Abstract
In the search for new therapeutic tools against neglected diseases produced by trypanosomatid parasites, and particularly against African Trypanosomiasis, whose etiological agent is Trypanosoma brucei, organoruthenium compounds with bioactive nitrofuran containing thiosemicarbazones (L) as co-ligands were obtained. Four ruthenium(ii) complexes with the formula [Ru2(p-cymene)2(L)2]X2, where X = Cl or PF6, were synthesized and the crystal structures of two of them were solved by X-ray diffraction methods. Two of the complexes show significant in vitro growth inhibition activity against Trypanosoma brucei brucei and are highly selective towards trypanosomal cells with respect to mammalian cells (J774 murine macrophages). These promising results make the title organoruthenium compounds good lead candidates for further developments towards potential antitrypanosomal organometallic drugs.
Introduction
Tropical diseases constitute a major health problem and offer big challenge for drug discovery.1 Among the infectious illnesses designated by the World Health Organization as neglected tropical diseases, those produced by protozoan parasites that belong to the trypanosomatid genus and kinetoplastid order constitute major health problems concentrated in the poorest tropical or subtropical regions of the planet.2–4 Particularly, Human African Trypanosomiasis, together with American Trypanosomiasis and Leishmaniasis, account for the highest rates of death among neglected tropical diseases.2 Human African Trypanosomiasis (Sleeping sickness), caused by Trypanosoma brucei (T. brucei) (Trypanosoma brucei gambiense and Trypanosoma brucei rhodesiense), is currently a resurgent disease with epidemic character in many regions of Africa that can be fatal if not treated. The lack of surveillance, health care and new treatments and the emergence of resistance to old drugs favored the reappearance of the disease.2,5 The drugs currently available for the treatment of Human African Trypanosomiasis, pentamidine, suramin, eflornithine and melarsoprol, show toxicity problems and variable efficacy depending on the type and stage of the disease.5,6
Attempts towards the development of trypanocidal metal-based compounds have been described.1,7–11 In particular, we have been successfully working on the development of potential anti-trypanosomal metal-based agents through different strategies.1,12–22 One of these approaches involves metal coordination of trypanocidal ligands. The design of antiparasitic compounds which combine ligands bearing anti-trypanosomal activity and pharmacologically active metals in a single chemical entity could provide drugs capable of modulating multiple targets simultaneously.23 This strategy aims to enhance efficacy and improve safety by lowering the therapeutic dose and/or circumventing drug resistance mechanisms.24 The metal could act as a carrier and stabilizer for the drug until it reaches its target, while at the same time the organic drug carries and protects the metal, preventing side reactions in its transit towards a second target of biological action. Metal–drug combined effects may result in an important enhancement of the activity of the drug and the appearance of new mechanisms of action.25 Applying this concept Sánchez-Delgado et al. studied ruthenium complexes of clotrimazole and ketoconazole for anti-trypanosome therapy and ruthenium chloroquine species as antimalarials. Synergistic effects were observed in most cases.1,7–10,25
We also used this approach to develop classic Pd(ii), Pt(ii), Ru(ii) and Ru(iii) coordination compounds of bioactive 5-nitrofuryl and 5-nitroacroleine containing thiosemicarbazones.12,15,17,18 These ligands had shown higher in vitro activity against T. cruzi, the etiological agent of American Trypanosomiasis (Chagas disease), than the reference drug nifurtimox. Their main mode of action, as for nifurtimox and other nitroheterocyclic antiparasitic agents, is related to the intracellular reduction of the nitro moiety followed by redox cycling, yielding reactive oxygen species (ROS) known to cause parasite damage.26 Moreover, some years ago the genomes of T. cruzi, T. brucei and Leishmania major were decoded,27 showing a high degree of similarity. Thus, T. cruzi and T. brucei could possess common enzymatic targets and metabolic pathways and therefore these thiosemicarbazone ligands could also be active on T. brucei.3 In this sense, the anti-T. cruzi drug nifurtimox (combined with eflornithine) was included in 2009 in the World Health Organization's Model List of Essential Medicines for the treatment of Human African Trypanosomiasis, which also supports our proposal.28
In the search for new metal-based antitrypanosomal agents, we expanded our previous work on classical coordination compounds through the development of organometallic compounds that include these potentially bioactive thiosemicarbazones as co-ligands. Organoruthenium compounds are currently emerging as innovative metal-based drugs, mainly as promising antitumor agents.29,30 In addition, organoruthenium and organoiron compounds had been previously identified as potential antimalarial agents.25,31,32 In particular, ferroquine is being developed by Sanofi-Aventis under phase II clinical trials.33 Nevertheless, organometallics against American or African Trypanosomiasis have not yet been described. Ruthenium-arene compounds including bioactive co-ligands could show potential as therapeutic tools against trypanosomatid parasites and, particularly, against T. brucei. In this work, we synthesized and characterized four new Ru-p-cymene compounds that include four bioactive 5-nitrofuryl containing thiosemicarbazones, shown in Fig. 1, as co-ligands. The novel organometallics and the free ligands were evaluated in vitro on T. brucei brucei. T. brucei brucei is the etiologic agent of Nagana disease of cattle and it is a suitable model of the species that cause Human African Trypanosomiasis. In addition, the selectivity of their antitrypanosomal action was evaluated by testing the cytotoxicity of the most active compounds on J774 macrophages as mammalian model cells.
Fig. 1.

Bioactive 5-nitrofuryl containing thiosemicarbazones selected as ligands.
Experimental
General considerations
All common laboratory chemicals were purchased from commercial sources and used without further purification. [Ru(p-cymene)Cl2]2 and the thiosemicarbazone ligands were prepared according to previously published procedures.23,26
Physical measurements
C, H and N analyses were performed with a Carlo Erba Model EA1108 elemental analyzer. A 500-MS Varian Ion Trap Mass Spectrometer was used to measure electrospray ionization mass spectra (ESI-MS) of methanol solutions of the complexes. Each spectrum was obtained as a combination of several scans for each sample. UV-vis spectra were recorded on a Shimadzu UV-1603 instrument. FTIR spectra (4000–400 cm−1) of the complexes and the free ligands were measured as KBr pellets with a Bomen FTIR model MB102 instrument. 1H-NMR spectra of the complexes were recorded at 400 MHz on a Bruker Avance instrument. Experiments were performed at 30 °C in DMSO-d6. Tetramethylsilane was used as the internal standard. Cyclic voltammetry experiments were carried out on ca. 1.0 × 10−3 MDMSO (spectroscopic grade) solutions using a Metrohm 693 VA instrument with a 694 VA Stand converter and a 693 VA Processor and a three-electrode cell under nitrogen atmosphere at room temperature with tetrabutyl ammonium perchlorate (TBAP) (ca. 0.1 M) as the supporting electrolyte. A hanging drop mercury electrode (HDME) was used as the working electrode, a platinum wire as the auxiliary electrode, and saturated calomel (SCE) as the reference electrode.
