Abstract
Autosomal recessive primary microcephaly (MCPH) is characterised by a significant reduction in prenatal human brain growth, without alteration of cerebral architecture. The genetic aetiology of MCPH is bi-allelic mutations in genes coding for a subset of centrosomal proteins1-10. While at least three of these proteins have been implicated in centrosome duplication11, the nature of centrosome dysfunction that underlies the neurodevelopmental defect in MCPH is unclear. Here we report a homozygous MCPH-causing mutation in the human CEP63 gene. CEP63 forms a complex with another MCPH protein, CEP152, a conserved centrosome duplication factor12-15. Together, they are essential for maintaining normal centrosome numbers in cells. Using super-resolution microscopy we find that CEP63 and CEP152 co-localise in a discrete ring around the proximal end of the parental centriole, a pattern specifically disrupted in CEP63-deficient patient-derived cells. This work suggests that the CEP152-CEP63 ring-like structure ensures normal neurodevelopment and its impairment particularly affects human cerebral cortex growth.
Keywords: centrosome, mitotic spindle, centrosome duplication, microcephaly, brain
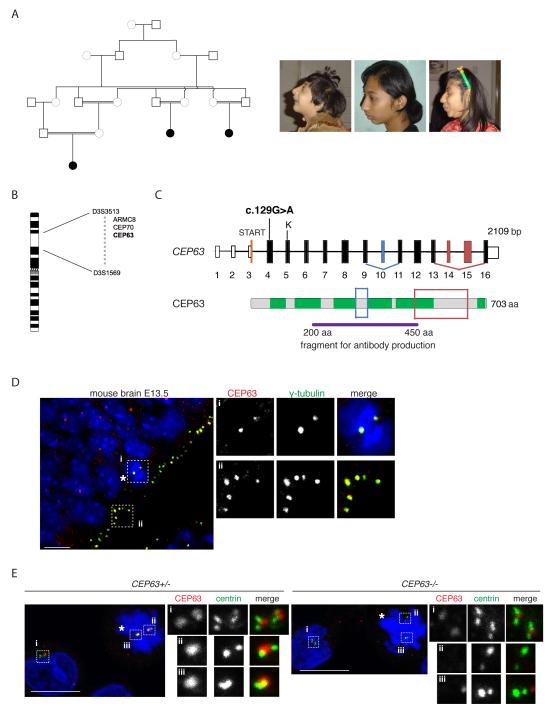
We ascertained a consanguineous Pakistani family in which three members had primary microcephaly at birth (head circumference −4SD to −6SD) and by five years showed mild/moderate mental retardation (Fig. 1 A and Supplementary Note). They were microcephalic and, to a lesser degree, had proportionate short stature, leading to a diagnosis of MCPH with growth retardation or mild Seckel syndrome 16. There was no genetic linkage to known MCPH or Seckel syndrome loci. Autozygosity mapping revealed a single, concordant homozygous locus of 24Mb on chromosome 3 shared by the patients (Fig. 1B). Within this genomic region we identified a pathogenic mutation in the centrosomal gene CEP63 17; c.129G>A causing an immediate STOP codon W43X in exon 4. Alternative splicing of CEP63 produces four products that are expressed in foetal brain (Fig. 1C, Supplementary Fig. 1A). Similar to other MCPH genes18-21, CEP63 underwent positive Darwinian selection during the evolutionary period that higher ape and human brains have grown in relative size (Supplementary Fig. 1B). An antibody recognising CEP63 stained centrosomes of apical neural precursors in mouse brain (Fig. 1D). CEP63 was enriched in centrosomes of B-lymphocytes derived from parent-of-patient (CEP63+/−) (Fig.1E and Supplementary Fig. 2A). A weak CEP63 signal was detectable in centrosomes of patient cells (CEP63−/−), implying that c.129G>A is a hypomorphic mutation. Indeed, CEP63 transcripts were present in patient cells (Supplementary Fig. 2B).
Fig.1. Identification of a MCPH-causing mutation in the CEP63 gene.
(A) Schematic diagram shows simplified pedigree. Filled in circles indicate individuals with MCPH. Only the closest link between family members is depicted. Pictures of three afflicted individuals are shown on right. (B) The three affected individuals share only one region that was concordantly homozygous in chromosome 3. The region was defined by heterozygous allele results for the markers D3S3513 at 137cM/121Mb and D3S1569 at 158cM/145Mb. The three candidate genes sequenced are shown within the linkage interval. (C) Exons of CEP63 gene are shown as bars; solid bars are translated. There is a single CpG island for the gene with transcription starting in exon 3. The position of the nonsense mutation c.129G>A is located in exon 4. A predicted Kozak sequence downstream of the mutation is shown (labelled as ‘K’). Alternatively spliced exons are shown in blue and red (see Supplementary Fig. 1A). CEP63 protein is shown with six predicted coiled-coil domains marked in green. Framed areas correspond to differentially spliced regions. The CEP63 antibody was raised against a recombinant fragment of CEP63 shown. (D) CEP63 protein expression in mouse cerebral cortex neuroepithelium at E13.5. Mitotic cell is marked with asterisk. Magnified views of framed areas are shown on right. Samples were co-stained with antibodies against CEP63 (red in merge) and the centrosomal protein γ-tubulin (green in merge). DNA is in blue. (E) CEP63 protein expression in parent-of-patient (CEP63+/−) and patient (CEP63−/−) lymphocytes. Mitotic cells are marked with asterisks. Magnified views of centrosomes are shown on right. Cells were co-stained with antibodies against CEP63 (red in merge) and the centriolar protein, centrin-3 (green in merge). DNA is in blue. Scale bars=10μm.
