsky BH, Long MY. Age-dependent chromosomal distribution of male-biased genes in Drosophila. Genome Res. 2010a;20:1526–1533. doi: 10.1101/gr.107334.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YE, Vibranovski MD, Landback P, Marais GAB, Long MY. Chromosomal redistribution of male-biased genes in mammalian evolution with two bursts of gene gain on the X chromosome. PLoS Biology. 2010b;8:e1000494. doi: 10.1371/journal.pbio.1000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Wang J, Huang L, Nie WH, Wang JH, Liu Y, Zhao XY, Yang FT, Wang W. Sex chromosomes in the black muntjac recapitulate incipient evolution of mammalian sex chromosomes. Genome Biol. 2008;9:R98. doi: 10.1186/gb-2008-9-6-r98. [DOI] [PMC free article] [PubMed] [Google Scholar]