Synthesis of the compounds
[RuII2(η6-p-cymene)2(L1)2]Cl2
[Ru(p-cymene)Cl2]2 (0.08 mmol, 50 mg) and L1 (0.16 mmol, 35 mg) were dissolved in 5 mL of dichloromethane. The reaction mixture was refluxed for 5 h. A red solid was isolated by filtration of the hot mixture. Yield: 23 mg, 58%. Found: C, 39.26; H, 3.95; N, 11.58. Calc. for Ru2Cl2C32H38N8O6S2: C, 39.62; H, 4.12; N, 11.62%. ESI-MS (MeOH) m/z [Found (Calcd)]: 449.1 (449.0) (100%) ([Ru2(p-cymene)2(L1)2]2+ ≡ [Ru2 C32H38N8O6S2]2+). UV-vis (phosphate buffer pH 7.04 2% DMSO) λmax (ε M−1cm−1): 413 (14.8 × 103), 291 nm. 1H-NMR: δ = 7.49 (2H, d, furan–H), 7.87 (2H, d, furan–H), 8.35 (2H, s, –CH=N), 7.94 (2H, br s, NH), 1.05 (6H, d, p-cym CH(CH3)2), 1.14 (6H, d, p-cym CH(CH3)2), 2.24 (6H, s, C–CH3 p-cym), 2.59 (2H, m, p-cym CH(CH3)2), 5.71 (2H, d, p-cym phenyl–H), 5.83 (2H, d, p-cym phenyl–H), 6.21 (2H, d, p-cym phenyl–H), 6.36 (2H, d, p-cym phenyl–H) ppm. Single crystals suitable for X-ray diffraction were obtained by slow evaporation of a dichloromethane solution.
[RuII2(η6-p-cymene)2(L2)2](PF6)2
Solutions of [Ru(p-cymene)Cl2]2 (0.08 mmol, 50 mg; 10 mL MeOH) and L2 (0.16 mmol, 38 mg; 10 mL MeOH) were mixed and stirred at room temperature for 24 h. The red reaction mixture was reduced to 10 mL on a rotary evaporator. Isopropanol (4 mL) and 5 mL of a NaPF6 methanolic solution (28 mg, 0.17 mmol) were sequentially added. A red solid precipitated. Yield: 43 mg, 86%. Found: C, 33.46; H, 3.51; N, 9.03. Calc. for Ru2P2F12C34H42N8O6S2: C, 33.55; H, 3.65; N, 9.29%. ESI-MS (MeOH) m/z [Found (Calcd)]: positive mode: 463.1 (463.0) (100%) ([Ru (p-cymene)2(L2)2]2+ ≡ [Ru2C34H42N8O6S2]2+); negative mode: 144.8 (144.9) (PF6−). UV-vis (phosphate buffer pH 7.04 2% DMSO) λmax (ε M−1cm−1): 414 (17.6 × 103), 288 nm. 1H-NMR: δ = 7.27 (2H, d, furan–H), 7.80 (2H, d, furan–H), 8.46 (2H, s, –CH=N), 8.43 (2H, br s, NH), 0.90 (6H, d, p-cym CH(CH3)2), 0.98 (6H, d, p-cym CH(CH3)2), 2.36 (6H, s, C–CH3 p-cym), 5.94 (2H, d, p-cym phenyl–H), 6.06 (2H, d, p-cym phenyl-H), 6.28 (2H, d, p-cym phenyl-H), 6.46 (2H, d, p-cym phenyl-H) ppm.
[RuII2(η6-p-cymene)2(L3)2](PF6)2
Solutions of [Ru(p-cymene)Cl2]2 (0.08 mmol, 50 mg; 10 mL MeOH) and L3 (0.16 mmol, 40 mg; 10 mL MeOH) were mixed and stirred at room temperature for 24 h. The red reaction mixture was reduced to 10 mL on a rotary evaporator. Isopropanol (4 mL) and 5 mL of a NaPF6 methanolic solution (28 mg, 0.17 mmol) were sequentially added. A red solid precipitated. Yield: 41 mg, 80%. Found: C, 34.80; H, 3.78; N, 8.84. Calc. for Ru2P2F12C36H46N8O6S2: C, 34.73; H, 3.86; N, 9.00%. ESI-MS (MeOH) m/z [Found (Calcd)]: positive mode: 477.1 (477.0) (100%) ([Ru2(p-cymene)2(L3)2]2+ ≡ [Ru2C36H46N8O6S2]2+); negative mode: 144.8 (144.9) (PF6−). UV-vis (phosphate buffer pH 7.04 2% DMSO) λmax (ε M−1cm−1): 417 (15.0 × 103), 287 nm. 1H NMR: δ = 1.03 (6H, t,–CH3), 7.21 (2H, d, furan–H), 7.79 (2H, d, furan–H), 8.43 (2H, s, –CH=N), 8.46 (2H, br s, NH), 0.92 (6H, d, p-cym CH(CH3)2), 0.95 (6H, d, p-cym CH(CH3)2), 2.39 (6H, s, C-CH3 p-cym), 2.73 (2H, m, p-cym CH(CH3)2), 2.93 (2H, m, p-cym CH(CH3)2), 5.96 (2H, d, p-cym phenyl-H), 6.10 (2H, d, p-cym phenyl-H), 6.32 (2H, d, p-cym phenyl-H), 6.39 (2H, d, p-cym phenyl-H) ppm. Suitable crystals for X-ray diffraction were obtained by slow evaporation of a methanol solution.
[RuII2(η6-p-cymene)2(L4)2]Cl2
Solutions of [Ru(pcymene)Cl2]2 (0.08 mmol, 50 mg; 10 mL MeOH) and L4 (0.16 mmol, 49 mg; 10 mL MeOH) were mixed and stirred at room temperature for 24 h. The red reaction mixture was reduced to 10 mL on a rotary evaporator. A red solid was isolated by filtration. Yield: 30 mg, 65%. Found: C, 47.16; H, 4.15; N, 9.89. Calc. for Ru2Cl2C44H46N8O6S2: C, 47.10; H, 4.28; N, 9.99%. ESI-MS (MeOH) m/z [Found (Calcd)]: positive mode: 525.2 (525.1) (100%) ([Ru2(p-cymene)2(L4)2]2+ ≡ [Ru2C44H46N8O6S2]2+). UV-vis (phosphate buffer pH 7.04 2% DMSO) λmax (ε M−1cm−1): 426 (17.1 × 103), 311 nm. 1H NMR: δ = 6.94 (2H, t, phenyl–H), 7.25 (4H, t, phenyl–H), 7.86 (2H, d, furan–H), 7.63 (4H, d, phenyl–H), 8.21 (2H, d, furan–H), 8.80 (2H, s, –CH=N), 9.70 (2H, br s, NH) 1.05 (6H, d, p-cym CH(CH3)2), 1.07 (6H, d, p-cym CH(CH3)2), 2.07 (6H, s, C–CH3p-cym), 5.59 (2H, d, p-cym phenyl-H), 5.62 (2H, d, p-cym phenyl-H), 5.71 (2H, d, p-cym phenyl-H), 5.73 (2H, d, p-cym phenyl-H) ppm.
X-ray crystallographic studies
Suitable crystals for X-ray diffraction studies were obtained as described above. Data were collected at 125(2) K by using graphite monochromated Mo-Kα radiation in a Bruker SMART APEX II CCD X-ray diffractometer. Structure resolution and refinement were performed using SHELX.34 Details are included in Table 1. Those H atoms not found in Fourier maps were included from models and constrained as riding on their bound atoms. Generally, we preferred the inclusion of experimentally found H atoms in the atomic list, and so some H atom distances are shorter than the conventional value 0.96Å.
Table 1.