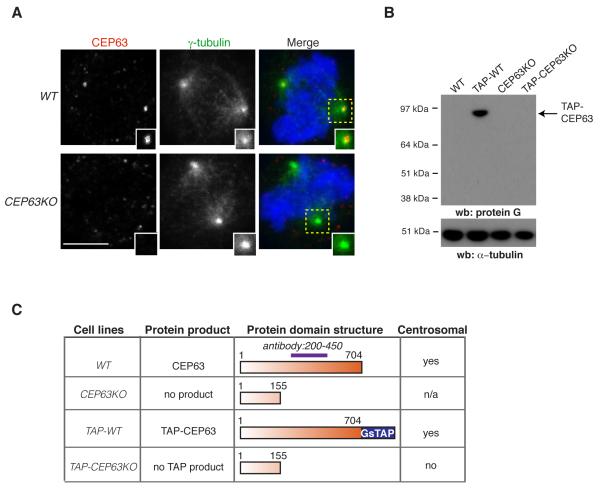
MCPH-linked genes encode proteins that function in mitotic spindle assembly and/or centriole biogenesis 10. Consistently, CEP63 has been implicated in mitotic entry and spindle formation 22,23. To address the role of CEP63 in the centrosome, we generated a CEP63 knockout in DT40 chicken B-lymphocytes using homologous gene-targeting. Exons 5-9 of CEP63 were replaced with drug resistance genes (CEP63KO; Supplementary Fig. 3). CEP63 antibody stained centrosomes of wild-type (WT), but not CEP63KO cells (Fig. 2A). Since the antibody epitope was largely disrupted in CEP63KO cells, these results were confirmed by labelling gene products with a GsTAP tag (comprising protein G and streptavidin-binding protein24) inserted in-frame to the 3′ ends of the CEP63 alleles of WT and CEP63KO cells (TAP-WT and TAP-CEP63KO cells, respectively; Supplementary Fig. 3). TAP-WT cells expressed a single protein product (TAP-CEP63) that localised to centrosomes throughout the cell cycle (Fig. 2B,C and Supplementary Fig. 4A). No product was detected in TAP-CEP63KO cells, indicating exons 6-16 were not translated. Homozygous CEP63KO clones displayed increased population doubling times (Supplementary Fig. 4B), but cell cycle profiles were similar between WT (G1:20±3%, S:51±5%, G2/M:25±6%) and CEP63KO (G1:20±2%, S:50±2%; G2/M:23±3%). However, CEP63KO cells exhibited mitotic spindle defects such as monopolar and multipolar spindles 22 (Fig. 3A). Time-lapse microscopy confirmed these phenotypes (Supplementary Fig. 5 and Videos 1,2).
Fig.2. Disruption of the centrosomal gene CEP63 in vertebrate cells.
(A) WT and CEP63KO DT40 cells were co-stained with antibodies against CEP63 (red in merge) and γ-tubulin (green in merge). Magnified views of framed areas are shown in insets. DNA is blue. Scale bar=5μm. (B) Western blots of cytoplasmic cell extracts (CCE) from WT and gene-disrupted DT40 cell lines were probed with protein G antibodies. TAP cell lines contain in-frame GsTAP tag in their CEP63 alleles. α-tubulin serves as loading control. (C) Summary of the localisation and expression of tagged and truncated CEP63 products in the DT40 cell lines specified. n/a: the antibody epitope is largely destroyed in predicted protein product.
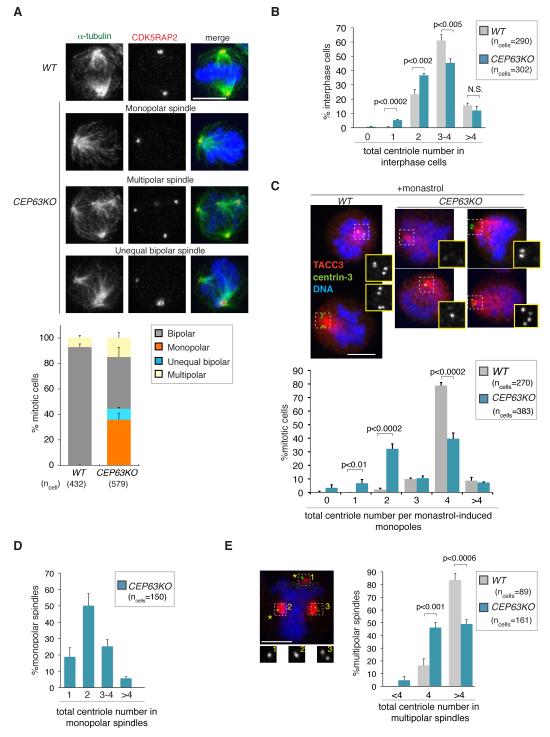
Fig.3. CEP63 is required for maintaining normal centrosome numbers.
(A) Examples for mitotic spindle defects of CEP63KO DT40 cells are shown. Cells are stained with antibodies against the centrosome marker CDK5RAP2 (red in merge) and α-tubulin (green in merge). DNA is in blue. Graph shows quantification of spindle phenotypes (n=3). Scale bars=5μm. (B) Graph shows number of centrioles in interphase WT and CEP63KO DT40 cells (n=3). Antibodies against centrin-3 were used for experiment. (C) Graph shows number of centrioles in monastrol-induced monopoles of WT and CEP63KO DT40 cells (n=3). Note that centriole numbers 1 and 3 could sometimes reflect insufficient spatial resolution of engaged centriole pairs. Examples for CEP63KO monopoles with different centriole numbers are shown; cells were co-stained with antibodies against the spindle pole marker TACC3 (red in merge) and centrin-3 (green in merge). DNA is in blue. Magnified views of centrin-3 staining in framed areas are shown in insets. Similar results were obtained using polyglutamylated tubulin as centriole marker; 0-2 centrioles were present in 42%, 3-4 centrioles in 52% and >4 centrioles in 6% of cells (100 CEP63KO mitotic cells). (D) Graph shows number of centrioles in untreated monopolar CEP63KO spindles (n=3). Antibodies against centrin-3 were used for experiment. (E) Graph shows number of centrioles in mitotic cells with multipolar spindles (n=3). On the left an example for CEP63KO multipolar spindle is shown with two singlet centrioles (asterisks) at spindle poles. Cell was co-stained with antibodies against the spindle pole marker TACC3 (red in merge) and centrin-3 (green in merge). DNA is in blue. Magnified views of centrin-3 staining in framed areas are shown in insets. Scale bars=5μm. Bars in graphs correspond to mean±standard deviation (s.d.). p values were obtained by two-tailed, unpaired Student t test. N.S.=not significant.
After mitosis a cell inherits a centrosome comprising a mother and daughter centriole embedded in the pericentriolar matrix (PCM). The centrosome duplicates in S-phase when a procentriole forms adjacent to each parental centriole engaging in an orthogonal configuration 25 (see schematic in Supplementary Fig. 6). The two centrosomes then mature by PCM expansion and separate to form the mitotic spindle poles. Monopolar spindles could therefore arise from defects in centrosome maturation, separation or duplication. Since PCM components γ-tubulin (Fig. 2A), CDK5RAP2 and TACC3 26 (Fig. 3) were present in CEP63KO spindle poles, CEP63 deficiency does not preclude centrosome maturation. Furthermore, centrosome separation appeared normal in CEP63KO cells (Supplementary Fig. 7, Videos 3-8). Thus, monopolar spindles must reflect a defect in centrosome duplication. Indeed, interphase CEP63KO cells contained fewer centrioles than WT (Fig. 3B). Since interphase cells normally contain 2-4 centrioles and in some cells multiple small centrin foci were visible, a more accurate quantification of centriole numbers was achieved in mitotic cells arrested with monastrol 27. Monastrol prevents centrosome separation, thereby converging centrioles into a monopole. Consistent with the presence of 2 duplicated centrosomes 80% of WT but only 40% of CEP63KO monopoles contained 4 centrioles (Fig. 3C). 40% of monastrol-induced and 70% of untreated CEP63KO monopolar spindles contained only 1-2 centrioles (Fig. 3 C,D).
If a mitotic cell contains only 2 centrioles, it could have defects in procentriole assembly or disengagement, a process whereby the tight link between parental and procentrioles is removed, allowing procentrioles to duplicate in the next cell cycle 28,29 (Supplementary Fig. 6). CEP63 is not a core centriole assembly factor, since CEP63KO cells do not become depleted of centrosomes over time (Supplementary Fig. 8A). Moreover, CEP63KO amplify centrosomes when treated with a DNA replication poison (Supplementary Fig. 8B). Since overduplication depends on recurring cycles of procentriole assembly and disengagement30, CEP63 is dispensable for these steps during an extended cell cycle arrest. Yet, during normal cell cycles CEP63 deficiency could cause a delay in procentriole assembly leading to a stochastic failure in centrosome duplication and thus monopolar spindles. Such a delay, however, does not explain the increase in multipolar spindles in CEP63KO cells. Since centriole pairs segregate with mitotic spindle poles, aberrant engagement between mother and procentrioles could cause asymmetric centriole segregation and multipolarity 31. Consistent with a potential defect in centriole engagement, nearly half of CEP63KO multipolar spindles contained 4 centrioles in total that were distributed as singlets between 3 or 4 spindle poles (Fig. 3E). This extreme phenotype occurred in only 7% of CEP63KO cells, but distances between mother and daughter centrioles were elevated in mitotic CEP63KO centrosomes (Supplementary Fig. 8C). Since a marker of centriole distal ends was used to evaluate distances (centrin-3), abnormal centriole structure or length could distort the data. However, transmission electron microscopy revealed no such anomalies (Supplementary Fig. 8D). In summary, abnormal centrosome numbers in CEP63KO cells could result from delayed procentriole assembly and impaired engagement.