Crystal data and structure refinement for [Ru2(p-cymene)2(L1)2]Cl2 and [Ru2(p-cymene)2(L3)2](PF6)2 complexes
| [Ru2(p-cymene)2(L1)2]Cl2·8H2O | [Ru2(p-cymene)2(L3)2](PF6)2·H2O | |
|---|---|---|
| Empirical formula | C32H38Cl2N8O14Ru2S2 | C36H48F12N8O7P2Ru2S2 |
| Crystal color | Dark red | Brown |
| Formula weight | 1095.88 | 1261.04 |
| Crystal System | Monoclinic | Monoclinic |
| Space group | P 21/c | Cc |
| Temperature K | 125(2) | 125(2) |
| Wavelength (Å) | 0.71073 | 0.71073 |
| a (Å) | 19.809(7) | 21.639(8) |
| b (Å) | 25.120(8) | 11.066(4) |
| c (Å) | 19.115(6) | 21.408(12) |
| β (°) | 108.286(4) | 113.206(4) |
| Volume (Å3) | 9031(5) | 4712(4) |
| Z, density (mg mm−3) | 8, 1.612 | 4, 1.778 |
| Absorption coefficient | 0.948 | 0.901 |
| Flack parameter | — | 0.03(2) |
| Crystal size (mm) | 0.17 × 0.17 × 0.13 | 0.10 × 0.08 × 0.08 |
| θ range data collection | 1.35, 24.41 | 2.05, 26.73 |
| Limiting indices | −23,23/−29,29/−22,22 | −27, 27/−14,14/−27,27 |
| Data collected/unique/Rint | 85246, 14865/0.0883 | 26397/9984/0.0653 |
| Max, min. Transmission | 0.86/0.83 | 0.93/0.91 |
| Refinement method | F2 | F2 |
| Refined data/parameters | 10119/1152 | 8246/677 |
| Goodness-of-fit on F2 | 1.002 | 0.988 |
| Final R,a Rw(all) [I > 2σ(I)] | 0.0457, 0.0842 | 0.0425, 0.0798 |
R1 = Σ||Fo|−|Fc||/Σ|Fo|, wR2 = [Σw(|Fo|2−|Fc|2)2/Σw(|Fo|2)2]1/2.
Biological studies
Activity onT. brucei brucei strain 427
The infective form of T. brucei brucei strain 427, cell line 449 (encoding one copy of the tet-repressor protein: PleoR;35), was aerobically cultivated in a humidified incubator at 37 °C with 5% CO2 in HMI-9 medium36 supplemented with 10% (v/v) fetal calf serum (FCS), 10 U mL−1 penicillin, 10 μg mL−1 streptomycin and 0.2 μg mL−1 phleomycin. 10 mM stock solutions of the test compounds and of the Ru precursor [Ru(p-cymene)Cl2]2 were prepared using DMSO as solvent and then diluted in culture medium to obtain five experimental concentrations (from 100 μM to 0.01 μM). Controls included compound vehicle (DMSO) at final concentrations ranging from 0.0001 to 1% (v/v) and culture medium (growth control). Each condition was tested in triplicate. The cytotoxic effect of the compounds towards trypanosomes was evaluated by absolute cell counting. Trypanosomes were grown to exponential phase, harvested by centrifugation at 2000 g for 10 min at room temperature and suspended at a density of 5 × 105 cells mL−1 in fresh culture medium. 1 mL of the cell suspension was plated into each well of a 24-well culture plate. Compounds or DMSO were immediately added at the concentrations described above and the culture plate was incubated at 37 °C with 5% CO2. After 24h, living parasites were counted with a Neubauer chamber under a light microscope or by flow cytometry. For the first counting technique, parasites in wells were suspended by pipetting, if necessary, diluted in phosphate buffered saline solution with 1% (w/v) glucose, and then loaded onto a Neubauer chamber. Living parasites were counted on both hemi-chambers with a light microscope. Lack of motility and abnormal morphology were criteria used to exclude dead parasites. For the flow cytometry analysis, 490 μL were taken from each well and 10 μL of Count Bright™ absolute counting beads (Invitrogen) were added as an internal counting standard. The mixture was homogenised by short vortexing and analyzed using a CyAn™ ADP (Dako) flow cytometer. The instrument parameters were adjusted to discriminate cells from debris by gating on the forward- and side-scatter signals, and to detect the microspheres according to their fluorescence (λex 488 nm). Analyses were performed on linear forward- vs. logarithmic side-scatter dot plots from approximately 1000 gated microspheres using the Summit 4.3 software (Dako). The cell density was calculated using the following formula: cells μL−1 = A/B × C/D, where A = number of cell events, B = number of beads events, C = assigned bead count of the lot (beads/10 μL), and D = sample volume (490 μL). For each compound concentration, cytotoxicity was calculated according to the following equation: Cytotoxicity (%) = (experimental value − DMSO control)/(growth control − DMSO control) × 100. The input value is the mean (n = 3) of the cell densities and corrected absorbance at 450 nm (Aic450), respectively. The data were plotted as percentage cytotoxicity versus drug concentration. IC50 values were obtained from dose response curves fitted to a sigmoidal equation (Boltzmann model) or extrapolated from non-linear fitting plots.
Cytotoxicity on murine macrophages
The J774 mouse macrophage cell line was cultivated in a humidified 5% CO2/95% air atmosphere at 37 °C in DMEM medium supplemented with 10% (v/v) FCS, 10 U mL−1 penicillin and 10 μg mL−1 streptomycin. Stock solutions of the compounds to be tested were prepared as described for anti-T. brucei activity tests. Each condition was tested in triplicate. The cytotoxic effect of the compounds towards macrophages was evaluated by colorimetric assay of cell viability with a tetrazolium salt (WST-1 reagent). For macrophages, cells grown to the mid-log phase were detached from the culture flask by trypsinization for 10 min at room temperature followed by gentle scraping. After centrifugation at 300 g for 5 min at room temperature, the cell pellet was resuspended in fresh culture medium at a cell density of 5 × 104 cells mL−1. 250 μL of the cell suspension was added per well to a 96-well flat bottom microculture plate. After 24 h incubation in a humidified atmosphere with 5% CO2 at 37 °C, the conditioned medium was replaced in each well by 200 mL fresh culture medium alone (growth control) or the added compounds or DMSO at different concentrations (see above). Following an additional 24 h incubation, the culture medium was removed and the wells were washed twice with 250 μL of prewarmed fresh culture medium. Cell viability was assessed using the reagent WST-1 (Roche) diluted 1 : 10 in culture medium. 110 mμL of the diluted reagent was added per well and incubated at 37 °C with 5% CO2. After 3 h, the absorbance of the formazan dye produced by metabolically active cells was measured at 450 nm (reference wavelength at 630 nm) with an EL 800 microplate reader (Biotek). Control wells (n = 12) containing cell proliferation reagent but lacking cells were included as blank. The corrected absorbance values at 450 nm were obtained by subtracting the corresponding absorbance value at 630 nm and the blank average (e.g. Aic450 = Ai450 − Ai630nm − Ablank450nm). Cytotoxicity was calculated and IC50 values were obtained as described for cytotoxicity on T. brucei brucei.
Calf thymus DNA interaction experiments
Complexes were preliminary tested for their DNA interaction ability using native calf thymus DNA (CT DNA) (Type I) by modification of a previously reported procedure.12,37 CT DNA (50 mg) was dissolved in water (30 mL) overnight. Solutions of the complexes in DMSO (spectroscopy grade) (1 mL, 10−3 M) were incubated at 37 °C with a solution of CT DNA (1 mL) for 96 h. DNA/complex mixtures were exhaustively washed to eliminate the unreacted complex. Quantification of bound metal was achieved by atomic absorption spectroscopy on a Perkin Elmer 5000 spectrometer. Standards were prepared by diluting a metal standard solution for atomic absorption spectroscopy. Final DNA concentration per nucleotide was determined by UV absorption spectroscopy using a molar absorption coefficient of 6000 M−1 cm−1 at 260 nm.