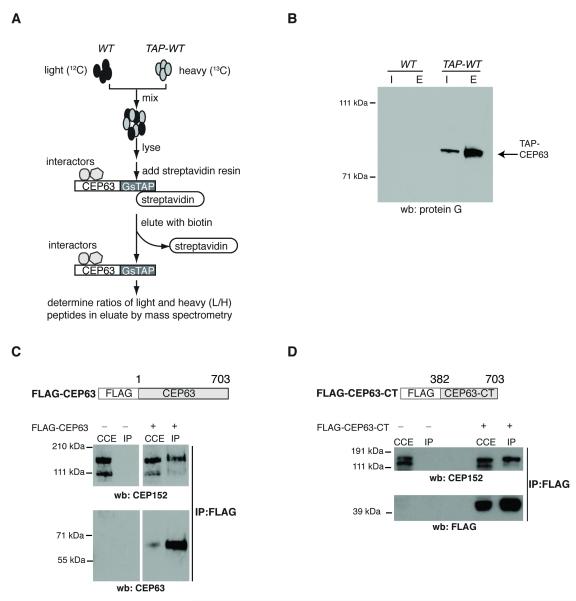
To examine the molecular mechanism of CEP63 action, interactors of CEP63 were identified by affinity-purifying GsTAP-CEP63 from biallelically tagged TAP-WT cells (Fig. 4A,B and Supplementary Fig. 9). WT and TAP-WT cells were differentially labelled with light (WT) or heavy (TAP-WT) forms of amino acids. Streptavidin-bound fractions were analysed by mass-spectrometry and ratios of light to heavy peptides (L/H) were determined using SILAC 32. Two proteins exhibited L/H ratios lower than 0.25: CEP63 and CEP152 (Supplementary Tables 1 and 2). Mammalian CEP63 was among nine proteins reported as CEP152 interactors by the Mitocheck consortium 33. Binding between CEP152 and FLAG-tagged CEP63 was confirmed in human cells (Fig. 4C), an interaction mediated by the C-terminal part of CEP63 (Fig. 4D).
Fig.4. CEP63 forms a protein complex with CEP152.
(A) Diagram illustrates experimental design to identify CEP63-interacting proteins. (B) Western blot shows an example for single step purification of GsTAP-tagged CEP63 protein. Cell lysates of WT or TAP-WT DT40 cells (Input=I) were incubated with streptavidin-agarose resin. Bound proteins were eluted from resin with biotin (Eluate=E). Immunoblot was probed with protein G antibodies to detect GsTAP-tagged CEP63. (C) Cytoplasmic cell extracts (CCE) were prepared from HeLa cells that were mock transfected (−) or transfected with FLAG-CEP63 (+). Anti-FLAG antibodies were used for immunoprecipitation (IP). Immunoblots were probed with antibodies against CEP63 and CEP152, as marked. (D) Cytoplasmic cell extracts (CCE) were prepared from HeLa cells that were mock transfected (−) or transfected with FLAG-CEP63-CT (+). Anti-FLAG antibodies were used for immunoprecipitation (IP). Immunoblots were probed with antibodies against CEP152 and FLAG, as marked.
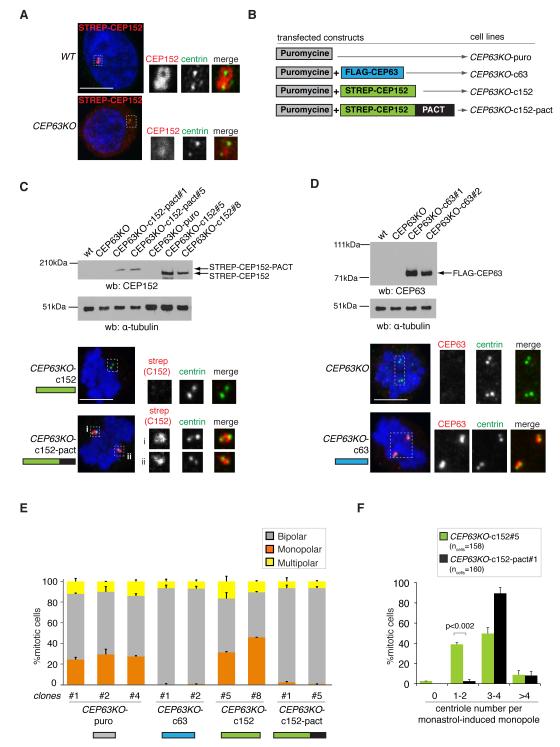
CEP152 is a key regulator of centriole biogenesis and centrosome function 12-15,34,35 and is mutated in MCPH and Seckel syndrome 9,36. We thus wondered if abnormal CEP152 function underlies the phenotypes of CEP63KO DT40 cells. Existing antibodies did not recognise chicken CEP152, so CEP152 localisation in DT40 cells was assessed with ectopically expressed STREP-tagged human CEP152. While STREP-CEP152 accumulated in WT centrosomes, it was weak in interphase and absent from mitotic CEP63KO centrosomes (Fig.5A). To restore CEP152 levels in CEP63KO centrosomes CEP152 was fused to PACT 37, a centrosome localisation signal that mediates CEP63-independent targeting of GFP (Fig. 5B, Supplementary Fig. 7). Indeed, unlike STREP-CEP152, STREP-CEP152-PACT localised to CEP63KO centrosomes throughout the cell cycle (Fig. 5C). To address whether phenotypes of CEP63KO cells were due to diminished centrosomal CEP152, stable CEP63KO clones were generated by introducing the following transgenes: puromycin resistance gene only or in combination with FLAG-CEP63, STREP-CEP152 or STREP-CEP152-PACT (Fig. 5B-D). Remarkably, expression of STREP-CEP152-PACT complemented the mitotic spindle phenotypes and restored centriole numbers in CEP63KO cells (Fig. 5E, F). STREP-CEP152-PACT complemented the phenotypes as efficiently as FLAG-CEP63, implying that CEP63 maintains centrosome numbers via centrosomal recruitment of CEP152.
Fig.5. CEP63-dependent centrosomal accumulation of CEP152 maintains normal centrosome numbers.
(A) Localisation of STREP-tagged human CEP152 in transfected WT and CEP63KO DT40 cells. Magnified views of framed areas are shown in panels below. Cells are co-stained with antibodies against centrin-3 (green in merge) and CEP152 (red in merge). (B) Outline of constructs that were transfected into CEP63KO DT40 cells to derive cell lines stably expressing transgenes. (C) Cytoplasmic cell extracts of CEP63KO-derived DT40 cell lines (nomenclature as in B) were immunoblotted with antibodies against CEP152 and as a loading control, α-tubulin. Below mitotic cells co-stained with antibodies against centrin-3 (green in merge) and strep-tag II (red in merge) are shown. Magnified views of framed areas are shown. DNA is in blue. (D) Cytoplasmic cell extracts of CEP63KO-derived DT40 cell lines (nomenclature as in B) were immunoblotted with antibodies against CEP63 and as a loading control, α-tubulin. Below mitotic cells co-stained with antibodies against centrin-3 (green in merge) and CEP63 (red in merge) are shown. Magnified views of framed areas are shown. DNA is in blue. (E) Graph depicts quantification of spindle phenotypes in CEP63KO-derived DT40 cell lines (nomenclature as in B; n=2; >350 mitotic cells per clone). (F) Graph depicts number of centrioles in monastrol-induced monopoles of CEP63KO-c152 or CEP63KO-c152-pact DT40 cell lines (nomenclature as in B; n=2). Bars in graphs correspond to mean±s.d. p values were obtained by two-tailed, unpaired Student t test. Scale bars=5μm.
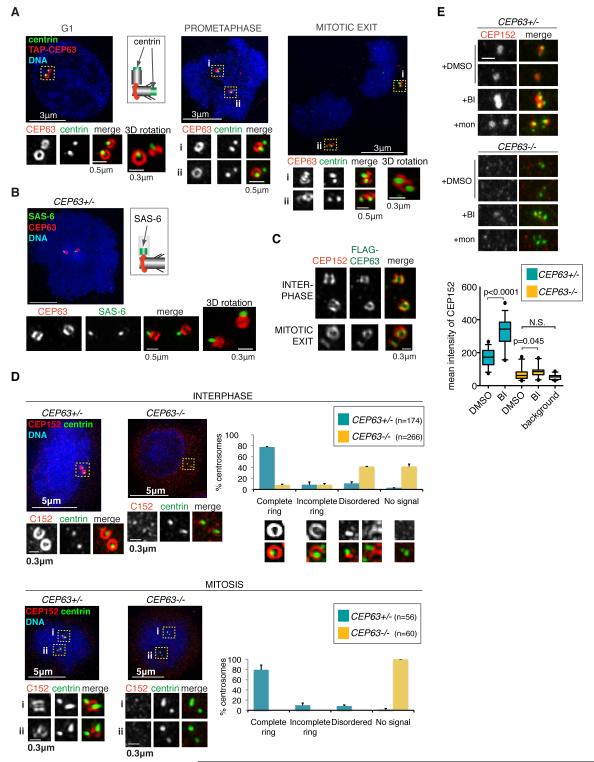
Super-resolution microscopy (3D-SIM) revealed that CEP63 formed a discrete centrosomal ring in TAP-WT DT40 cells (Fig. 6A). The diameter of the ring was approximately 280 nm, similar to centriole diameter. Its position relative to centrin-3 indicated a proximal location for CEP63. Partial overlap was seen with polyglutamylated tubulin antibody marking centriole walls (Supplementary Fig. 10A). The PCM component γ-tubulin was organised around the circumference of the ring (Supplementary Fig. 10B). CEP63 thus occupies a position between the proximal end of parental centriole wall and the PCM. From G1 until mitotic exit only parental centrioles carried a ring. Procentrioles acquired CEP63 signal in late mitosis, but their rings were completed only after cytokinesis, coinciding with centriole disengagement and PCM recruitment (Fig. 6A) 28,31. The ring was also present in humans (Supplementary Fig. 10C) and appeared lateral to SAS-6 found at the proximal end of procentrioles 38 (Fig. 6B).
Fig.6. CEP63 and CEP152 form a ring around parental centrioles, a structure disrupted in CEP63−/− patient cells.
(A) 3D-SIM images are shown of TAP-WT DT40 cells co-stained with antibodies against protein G (recognises TAP-CEP63; red in merge) and centrin-3 (green in merge) antibodies. Each centrosome is shown at high magnification with separate images depicting protein G (top panels) and centrin-3 (middle panels) stainings. Specific cell cycle stages are stated. All images are maximum projections apart from those referred to as 3D rotations. The latter represent alternative views of centrioles (generated by rotations of 3D volumes) to highlight particular features. Schematic shows relative positions of centrin and CEP63 staining. (B) 3D-SIM image of CEP63+/− human lymphocyte co-stained with antibodies against CEP63 (green in merge) and the daughter centriole marker, SAS-6 (red in merge). Schematic shows relative positions of SAS-6 and CEP63 staining. (C) HeLa cells stably expressing FLAG-CEP63 were co-stained with antibodies against FLAG (green in merge) and CEP152 (red in merge). 3D-SIM images of centrosomes are shown. (D) 3D-SIM images of interphase (top) and mitotic (bottom) CEP63+/− and CEP63−/− human lymphocytes are shown. Cells were co-stained with antibodies against CEP152 (red in merge) and centrin-3 (green in merge). DNA is in blue. Framed areas are shown in higher magnification with corresponding CEP152 (left panels) and centrin-3 (middle panels) signal. Graphs on the right show percentages of CEP63+/− and CEP63−/− centrosomes with specific CEP152 localisation pattern (n=2). (E) CEP63+/− and CEP63−/− human lymphocytes were treated with DMSO, PLK1 inhibitor BI-2536 (BI) or monastrol (mon) for 90 minutes prior to fixation. Cells were co-stained with antibodies against CEP152 (red in merge) and centrin-3 (green in merge). Examples for centrosomes from individual mitotic cells are shown. Note that mon and BI induce monoastral spindle, whereas those in DMSO remain bipolar. Scale bar=0.5μm. Distribution of mean intensities of centrosomal CEP152 signal in patient cells is illustrated in graph. Number of centrosomes scored: CEP63+/−: n=30 DMSO- and n=38 BI-treated; CEP63−/−: n=26 DMSO- and n=24 BI-treated. Background CEP152 signal was measured in areas of CEP63−/− cells that do not contain centrosomes (n=26). Box plots: length of whiskers is at 5th and 95th percentiles, the box shows interquartile (25-75) range and horizontal line represents the median. p values were obtained by two-tailed, unpaired Student t test. N.S.=not significant.