Results and discussion
Syntheses and characterization of the new ruthenium complexes
Four Ru(II)-p-cymene dimeric complexes including bioactive 5-nitrofuryl containing thiosemicarbazones as ligands were synthesized with high purity and reasonable yields. Analytical, ESI mass spectrometry and FTIR and 1H-NMR spectroscopic results are in agreement with the formula [Ru2(p-cymene)2(L)2]X2, where X = Cl or PF6 depending on the thiosemicarbazone ligand. In all cases ESI-MS spectra in the positive mode showed the most intense peak at a m/z value that is assigned by simulation of the spectra to the [Ru2(p-cymene)2 (L)2]2+ dimer. The isotope pattern for this signal is consistent with this assignment. In addition, the L2 and L3 complexes showed in the negative mode the peak corresponding to m/z of the hexafluorophosphate counterion. As a general trend for all the compounds ESI(+)-MS spectra for 24 h-aged solutions still exhibited the major signal ascribed to the dimeric parent compound with overall charge +2, indicating that the parent complex was still present in solution.
Infrared spectroscopic results
Although the complexity of the IR spectra of the thiosemicarbazone ligands and, consequently, of their ruthenium complexes increases as the R-substituent complexity increases, this family of ruthenium compounds shows a common spectral pattern. The IR vibrational spectroscopic behavior of the four ruthenium p-cymene compounds was compared to that previously reported for the free ligands and their palladium(II), platinum(II), ruthenium(II) and ruthenium(III) coordination compounds.12,15,17,18,38 Significant vibration bands of the ruthenium compounds, diagnostic of the coordination mode of the ligands, could be tentatively assigned (Table 2). After coordination, the ν(C=N) bands of the thiosemicarbazone free ligands, at approximately 1500–1600 cm−1, shift to higher frequencies. In addition, ν(C=S) bands at around 820–850 cm−1, should shift to lower wavenumbers when thiosemicarbazones act as N, S bidentate ligands but, as has been previously stated, they could not be unambiguously assigned for the metal compounds due to the complexity of the spectra in this region.38 Nevertheless, significant changes are observed in this region after coordination. These modifications are consistent with coordination of the thiosemicarbazone ligands through the thiocarbonylic sulfur and the azomethynic nitrogen. The ν(NH) band, at approximately 3120–3150 cm−1, is not observed in the complexes due to deprotonation of the ligands.
Table 2.
Significant vibration bands of the [Ru2(p-cymene)2(L1)2]X2 complexes, useful for determining the mode of coordination of the ligands
| Compound | ν(C=N) | νs(NO2) | ν(N–N) | δ(NO2) + furana | Other bands |
|---|---|---|---|---|---|
| HL1b | 1602 | 1356 | 1108 | 811 | |
| [Ru2(p-cymene)2(L1)2]Cl2 | 1605 | 1349 | 1160 | 809 | |
| HL2b | 1599 | 1354 | 1114 | 808 | |
| [Ru2(p-cymene)2(L2)2](PF6)2 | 1588 | 1351 | 1168 | 813 | νa(PF) 847; δa(FPF) 558 |
| HL3b | 1602 | 1352 | 1104 | 805 | |
| [Ru2(p-cymene)2(L3)2](PF6)2 | 1586 | 1351 | 1172 | 813 | νa(PF) 846; δa(FPF) 558 |
| HL4b | 1595 | 1344 | 1104 | 811 | |
| [Ru2(p-cymene)2(L4)2]Cl2 | 1602 | 1338 | 1166 | 809 |
ν: stretching; δ: bending; a: asymmetric;
δ(NO2) + furan modes or furan hydrogen wagging symmetric modes;
From reference 38.
The strong νa(PF) and δa(FPF) bands of the PF6− counterion are also observed, at around 840 and 560 cm−1, for the Ru-p-cymene L2 and L3 hexafluorophosphate salts.39
1H-NMR results
NMR experiments confirmed the structures of the synthesized complexes and the results are in agreement with those of the other spectroscopies. The 1H-NMR experiments showed narrow signals, typical for Ru(II) diamagnetic complexes. 1H-NMR integrations and signal multiplicities are in agreement with the proposed formulae. Signals corresponding to the p-cymene moiety are duplicated due to non equivalent p-cymene molecules in the dimer. The complexes showed similar 1H chemical shifts for the common nitrofuryl thiosemicarbazone portions and also for the p-cymene moiety. Relevant results are shown in Table 3. The attached figure shows the numbering scheme of the free thiosemicarbazone ligands. When the thiosemicarbazone ligand is coordinated, the effect of both the metal coordination and the ligand deprotonation is apparent for the protons that are located close to the coordinating atoms i.e. the azomethynic nitrogen and the thiocarbonylic sulfur. Upon coordination, the most distinguishing feature of the spectra is the change in chemical shift of the ilydenic proton, numbered 3. In all complexes, the signal of this proton is significantly displaced downfield. The absence of a signal corresponding to proton 4 is in accordance with the coordination of the ligands in a monodeprotonated form.
Table 3.
Relevant 1H-NMR chemical shift values (δ) in ppm of HL and the L portion in [Ru2(p-cymene)2(L)2]X2 at 303 K in DMSO-d6. [Ru2(p-cymene)2(L)2]X2 compounds are designated as Ru-L for simplicity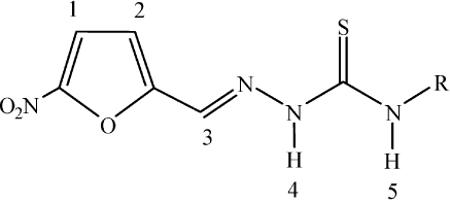
| H | HL1b | Ru–L1 | Δ δ aRuL1 | HL2b | Ru–L2 | Δ δ aRuL2 | HL3b | Ru–L3 | Δ δ aRuL3 | HL4b | Ru–L4 | Δ δ aRuL4 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 7.76 | 7.87 | 0.11 | 7.79 | 7.80 | 0.01 | 7.79 | 7.79 | 0.00 | 7.82 | 8.21 | 0.39 |
| 2 | 7.37 | 7.49 | 0.12 | 7.31 | 7.27 | −0.04 | 7.31 | 7.21 | −0.04 | 7.49 | 7.86 | 0.37 |
| 3 | 8.00 | 8.35 | 0.35 | 7.98 | 8.46 | 0.48 | 7.98 | 8.43 | 0.45 | 8.08 | 8.80 | 0.72 |
| 4 | 11.87 | — | — | 11.87 | — | — | 11.82 | — | — | 12.20 | — | — |
| 5 | 8.52 | 7.94 | −0.58 | 8.52 | 8.43 | −0.09 | 8.54 | 8.46 | −0.08 | 10.16 | 9.70 | −0.46 |
Δδ = (δComplex − δLigand);
From reference 26.