Reminiscent of the CEP63 ring, CEP152 was found to localise to the proximal end of the mother centriole wall 13,15. Indeed, CEP152 and FLAG-CEP63 co-localised in HeLa cells (Fig. 6C). Similar to CEP63KO DT40 cells, siRNA-mediated depletion of CEP63 prevented centrosomal accumulation of CEP152 in human cells (Supplementary Fig. 11). This prompted us to investigate the localisation and sub-centrosomal distribution of CEP152 in CEP63+/− and CEP63−/− patient lymphocytes. Despite CEP63+/− and CEP63−/− cells expressing similar amounts of CEP152, protein levels were reduced in CEP63−/− centrosomes (Supplementary Figs. 2A and 12A, B). While most interphase CEP63+/− centrosomes contained CEP152 rings, these were rare in CEP63−/− centrosomes (Fig. 6D). Like CEP63KO DT40 centrosomes, mitotic CEP63−/− centrosomes were devoid of CEP152, a defect complemented by expression of FLAG-CEP63 (Supplementary Fig. 12C). CEP63−/− cells exhibited no mitotic spindle abnormalities, but a small increase in centriole distances was detected in mitotic cells (Supplementary Fig. 12D).
Why is CEP152 localisation particularly affected by CEP63 deficiency during mitosis? We hypothesised that CEP63 opposes an activity that removes/modifies centrosomal CEP152. A good candidate is POLO-LIKE KINASE 1 (PLK1) that links centriole biogenesis to cell cycle progression28,30,31,39. Indeed, treatment with the PLK1 inhibitor BI-253640 dramatically increased centrosomal CEP152 levels in mitotic CEP63+/− cells (Fig. 6E). This was not due to mitotic arrest imparted by BI-2536, since monastrol had no effect. While we cannot exclude that PLK1 modifies the epitope of the CEP152 antibody, our results nonetheless implicate PLK1 in regulating CEP152. BI-2536 treatment produced a small increase in CEP152 at CEP63−/− centrosomes, indicating that PLK1 inhibition can only partially override the defect in CEP152 recruitment caused by CEP63 deficiency.
CEP152 scaffolds procentriole formation by promoting centrosomal accumulation of CPAP and POLO-LIKE KINASE 4 (PLK4) 13-15, a kinase that controls procentriole formation 25,29,41-43. In both chicken CEP63KO and patient CEP63−/− cells CEP152 was absent from mitotic centrosomes, but residual CEP152 protein was detectable in interphase. Since PLK4 activity peaks in mitosis 44, depending on the window of PLK4 activity and the kinetics of CEP63-independent accumulation of CEP152, a delay in CEP152 recruitment could preclude procentriole formation especially in rapidly proliferating cells like DT40 with 8 or apical progenitors with 10-14 hour cycles 45. In contrast, CEP63−/− patient lymphocytes have a 24-hour doubling time that could explain lack of centrosome duplication defects in these cells. Our finding that a CEP152-PACT fusion product complemented centrosome phenotypes of CEP63KO cells suggests that the role of CEP63 in centrosome duplication is to maintain CEP152 at centrosomes. How can PACT mimic the action of CEP63? While PACT is derived from the PCM component AKAP45037, it accumulates on centrioles46 and therefore STREP-CEP152-PACT is probably recruited near the centriole wall in CEP63KO cells.
The position of the CEP63-CEP152 ring coincides with the site where a cohesive structure is predicted to engage parental and procentrioles. A limited role for CEP63 in engagement is supported by the co-existence of mono- and multipolar spindles in CEP63KO populations. This function is consistent with findings in Xenopus, where DNA damage-induced removal of CEP63 from the centrosome caused multipolar spindle formation22. Further support for a link between MCPH and centriole engagement emerged from studies implicating MCPH7/STIL/ana2 47 and MCPH3/CDK5RAP2 48,49 in this process. During neurogenic mitoses of the developing mouse neocortex, progenitors retain the centrosome containing the old mother centriole 50, whereas newborn neurons inherit the new mother centriole. Since this asymmetry contributes to cell-fate determination50, delayed procentriole assembly and impaired engagement between mother and daughter centrioles can alter stem cell fate. In summary we show that the CEP63-CEP152 MCPH complex is an essential part of the molecular machinery controlling centrosome numbers, and defects in either component result in a diminished pool of neural precursors that cannot provide for the enlarged human brain.
Supplementary Material
Acknowledgments
We would like to thank Drs Munro, Pines and Rios for constructs and reagents, Drs Cox, Ebrahimi, Thornton for help, Drs Lakshminarasimhan, Patel, Rios for the critical reading of the manuscript and Dr Bornens and the Gergely lab for helpful suggestions. The authors would like to thank the families for their participation. We are grateful for the support provided by the microscopy and proteomics core facilities at CRI. This work was made possible by funding from the Wellcome Trust to AKN, CGW, MK, OPC, from Cancer Research UK to ARB, CDS, FG, JHS, SR and a Royal Society University Research Fellowship to FG.
Abbreviations
- BrdU
Bromodeoxyuridine
- CEP
centrosomal protein
- GFP
green fluorescent protein
- MCPH
autosomal primary recessive microcephaly
- PCM
pericentriolar matrix
Footnotes
Author Contribution J-H.S. performed most experiments presented in manuscript. A.R.B. generated patient cell lines and performed initial cell biology analysis. A.K.N. performed molecular genetic mapping and gene mutation identification. O.P.C. performed gene expression analysis in patient cells. M.K carried out embryonic brain immunohistochemistry. A.S. and S.R. provided support with super-resolution microscopy. C.D’S. helped with generation and analysis of proteomic data. C.G.W. performed patient ascertainment, clinical studies and gene identification strategy. C.G.W. and F.G. designed the study and wrote paper with comments from all authors.
Accession codes Human CEP63: NM_025180.3 (NCBI)
Human CEP152: NM_014985.3 (NCBI)
Chicken CEP63: ENSGALT00000010715 (Ensembl)
References
- 1.Guernsey DL, et al. Mutations in centrosomal protein CEP152 in primary microcephaly families linked to MCPH4. Am J Hum Genet. 87:40–51. doi: 10.1016/j.ajhg.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson AP, et al. Identification of microcephalin, a protein implicated in determining the size of the human brain. Am J Hum Genet. 2002;71:136–42. doi: 10.1086/341283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bond J, et al. ASPM is a major determinant of cerebral cortical size. Nat Genet. 2002;32:316–20. doi: 10.1038/ng995. [DOI] [PubMed] [Google Scholar]
- 4.Bond J, et al. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat Genet. 2005;37:353–5. doi: 10.1038/ng1539. [DOI] [PubMed] [Google Scholar]
- 5.Kumar A, Girimaji SC, Duvvari MR, Blanton SH. Mutations in STIL, encoding a pericentriolar and centrosomal protein, cause primary microcephaly. Am J Hum Genet. 2009;84:286–90. doi: 10.1016/j.ajhg.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicholas AK, et al. WDR62 is associated with the spindle pole and is mutated in human microcephaly. Nat Genet. 2010;42:1010–4. doi: 10.1038/ng.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu TW, et al. Mutations in WDR62, encoding a centrosome-associated protein, cause microcephaly with simplified gyri and abnormal cortical architecture. Nat Genet. 2010;42:1015–20. doi: 10.1038/ng.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bilguvar K, et al. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature. 2010;467:207–10. doi: 10.1038/nature09327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guernsey DL, et al. Mutations in centrosomal protein CEP152 in primary microcephaly families linked to MCPH4. Am J Hum Genet. 2010;87:40–51. doi: 10.1016/j.ajhg.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thornton GK, Woods CG. Primary microcephaly: do all roads lead to Rome? Trends Genet. 2009;25:501–10. doi: 10.1016/j.tig.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bettencourt-Dias M, Hildebrandt F, Pellman D, Woods G, Godinho SA. Centrosomes and cilia in human disease. Trends Genet. 2011;27:307–15. doi: 10.1016/j.tig.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blachon S, et al. Drosophila asterless and vertebrate Cep152 Are orthologs essential for centriole duplication. Genetics. 2008;180:2081–94. doi: 10.1534/genetics.108.095141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cizmecioglu O, et al. Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J Cell Biol. 2010;191:731–9. doi: 10.1083/jcb.201007107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dzhindzhev NS, et al. Asterless is a scaffold for the onset of centriole assembly. Nature. 2010;467:714–8. doi: 10.1038/nature09445. [DOI] [PubMed] [Google Scholar]
- 15.Hatch EM, Kulukian A, Holland AJ, Cleveland DW, Stearns T. Cep152 interacts with Plk4 and is required for centriole duplication. J Cell Biol. 2010;191:721–9. doi: 10.1083/jcb.201006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roberts E, et al. Autosomal recessive primary microcephaly: an analysis of locus heterogeneity and phenotypic variation. J Med Genet. 2002;39:718–21. doi: 10.1136/jmg.39.10.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersen JS, et al. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 18.Evans PD, et al. Microcephalin, a gene regulating brain size, continues to evolve adaptively in humans. Science. 2005;309:1717–20. doi: 10.1126/science.1113722. [DOI] [PubMed] [Google Scholar]
- 19.Mekel-Bobrov N, et al. Ongoing adaptive evolution of ASPM, a brain size determinant in Homo sapiens. Science. 2005;309:1720–2. doi: 10.1126/science.1116815. [DOI] [PubMed] [Google Scholar]
- 20.Evans PD, Vallender EJ, Lahn BT. Molecular evolution of the brain size regulator genes CDK5RAP2 and CENPJ. Gene. 2006;375:75–9. doi: 10.1016/j.gene.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 21.Tang BL. Molecular genetic determinants of human brain size. Biochem Biophys Res Commun. 2006;345:911–6. doi: 10.1016/j.bbrc.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 22.Smith E, et al. An ATM- and ATR-dependent checkpoint inactivates spindle assembly by targeting CEP63. Nat Cell Biol. 2009;11:278–85. doi: 10.1038/ncb1835. [DOI] [PubMed] [Google Scholar]