Crystal structures
The title compounds are binuclear dicationic complexes having a central core formed by two Ru and two S atoms (Fig. 2 and Fig. S1, ESI†). This 4-membered ring is not planar as the dihedral angle between the Ru1–S1–S2 and Ru2–S1–S2 planes ranges between 7.5° and 13.7° (Table 4). The metal coordination polyhedron is piano-stool shaped, the legs of which are defined by one N and two S atoms. There are some compounds containing the dinuclear arene-RuS moiety in the CSD, and about 40% of them show bonding interactions between metal centers (Ru–Ru distances within 3Å). The second group has Ru–Ru separations within the range 3.30–3.70Å; the 2 complexes here reported fall within this group with Ru–Ru separations longer than 3.53Å. As in our complexes, this latter set having large separation between metals is also associated with small dihedral angles between the Ru1–S1–S2 and Ru2–S1–S2 planes in the CSD. There is marked asymmetry of the Ru–S bond distances in the title compounds; the shorter bond is about 2.35Å and the longer one about 2.43 Å. Stacking of the thiosemicarbazone ligand in the title compounds is seen, defining a sort of pentagonal prism with 2 N atoms within graphite interaction of 3.401(6) Å for the L3 complex (Table 4, Fig. 3 for Ru–L3 complex). Both independent molecules in the L1 complex show related values of 3.285(9) Å and 3.387(8) Å. To our knowledge, the only reported structure showing a similar feature is bis(μ2-N-cyano-N′-methylthiourea)-bis(p-cymene)-di-ruthenium(II),40 where a ring of sequence Ru–S–C–N replaces our 5 membered ring Ru–S–C–N–N moiety. In this case, the corresponding prism has four faces made by the two Ru–S bonds plus two C---N separations of 3.01 and 3.04 Å. Coplanarity between the furyl and nitro moieties within 13° is seen as shown by the dihedral angle between both planes. Also both furyl rings in each complex are almost co-planar (Table 4), although these rings are not above each other.
Fig. 2.
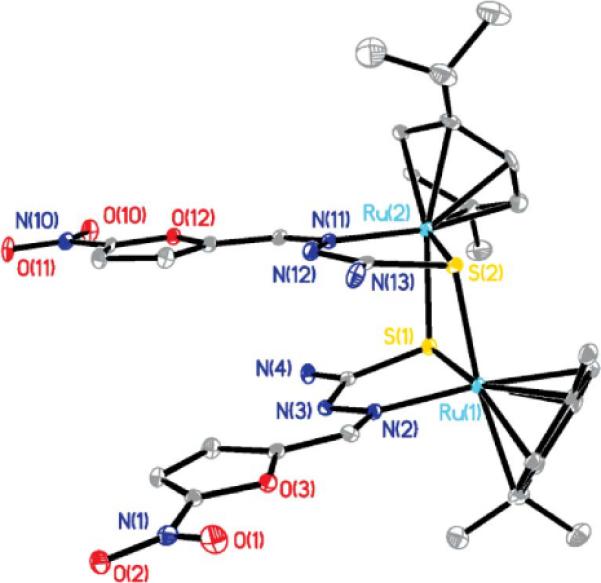
The cationic [Ru2(p-cymene)2(L1)2]2+ in [Ru2(p-cymene)2(L1)2]Cl2 showing ellipsoids; H atoms, water molecules and chloride are omitted.
Table 4.
Selected geometrical parameters including bond distances, separation for non bonded atoms, dihedral angles and bond angles for [Ru2(p-cymene)2(L1)2]Cl2 and [Ru2(p-cymene)2(L3)2](PF6)2 complexes
| [Ru2(p-cymene)2(L1)2]Cl2 8H2O | [Ru2(p-cymene)2(L3)2](PF6)2·H2O | |
|---|---|---|
| Ru1–Ru2 | 3.565(1), [3.537(1)] | 3.537(1) |
| Ru1–S1, Ru1–S2 | 2.350(1), 2.428(2) (Ru3) [2.344(2), 2.417(2)] | 2.3521(15), 2.4313(16) |
| Ru2–S1, Ru2–S2 | 2.418(2), 2.347(2) (Ru4) [2.420(2), 2.349(1)] | 2.3495(15), 2.4009(16) |
| Ru–N | 2.077(5), 2.077(5) (Ru3) [2.081(5), 2.093(5)] (Ru4) | 2.085(4), 2.092(4) |
| Ru–C(aromatic) - range | 2.162(9)–2.275(5) | 2.178(5)–2.277(5) |
| Ru–arene centroid | 1.701(7), 1.707(7) (Ru3) [1.719(7) 1.706(7)] (Ru4) | 1.711(6), 1.705(6) |
| N′–N″ stacking | 3.285(9), [3.387(8)] | 3.401(6) |
| Ru1–S1–S2 Dihedral, Ru2–S1–S2 | 14.0, [12.1] | 12.7 |
| furyl–furyl dihedral | 7.5, [13.7] | 13.9(2) |
Distances in Å. Values in square brackets are a selection of geometrical data for the 2nd molecule in the asymmetric unit of [Ru2(p-cymene)2(L1)2]Cl2, for instance Ru3–Ru4 separation of 3.537(1) Å, is shown next to that of Ru1–Ru2.
Fig. 3.
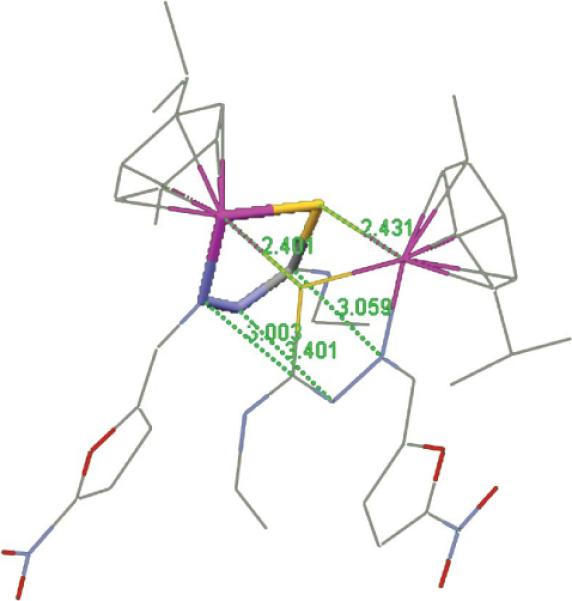
The pentagonal prism for the [Ru2(p-cymene)2(L3)2]2+ cation (only one base is stick style), whose lateral faces are defined by two Ru–S bond distances (2.401 Å, 2.431 Å) and three additional separations (C---N, N---N, and N---C), 3.509 Å, 3.401 Å and 3.003 Å, respectively; H atoms omitted.
Comparing our our 2 complexes, it is seen that the pair of Ru–S bond distances in each complex is not sensitive to the substituents in the thiosemicarbazone ligand since one is systematically longer (2.35 and 2.43Å). The same equivalence is seen in the Ru–N bond lengths, about 2.08Å, and the Ru–arene–centroid distance, about 1.71Å. However, a difference is seen in the stacking N---N separation, from 3.28 to 3.40Å, which may be due to packing forces.
Geometric data for both molecules in the asymmetric unit of [Ru2(p-cymene)2(L1)2]Cl2 are mostly equivalent. Exceptions are the Ru---Ru separation in one complex, 3.565(1)Å (Ru1---Ru2), which is slightly longer than for Ru3---Ru4, 3.537(1)Å. Also, the stacking distance between N atoms differs between both dinuclear complexes, 3.285(9)Å (N3---N12), and 3.387(8)Å (N22---N32), respectively, as do the dihedral angles between furyl rings or within the 4-membered ring Ru-S-Ru-S (Table 4). The packing of the [Ru2(p-cymene)2(L1)2]Cl2 complex shows a large amount of water molecules within a channel (Fig. 4), occupying the central part of the crystal lattice along the a direction. Some of these water molecules establish H-bonds with N23, the nitro group (O31) and the chloride. There are 5 dinuclear Ru octahydrates found in the CSD. One of them, (μ2-oxo)-bis((oxalato)-(2,2′,6′,2″-terpyridine))-di-ruthenium(III) octahydrate,41 shows a water channel with diameter of about 8Å through the center of the crystallographic cell. This is narrower than the one found in our [Ru2(p-cymene)2(L1)2]Cl2·8H2O complex, having a water channel diameter of about 12Å.
Fig. 4.

Ellipsoid style for packing of [Ru2(p-cymene)2(L1)2]Cl2 showing the channel of disordered water (red oxygen) parallel to the a axis; H atoms omitted.
Biological studies
In vitro activity on T. brucei brucei strain 427
All the new ruthenium compounds showed growth inhibitory activity on T. brucei brucei, inducing a dose-dependent antiproliferative effect on parasites treated for 24 h (Table 5). [Ru2(p-cymene)2(L1)2]Cl2 and [Ru2(p-cymene)2(L4)2]Cl2 are the most active, with IC50 values of 0.5 μM and 2.9 μM, respectively (Table 5). The growth inhibitory activity of Ru–L2 and Ru–L3 compounds against T. brucei brucei was 6- to 30-fold lower than those of the Ru–L1 and Ru–L4 derivatives. Interestingly, in contrast to the other Ru complexes, exposure of the parasites to 1 μM Ru–L4 for an additional 24 h caused a significant cell death (drop of two Log10 in cell number, not shown). This data suggests a mechanism of action for Ru–L4 that might involve slow metabolism of the compound to more active byproducts, or accumulative and irreversible cellular damage. It is interesting to emphasize that [Ru2(p-cymene)2(L4)2]Cl2 showed a very significant increase in growth inhibitory activity with respect to the L4 ligand (IC50 > 100 μM). The Ru-p-cymene starting compound, [Ru(p-cymene)Cl2]2, did not show growth inhibitory activity on the parasite (IC50 > 100 μM, Table 5). This fact demonstrates the relevance of including the bioactive thiosemicarbazone ligand in the Ru coordination sphere. The thiosemicarbazone ligands L1–L3 per se exerted a cytotoxic effect comparable to their Ru-p-cym complexes (IC50 values of 1.1 μM, 11.0 μM, 17.3 μM for L1, L2, L3 and 2.9 μM, 10.6 μM and 17.3 μM for Ru–L1, Ru–L2 and Ru–L3, respectively).
Table 5.
In vitro activity against T. brucei brucei, cytotoxicity on murine macrophages and selectivity index (SI) values of the 5-nitrofuryl thiosemicarbazone ligands and their Ru p-cymene derivatives
| Compound | Macrophages IC50/μM | T. brucei IC50/μM | SI (fold)a |
|---|---|---|---|
| [Ru2(p-cymene)2(L1)2]Cl2 | >100 | 2.9 | >35 |
| [Ru2(p-cymene)2(L2)2](PF6)2 | ND | 10.6 | ND |
| [Ru2(p-cymene)2(L3)2](PF6)2 | ND | 17.3 | ND |
| [Ru2(p-cymene)2(L4)2]Cl2 | 26 | 0.5 | 52 |
| L1 | 64 | 1.1 | 58 |
| L2 | ND | 11.0 | ND |
| L3 | ND | 17.3 | ND |
| L4 | ND | >100 μM | ND |
| [Ru(p-cymene)Cl2]2 | ND | >100 μM | ND |
SI: IC50 macrophages/IC50 T. brucei brucei. ND: not determined. Results are the means of three different experiments.
Cytotoxicity on J774 murine macrophages
The specificity of the antitrypanosomal activity of the most active Ru compounds (Ru–L1 and Ru–L4) and thiosemicarbazone ligand L1 was evaluated by analyzing their cytotoxicity against a murine macrophage-like cell line (J774). The cytotoxicity observed was, in ascending order: Ru–L1 (IC50 > 100 μM) > L1 (IC50 64 μM) > Ru-L4 (IC50 26 μM). Comparison of the selectivity indices (SI) reveals that L1 (SI 58) and the Ru–L4 complex (SI 52) are about 50-fold more potent on bloodstream trypanosomes than on the mammalian cell line. On the same order of magnitude but 1.5-fold lower is the SI for Ru–L1 (SI 35).
The Special Programme for Research and Training in Tropical Diseases (TDR), which performs leading research on lead discovery against neglected diseases, has recently defined the following criteria to consider a compound as a drug hit for African trypanosomiasis: IC50 of ≤ 0.2 μg mL−1 against whole organism and SI > 100.43 Based on these criteria, none of the compounds tested qualifies as a hit. However, the biological activity displayed by [Ru2(p-cymene)2(L4)2]Cl2 (IC50 on T. brucei brucei of 0.5 μM, SI 52) and [Ru2(p-cymene)2(L1)2]Cl2 (IC50 on T. brucei brucei of 2.9 μM, SI > 35) merits further investigation to draw final conclusions in this respect. For instance, potential synergistic combinations of these compounds with drugs currently employed to treat African trypanosomiasis might be explored with the goal of reducing the chemotherapeutic dosage, and the associated side effects, as well as preventing or delaying the appearance of drug resistance.
Other biologically relevant results
The compounds were designed to show a dual mechanism of action by affecting different targets or biologically significant processes in the parasite. The compounds could potentially act by intra-parasite generation of toxic free radicals due to the 5-nitrofuryl thiosemicarbazone ligand. Additionally, they could interact with DNA through the Ru-arene moiety. Accordingly, two preliminary studies, redox behavior by cyclic voltammetry and CT-DNA covalent interaction studies, were performed to evaluate the potential of these organometallics to act through such a dual mechanism of action.
Cyclic voltammetry results
All ruthenium p-cymene compounds displayed comparable voltammetric behavior in DMSO solution. The metal complexes showed two well-defined waves at around −1.6 and −0.7 V vs. SCE (Fig. 5). Couple I could be assigned to a metal centered reduction process as reported for other Ru-p-cymene complexes.44 No significant changes in the corresponding peak potential were observed for the different compounds studied. As previously observed for the thiosemicarbazone ligands and their palladium, platinum and ruthenium complexes, couple II corresponds to a quasireversible process involving a one-electron transfer that had been assigned to the generation of the anion radical RNO2˙- by reduction of the nitro group.12,15,18
Fig. 5.
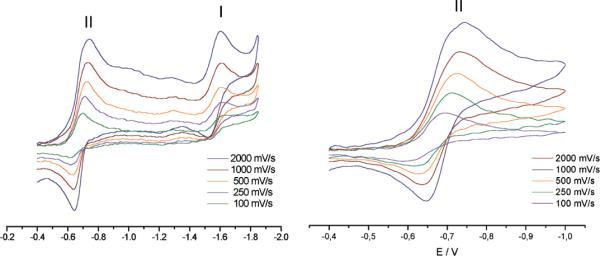
Cyclic voltammetric results for [Ru2(p-cymene)2(L4)2]Cl2. Experimental conditions: hanging drop Hg working electrode, Pt wire auxiliary electrode, saturated calomel (SCE) reference electrode, supporting electrolyte 0.1 M TBAP, 1 mM DMSO.
The biological significance of the reduction potential of the nitro moiety present in the thiosemicarbazone active ligand, in relation to the ability of the compounds to be bioreduced in the parasite leading to the toxic nitro anion radical species, has been previously stated.12,42 Therefore, the effect of ruthenium complexation on this peak potential was studied. Table 6 shows the reduction potentials of the nitro moiety for the four ruthenium p-cymene compounds. The potentials previously reported for the corresponding ligands and some ruthenium 5-nitrofuryl thiosemicarbazone coordination compounds are included for comparison.18 Small changes were observed in the reduction potentials of the nitro moiety of the ligands as a consequence of ruthenium complexation. The new Ru-p-cymene compounds showed lower values than those observed for the corresponding ligands which could have a positive effect on the biological activity of the new compounds as they would be more easily bioreduced. In this aspect the generation of these novel Ru-p-cymene dimers including 5-nitrofuryl thiosemicarbazones as ligands could lead to a favorable change in bioreduction behavior, with respect to the previously reported Ru 5-nitrofuryl thiosemicarbazone coordination compounds that showed higher reduction potential values than was observed for the corresponding ligands.18
Table 6.