- 23.Loffler H, et al. Cep63 Recruits Cdk1 to the Centrosome: Implications for Regulation of Mitotic Entry, Centrosome Amplification, and Genome Maintenance. Cancer Res. 2011;71:2129–2139. doi: 10.1158/0008-5472.CAN-10-2684. [DOI] [PubMed] [Google Scholar]
- 24.Burckstummer T, et al. An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat Methods. 2006;3:1013–9. doi: 10.1038/nmeth968. [DOI] [PubMed] [Google Scholar]
- 25.Bettencourt-Dias M, Glover DM. Centrosome biogenesis and function: centrosomics brings new understanding. Nat Rev Mol Cell Biol. 2007;8:451–63. doi: 10.1038/nrm2180. [DOI] [PubMed] [Google Scholar]
- 26.Barr AR, Kilmartin JV, Gergely F. CDK5RAP2 functions in centrosome to spindle pole attachment and DNA damage response. J Cell Biol. 2010;189:23–39. doi: 10.1083/jcb.200912163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayer TU, et al. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286:971–4. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- 28.Tsou M-FB, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006;442:947–951. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- 29.Nigg EA. Centrosome duplication: of rules and licenses. Trends in Cell Biology. 2007;17:215–221. doi: 10.1016/j.tcb.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 30.Loncarek J, Hergert P, Khodjakov A. Centriole reduplication during prolonged interphase requires procentriole maturation governed by Plk1. Curr Biol. 2010;20:1277–82. doi: 10.1016/j.cub.2010.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang WJ, Soni RK, Uryu K, Tsou M.F. Bryan. The conversion of centrioles to centrosomes: essential coupling of duplication with segregation. J Cell Biol. 2011;193:727–39. doi: 10.1083/jcb.201101109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–86. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 33.Hutchins JR, et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328:593–9. doi: 10.1126/science.1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonaccorsi S, Giansanti MG, Gatti M. Spindle self-organization and cytokinesis during male meiosis in asterless mutants of Drosophila melanogaster. J Cell Biol. 1998;142:751–61. doi: 10.1083/jcb.142.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Varmark H, et al. Asterless is a centriolar protein required for centrosome function and embryo development in Drosophila. Curr Biol. 2007;17:1735–45. doi: 10.1016/j.cub.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 36.Kalay E, et al. CEP152 is a genome maintenance protein disrupted in Seckel syndrome. Nat Genet. 2011;43:23–6. doi: 10.1038/ng.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gillingham AK, Munro S. The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. 2000;1:524–9. doi: 10.1093/embo-reports/kvd105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leidel S, Delattre M, Cerutti L, Baumer K, Gonczy P. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat Cell Biol. 2005;7:115–25. doi: 10.1038/ncb1220. [DOI] [PubMed] [Google Scholar]
- 39.Tsou MF, et al. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev Cell. 2009;17:344–54. doi: 10.1016/j.devcel.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lenart P, et al. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr Biol. 2007;17:304–15. doi: 10.1016/j.cub.2006.12.046. [DOI] [PubMed] [Google Scholar]
- 41.Bettencourt-Dias M, et al. SAK/PLK4 is required for centriole duplication and flagella development. Curr Biol. 2005;15:2199–207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 42.Kleylein-Sohn J, et al. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 43.Puklowski A, et al. The SCF-Fbxw5 E3-ubiquitin ligase is regulated by Plk4 and targets HsSAS-6 to control centrosome duplication. Nat Cell Biol. 2011 doi: 10.1038/ncb2282. [DOI] [PubMed] [Google Scholar]
- 44.Rogers GC, Rusan NM, Roberts DM, Peifer M, Rogers SL. The SCF Slimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J Cell Biol. 2009;184:225–39. doi: 10.1083/jcb.200808049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Calegari F, Haubensak W, Haffner C, Huttner WB. Selective lengthening of the cell cycle in the neurogenic subpopulation of neural progenitor cells during mouse brain development. J Neurosci. 2005;25:6533–8. doi: 10.1523/JNEUROSCI.0778-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keryer G, et al. Dissociating the centrosomal matrix protein AKAP450 from centrioles impairs centriole duplication and cell cycle progression. Mol Biol Cell. 2003;14:2436–46. doi: 10.1091/mbc.E02-09-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stevens NR, Roque H, Raff JW. DSas-6 and Ana2 coassemble into tubules to promote centriole duplication and engagement. Dev Cell. 2010;19:913–9. doi: 10.1016/j.devcel.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barrera JA, et al. CDK5RAP2 regulates centriole engagement and cohesion in mice. Dev Cell. 2010;18:913–26. doi: 10.1016/j.devcel.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lizarraga SB, et al. Cdk5rap2 regulates centrosome function and chromosome segregation in neuronal progenitors. Development. 2010;137:1907–17. doi: 10.1242/dev.040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X, et al. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature. 2009;461:947–55. doi: 10.1038/nature08435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gretchen J. Lymphocyte Isolation and Culture. Cold Spring Harb Protoc. 2006 [Google Scholar]
- 52.Lewis AE, et al. Identification of nuclear phosphatidylinositol 4,5-bisphosphate-interacting proteins by neomycin extraction. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.003376. M110 003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Finch AJ, et al. Uncoupling of GTP hydrolysis from eIF6 release on the ribosome causes Shwachman-Diamond syndrome. Genes Dev. 2011;25:917–29. doi: 10.1101/gad.623011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.