Reduction potentials of the nitro moiety for the obtained ruthenium p-cymene compounds and the corresponding ligands measured in DMSO at 2000 mV s−1. Reduction potentials for previously reported ruthenium 5-nitrofurylthiosemicarbazone coordination compounds are included for comparison18
| Compound | Epcc | Epad |
|---|---|---|
| HL1a | −0.84 | −0.75 |
| [Ru2(p-cymene)2(L1)2]Cl2 | −0.80 | −0.66 |
| [RuCl2(HL1)2]b | −0.98 | −0.77 |
| [RuCl3(dmso)(HL1)]b | −0.99 | −0.74 |
| [RuCl(PPh3)(L1)2]b | −1.09 | −0.72 |
| HL2a | −0.79 | −0.71 |
| [Ru2(p-cymene)2(L2)2](PF6)2 | −0.84 | −0.68 |
| [RuCl2(HL2)2]b | −1.02 | −0.77 |
| [RuCl3(dmso)(HL2)]b | −0.92 | −0.73 |
| HL3a | −0.79 | −0.71 |
| [Ru2(p-cymene)2(L3)2](PF6)2 | −0.84 | −0.68 |
| [RuCl2(HL3)2]b | −1.12 | −0.69 |
| [RuCl3(dmso)(HL3)]b | −0.96 | −0.69 |
| [RuCl(PPh3)(L3)2]b | −0.87 | −0.73 |
| HL4a | −0.78 | −0.70 |
| [Ru2(p-cymene)2(L4)2]Cl2 | −0.74 | −0.65 |
| [RuCl2(HL4)2]b | −0.94 | −0.77 |
| [RuCl3(dmso)(HL4)]b | −1.01 | −0.72 |
| [RuCl(PPh3)(L4)2]b | −1.03 | −0.72 |
From reference 12
From reference 18.
Cathodic peak potential
Anodic peak potential.
Potentials are reported in Volts vs. saturated calomel electrode.
Calf thymus DNA interaction experiments
In order to initially address if interaction with DNA could be part of the mode of action of the complexes, experiments with CT DNA were carried out. Binding of the Ru compounds to CT-DNA was studied by combining atomic absorption determinations (for the metal) and electronic absorption measurements for DNA quantification. The new compounds are very poor binding agents for CT DNA (Table 7). The observed ruthenium–DNA binding levels were almost 10-fold lower than those of the palladium(II) 5-nitrofuryl thiosemicarbazone complexes previously developed.12 These results are in agreement with the structure of the complexes that seems not to allow covalent binding to DNA bases due to the lack of labile bonds such as Ru–Cl. Nevertheless, other DNA interaction types should be evaluated in further studies.
Table 7.
Interaction of the Ru compounds with CT DNA after 96 h of incubation at 37 °C
| Compound | nmol Ru/mg DNA base | Ru/basea | base/Ru |
|---|---|---|---|
| [Ru2(p-cymene)2(L1)2]Cl2 | 42.7 | 0.014 | 71 |
| [Ru2(p-cymene)2(L2)2](PF6)2 | 28.2 | 0.009 | 108 |
| [Ru2(p-cymene)2(L3)2](PF6)2 | 22.7 | 0.008 | 134 |
| [Ru2(p-cymene)2(L4)2]Cl2 | 37.5 | 0.013 | 80 |
ruthenium (mol) per DNA base (mol).
Conclusions
Four new dimeric ruthenium arene compounds of the formula [Ru2(p-cymene)2(L)2]X2, where X = Cl or PF6, with bioactive nitrofuryl containing thiosemicarbazones as co-ligands were synthesized and fully characterized. Two of them showed in vitro activity against T. brucei brucei and high selectivity indices (IC50 mammalian cells/IC50 parasite). Preliminary studies suggest the potential of the complexes to act as dual parasite inhibitors. Further studies are needed to obtain insight into the probable mechanism of action of the new Ru p-cymene compounds and are currently in progress. The promising results obtained herein make these organoruthenium compounds good lead candidates for further developments towards potential antitrypanosomal organometallic drugs.
Acknowledgements
Authors wish to thank CYTED through network 209RT0380, Dr Isabel Tomaz (Centro de Ciências Moleculares e Materiais, Faculdade de Ciências, Lisbon University, Portugal) for performing ESI-MS experiments, the Portuguese National Mass Spectroscopy Network (RNEM, IST-UTL Center) and M.C. Oliveira (CQE-IST-TU Lisbon) for providing access to the MS facilities. Authors also thank the US National Science Foundation through grant 0521237 for the X-ray diffractometer and the NIH through Grant # SC1GM089558-01A1 (to R.A. S.-D). The technical assistance of MD Valentina Porro (Cell Biology Unit, Institut Pasteur de Montevideo) during flow cytometry analysis is gratefully acknowledged.
Footnotes
Electronic supplementary information (ESI) available. CCDC reference numbers 819342-819343. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c1dt11519g
References
- 1.Navarro M, Gabbiani G, Messori L, Gambino D. Drug Discovery Today. 2010;15:1070–1077. doi: 10.1016/j.drudis.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Hotez PJ, Molyneux DH, Fenwick A, Kumaresan J, Ehrlich Sachs S, Sachs JD, Savioli L. N. Engl. J. Med. 2007;357:1018–1027. doi: 10.1056/NEJMra064142. [DOI] [PubMed] [Google Scholar]
- 3.Ribeiro I, Sevcsik AM, Alves F, Diap G, Don R, Harhay MO, Chang S, Pecoul B. PLoS Neglected Trop. Dis. 2009;3:e484. doi: 10.1371/journal.pntd.0000484. DOI: 10.1371/journal.pntd.0000484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Urbina J. Expert Opin. Ther. Pat. 2003;13:661–669. [Google Scholar]
- 5.Delespaux V, de Koning HP. Drug Resist. Updates. 2007;10:30–50. doi: 10.1016/j.drup.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 6.Croft S, Barret M, Urbina J. Trends Parasitol. 2005;21:508–512. doi: 10.1016/j.pt.2005.08.026. [DOI] [PubMed] [Google Scholar]
- 7.Sánchez-Delgado RA, Anzellotti A. Mini-Rev. Med. Chem. 2004;1:23–30. doi: 10.2174/1389557043487493. [DOI] [PubMed] [Google Scholar]
- 8.Sánchez-Delgado RA, Anzellotti A, Suárez L. Metal ions in Biological Systems (H. Sigel and A. Sigel, ed.), 41: Metal Ions and Their Complexes in Medication. Marcel Dekker; New York: 2004. pp. 379–419. [PubMed] [Google Scholar]
- 9.Magalhães Moreira DR, Lima Leite AC, Ribeiro dos Santos R, Soares MBP. Curr. Drug Targets. 2009;10:212–231. doi: 10.2174/138945009787581140. [DOI] [PubMed] [Google Scholar]
- 10.Cavalli A, Bolognesi ML. J. Med. Chem. 2009;52:7339–7359. doi: 10.1021/jm9004835. [DOI] [PubMed] [Google Scholar]
- 11.Fricker SP, Mosi RM, Cameron BR, Baird I, Zhu Y, Anastassov V, Cox J, Doyle PS, Hansell E, Lau G, Langille J, Olsen M, Qin L, Skerlj R, Wong RSY, Santucci Z, McKerrow JH. J. Inorg. Biochem. 2008;102:1839–1845. doi: 10.1016/j.jinorgbio.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 12.Otero L, Vieites M, Boiani L, Denicola A, Rigol C, Opazo L, Olea-Azar C, Maya JD, Morello A, Krauth-Siegel RL, Piro OE, Castellano E, González M, Gambino D, Cerecetto H. J. Med. Chem. 2006;49:3322–3331. doi: 10.1021/jm0512241. [DOI] [PubMed] [Google Scholar]
- 13.Urquiola C, Vieites M, Aguirre G, Marín A, Solano B, Arrambide G, Lavaggi ML, Torre MH, González M, Monge A, Gambino D, Cerecetto H. Bioorg. Med. Chem. 2006;14:5503–5509. doi: 10.1016/j.bmc.2006.04.041. [DOI] [PubMed] [Google Scholar]
- 14.Vieites M, Smircich P, Parajón-Costa B, Rodríguez J, Galaz V, Olea-Azar C, Otero L, Aguirre G, Cerecetto H, González M, Gómez-Barrio A, Garat B, Gambino D. JBIC, J. Biol. Inorg. Chem. 2008;13:723–735. doi: 10.1007/s00775-008-0358-7. [DOI] [PubMed] [Google Scholar]
- 15.Vieites M, Otero L, Santos D, Gajardo D, Toloza J, Figueroa R, Norambuena E, Olea-Azar C, Aguirre G, Cerecetto H, González M, Morello A, Maya JD, Garat B, Gambino D. J. Inorg. Biochem. 2008;102:1033–1043. doi: 10.1016/j.jinorgbio.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Vieites M, Smircich P, Guggeri L, Marchán E, Gómez-Barrio A, Navarro M, Garat B, Gambino D. J. Inorg. Biochem. 2009;103:1300–1306. doi: 10.1016/j.jinorgbio.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 17.Vieites M, Otero L, Santos D, Olea-Azar C, Norambuena E, Aguirre G, Cerecetto H, González M, Kemmerling U, Morello A, Maya JD, Gambino D. J. Inorg. Biochem. 2009;103:411–418. doi: 10.1016/j.jinorgbio.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 18.Pagano M, Demoro B, Toloza J, Boiani L, González M, Cerecetto H, Olea-Azar C, Norambuena E, Gambino D, Otero L. Eur. J. Med. Chem. 2009;44:4937–4943. doi: 10.1016/j.ejmech.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 19.Benítez J, Guggeri L, Tomaz I, Costa Pessoa J, Moreno V, Lorenzo J, Avilés FX, Garat B, Gambino D. J. Inorg. Biochem. 2009;103:1386–1394. doi: 10.1016/j.jinorgbio.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 20.Benítez J, Guggeri L, Tomaz I, Arrambide G, Navarro M, Costa Pessoa J, Garat B, Gambino D. J. Inorg. Biochem. 2009;103:609–616. doi: 10.1016/j.jinorgbio.2008.10.018. [DOI] [PubMed] [Google Scholar]
- 21.Demoro B, Caruso F, Rossi M, Benítez D, Gonzalez M, Cerecetto H, Parajón-Costa B, Castiglioni J, Gallizi M, Docampo R, Otero L, Gambino D. J. Inorg. Biochem. 2010;104:1252–1258. doi: 10.1016/j.jinorgbio.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benítez J, Becco L, Correia I, Leal SM, Guiset H, Costa Pessoa J, Lorenzo J, Tanco S, Escobar P, Moreno V, Garat B, Gambino D. J. Inorg. Biochem. 2011;105:303–311. doi: 10.1016/j.jinorgbio.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 23.Morphy R, Rankovic Z. J. Med. Chem. 2005;48:6523–6543. doi: 10.1021/jm058225d. [DOI] [PubMed] [Google Scholar]
- 24.Chibale K. ARKIVOC IX. 2002:93–98. [Google Scholar]
- 25.Rajapakse CSK, Martinez A, Naoulou B, Jarzecki AA, Suarez L, Deregnaucourt C, Sinou V, Schrevel J, Musi E, Ambrosini G, Schwartz GK, Sanchez-Delgado RA. Inorg. Chem. 2009;48:1122–1131. doi: 10.1021/ic802220w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aguirre G, Cerecetto H, González M, Gambino D, Otero L, Olea-Azar C, Rigol C, Denicola A. Bioorg. Med. Chem. 2004;12:4885–4893. doi: 10.1016/j.bmc.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 27.El-Sayed NM, et al. Science. 2005;309:404–409. doi: 10.1126/science.1112181. [DOI] [PubMed] [Google Scholar]
- 28. http://www.who.int/trypanosomiasis_african/research/combination_treatment/en/index.html.
- 29.van Rijt SH, Sadler PJ. Drug Discovery Today. 2009;23:1089–1097. doi: 10.1016/j.drudis.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dyson PJ, Sava G. Dalton Trans. 2006:1929–1933. doi: 10.1039/b601840h. [DOI] [PubMed] [Google Scholar]
- 31.Biot C. Curr. Med. Chem.: Anti-Infect. Agents. 2004;3:135–147. [Google Scholar]
- 32.Dubar F, Khalife J, Brocard J, Dive D, Biot C. Molecules. 2008;13:2900–2907. doi: 10.3390/molecules13112900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dive D, Biot C. ChemMedChem. 2008;3:383–391. doi: 10.1002/cmdc.200700127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheldrick GM. Acta Crystallogr. 2008;A64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 35.Biebinger S, Wirtz LE, Lorenz P, Clayton C. Mol. Biochem. Parasitol. 1997;85:99–102. doi: 10.1016/s0166-6851(96)02815-0. [DOI] [PubMed] [Google Scholar]
- 36.Hirumi H, Hirumi K. J. Parasitol. 1989;75:985–989. [PubMed] [Google Scholar]
- 37.Mahnken RE, Billadeau MA, Nikonowicz EP, Morrison H. J. Am. Chem. Soc. 1992;114:9253–9265. [Google Scholar]
- 38.Gambino D, Otero L, Vieites M, Boiani M, González M, Baran EJ, Cerecetto H. Spectrochim. Acta, Part A. 2007;68:341–348. doi: 10.1016/j.saa.2006.11.043. [DOI] [PubMed] [Google Scholar]
- 39.Heyns AM. Spectrochim. Acta, Part A. 1977;33:315–322. [Google Scholar]
- 40.Henderson W, Nicholson BK, Dinger MB, Bennett RL. Inorg. Chim. Acta. 2002;338:210–218. [Google Scholar]
- 41.Lebeau EL, Ajao Adeyemi S, Meyer TJ. Inorg. Chem. 1998;37:6476–6484. doi: 10.1021/ic970908z. [DOI] [PubMed] [Google Scholar]
- 42.Otero L, Folch C, Barriga G, Rigol C, Opazo L, Vieites M, Gambino D, Cerecetto H, Norambuena E, Olea-Azar C. Spectrochim. Acta, Part A. 2008;70:519–523. doi: 10.1016/j.saa.2007.07.045. [DOI] [PubMed] [Google Scholar]
- 43. http://apps.who.int/tdr/documents/bl-2008-business-plan-2008.pdf.
- 44.Sumiyoshi T, Gunnoe TB, Petersen JL, Boyle PD. Inorg. Chim. Acta. 2008;361:3254–3262. [Google Scholar]


